Abstract
We developed a method for restricted enzymatic proteolysis using the outer membrane protease T (OmpT) to produce large peptides (> 6.3 kDa on average) for mass spectrometry-based proteomics. Using this approach to analyze prefractionated high-mass HeLa proteins we identified 3,697 unique peptides from 1,038 proteins. We demonstrated the ability of large OmpT peptides to differentiate closely related protein isoforms and to enable the detection of many post-translational modifications.
The “bottom-up” and “top-down” approaches represent two strategies for proteomic studies using mass spectrometry. Bottom-up proteomics relies upon enzymatic protein digestions prior to on-line liquid chromatography-coupled tandem mass spectrometry analysis (LC-MS/MS)1. Top-down proteomics omits proteolysis and focuses on complete characterization of intact proteins and their post-translational modifications (PTMs)2. While both approaches continue to mature, they each have limitations. The tryptic peptides used in the bottom-up approach are the primary unit of measurement, but their relatively small size (typically ~8–25 residues long) leads to problems such as sample complexity, difficulties in assigning peptides to specific gene products rather than protein groups4 and loss of single and combinatorial PTM information. The top-down approach handles these issues by characterizing intact proteins, but becomes less successful in the high-mass region. Therefore, a hybrid approach based on 2–20 kDa peptides could marry positive aspects of both bottom-up and top-down proteomics.
We previously proposed a generic approach to “middle-down” proteomics to interrogate high-mass proteome with two essential features: a size-dependent protein fractionation technique and a robust but restricted proteolysis method5 (Fig. 1a). A continuous tube-gel electrophoresis technique can now provide the size-dependent fractionation of a complex proteome6. Previous efforts to explore restricted proteolysis options included using alternative enzymes to trypsin (for example, Lys-C7 and Lys-N8) and chemical methods (for example, microwave-assisted acid hydrolysis9). Nonetheless, these methods produced peptides only marginally longer than tryptic peptides in large-scale proteomic studies.
Figure 1.
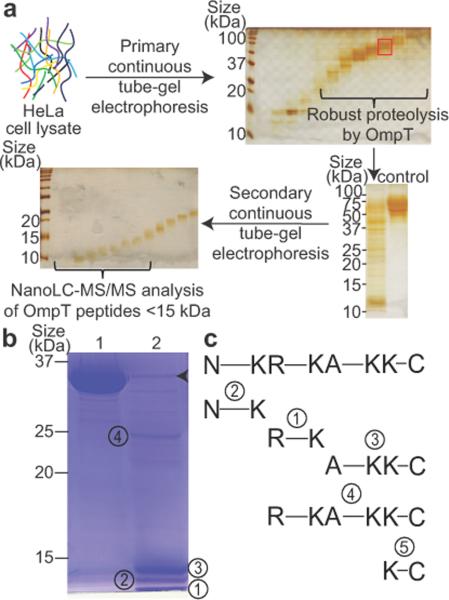
OmpT-based platform for middle-down proteomics and characterization of OmpT peptides from digestion of a standard protein. (a) The middle-down workflow was illustrated on proteins from a HeLa cell lysate sorted into narrow size ranges by molecular-weight based pre-fractionation (see silver stained gel, top row). A representative OmpT digestion of a fraction containing 50–75 kDa proteins (highlighted in the red box) was visualized by silver staining (left lane, bottom right) along with the control sample with no digestion (right lane). The digested samples were separated further and fractions below ~15 kDa were subjected to nanoLC-MS/MS analysis. (b) Peptide products from digestion of glyceraldehyde 3-phosphate dehydrogenase (GAPDH, 36 kDa) by OmpT were visualized on a Coomassie stained SDS-PAGE gel. Lane 1, GAPDH incubated without OmpT. Lane 2, GAPDH after OmpT digestion. Major peptide products are numbered from 1 through 4. Arrowhead indicates the intact OmpT enzyme. (c) Alignment of identified OmpT peptides by nanoLC-MS/MS with the original GAPDH sequence on top. Peptide cleavage sites are illustrated and N and C represent the protein N and C termini.
Here we present the protease OmpT to achieve a robust, yet restricted, proteolysis of a complex proteome (with its benefits described in Supplementary Fig. 1). OmpT is known to cleave between two consecutive basic amino acid residues (K/R–K/R) and is reported to have favorable kinetics with its kcat/Km in the 104 – 108 s−1M−1 range. Derived from the Escherichia coli K12 outer membrane, OmpT belongs to the novel omptin protease family10. In this study, we developed OmpT into an efficient reagent to generate > 2 kDa peptides for middle-down proteomics.
