Abstract
Rickettsia typhi, the causative agent of murine (endemic) typhus, is an obligate intracellular pathogen with a life cycle involving both vertebrate and invertebrate hosts. In this study, we characterized a gene (RT0218) encoding a C-terminal ankyrin repeat domain-containing protein, named Rickettsia ankyrin repeat protein 1 (RARP-1), and identified it as a secreted effector protein of R. typhi. RT0218 showed differential transcript abundance at various phases of R. typhi intracellular growth. RARP-1 was secreted by R. typhi into the host cytoplasm during in vitro infection of mammalian cells. Transcriptional analysis revealed that RT0218 was cotranscribed with adjacent genes RT0217 (hypothetical protein) and RT0216 (TolC) as a single polycistronic mRNA. Given one of its functions as a facilitator of extracellular protein secretion in some Gram-negative bacterial pathogens, we tested the possible role of TolC in the secretion of RARP-1. Using Escherichia coli C600 and an isogenic tolC insertion mutant as surrogate hosts, our data demonstrate that RARP-1 is secreted in a TolC-dependent manner. Deletion of either the N-terminal signal peptide or the C-terminal ankyrin repeats abolished RARP-1 secretion by wild-type E. coli. Importantly, expression of R. typhi tolC in the E. coli tolC mutant restored the secretion of RARP-1, suggesting that TolC has a role in RARP-1 translocation across the outer membrane. This work implies that the TolC component of the putative type 1 secretion system of R. typhi is involved in the secretion process of RARP-1.
INTRODUCTION
Rickettsia spp. (belonging to the Alphaproteobacteria, Rickettsiales, Rickettsiaceae) are Gram-negative, aerobic, coccobacilli, with all described members having an obligate intracellular lifestyle. Phylogenetic analyses have classified Rickettsia spp. into four groups: ancestral group (AG), typhus group (TG), transitional group (TRG), and spotted fever group (SFG) rickettsiae, with additional studies suggesting even further classification within the Rickettsia tree (28). Rickettsia typhi, a species of TG rickettsiae and the etiological agent of murine typhus, is internalized into eukaryotic host cells via induced phagocytosis. The bacteria rapidly escape from the phagosome and multiply within the host cell cytoplasm (62).
Compared to free-living bacterial species, rickettsiae have reductive genomes (∼1.3 Mb), a consequence of adapting to an intracellular niche (4, 30). Relative to other Rickettsia spp., TG rickettsiae have the smallest genomes (1.1 Mb), encoding few mobile genetic elements and the most reduced cellular and metabolic pathways (30). The genome sequence of R. typhi revealed many genes encoding hypothetical proteins (HP) that are conserved in Rickettsia spp. but lack homologs in other bacterial genomes (39). These genes, some of which encode domains typical of eukaryotic proteins, are important candidates for advancing the knowledge of the mechanisms underlying R. typhi pathogenesis and intracellular colonization by rickettsiae, as well as for identifying potential therapeutic targets against rickettsial infection.
R. typhi has been reported to contain a gene (RT0218) encoding a putative ankyrin (ANK) repeat-containing protein (Ank; 586 amino acids [aa]), here named RARP-1 (Rickettsia ankyrin-repeat protein 1) (39). The predicted RARP-1 is conserved in all sequenced Rickettsia genomes. The ankyrin repeat domain, a motif found commonly in a wide range of eukaryotic protein families, has been identified in proteins encoded within many prokaryotic and viral genomes (2, 57). The ANK repeat (∼33 amino acids) is a motif that folds into a helix-loop-helix structure with antiparallel α-helices followed by an outer membrane β-hairpin/loop region (41). The Pfam database (23) indicates that the number of ANK repeats per protein varies from 1 to 34; however, the majority of proteins contain fewer than 6 repeats (40). Bacterial Anks are typically involved in mediating protein-protein or protein-DNA interactions in host cells for a wide variety of functions, i.e., transcription regulation, signal transduction, inflammatory response, and protein transport mechanisms (2, 25, 35, 68). In contrast to other repeat motifs involved in protein-protein interactions, ANK repeats have not been shown to recognize any signature DNA or amino acid sequence or any specific molecular structure (35). Anks are encoded within the genomes of various obligate and facultative intracellular bacteria including Rickettsia spp. (2), Anaplasma phagocytophilum (25, 54), Ehrlichia chaffeensis (61, 68), Orientia tsutsugamushi (12), Legionella pneumophila (47), Coxiella burnetii (58), and Wolbachia pipientis (63). Some of these intracellular pathogens deliver Anks into eukaryotic cells via a type IV secretion system (T4SS), with these “effectors” functionally mimicking or interfering with host cell processes to aid in the infection process (1, 36, 44, 54). A recent report has identified the role of T1SS for extracellular secretion of E. chaffeensis Ank200, also suggesting that the T1SS is functional across Rickettsiales (59).
Rickettsia genomes encode various secretion systems, including the Sec translocon, elements of the twin-arginine translocation pathway, the Rickettsiales vir homolog T4SS, the autotransporter or type V secretion system (T5SS), and the type I secretion system (T1SS) (30). Our previous work on R. typhi protein secretion has focused mainly on the characterization of the Sec pathway (49–51) and the rvh T4SS (26, 27). Little is known about the function of the rickettsial T1SS, despite the presence of genes encoding the T1SS components within all sequenced Rickettsia genomes. The T1SS consists of three major protein components: an inner membrane (IM) ATP binding cassette transporter (ABC), a periplasmic membrane fusion protein (MFP), and a multifunctional outer membrane (OM) protein of the TolC family. The IM and periplasmic components are substrate specific; however, the OM TolC protein can couple with many different IM/periplasmic proteins to transport a variety of molecules. The complete channel exports virulence-associated proteins, antibiotics, detergents, colicin V, and organic solvents (67). The TolC protein plays an important role in the virulence of many Gram-negative bacterial pathogens (7, 11, 13, 22, 32, 46). Thus, TolC should not be overlooked for its possible role in protein secretion in rickettsiae.
In this study, we characterized RARP-1 and demonstrated its extracellular secretion into host cells. Additionally, we showed that RT0218 (encoding RARP-1) is cotranscribed with RT0216 (encoding an R. typhi TolC homolog) and present evidence for the role of TolC in RARP-1 secretion.
MATERIALS AND METHODS
Bacterial strains, host cell culture, and growth conditions.
R. typhi strain Wilmington (ATCC VR-144; Manassas, VA) was propagated in Vero76 (African green monkey kidney cells, ATCC CRL-1587) cells. Vero76 and HeLa (human epithelial cervical adenocarcinoma, ATCC CCL-2) cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 4.5 g of glucose/liter with glutamine (Biofluids Inc., MD) and 10% fetal bovine serum (FBS; Gemini, CA) at 34°C with 5% CO2. Escherichia coli C600 (wild type [WT]) and tolC mutant (C600 tolC::Tn5) strains used in heterologous secretion assays are derivatives of the E. coli K-12 strain and were kindly provided by Philippe Delepelaire from the Institute Pasteur, Paris, France. E. coli Top10, C600, and the tolC mutant strains were routinely grown in lysogeny broth (LB) medium at 37°C. The tolC mutant has a kanamycin resistance cassette (Kmr) inserted in the tolC gene. The tolC mutant strains containing the plasmid constructs were maintained with 50 μg/ml kanamycin and 100 μg/ml ampicillin. E. coli genomic DNA was extracted using a Wizard Genomic DNA Purification Kit (Promega, WI) according to the manufacturer's instructions.
R. typhi infection in Vero76 cells.
R. typhi cells were partially purified from infected Vero76 cells. The infected cells were harvested in 5 ml of DMEM supplemented with 5% FBS and disrupted by mild sonication for 15 s by using a sonic dismembrator (Fisher Scientific, PA). The disrupted host cells were centrifuged at 1,000 × g for 10 min at 4°C to remove host cell debris or any remaining intact host cells. The supernatant was filtered through a 5-μm-pore-size filtering unit (Millipore). The filtrate (0.5 ml) was used to infect confluent monolayers of Vero76 cells (2.1 × 106 cells), which were incubated at 37°C with 5% CO2 for 4 days. For whole-cell lysate preparation, the infected or uninfected cells were harvested and washed with ice-cold phosphate-buffered saline (PBS) supplemented with complete mini-EDTA-free protease inhibitor (Roche, IN). The host cells were resuspended in PBS supplemented with protease inhibitor and lysed by ultrasonication treatment of cells for 20 s on ice. For genomic DNA isolation, the partially purified rickettsiae were further centrifuged at 14,000 × g for 10 min at 4°C. Genomic DNA of R. typhi was extracted from the pellets by using either an All Prep DNA/RNA Mini Kit (Qiagen, CA) or a Wizard genomic DNA purification kit as previously described (51).
Isolation of RNA and RT-PCR.
Total RNA was isolated from R. typhi-infected Vero76 and HeLa cells (48 h postinfection) using TRIzol reagent (Invitrogen, CA) and treated with RQ1 RNase-free DNase (Promega). The DNase (Qiagen)-treated total RNA was then purified with an RNeasy MinElute Cleanup Kit (Qiagen). For the E. coli C600 strain harboring pTrc-RT0216-RT0217-RT0218, RNA was isolated using an All Prep DNA/RNA Mini Kit. Briefly, the pellet from isopropyl-β-d-thiogalactopyranoside (IPTG)-induced culture was resuspended in RLT Plus buffer containing 2-mercaptoethanol (Invitrogen), and RNA was isolated according to the manufacturer's recommendations. To remove any remaining contaminating DNA, RNA was treated with RNase-free DNase (Qiagen) as well as with Turbo DNase (Ambion, CA). Reverse transcription-PCR (RT-PCR) was performed with 200 ng of total RNA, using the primer pairs RT0216-F1 and RT0218-R1 for RT0216 transcript, RT0218 RT-F1 and RT0218 RT-R1 for RT0218, or RT0218 RT-F1 and RT0218 RT-R2 for RT0216-RT0217-RT0218, in 25-μl reaction volumes by using SuperScript III One-Step RT-PCR with Platinum Taq (Invitrogen). The thermal cycling conditions consisted of one cycle at 50°C for 1 h and at 94°C for 2 min, followed by 39 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min, with a final extension step of 72°C for 5 min. To confirm the absence of DNA in purified RNA samples, reverse transcriptase was omitted from the reaction mixture, and samples were checked by PCR using standard conditions.
Isolation of RNA and real-time qRT-PCR.
The titers of partially purified R. typhi from Vero76 cells were determined using a Live/Dead BacLight bacterial viability assay (Molecular Probes, OR), according to the manufacturer's instructions. Rickettsiae were counted with a hemacytometer using fluorescence microscopy (excitation, 480 nm; Nikon H550L). The Vero76 cells (∼1 × 106 cells) were infected with R. typhi at a multiplicity of infection (MOI) of ∼100 and incubated at 37°C with 5% CO2 for 15 min, 1 h, 4 h, 8 h, and 48 h. At each time point, culture medium was removed, and rickettsia-infected cells were washed with fresh growth medium. Total RNA was isolated from infected and uninfected cells using TRIzol reagent and treated with RQ1 RNase-free DNase as described previously (48). RT0218 expression profiles were determined by one-step real-time quantitative reverse transcription-PCR (qRT-PCR). qRT-PCR was performed with 200 ng of total RNA isolated from R. typhi-infected Vero76 cells using a Brilliant II SYBR green qRT-PCR Master Mix kit (Stratagene, CA) according to the manufacturer's instructions. The specific primers (listed in Table S1 in the supplemental material) for R. typhi genes used in qRT-PCR were AZ5194 and AZ5195 for RT0218 and AZ4923 and AZ4924 for RT0119 (rickettsial housekeeping gene, rpsL). PCR conditions were as follows: reverse transcription at 50°C for 30 min and then at 95°C for 10 min; 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 30 s; and a final cycle for generation of the dissociation curve. The qRT-PCR amplification and detection were performed on an MX3005P thermal cycler (Stratagene) as described previously (48). The expression data generated by real-time qRT-PCR were analyzed using the Q-Gene program with amplification efficiency correction (42). The amplification efficiency for each primer pair was calculated using the LinReg PCR software tool (53). To calculate the normalized expression of the target gene, efficiency-corrected threshold cycle (CT) values for RT0218 were normalized with CT values of RT0119 (rpsL).
Raising antibodies against RARP-1 peptide antigen.
For immunization purposes, the peptide sequence was chosen based on the predicted immunogenicity using Open Biosystems Antigen Profiler software (Open Biosystems, Rockford, IL). Rabbit polyclonal antibodies were generated against the peptide EKGQTKAIHKESQQIDPRE conjugated to keyhole limpet hemocyanin (KLH) corresponding to 76 to 94 amino acid residues of RARP-1. The antigen preparation, immunization, and purification were performed by Open Biosystems (Thermo Scientific, IL), according to the company's 70-day rabbit immunization protocol. For protein expression assays, the affinity-purified anti-RT0218 antibodies were preadsorbed with Vero76 cell lysate by incubation for 1 h at room temperature, and IgGs were purified using a MelonG IgG purification kit (Thermo Scientific).
Immunofluorescence microscopy.
R. typhi-infected Vero76 and HeLa cells were fixed with 4% paraformaldehyde (in PBS) at room temperature for 10 min, stored at 4°C overnight, and then washed three times with PBS before labeling. The blocking and permeabilization were performed using PBS containing 10% FBS for 30 min at room temperature and PBS containing 0.01% Triton X-100 for 5 min at room temperature, respectively. For double immunofluorescence labeling, cells were probed with rabbit anti-RT0218 antibodies and rat anti-R. typhi serum as primary antibodies for 1 h at room temperature and then with goat anti-rabbit Alexa Fluor 594-conjugated IgG and goat anti-rat Alexa Fluor 488-conjugated IgG (Molecular Probes) secondary antibodies (see Table S4 in the supplemental material). The cells were washed with PBS and mounted with Vectashield mounting medium containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories, CA). Samples were analyzed under a Nikon Eclipse E600 fluorescence microscope. Images were captured using QCapture Pro, version 5.1, imaging and analysis software and processed with Adobe Photoshop CS2, version 9.0.2.
Western blot analyses.
The samples were mixed with 2× Tris-glycine-SDS sample loading buffer (Invitrogen) containing 2-mercaptoethanol and boiled for 5 min at 95°C. The proteins were separated on Novex 4 to 20% Tris-glycine or Novex 4 to 12% Tris-glycine-polyacrylamide gels (Invitrogen) at 130 V using Tris-glycine-SDS running buffer (Bio-Rad) under denaturing conditions and transferred to a polyvinylidene difluoride (PVDF) membrane using an iBlot gel transfer system (Invitrogen). The membranes were blocked in StartingBlock (Tris-buffered saline [TBS]) blocking buffer (Thermo Scientific) containing Tween 20, pH 7.4 (TBST), for 30 min at room temperature and then incubated with primary antibody diluted in blocking buffer, either for 1 h at room temperature or overnight at 4°C. Following incubation with primary antibodies, the membranes were washed four times in TBST (5-min washes at room temperature) and then incubated with the appropriate secondary antibodies (conjugated to horseradish peroxidase) diluted in blocking buffer for 1 h at room temperature. The membranes were washed four times in TBST and developed using a SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Scientific). The membranes were then imaged using the G:BOX chemiluminescence imaging system and the associated GeneSnap acquisition software (Syngene). The antibodies used in this study are listed in Table S4 in the supplemental material.
RARP-1 translocation assay.
A confluent monolayer of Vero76 cells was infected with R. typhi (MOI of ∼100) for 48 h at 37°C with 5% CO2. The cells were washed once with PBS to remove extracellular bacteria, harvested by trypsinization, and washed again with ice-cold PBS supplemented with complete mini-EDTA-free protease inhibitor. The host cell cytoplasmic and nuclear proteins were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific), according to the manufacturer's instructions. The remaining pellet (containing rickettsiae) was collected after extraction of cytoplasmic and nuclear protein and resuspended in PBS containing protease inhibitor. The rickettsiae in the pellet were lysed by ultrasonication treatment of cells for 20 s on ice. The uninfected Vero76 cells were also processed as described above. The protein fractions were separated on 4 to 20% Tris-glycine precast gels (Invitrogen) and probed by Western blot analysis as described above. The membranes were probed with rabbit polyclonal anti-RT0218 peptide antibodies or with antibodies against rickettsial cytoplasmic elongation factor (EF) thermostable protein (rabbit anti-EF-Ts antibodies) (48). The mouse monoclonal antibodies against glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Abcam, MA) were used as a host cell cytoplasmic protein loading control.
Cloning and secretion assay of RARP-1 in E. coli.
A 1,760-bp gene fragment (full-length RT0218) encoding RARP-1 (586 amino acids) was amplified from the genomic DNA of R. typhi by PCR using the primer pair RT0218-SS F and RT0218-R2. For cloning RT0218 derivatives, PCR was performed using the primer pair RT0218-F1 and RT0218-R2 for amplification of RT0218 without the N-terminal signal peptide or with RT0218_SS F and RT0218-R3 for amplification of RT0218 without the C-terminal ANK domain. The 3,672-bp fragment, spanning the three genes RT0216-RT0217-RT0218, was amplified from R. typhi genomic DNA by PCR using the primers RT0216-F2 and RT0218-R2. The primers are listed in Table S1 in the supplemental material. The amplicons were ligated into pTrcHis2-TOPO vector under the control of the trc promoter (Invitrogen), and recombinant plasmids were chemically transformed into either the WT C600 or C600 ΔtolC strain. The sequences of all the fragments were confirmed by sequencing. The sequences and deduced amino acid sequences were analyzed using MacVector, version 7.1.1 software (Genetics Computer Group, Inc. Madison, WI).
For recombinant protein expression, bacteria were grown overnight at 37°C in LB medium containing appropriate antibiotics. The following day, the culture was diluted in fresh LB medium and grown at 37°C to mid-log phase (optical density at 600 nm [OD600] of ∼0.3 to 0.5). Protein expression was induced by the addition of 1 mM isopropyl-beta-d-thiogalactopyranoside (IPTG) at 16°C with incubation overnight. The culture was collected the following day and centrifuged at 10,000 × g for 20 min to separate secreted supernatant proteins from the bacterial cell-containing pellet fraction. The culture supernatants were filtered using a 0.22-μm-pore-size filter (Millipore, MA), and proteins were precipitated to concentrate the supernatant (100- to 150-fold) using prechilled 20% trichloroacetic acid in acetone for 1 h at 4°C. The precipitated proteins were collected by centrifugation at 13,000 × g for 15 min at 4°C. The pellets were washed three times in ice-cold acetone and air dried for 10 min. The pellets containing bacterial cells or precipitated supernatant proteins were resuspended in PBS (containing complete mini-protease inhibitor cocktail) with 2× Tris-glycine-SDS sample loading buffer, and amounts equivalent to 1 OD600 unit were separated on Tris-glycine-SDS gels and immunoblotted with RT0218-specific antibodies.
Recombinant protein purification.
The E. coli cells expressing recombinant protein were harvested by centrifugation at 8,000 × g and suspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0). The cells were incubated with lysozyme (1 mg/ml) for 20 min on ice and then lysed by a French press cell disrupter (Thermo Scientific). The cell lysate was cleared by centrifugation at 50,000 × g for 10 min at 4°C. Recombinant proteins were purified by affinity chromatography under native conditions using nickel-nitrilotriacetic acid resin (Ni-NTA) spin or superflow columns (Qiagen) according to the manufacturer's instructions. Briefly, 1 ml of Ni-NTA resin was added to 3 ml of the cleared cell lysate containing soluble proteins and incubated at 4°C for 2 h. The column was packed and washed three times with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) to remove nonspecifically bound proteins. The fusion protein was eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). Purified protein was dialyzed using Slide-A-Lyzer dialysis cassettes (Thermo Scientific) and concentrated with Amicon Ultra Centrifugal Filter Devices (Millipore).
SDS sensitivity assays.
The growth of E. coli cells was examined in the presence of SDS. In the first assay, sensitivity to SDS was checked on solid medium, and single colonies were streaked onto LB agar plates supplemented with antibiotics, IPTG (1 mM), and SDS at various concentrations (0, 0.1, and 1%); growth of colonies was recorded after overnight incubation at 37°C. In the second assay, growth was tested using liquid cultures. Cells were grown to mid-log phase at 37°C, and then the cultures were diluted with LB medium supplemented with IPTG (1 mM) and SDS at various concentrations and incubated overnight at 16°C. The cultures were grown for 24 h before the OD600 was determined.