We first performed in silico digestions of the human proteome using various enzymatic or chemical approaches to create histograms of their predicted peptide masses (Supplementary Fig. 2). Most traditional enzymatic approaches generated predominantly small peptides (< 2 kDa). OmpT, which cleaves between less common dibasic sites, produced a distribution with a greater number of peptides > 3 kDa.
We overexpressed and refolded OmpT to obtain active enzyme (see Online Methods), then optimized its digestion of four standard protein substrates. Optimal conditions for OmpT digestion were at pH 6.0 in 2–3 M urea at 22°C (Supplementary Fig. 3). We used urea to reduce the higher-order structure present in large protein substrates, an important step for the cleavage efficiency of OmpT11. Characterization of digestion products from the 36 kDa GAPDH standard is shown as an example (Fig. 1b–c and Supplementary Fig. 4). In addition to the predicted dibasic cleavages, we also observed a K-A cleavage, corroborating previous reports that OmpT can still cleave with aliphatic amino acids in its P1' position12, especially under strongly denaturing conditions13. Although the GAPDH sequence contains a K-K site (Supplementary Fig. 3g), the cleaved product at this site (peptide 5 in Fig. 1c) was barely observable upon LC-MS/MS analysis (data not shown). This is likely because the flanking amino acids are two aspartic acids whose negative charges may prevent the binding of the nearby K-K site to the negatively charged OmpT active site14. We also characterized three other proteins digested with OmpT in detail (Supplementary Fig. 3b,d–f,h).
We established an OmpT-based middle-down platform to analyze complex mixtures pre-sorted by protein size (Fig. 1a). Integrating data from the middle-down workflow applied to ~20–100 kDa proteins fractionated from the HeLa cell proteome, we identified 3,697 unique peptides (average size: 6.3 kDa) from 1,038 unique proteins (26% average sequence coverage at an estimated 1% false discovery rate (FDR)2 (Supplementary Table 1). Two database search modes were used: biomarker and absolute mass (explained further below and in Online Methods). Both the forward and decoy databases for biomarker and absolute mass searches are available online using ProSightPTM 2.0 (http://prosightptm2.northwestern.edu). ProSightPC users can also download these databases using the link (ftp://prosightftp:gsX1gON@prosightpc.northwestern.edu/) and run both search modes locally. Results from an individual LC-MS/MS analysis of fractionated OmpT peptides from the middle-down workflow are provided as an example (Supplementary Fig. 5). We also performed a negative control treatment of substrate proteins in the absence of OmpT, which showed no sample auto-degradation (data not shown).
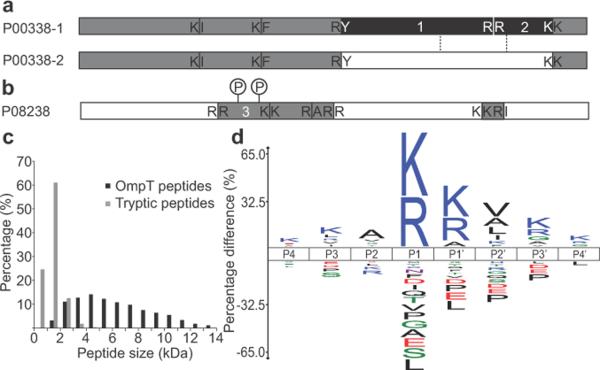
Proteotypic OmpT peptides can allow differentiation of specific protein isoforms. Detailed sequence alignments between protein isoforms revealed high sequence identity, while OmpT peptides, owing to their desirably large size, covered unique regions where isoform sequences differed (Fig. 2a and Supplementary Fig. 6a–b). Long peptides can also prove beneficial for detection and identification of modified peptides. In this study, ~25% of OmpT peptides were identified with PTMs (using annotated modifications from the UniProt database2) and several examples of multiply modified peptides were found (Fig. 2b and Supplementary Fig. 6c–e). An additional 8% of unique peptides with unexpected mass discrepancies were confidently identified in error tolerant searching. Together, these data imply that the OmpT-based workflow can provide isoform-specific assignments, characterization of modified peptides and combinatorial PTM information complementary to traditional protease-based proteomic approaches.
Figure 2.