Bioinformatics analysis.
Gene and protein sequences for R. typhi (i) RT0216, (ii) RT0217, and (iii) RT0218 were obtained from the genome sequence at NCBI (NC_006142). Homologs to these sequences were compiled from BLASTP searches against the Rickettsia database (taxonomy identifier [taxid] 780), as well as from conserved protein families compiled at the Pathosystems Resource Integration Center (PATRIC) website (29). The operon structures of these genes across 18 Rickettsia genomes were analyzed using the Compare Region Viewer tool at PATRIC. For each protein family (TolC, PopZ homolog, and RARP-1), the presence of a signal sequence and cellular localization were evaluated by using SignalP (45), and SecretomeP, version 2.0 (8), respectively. All gene and protein alignments were performed using MUSCLE, version 3.6 (21), with default parameters.
(i) RT0216.
The R. typhi TolC homolog was compared to Escherichia coli TolC proteins in two ways. First, a sequence alignment was constructed with mapping of the secondary structure over the comparison. Second, R. typhi TolC was threaded to the structure of E. coli TolC (Protein Data Bank [PDB] code 1ek9C). Using RT0216 as a query, the E. coli structure (best match using PSI-BLAST [3]) was retrieved in a template search using SWISS-MODEL (37). The SWISS-MODEL DeepView program was used to visualize the structural modeling.
(ii) RT0217.
Non-Rickettsia homologs of the R. typhi RT0217 sequence were searched for in the Rickettsiales (taxid 766) and Alphaproteobacteria (taxid 28211) databases at NCBI. Homologs found in other Rickettsiales genomes (all major genera except Neorickettsia) were compiled and analyzed as described above. Putative RT0217 homologs within 122 alphaproteobacterial genomes were also compiled and analyzed as described above, with sequence logos generated for the conserved N- and C-terminal regions using WebLogo (15).
(iii) RT0218.
BLASTP searches against the nonredundant protein sequences at NCBI revealed that an RARP-1 protein homolog is present in all 18 sequenced Rickettsia genomes. HHrepID, version 2.16.1 (9), was used to detect repeat regions between the predicted signal sequence and the C-terminal ANK domain. The three ANK repeats at the C terminus were identified by PSI-BLAST matches to ANK domains from other bacterial and eukaryotic proteins, with structural features within each repeat determined using a conserved ANK model (40).
MS.
Purified proteins and supernatant proteins from E. coli were separated on 4 to 12% SDS gels and visualized with Coomassie R250-based Imperial Protein Stain (Thermo Scientific). A protein band of ∼80 kDa in the purified-protein fraction from C600 harboring pTrc-RT0218 and the tolC mutant strain harboring pTrc-RT0216-RT0217-RT0218 and a similarly sized band in the supernatant of C600/pTrc-RT0218 and cell lysate as well as the supernatant of C600/pTrc-lacZ were excised and treated by in-gel digestion with trypsin and analyzed by mass spectrometry (MS) protein analyses at the protein core facility at the Center for Vascular and Inflammatory Diseases (University of Maryland, Baltimore, MD). The Coomassie-stained bands were washed; disulfide bonds were reduced and alkylated. Proteins were digested with methylated porcine trypsin overnight at 37°C. Peptides were extracted and desalted with Empore C18 discs, dried, and dissolved in 0.1% formic acid. Peptides were loaded onto a 0.150-mm by 250-mm capillary column packed with TARGA C18 (5-μm particle size; Nest Group, Soughborough, MA) and eluted with a gradient from 0 to 30% acetonitrile over 50 min directly into a Captive Spray ion source (Bruker-Michrom, Auburn, CA) which was fitted to the inlet of an LCQ (liquid chromatography quadrupole) Advantage ion trap mass spectrometer (Thermo Scientific). Data were acquired in the data-dependent scan mode consisting of successive rounds where the three most abundant ions were detected, selected for fragmentation, and scanned out. These ions were then excluded from further analysis for 60 s to allow for analysis of less abundant ions in subsequent scan cycles. For database searches, the raw data files were first converted to the mzXML file format and processed using the Trans-Proteomic Pipeline (17). The search engine Tandem (14) was used to search a protein sequence database consisting of both E. coli K-12 (UniprotKB) and R. typhi Wilmington (NCBI).
RESULTS
RT0218 bioinformatics analysis.
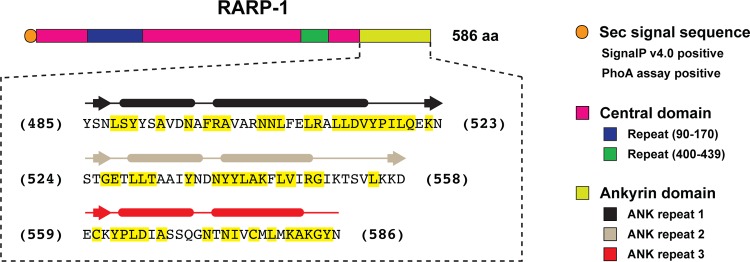
In silico analysis of the genome sequence of R. typhi strain Wilmington revealed a gene (RT0218, 1,760 bp), encoding an HP (586 aa; GenBank accession number YP_067182.1) with an N-terminal signal peptide, a central domain of unknown function, and a C-terminal ANK repeat domain (Fig. 1). The encoded protein, RARP-1, was predicted to contain three ANK repeat regions (∼33 residue repeats) (see Fig. S1 in the supplemental material for a detailed description). SignalP, version 4.0 (45), predicted a signal peptidase cleavage site between Ala23 and Ser24. Alignment of RARP-1 with homologs encoded in 17 other Rickettsia genomes (see Table S2 in the supplemental material) indicates that the protein sequence is conserved, with amino acid identity ranging from 79% (Rickettsia prowazekii strains) to 39% (Rickettsia bellii strain RML369-C). Importantly, the majority of sequence divergence is localized within the central domain, with strong sequence conservation in the ANK repeat domain, with amino acid identity ranging from 89% (R. prowazekii) to 74% (R. bellii strain RML369-C) (see Fig. S2).
Fig 1.
Domain organization of R. typhi RARP-1. The N-terminal signal peptide (orange; residues 1 to 23) was predicted with SignalP, version 4.0 (45). The large central domain (magenta; residues 24 to 484) contains two repeat regions (blue and green), predicted using HHrepID, version 2.16.1 (9), with no significant matches to any previously defined conserved domain. The three ankyrin (ANK) repeats at the C terminus (light green; residues 485 to 586) were identified by PSI-BLAST (3) matches to ANK domains from other bacterial and eukaryotic proteins, with structural features within each repeat determined using a conserved ANK model (40). Residues within ANK repeats highlighted in yellow depict invariant residues in RARP-1 homologs encoded within 17 other Rickettsia genomes (see Fig. S1 in the supplemental material for more details).
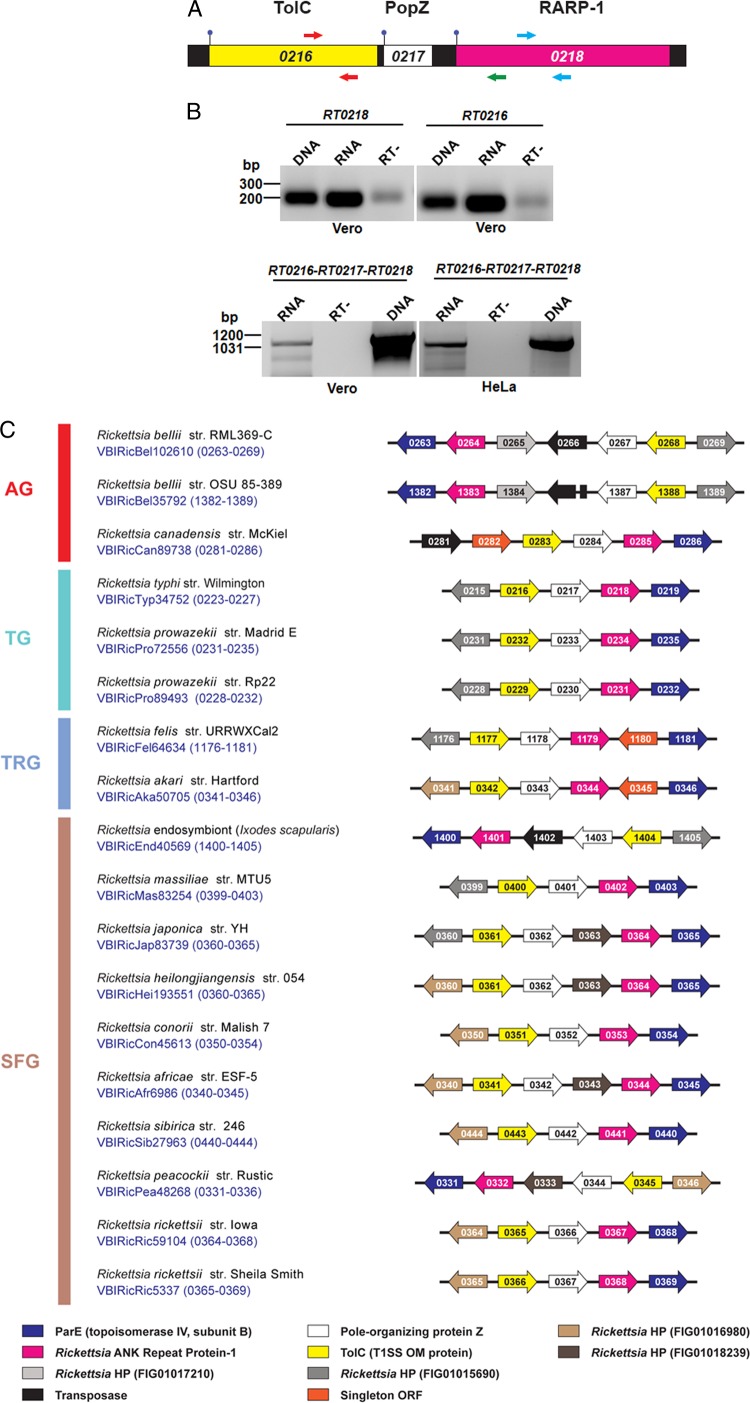
RT0218 is cotranscribed with RT0216 and RT0217 as a single transcript.
The gene locus of RT0218 (RARP-1) is adjacent to RT0217 (small HP) and RT0216 (TolC homolog) on the R. typhi chromosome (Fig. 2A). To examine whether RT0218 is cotranscribed with RT0216 and RT0217, RT-PCR was performed using total RNA isolated from Vero76 and HeLa cells infected with R. typhi (48 h postinfection). The specific primers designed to amplify the fragment across all three contiguous genes yielded a product of the expected size (1,069 bp) (Fig. 2B). We confirmed the transcription of RT0218 and RT0216 individually in Vero76 cells (Fig. 2B) as well as in HeLa cells (data not shown). The nucleotide sequence of the RT-PCR amplicon indicating the cotranscription of genes was confirmed to be identical to the R. typhi genome sequence spanning a specific region across all three genes. Thus, we confirmed that these three genes are cotranscribed by R. typhi as a polycistronic mRNA during infection in both Vero76 and HeLa cells. The homologs of these three genes are also clustered in other Rickettsia genomes, with minor differences due to genes encoding transposases and HP inserted into the operon (Fig. 2C), suggesting a conserved function for these genes in rickettsial biology.
Fig 2.
Cotranscription of RT0216-RT0217-RT0218. (A) Structure of three genes, RT0216-RT0217-RT0218, in the operon. Genes encoding TolC (genome coordinates 278261 to 279725), HP RT0217 (279757 to 280140) (see Fig. S3 in the supplemental material), and RARP-1 (280273 to 282033) are diagrammed at the top according to open reading frame lengths. DNA schema depicts coding sequences for RT0216 (yellow), RT0217 (white), and RT0218 (magenta) and intergenic regions (black). Arrows depict RT0216-specific (red) and RT0218-specific (blue) primers, with an additional reverse primer (green) used in conjunction with the RT0216 forward primer to amplify a fragment across all three genes. The ball-and-stick symbol indicates the predicted start codon for each open reading frame. (B) RT-PCR analysis of total RNA isolated from R. typhi-infected Vero76 and HeLa cells at 48 h postinfection using primers designed for targeted genes (as shown in panel A). The anticipated size (1,069 bp) of the RT0216-RT0217-RT0218 amplicon was identified. Reverse transcriptase was omitted (RT−) in one of the reaction mixtures to confirm the absence of DNA. As a positive control, PCR was performed with R. typhi DNA using Taq polymerase. The molecular mass standards (in kDa) are shown on the left. (C) RT0216-RT0217-RT0218 map across Rickettsia genomes. Gene structure within each genome was drawn with reference to 18 genomes displayed in the Compare Region Viewer tool at PATRIC (29). Locus tags refer to the PATRIC annotation, which can be translated to NCBI Refseq numbers using the ID Mapping tool at PATRIC. Genes pointing right are carried by the forward strand; genes pointing left are carried by the reverse strand. Colors for all genes are described at bottom. At right, the genomes are classified into four rickettsial groups based on phylogenomic analysis (30), with rickettsial groups as follows: red, ancestral group (AG); aquamarine, typhus group (TG); blue, transitional group (TRG); brown, spotted fever group (SFG). The Refseq protein numbers (RT0215 to RT0219) are shown for the R. typhi operon for consistency. str, strain.
Bioinformatics analysis of RT0216 and RT0217.
RT0216, which is comprised of 1,364 nucleotides, encodes a protein (TolC) of 454 aa (GenBank accession number YP_067180.1). R. typhi TolC contains a predicted N-terminal signal peptide sequence with the cleavage site located between residues Ala19 and Val20 (45). Multiple sequence alignment of R. typhi TolC with homologs encoded within 17 other Rickettsia genomes (see Table S3 in the supplemental material) revealed strong conservation, with amino acid identity ranging from 95% (R. prowazekii strains) to 78% (R. belli strains) (see Fig. S2 in the supplemental material). This indicates a highly conserved function for TolC across members of the genus. The R. typhi TolC amino acid sequence displays approximately 23% identity and 43% similarity to the E. coli TolC homolog. Based on in silico analyses, R. typhi TolC is predicted to contain three domains: a β-barrel domain that inserts into the OM, a 100-Å-long α-helical barrel domain that protrudes into the periplasm, and an α/β domain (equatorial domain) that forms the middle section of the α-helical barrel (see Fig. S3). The R. typhi TolC contains all the key amino acid residues that are critical for preserving the structural integrity of the TolC channel. In particular, the conserved Asp residues at positions 370 and 373 in R. typhi TolC, which correspond to Asp371 and Asp374 of E. coli TolC (see Fig. S3), form a ring structure along with the two aspartate residues of other monomers that comprises the tapered periplasmic entrance of the channel, which functions as a regulatory gate (6). The overall conservation of sequence, predicted structure, and essential amino acids suggests that TolC is functionally conserved in R. typhi.
Comparative sequence analysis of RT0217 (413 bp) with other rickettsial homologs indicated that this small encoded HP (137 aa; GenBank accession number AAU03699.1) is conserved across Rickettsia spp., with amino acid identity ranging from 91% (R. prowazekii strains) to 75% (R. bellii strains) (see Fig. S2 in the supplemental material). A search against the NCBI Rickettsiales database revealed a divergent homolog encoded within the genomes of all major Rickettsiales genera (Midichloria, Orientia, Wolbachia, Anaplasma, and Ehrlichia) except Neorickettsia. Despite extensive divergence in sequence length and composition, with proteins raging from 102 aa (“Candidatus Midichloria mitochondrii strain IricVA”) to 242 aa (Ehrlichia canis strain Jake), two conserved regions were identified at the N and C termini (see Fig. S4 in the supplemental material). In BLASTP searches, RT0217 scored best to COG3827 (E-value, 4.22e−46), an uncharacterized protein conserved in most Alphaproteobacteria. Another profile also scored well (E-value, 6.51e−21), pfam10691, which includes a domain of unknown function (DUF2497) also specific to the Alphaproteobacteria. Inspection of literature related to all of the proteins within COG3827 and pfam10691 revealed one characterized protein, pole-organizing protein Z (PopZ) of Caulobacter crescents (24.9% identity and 42.2% similarity to RT0217), which was demonstrated to be critical for cell division and chromosomal segregation (20).
Expression of RT0218 during eukaryotic host cell infection.
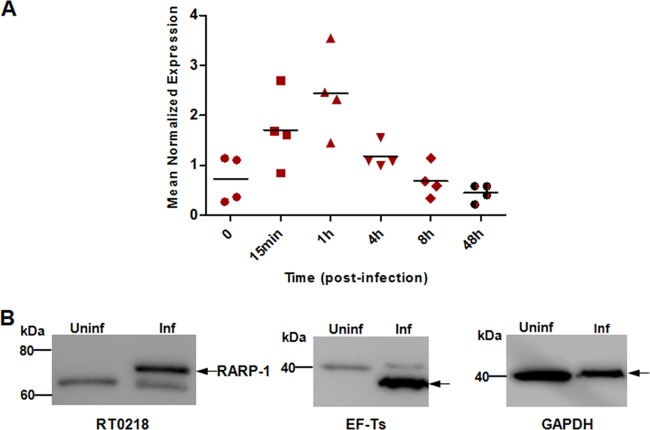
To examine how RT0218 expression is modulated at various stages of R. typhi growth in Vero76 cells, quantitative real-time RT-PCR was performed using total RNA purified from Vero76 cells at different time points postinfection (0 to 48 h). Our data show that RT0218 was transcribed throughout the infection cycle and that expression was upregulated at around 1 h postinfection (Fig. 3A), suggesting that this gene may play a role during early infection of host cells.
Fig 3.
RT0218 transcriptional profile and protein expression during infection. (A) Vero76 cells were infected with R. typhi, and total RNA was isolated at the indicated time points. Real-time quantitative RT-PCR was performed using total RNA for each time point, and RT0218 mean normalized expression was calculated relative to the rickettsial housekeeping gene (rpsl) transcript abundance. The dots represent data from four independent experiments. (B) Cell lysate was prepared from R. typhi-infected ([Inf] MOI of ∼100) or uninfected (Uninf) Vero76 cells, and proteins were electrophoresed by SDS-PAGE. The membranes were probed either with RT0218-specific antibodies preadsorbed with Vero76 cell lysate or with anti-EF-Ts antibodies to detect rickettsial housekeeping cytoplasmic EF-Ts protein (expected molecular mass of ∼34 kDa). The antibody against the host cell cytoplasmic GAPDH protein (∼37 kDa) was used as the host cell protein loading control. The arrows on the right indicate the expected positions of the proteins.
In order to determine if RARP-1 was expressed during R. typhi infection of mammalian cells, Vero76 cells were infected with R. typhi, and the expression of endogenous RARP-1 was assessed by Western blot analysis using polyclonal antibodies generated against the peptide antigen of RARP-1, preadsorbed with Vero76 cell lysate. A unique band was observed in the infected cell lysate corresponding to the predictive molecular mass of RARP-1 (∼66 kDa) (Fig. 3B), indicating the expression of RARP-1 during the infection in Vero76 cells. Another band (above 60 kDa) appeared in both uninfected and infected cell lysates; however, the intensity of this band was decreased upon preadsorption of anti-RT0218 antibodies with Vero76 cell lysate (data not shown), suggesting that this band is present due to nonspecific interaction of the RT0218 antiserum with an unknown host cell protein. The antibodies against rickettsial EF-Ts (∼34 kDa) indicated the high levels of R. typhi in infected Vero76 cells.