Proteotypic OmpT peptides, peptide size distribution and iceLogo of OmpT recognition site. (a) Peptides 1 and 2 (10.8 kDa and 5.4 kDa respectively) cover a proteotypic sequence region of 37 kDa L-lactate dehydrogenase A chain isoform 1 (Uniprot number: P00338-1, 87% identify to isoform 2). Cleavage sites for OmpT peptides are shown. The schematic isoform alignment (detailed sequence alignment in Supplementary Fig. 6a), marks the region where the two isoform sequences differ between dashed lines. Peptides covering the distinct part of a certain isoform are shaded in black; peptides covering the common regions of all isoforms are in grey. (b) 84 kDa heat shock protein HSP 90-beta (Uniprot accession number: P08238) identified by peptides in grey; phosphorylation sites in peptide 3 (8.9 kDa) are indicated (survey spectrum of the singly and doubly modified species in Supplementary Fig. 6c). (c) Mass distribution of identified OmpT peptides (below ~15 kDa) in comparison with tryptic peptides16. (d) IceLogo of OmpT recognition sequences from P4 through P4' sites. OmpT cleaves between P1 and P1'. The y axis displays the percentage difference of amino acid frequencies between the experimental set and the reference set at each position.
The OmpT peptide size distribution was plotted in comparison with tryptic peptides (Fig. 2c). Because we only analyzed fractions below ~15 kDa, the average size of peptides identified here was 6.3 kDa, but OmpT peptides above 15 kDa were readily visible on gels (Fig. 1a and Supplementary Fig. 5c). We compared the performance between collision induced dissociation (CID) and electron transfer dissociation (ETD) using OmpT peptides (Supplementary Fig. 7 and Supplementary Table 2). The low degree of overlap between the methods indicates that both CID and ETD can serve as highly complementary fragmentation approaches to identify and characterize OmpT peptides.
Although the substrate specificity of OmpT has been extensively studied, previous model substrates were mostly short peptides and unstructured protein linker regions12,13,15. This study helped to improve our understanding of OmpT's sequence preference under denaturing conditions (3 M urea) where whole proteins were the substrates. We searched the entire dataset in “biomarker” mode against an intact protein database. A biomarker search assumes no specific proteolytic cleavage, but rather queries every possible sub-sequence in the database within tolerance from an observed peptide mass. Confident biomarker peptide hits were then used to extract the P4 through P4' recognition sites of OmpT for the generation of an unbiased consensus sequence. From these data, we generated an iceLogo that normalizes observed amino acid frequencies at each site to a reference set of proteomic amino acid frequencies (Fig. 2d and Supplementary Fig. 8a–b). We also made a WebLogo for comparison, which illustrates amino acid frequencies at each site solely based on the input sequences without normalization (Supplementary Fig. 8c).
As shown in iceLogo and WebLogo representations, the P1 site was restricted almost exclusively to lysine and arginine, while the P1' site was more permissive, allowing predominantly lysine and arginine, but also alanine and serine. The relative promiscuity of OmpT at the P1' position may be attributed to the location of P1'-substrate binding site near the loops on top of the beta-barrel14, which could have increased flexibility under denaturing conditions. Because of OmpT's broader specificity at the P1' site, we defined the “major cleavage sites” as K/R–K/R/A/S and performed another in silico digestion of human proteome at all these major sites assuming 0 and 2 missed cleavages (Supplementary Fig. 2). The resultant peptide size distributions strongly resembled the distributions assuming only K/R–K/R cleavages.
In addition to selectivities at the P1 and P1' sites, the P2' site also had a slight preference for aliphatic amino acids. Overall, OmpT favored positively charged residues across its recognition sites (with the exception of P2) and resisted negatively charged and proline residues. Selectivities outside P1–P1' have been previously reported12,15 and might explain the average number of observed missed cleavages (0.99 ± 1.29) at the major sites. In spite of these preferences, OmpT is still a stringent protease with well-defined substrate specificities, which will be better understood with future experimentation and data mining.
The stable beta-barrel structure of this membrane endopeptidase endows it with a remarkable resistance to both denaturants and surfactants, allowing extensive denaturation of large protein substrates under strongly solubilizing conditions for robust proteolysis. The analysis of widely-distributed OmpT peptides across a broad mass range will necessitate adjustments in separation protocols and LC-MS/MS methods accordingly. New and next generation instruments will further increase the routine size range accessible for sequencing, and many proteomics search engines will require modification to identify these larger peptides. With a demonstrated capacity for robust and restricted proteolysis, OmpT is as an attractive option for mass spectrometry-based interrogation of protein primary structure.