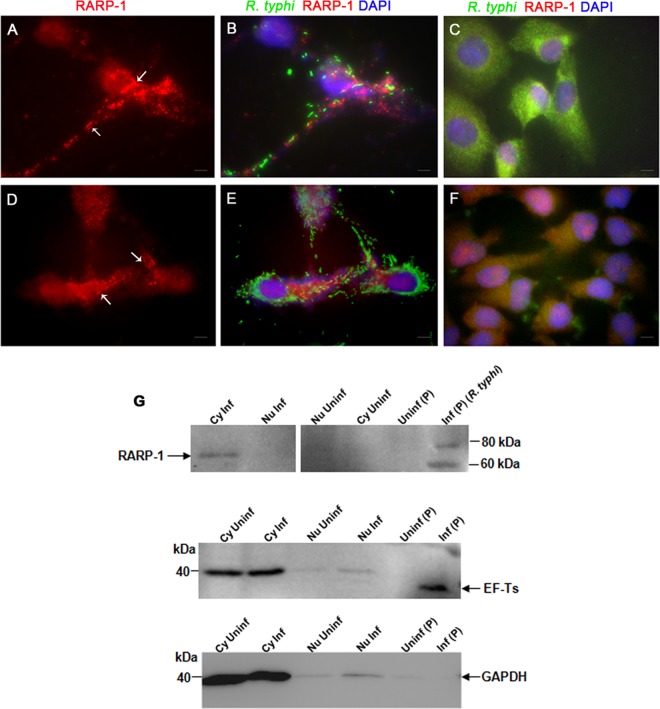
R. typhi secretes RARP-1 into host cells.
Secreted bacterial Anks are known to localize to distinct compartments of the eukaryotic cells in order to manipulate host cell processes (44, 52, 58). In order to find out if RARP-1 is a secreted effector and to obtain clues about the function of RARP-1, we analyzed the subcellular localization of RARP-1 in host cells during R. typhi infection using immunofluorescence and immunoblotting assays. Vero76 and HeLa cells were infected with R. typhi, followed by immunolabeling with anti-RT0218 antibodies and anti-R. typhi antibodies. RARP-1 was observed throughout the host cell cytoplasm as specific punctate forms in both Vero76 (Fig. 4A and B) and HeLa (Fig. 4D and E) cells. The uninfected Vero76 and HeLa cells showed the absence of immunostaining with RT0218-specific antibodies (Fig. 4C and F). To further confirm the cytoplasmic localization of RARP-1 in host cells, we isolated cytoplasmic and nuclear proteins from Vero76 cells at 48 h postinfection. Immunoblotting of nuclear and cytoplasmic proteins from uninfected and infected cells with anti-RT0218 antibodies showed a band above 60 kDa in the cytoplasmic fraction of infected cells. In contrast, this band was absent in the nuclear protein fraction, as well as in uninfected subcellular fractions, indicating that RARP-1 was translocated into host cells and subsequently localized to the host cell cytoplasm (Fig. 4G). Noticeably, the RARP-1 band identified in the cytoplasmic fraction was slightly smaller than the band detected in the pellet fraction containing rickettsial whole-cell lysate (Fig. 4G), which suggests that an unknown processing of RARP-1 due to posttranslational modifications may have occurred during the secretion process. EF-Ts was not detected in the infected cytoplasmic protein fraction, indicating that rickettsiae remained intact during this assay. Taken together, these data indicate that RARP-1 is secreted from the bacteria into the host cell cytoplasm during infection, where it may interact with a host cytoplasmic protein(s) to manipulate its function.
Fig 4.
Translocation of RARP-1 into host cells. Vero76 cells (A and B) and HeLa cells (D and E) were fixed with 4% paraformaldehyde at 24 h postinfection and immunolabeled using anti-RT0218 antibodies and anti-R. typhi rat serum as primary antibodies. The anti-rat-Alexa Fluor-488 (green) and anti-rabbit-Alexa Fluor-594 (red) were used as secondary antibodies. The cell nuclei are stained with DAPI (blue). The immunolabeled uninfected Vero76 and HeLa cells are shown in panels C and F, respectively. The white arrows show punctate structures indicating RARP-1 in the host cell cytoplasm. Scale bar, 5 μm. (G) Immunoblotting of the subcellular protein fractions (cytoplasmic [Cy], nuclear [Nu], and pellet [P]) of infected (MOI of ∼100) or uninfected Vero76 cells using anti-RT0218 antibodies. Controls included a blot probed with anti-EF-Ts (rickettsial housekeeping cytoplasmic protein) antibodies or monoclonal anti-GAPDH antibodies. The arrow indicates the secreted RARP-1 band with a molecular mass slightly smaller than the band detected in the pellet fraction containing rickettsial cell lysate, due to either nonspecific degradation or proteolytic processing of the protein. The faint band of EF-Ts (∼34 kDa) in the nuclear protein fraction indicates a minor carryover from the pellet fraction containing rickettsiae. The presence of GAPDH (∼36 kDa) in the nuclear fractions may be due to the presence of residual cytoplasmic proteins.
RARP-1 is secreted via outer membrane protein TolC.
The data presented above showed the secretion of RARP-1 into the host cell cytoplasm; however, the mechanism of secretion remained unknown. The conserved synteny of RT0218 and RT0216 across Rickettsia genomes and their cotranscription as a polycistronic message in R. typhi suggested a functional linkage of RARP-1 and TolC. Our previous work demonstrated that the RT0216 transcript was expressed during R. typhi infection of HeLa cells, with its corresponding N-terminal putative signal peptide sequence mediating TolC translocation into the periplasm, as demonstrated by an E. coli PhoA-based gene fusion assay (5).
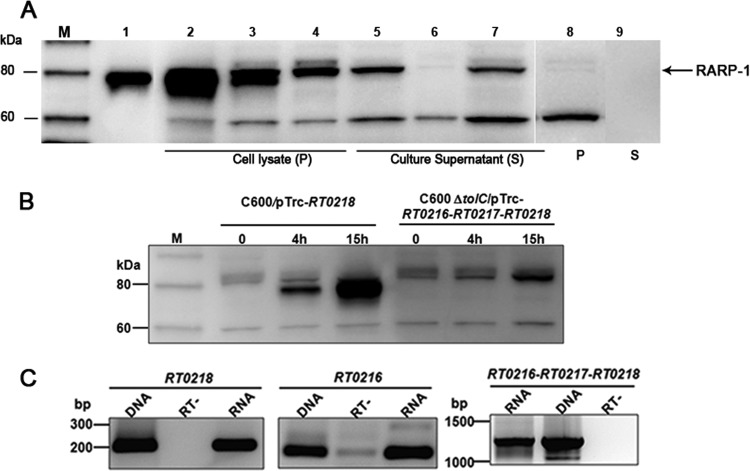
Therefore, we investigated whether RARP-1 is secreted by the bacteria into the extracellular milieu via a TolC-dependent secretion channel. Based on the sequence and predicted structural similarity in TolC homologs of R. typhi and E. coli (see Fig. S3 in the supplemental material), we used E. coli as a heterologous system for secretion assays. Full-length RT0218 cloned into pTrcHis2-TOPO vector was transformed into WT E. coli C600 and an isogenic tolC insertion mutant strain. Western blot analysis using polyclonal antibodies against RT0218 revealed that full-length RARP-1 was expressed by both the WT C600 and the tolC mutant strains harboring pTrc-RT0218, as shown by a unique band around ∼80 kDa in the cellular lysates (Fig. 5A, lanes 2 and 3, respectively). The presence of a second RARP-1 band below ∼80 kDa could be due to either nonspecific degradation or posttranslational modification of the protein. The analysis of the supernatant proteins of these strains showed a band of ∼80 kDa in the supernatant of the WT C600 strain, corresponding to RARP-1 (Fig. 5A, lane 5), whereas this band was absent in the culture supernatant of the tolC mutant strain (Fig. 5A, lane 6), suggesting the need of a functional TolC for secretion of RARP-1.
Fig 5.
RARP-1 is secreted into extracellular milieu by E. coli in a TolC-dependent manner, and R. typhi TolC restores RARP-1 secretion in tolC deficient E. coli strain. (A) Wild-type E. coli C600 and an isogenic tolC mutant harboring the indicated plasmids were grown to mid-log phase, and protein expression was induced by the addition of IPTG to a final concentration of 1 mM at 16°C. Following centrifugation of culture aliquots, proteins in the supernatants were precipitated with trichloroacetic acid. The total cell lysate equivalent to approximately 1.0 OD600 unit and concentrated (100-fold) culture supernatant equivalent to 2 ml of culture supernatant prior to concentration were loaded on each lane and subjected to SDS-PAGE. RARP-1 was detected by Western blotting using anti-RT0218 antibodies. The recombinant RARP-1 with a C-terminal His6 tag was purified from the total cell lysate of C600/pTrc-RT0218 using Ni-NTA agarose and used as a positive control. Lane 1, recombinant purified RARP-1; lane 2, cell lysate of C600/pTrc-RT0218; lane 3, cell lysate of C600 ΔtolC/pTrc-RT0218; lane 4, cell lysate of C600 ΔtolC/pTrc-RT0216-RT0217-RT0218; lane 5, supernatant of C600/pTrc-RT0218; lane 6, supernatant of C600 ΔtolC/pTrc-RT0218; lane 7, supernatant of C600 ΔtolC/pTrc-RT0216-RT0217-RT0218; lane 8, cell lysate of C600/pTrc-lacZ (for control plasmid); lane 9, supernatant of C600/pTrc-lacZ. Results are representative of three independent experiments. The arrow indicates the expected position of RARP-1. The molecular mass markers (lane M) are shown in kDa on the left. (B) C600/pTrc-RT0218 or C600 ΔtolC/pTrc-RT0216-RT0217-RT0218 was grown to mid-log phase and induced by the addition of IPTG for expression of a full-length RARP-1 fusion protein with His6 tag at the C terminus. The bacterial cell lysates were prepared by collecting culture at indicated time points, and the protein was detected by Western blotting using anti-RT0218 antibodies. The molecular mass markers (M) are shown in kDa on the left. (C) Total RNA was isolated from the tolC-complemented strain after protein expression was induced overnight at 16°C. One-step RT-PCR was performed using primers designed for targeted genes (as described in the legend of Fig. 2A). Controls included the PCR analysis of the plasmid DNA isolated from the complemented strain and a no-RT reaction. The anticipated sizes of the RT0216, RT0218, and RT0216-RT0217-RT0218 amplicons were identified, indicating the expression of RT0218 and RT0216 individually and the cotranscription of all three genes of the operon.
To confirm that the immunogenic ∼80-kDa band is RARP-1, the purified recombinant protein isolated from the cell lysate of WT C600 harboring pTrc-RT0218 (see Fig. S5 in the supplemental material) and the corresponding band in the crude culture supernatant were identified as RARP-1 by MS analyses (see Table S5). The corresponding regions of the gel in the cell lysate and concentrated culture supernatant of E. coli harboring pTrc-lacZ (negative control) (Fig. 5A, lanes 8 and 9, respectively) were also analyzed by MS and showed no peptides matching RARP-1. These data indicate that RARP-1 is secreted specifically into the culture supernatant in a TolC-dependent manner.
In spite of the observation that the tolC mutant carrying pTrc-RT0218 showed the same growth as the WT C600 harboring pTrc-RT0218, the expression of RARP-1 by the tolC mutant was relatively less than that of the WT strain, likely due to the pleiotropic effects of the tolC mutation (18). The immunogenic RARP-1 band of ∼80 kDa present in the supernatant from the WT strain was not detected in the supernatant from the tolC mutant strain expressing full-length RARP-1 when equal amounts of supernatant proteins from the WT and tolC mutant strains were loaded (i.e., 3-fold more supernatant proteins from the tolC mutant strain than WT supernatant proteins) on SDS gels and visualized by immunoblotting (data not shown). This suggests that RARP-1 occurrence in the culture supernatant is not due to cellular lysis.
R. typhi TolC restores RARP-1 secretion in an E. coli tolC mutant.
In order to examine if R. typhi TolC is a functional homolog of E. coli TolC and is involved in RARP-1 secretion, we performed R. typhi tolC complementation in an E. coli tolC mutant and examined if it could restore the RARP-1 secretion phenotype. To monitor RARP-1 secretion by the complemented strain, we made a construct (pTrc-RT0216-RT0217-RT0218) containing the three genes of the conserved operon and transformed it into the tolC mutant strain instead of inserting RT0218 and RT0216 separately into the expression vector. An E. coli tolC mutant harboring pTrc-RT0216-RT0217-RT0218 (tolC-complemented strain) synthesized the recombinant RARP-1, as shown by an immunogenic band of ∼80 kDa in the cellular lysates (Fig. 5A, lane 4). The RARP-1 band was detected in the culture supernatant of the tolC-complemented strain, with the protein secretion pattern similar to that of the WT C600 expressing RARP-1 (Fig. 5A, lane 7). In contrast, the secreted RARP-1 was noticeably absent in the culture supernatant of the tolC mutant harboring pTrc-RT0218 (Fig. 5A, lane 6). These data suggest that R. typhi TolC functionally complemented the tolC mutant strain and thus restored the secretion of RARP-1.
The tolC-complemented strain showed the absence of the second RARP-1 band below ∼80 kDa, suggesting that the protein expressed by the complemented strain is more stable than the protein expressed by the WT and the tolC mutant harboring pTrc-RT0218 (Fig. 5B). To confirm the identity of the recombinant RARP-1 from the tolC-complemented strain, the protein was purified from the cell lysate using Ni-NTA agarose (see Fig. S5 in the supplemental material), and the ∼80-kDa band was identified as RARP-1 by MS analysis (see Table S5). We also confirmed the transcription of all three genes in the complemented strain by RT-PCR (Fig. 5C).
To determine if R. typhi TolC restores the SDS resistance phenotype in the E. coli tolC mutant strain, we checked the growth of the tolC-complemented strain in the presence of SDS in either LB agar or liquid medium. As expected, WT E. coli harboring pTrc-RT0218 showed resistance to SDS, and the tolC mutant harboring pTrc-RT0218 was unable to grow in the presence of SDS. However, the tolC-complemented strain was sensitive to SDS under the growth conditions tested (data not shown).
Deletion of the N-terminal signal peptide or the C-terminal ANK domain abolished secretion of RARP-1.
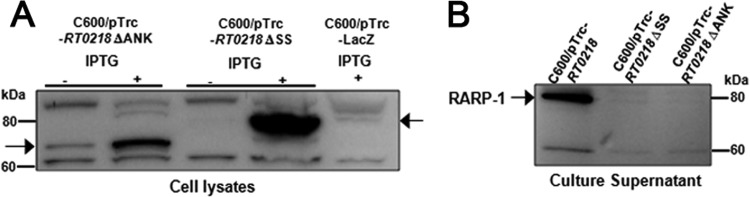
Previous work from our laboratory demonstrated that the RARP-1 signal peptide sequence fused to an E. coli alkaline phosphatase protein (PhoA), lacking its own N-terminal signal peptide, targets the protein into the periplasm, suggesting that RARP-1 is possibly secreted across the IM to the periplasm through the Sec translocon during the infection process (5). The presence of essential components of the Sec translocon, including the type 1 (LepB) and type 2 (LspA) signal peptidases and a SecA homolog, indicates the functionally active Sec translocon in R. typhi (49–51). To analyze the role of the signal peptide sequence in the RARP-1 secretion process, the RT0218 derivative without the N-terminal signal peptide sequence (ΔSS) was cloned into pTrcHis2 Topo vector and transformed into the C600 strain (C600/pTrc-RT0218 ΔSS). Western immunoblot analysis with RT0218-specific antibodies identified a band below ∼80 kDa in the cell lysate, indicating the expression of recombinant RARP-1 lacking its signal peptide (RARP-1 ΔSS; expected molecular mass, ∼67 kDa) (Fig. 6A), whereas the corresponding immunogenic band was not detected in the culture supernatant of this strain (Fig. 6B). These data indicate that RARP-1 ΔSS was not secreted by WT E. coli, thus arresting the protein in the bacterial cytosol, which suggests that the protein is translocated via the Sec-dependent pathway. To confirm that the lack of the protein secretion by this strain is not due to the formation of inclusion bodies in E. coli, pTrc-RT0218 ΔSS was transformed in E. coli Top10 cells, and soluble protein was purified from cleared cell lysates under native conditions using Ni-NTA-agarose (see Fig. S5 in the supplemental material).
Fig 6.
RARP-1 is not secreted in the absence of the N-terminal signal peptide or the C-terminal ankyrin repeat. The WT E. coli C600 strain harboring pTrc-RT0218 ΔSS or pTrc-RT0218 ΔANK was grown to mid-log phase and induced with IPTG for recombinant protein expression. (A) The whole-cell lysate of uninduced and induced cultures equivalent to an OD600 of 1.0 was loaded in each lane and subjected to SDS-PAGE for subsequent analysis by immunoblotting using anti-RT0218 antibodies. The cell lysate of WT E. coli containing pTrc-lacZ was used as a negative control. (B) The culture supernatants were precipitated with trichloroacetic acid, and a concentrated supernatant sample equivalent to 1.5 ml of culture medium was loaded in each lane and analyzed for the presence of RARP-1 by immunoblot analysis using anti-RT0218 antibodies. The supernatant from the C600 strain carrying pTrc-RT0218 was used as a positive control. Numbers indicate molecular masses (in kDa) of the ladder. The arrows indicate the expected positions of the proteins.
Typically, the signal sequence of T1SS protein substrates is located at the C terminus (27 to 100 aa) and interacts with an IM ABC transporter to trigger the assembly of the functional export complex (10). Therefore, we tested the role of the C-terminal ANK repeats of RARP-1 in the secretion process. The WT C600 transformed with a plasmid containing RT0218 without the C-terminal ANK domain (C600/pTrc-RT0218 ΔSS) showed a band above 60 kDa in the cellular lysate (expected molecular mass, ∼58 kDa), indicating the expression of RARP-1 without its ANK domain (RARP-1 ΔANK) (Fig. 6A). In contrast, the immunogenic protein band was not detected in the culture supernatant of this strain (Fig. 6B), suggesting that the ANK domain is required for RARP-1 secretion. Collectively, these data indicate that deletion of either the N-terminal signal peptide or the C-terminal ANK domain abolished the secretion of RARP-1.
DISCUSSION
Anks of many intracellular pathogens are translocated into host cells and localize to distinct subcellular compartments in order to subvert host cell processes (44, 52, 54, 68). The presence of a conserved RT0218 gene encoding RARP-1 within all sequenced Rickettsia genomes prompted us to characterize this protein. The molecular mechanisms involved in protein secretion across the rickettsial IM and OM remain an important area of rickettsial research. In this study, we focused on RARP-1 and an OM protein, TolC, located within the same operon on the R. typhi chromosome. First, we showed that RARP-1 was secreted into eukaryotic host cells during the R. typhi infection process. Second, our data suggest that RARP-1 is secreted by the bacterium into the extracellular milieu in a TolC-dependent manner, and that requires its N-terminal signal peptide and the C-terminal ANK domain. Since stable genetic manipulation including insertion, deletion, and complementation of genes has not yet been feasible in R. typhi, we utilized an E. coli heterologous system (C600 and an isogenic tolC mutant strain) to monitor the expression and secretion of RARP-1.
The RT0218 transcript was expressed throughout the infection cycle within eukaryotic host cells, with elevated expression at early time points of infection, suggesting the role of RT0218 during early stages of the rickettsial life cycle. RARP-1 was detected in Vero76 cells infected with R. typhi, indicating that the protein is expressed during the infection process. Fluorescence microscopy analyses of antibody-stained RARP-1 in infected Vero76 and HeLa cells, as well as immunoblotting of subcellular proteins isolated from R. typhi-infected Vero76 cells, show that RARP-1 is secreted into host cells and localizes to the host cell cytoplasm. The possible role of Anks in pathogenesis of intracellular bacteria has been previously reported (1, 2). L. pneumophila AnkX, a secreted dot/icm T4SS effector, localized to the host cell cytoplasm and prevented the maturation of the Legionella-containing phagosome during infection in macrophages by interfering with the microtubule-dependent vesicular transport (44). AnkA of A. phagocytophilum was shown to be translocated via the rvh T4SS into the host cell cytoplasm and subsequently to the nucleus, where it binds with the CYBB (cytochrome b beta chain) locus promoter by forming complexes with AT-rich DNA sequences to downregulate the transcription of CYBB and other host defense genes (25). While the functions of RARP-1 and its interacting host factors are unknown, our data suggest that RARP-1 may have a role in interacting with host cytoplasmic molecules for manipulating the host cell processes to enhance the survival and growth of rickettsiae in eukaryotic host cells.