Supplementary Material
ACKNOWLEDGEMENTS
We sincerely thank current and former members of the Kelleher and Sweedler research groups, especially B. Evans, P. Chu, P. Compton, A. Catherman, S. Sweet, I. Ntai, J. Lee, A. Vellaichamy and K. Catherman for technical support and insightful suggestions, P. Yau from the Protein Sciences Facility at the University of Illinois for the synthesis of the fluorogenic substrate, M. Burke for the access to a fluorimeter. The project described was supported by the National Institutes of Health through awards R01 GM067193, P30 DA018310 and F30 DA026672, by the National Science Foundation through award DMS 0800631, and by the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust.
Footnotes
AUTHOR CONTRIBUTIONS C.W. designed research, conducted experiments, analyzed data and wrote the paper; J.C.T., L. Z., K.R.D. and B.P.E. analyzed data; M.L., K.R.D. and D.R.A. conducted cell culture; P.M.T. analyzed data and wrote the paper; J.V.S interpreted data and wrote the paper; N.L.K designed research, interpreted data and wrote the paper.
References
- 1.de Godoy LM, et al. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 2.Tran JC, et al. Nature. 2011;480:254–258. doi: 10.1038/nature10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chait BT. Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 4.Nesvizhskii AI, Aebersold R. Mol. Cell. Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 5.Forbes AJ, Mazur MT, Patel HM, Walsh CT, Kelleher NL. Proteomics. 2001;1:927–933. doi: 10.1002/1615-9861(200108)1:8<927::AID-PROT927>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 6.Tran JC, Doucette AA. Anal. Chem. 2008;80:1568–1573. doi: 10.1021/ac702197w. [DOI] [PubMed] [Google Scholar]
- 7.Wu SL, Kim J, Hancock WS, Karger B. J. Proteome. Res. 2005;4:1155–1170. doi: 10.1021/pr050113n. [DOI] [PubMed] [Google Scholar]
- 8.Taouatas N, Drugan MM, Heck AJ, Mohammed S. Nat. Methods. 2008;5:405–407. doi: 10.1038/nmeth.1204. [DOI] [PubMed] [Google Scholar]
- 9.Cannon J, et al. J. Proteome. Res. 2010;9:3886–3890. doi: 10.1021/pr1000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramer RA, Zandwijken D, Egmond MR, Dekker N. Eur. J. Biochem. 2000;267:885–893. doi: 10.1046/j.1432-1327.2000.01073.x. [DOI] [PubMed] [Google Scholar]
- 11.White CB, Chen Q, Kenyon GL, Babbitt PC. J. Biol. Chem. 1995;270:12990–12994. doi: 10.1074/jbc.270.22.12990. [DOI] [PubMed] [Google Scholar]
- 12.Dekker N, Cox RC, Kramer RA, Egmond MR. Biochemistry. 2001;40:1694–1701. doi: 10.1021/bi0014195. [DOI] [PubMed] [Google Scholar]
- 13.Okuno K, et al. Biosci. Biotechnol. Biochem. 2002;66:127–134. doi: 10.1271/bbb.66.127. [DOI] [PubMed] [Google Scholar]
- 14.Vandeputte-Rutten L, et al. EMBO J. 2001;20:5033–5039. doi: 10.1093/emboj/20.18.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarter JD, et al. J. Bacteriol. 2004;186:5919–5925. doi: 10.1128/JB.186.17.5919-5925.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsen JV, et al. Mol. Cell. Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dekker N, Merck K, Tommassen J, Verheij HM. Eur. J. Biochem. 1995;232:214–219. doi: 10.1111/j.1432-1033.1995.tb20801.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee JE, et al. J. Am. Soc. Mass. Spectrom. 2009;20:2183–2191. doi: 10.1016/j.jasms.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran JC, Doucette AA. Anal. Chem. 2009;81:6201–6209. doi: 10.1021/ac900729r. [DOI] [PubMed] [Google Scholar]
- 20.Kramer RA, et al. Eur. J. Biochem. 2002;269:1746–1752. doi: 10.1046/j.1432-1327.2002.02820.x. [DOI] [PubMed] [Google Scholar]
- 21.Wessel D, Flugge UI. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 22.Elias JE, Gygi SP. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 23.Meng F, et al. Nat. Biotechnol. 2001;19:952–957. doi: 10.1038/nbt1001-952. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. J. R. Stat. Soc. Ser. B-Methodol. 1995;57:289–300. [Google Scholar]
- 25.Storey JD, Tibshirani R. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.