The cotranscription of RT0218 (RARP-1) with adjacent genes RT0217 (HP) and RT0216 (TolC) as a single polycistronic mRNA during R. typhi infection in eukaryotic host cells suggests a possible functional linkage of these genes. Bioinformatics analysis shows that RT0216 encodes a TolC homolog with extensive structural similarity to E. coli TolC. Based on these observations, we hypothesized that RARP-1 is secreted by the bacteria into the extracellular milieu in a TolC-dependent manner. Using E. coli as a heterologous system, we demonstrated that RARP-1 was secreted by WT E. coli (C600 strain) into the culture medium, whereas the isogenic tolC mutant was unable to secrete RARP-1, suggesting the role of TolC in the transport of RARP-1 into the extracellular milieu. However, the secretion by WT E. coli was not so efficient, as most of the protein remained inside the cells, suggesting the limited secretion of the protein by the heterologous transporter. The identified immunogenic protein band was confirmed as RARP-1 by MS analyses. The predicted molecular mass of recombinant RARP-1 is 70 kDa; however, our data indicated that RARP-1 electrophoretically migrated on SDS-PAGE with an apparent mass of ∼80 kDa, possibly due to the acidic nature of the protein (pI 5.3), as it is not unusual that acidic proteins migrate with altered masses (60).
Previous studies have shown the interchangeability of the T1SS genes between various organisms (19, 56). Another study on the twin-arginine translocation (Tat) pathway of Anaplasma marginale also shows the ability of its component genes to retain function in cross-species complementation using E. coli (43). RT0216 transcript expression during R. typhi infection in HeLa cells suggests that its encoded protein may play a role in R. typhi growth and pathogenesis (5). Interestingly, when R. typhi TolC was expressed along with RARP-1 in the E. coli tolC mutant strain, RARP-1 was detected in the culture supernatant, suggesting that R. typhi tolC complemented the tolC mutation in E. coli and restored the RARP-1 secretion phenotype, similar to WT E. coli. Noticeably, the tolC-complemented strain showed the presence of a single RARP-1 band in the cell lysate compared to two RARP-1 bands observed in the tolC mutant strain carrying RT0218 solely. This is likely due to increased stability of the protein in the presence of native R. typhi TolC. We corroborated the expression of RARP-1 by MS analysis and of the RT0216, RT0217, and RT0218 genes by RT-PCR in the tolC-complemented E. coli strain.
In E. coli, TolC interacts both with the AcrA/AcrB or HlyB/HlyD complex to constitute an efflux pump that expels detergents and drugs (e.g., SDS and acriflavine) or proteins/toxins (e.g., hemolysin A) from the cell cytoplasm, respectively (32, 33). However, the R. typhi TolC did not restore resistance to SDS in the tolC-complemented strain, probably due to either the specificity of the E. coli TolC protein for this substrate or the inability of R. typhi TolC to function with the AcrA/AcrB efflux complex. Taken together, these data show that R. typhi TolC restored the RARP-1 secretion in the tolC-complemented strain; however, it failed to grow in SDS-containing medium, indicating that it partially complemented the function of TolC in E. coli.
Typically, T1SS protein substrates lack a cleavable N-terminal signal peptide, with proteins secreted directly from the bacterial cytoplasm to the extracellular medium. In Gram-negative bacteria, N-terminal signal peptide-containing proteins are formed as inactive preproteins in the cytoplasm that are targeted to the IM SecYEG complex, where signal peptides are cleaved by signal peptidases during protein translocation to the periplasm (55). A previous study from our laboratory has shown that the N-terminal signal peptide of RARP-1 fused to E. coli PhoA directed the protein to the periplasm, indicating that RARP-1 is a Sec-dependent extracytoplasmic protein (5). In this study, we further demonstrate that WT E. coli expressing RARP-1 ΔSS is unable to secrete the protein into culture supernatant, verifying that the N-terminal signal peptide is required for translocation of RARP-1 across the IM via the Sec-dependent pathway to further engage the TolC-dependent channel for secretion. Interestingly, a nonclassical mechanism of protein secretion involving both the Sec translocon and T1SS, wherein the secreted proteins in the periplasm are further secreted extracellularly in a TolC-dependent manner, has been previously reported for signal peptide-containing heat-stable enterotoxins STI and STII produced by enterotoxigenic E. coli (24, 65, 66). Additionally, in silico analysis of RARP-1 with SecretomeP, version 2.0 (8), also identified it to be a nonclassically secreted protein.
Although the secretion signal of T1SS substrates is not clearly understood, it has been shown that the proximal 100 aa residues of the C terminus determine their secretion (16, 34). However, another report has indicated that multiple regions throughout HasA, a hemophore secreted by Serratia marcescens via the T1SS, are involved in the assembly, stabilization, and function of the T1SS (38). It has been previously reported that the last 50 amino acid residues of T1SS substrates are usually rich in LDAVTSIF amino acids and contain very few Cys residues (16). For RARP-1, 48% of the last 50 aa residues consist of signature amino acid residues LDAVTSIF and contain only three Cys residues, suggesting that a T1SS signal could be present within the ANK repeat-containing domain of RARP-1. Our data indicate that deletion of the C-terminal ANK repeat region (102 aa) abolished secretion of RARP-1 into the culture supernatant. This suggests that the C-terminal region is essential for the export of RARP-1 across the OM through a TolC channel, functioning either through direct structural involvement of ANK repeats in the secretion process or by acting as a signal sequence for targeting RARP-1 to the TolC-dependent channel. Further studies are under way to identify the minimum region of the ANK domain required for RARP-1 secretion.
The molecular mechanisms of targeting the periplasmic RARP-1 to the TolC channel and subsequent secretion remain unknown. The structural similarity of R. typhi TolC with E. coli TolC suggests its possible involvement in secreting proteins as a tripartite transport system in combination with IM ATPase and MFP. In enterotoxigenic E. coli, MacA/MacB, an ABC transporter, and an MFP, have been reported to be essential for the translocation of STII from the periplasm to the cell exterior (64). In silico analyses indicated the presence of T1SS components in the R. typhi genome: AprD (RT0305, a putative alkaline proteinase secretion ATP-binding protein) and AprE (RT0304, a putative alkaline proteinase secretion protein) that have 21% and 24% identity to E. coli HlyB and HlyD, respectively, and 42.1% and 28.2% identity with Neorickettsia risticii AprD (NRI_0040, type I secretion system ATPase) and AprE (NRI_0899, secretion protein, HlyD family), respectively. Also, an ortholog of the E. coli multidrug transporter protein EmrB (RT0146) and two copies of the Bcr (RT0591 and RT0693) are annotated in the R. typhi genome (39). Alternatively, it is tempting to speculate that RARP-1 is secreted via the novel Sec-TolC pathway, analogous to the type Vb (autotransporter) pathway, where the accessory pore-forming β-barrel protein analogous to the OM structure of TolC is required for the protein secretion (31).
In conclusion, we propose a model for the secretion pathway of RARP-1 (Fig. 7). The precursor RARP-1 is synthesized in the cytoplasm and translocated across the IM via the Sec translocase, with the N-terminal signal peptide cleaved during this process. The periplasmic protein, potentially by interacting with unknown periplasmic and IM proteins of the T1SS, triggers the recruitment of TolC to assemble the functional secretion channel and is secreted through the TolC across the OM into the extracellular environment. Further research will identify the role of additional proteins involved in the secretion process, as well as the function of RARP-1 in host cells. As the RT0218 and RT0216 locations are conserved in the genome of Rickettsia spp., we speculate that RARP-1 orthologs from other rickettsial species may also be secreted through a TolC-dependent secretion system and that this pathway may be utilized for the secretion of potential virulence-associated proteins involved in the intracellular survival of rickettsiae.
Fig 7.
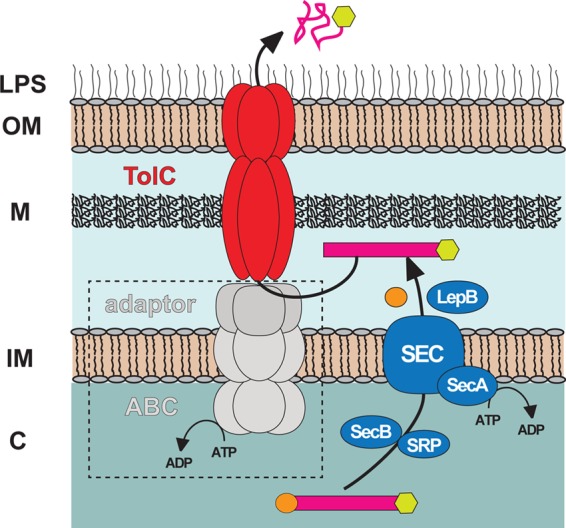
Schematic model depicting the proposed mechanism of RARP-1 secretion. The model proposed here shows the translocation of RARP-1 from the bacterial cytoplasm to the periplasm via the Sec translocon (blue), with subsequent engagement to the TolC channel (red) for secretion through the outer membrane to the cell exterior. In a canonical T1SS mechanism, TolC interacts with the IM ABC and periplasmic adaptor proteins (gray) to secrete substrates directly from the cytoplasm to the cell exterior. The role of these proteins (dashed box) in the secretion of RARP-1 is currently unknown. LPS, lipopolysaccharide; OM, outer membrane; M, murein layer; IM, inner membrane; C, cytoplasm.
Supplementary Material
ACKNOWLEDGMENTS
The project was supported by award numbers AI017828 and AI059118 from the National Institute of Allergy and Infectious Diseases (NIAID). J.J.G. acknowledges support from NIAID award HHSN272200900040C (to Bruno W. Sobral, Virginia Bioinformatics Institute, Virginia Tech).
We thank Philippe Delepalaire from the Institut Pasteur (Paris, France) for E. coli strains C600 and C600 tolC::Tn5. We thank Prarthana Vasudevan for technical help and Brian Hampton for help with mass spectrometry.
The content is solely the responsibility of the authors and does not necessarily represent official views of the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
Footnotes
Published ahead of print 6 July 2012
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Al-Khodor S, Price CT, Habyarimana F, Kalia A, Abu Kwaik Y. 2008. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70:908–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. 2010. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol. 18:132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ammerman NC, Gillespie JJ, Neuwald AF, Sobral BW, Azad AF. 2009. A typhus group-specific protease defies reductive evolution in rickettsiae. J. Bacteriol. 191:7609–7613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ammerman NC, Rahman MS, Azad AF. 2008. Characterization of Sec-translocon-dependent extracytoplasmic proteins of Rickettsia typhi. J. Bacteriol. 190:6234–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andersen C, Koronakis E, Hughes C, Koronakis V. 2002. An aspartate ring at the TolC tunnel entrance determines ion selectivity and presents a target for blocking by large cations. Mol. Microbiol. 44:1131–1139 [DOI] [PubMed] [Google Scholar]
- 7. Barabote RD, et al. 2003. Erwinia chrysanthemi tolC is involved in resistance to antimicrobial plant chemicals and is essential for phytopathogenesis. J. Bacteriol. 185:5772–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bendtsen JD, Kiemer L, Fausboll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol. 5:58 doi:10.1186/1471-2180-5-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Biegert A, Soding J. 2008. De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics 24:807–814 [DOI] [PubMed] [Google Scholar]
- 10. Binet R, Letoffe S, Ghigo JM, Delepelaire P, Wandersman C. 1997. Protein secretion by Gram-negative bacterial ABC exporters—a review. Gene 192:7–11 [DOI] [PubMed] [Google Scholar]
- 11. Buckley AM, et al. 2006. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell Microbiol. 8:847–856 [DOI] [PubMed] [Google Scholar]
- 12. Cho NH, et al. 2007. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc. Natl. Acad. Sci. U. S. A. 104:7981–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cosme AM, et al. 2008. The outer membrane protein TolC from Sinorhizobium meliloti affects protein secretion, polysaccharide biosynthesis, antimicrobial resistance, and symbiosis. Mol. Plant Microbe Interact. 21:947–957 [DOI] [PubMed] [Google Scholar]
- 14. Craig R, Beavis RC. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20:1466–1467 [DOI] [PubMed] [Google Scholar]
- 15. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Delepelaire P. 2004. Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694:149–161 [DOI] [PubMed] [Google Scholar]
- 17. Deutsch EW, et al. 2010. A guided tour of the trans-proteomic pipeline. Proteomics 10:1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dhamdhere G, Zgurskaya HI. 2010. Metabolic shutdown in Escherichia coli cells lacking the outer membrane channel TolC. Mol. Microbiol. 77:743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duong F, Lazdunski A, Murgier M. 1996. Protein secretion by heterologous bacterial ABC-transporters: the C terminus secretion signal of the secreted protein confers high recognition specificity. Mol. Microbiol. 21:459–470 [DOI] [PubMed] [Google Scholar]
- 20. Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134:956–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferhat M, et al. 2009. The TolC protein of Legionella pneumophila plays a major role in multi-drug resistance and the early steps of host invasion. PLoS One 4:e7732 doi:10.1371/journal.pone.0007732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finn RD, et al. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Foreman DT, Martinez Y, Coombs G, Torres A, Kupersztoch YM. 1995. TolC and DsbA are needed for the secretion of STB, a heat-stable enterotoxin of Escherichia coli. Mol. Microbiol. 18:237–245 [DOI] [PubMed] [Google Scholar]
- 25. Garcia-Garcia JC, Rennoll-Bankert KE, Pelly S, Milstone AM, Dumler JS. 2009. Silencing of host cell CYBB gene expression by the nuclear effector AnkA of the intracellular pathogen Anaplasma phagocytophilum. Infect. Immun. 77:2385–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gillespie JJ, et al. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4:e4833 doi:10.1371/journal.pone.0004833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gillespie JJ, et al. 2010. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect. Immun. 78:1809–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gillespie JJ, Nordberg EK, Azad AF, Sobral BW. Phylogeny and comparative genomics: the shifting landscape in the genomics era. In Palmer GH, Azad AF. (ed), Intracellular pathogens II: Rickettsiales, in press ASM Press, Washington, DC [Google Scholar]
- 29. Gillespie JJ, et al. 2011. PATRIC: the comprehensive bacterial bioinformatics resource with a focus on human-pathogenic species. Infect. Immun. 79:4286–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gillespie JJ, et al. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One 3:e2018 doi:10.1371/journal.pone.0002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koronakis V. 2003. TolC—the bacterial exit duct for proteins and drugs. FEBS Lett. 555:66–71 [DOI] [PubMed] [Google Scholar]
- 33. Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914–919 [DOI] [PubMed] [Google Scholar]
- 34. Letoffe S, Delepelaire P, Wandersman C. 1991. Cloning and expression in Escherichia coli of the Serratia marcescens metalloprotease gene: secretion of the protease from E. coli in the presence of the Erwinia chrysanthemi protease secretion functions. J. Bacteriol. 173:2160–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li J, Mahajan A, Tsai MD. 2006. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45:15168–15178 [DOI] [PubMed] [Google Scholar]
- 36. Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 9:2644–2657 [DOI] [PubMed] [Google Scholar]
- 37. Marchler-Bauer A, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Masi M, Wandersman C. 2010. Multiple signals direct the assembly and function of a type 1 secretion system. J. Bacteriol. 192:3861–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McLeod MP, et al. 2004. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J. Bacteriol. 186:5842–5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13:1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mosavi LK, Minor DL, Jr, Peng ZY. 2002. Consensus-derived structural determinants of the ankyrin repeat motif. Proc. Natl. Acad. Sci. U. S. A. 99:16029–16034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Muller PY, Janovjak H, Miserez AR, Dobbie Z. 2002. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32:1372–1374 1376:1378–1379 [PubMed] [Google Scholar]
- 43. Nunez PA, Soria M, Farber MD. 2012. The Twin-arginine translocation pathway in Alphaproteobacteria is functionally preserved irrespective of genomic and regulatory divergence. PLoS One 7:e33605 doi:10.1371/journal.pone.0033605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 46. Platz GJ, et al. 2010. A tolC mutant of Francisella tularensis is hypercytotoxic compared to the wild type and elicits increased proinflammatory responses from host cells. Infect. Immun. 78:1022–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Price CT, Al-Khodor S, Al-Quadan T, Abu Kwaik Y. 2010. Indispensable role for the eukaryotic-like ankyrin domains of the ankyrin B effector of Legionella pneumophila within macrophages and amoebae. Infect. Immun. 78:2079–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rahman MS, Ammerman NC, Sears KT, Ceraul SM, Azad AF. 2010. Functional characterization of a phospholipase A2 homolog from Rickettsia typhi. J. Bacteriol. 192:3294–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rahman MS, Ceraul SM, Dreher-Lesnick SM, Beier MS, Azad AF. 2007. The lspA gene, encoding the type II signal peptidase of Rickettsia typhi: transcriptional and functional analysis. J. Bacteriol. 189:336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rahman MS, Simser JA, Macaluso KR, Azad AF. 2005. Functional analysis of secA homologues from rickettsiae. Microbiology 151:589–596 [DOI] [PubMed] [Google Scholar]
- 51. Rahman MS, Simser JA, Macaluso KR, Azad AF. 2003. Molecular and functional analysis of the lepB gene, encoding a type I signal peptidase from Rickettsia rickettsii and Rickettsia typhi. J. Bacteriol. 185:4578–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ramabu SS, et al. 2011. Expression of Anaplasma marginale ankyrin repeat-containing proteins during infection of the mammalian host and tick vector. Infect. Immun. 79:2847–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62–66 [DOI] [PubMed] [Google Scholar]
- 54. Rikihisa Y, Lin M. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr. Opin. Microbiol. 13:59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saier MH., Jr 2006. Protein secretion and membrane insertion systems in gram-negative bacteria. J. Membr. Biol. 214:75–90 [DOI] [PubMed] [Google Scholar]
- 56. Scheu AK, et al. 1992. Secretion of the Rhizobium leguminosarum nodulation protein NodO by haemolysin-type systems. Mol. Microbiol. 6:231–238 [DOI] [PubMed] [Google Scholar]
- 57. Sonnberg S, Seet BT, Pawson T, Fleming SB, Mercer AA. 2008. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc. Natl. Acad. Sci. U. S. A. 105:10955–10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Voth DE, et al. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J. Bacteriol. 191:4232–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wakeel A, den Dulk-Ras A, Hooykaas PJJ, McBride JW. 2011. Ehrlichia chaffeensis tandem repeat proteins and Ank200 are type 1 secretion system substrates related to the repeats-in-toxin exoprotein family. Front. Cell. Infect. Microbiol. 1:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wakeel A, Zhang X, McBride JW. 2010. Mass spectrometric analysis of Ehrlichia chaffeensis tandem repeat proteins reveals evidence of phosphorylation and absence of glycosylation. PLoS One 5:e9552 doi:10.1371/journal.pone.0009552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wakeel A, Zhu B, Yu XJ, McBride JW. 2010. New insights into molecular Ehrlichia chaffeensis-host interactions. Microbes Infect. 12:337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Walker DH, Feng HM, Popov VL. 2001. Rickettsial phospholipase A2 as a pathogenic mechanism in a model of cell injury by typhus and spotted fever group rickettsiae. Am. J. Trop. Med. Hyg. 65:936–942 [DOI] [PubMed] [Google Scholar]
- 63. Walker T, et al. 2007. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol. 5:39 doi:10.1186/1741-7007-5-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yamanaka H, Kobayashi H, Takahashi E, Okamoto K. 2008. MacAB is involved in the secretion of Escherichia coli heat-stable enterotoxin II. J. Bacteriol. 190:7693–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yamanaka H, Nomura T, Fujii Y, Okamoto K. 1997. Extracellular secretion of Escherichia coli heat-stable enterotoxin I across the outer membrane. J. Bacteriol. 179:3383–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yamanaka H, Nomura T, Fujii Y, Okamoto K. 1998. Need for TolC, an Escherichia coli outer membrane protein, in the secretion of heat-stable enterotoxin I across the outer membrane. Microb. Pathog. 25:111–120 [DOI] [PubMed] [Google Scholar]
- 67. Zgurskaya HI, Krishnamoorthy G, Ntreh A, Lu S. 2011. Mechanism and function of the outer membrane channel TolC in multidrug resistance and physiology of enterobacteria. Front. Microbiol. 2:189 doi:10.3389/fmicb.2011.00189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhu B, et al. 2009. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect. Immun. 77:4243–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.