Abstract
Significance: In recent years, bacterial iron–sulfur cluster proteins that function as regulators of gene transcription have emerged as a major new group. In all cases, the cluster acts as a sensor of the environment and enables the organism to adapt to the prevailing conditions. This can range from mounting a response to oxidative or nitrosative stress to switching between anaerobic and aerobic respiratory pathways. The sensitivity of these ancient cofactors to small molecule reactive oxygen and nitrogen species, in particular, makes them ideally suited to function as sensors. Recent Advances: An important challenge is to obtain mechanistic and structural information about how these regulators function and, in particular, how the chemistry occurring at the cluster drives the subsequent regulatory response. For several regulators, including FNR, SoxR, NsrR, IscR, and Wbl proteins, major advances in understanding have been gained recently and these are reviewed here. Critical Issues: A common theme emerging from these studies is that the sensitivity and specificity of the cluster of each regulatory protein must be exquisitely controlled by the protein environment of the cluster. Future Directions: A major future challenge is to determine, for a range of regulators, the key factors for achieving control of sensitivity/specificity. Such information will lead, eventually, to a system understanding of stress response, which often involves more than one regulator. Antioxid. Redox Signal. 17, 1215–1231.
Introduction
Iron–sulfur clusters, which contain iron and inorganic sulfide, are among the most ancient of protein cofactors. Since their discovery in the 1960s (5, 13), it has become clear that they are extremely widespread in biology, exhibiting a remarkable breadth and flexibility in the roles they perform, from electron transfer and storage, to redox and nonredox chemical catalysis (12, 14, 83). Because clusters of different nuclearity and shape are able to interconvert, they can act as powerful drivers of protein conformational change. Here, we focus on their roles in transcriptional regulatory proteins, in which the cluster senses environmental change, leading directly to changes in DNA binding affinity.
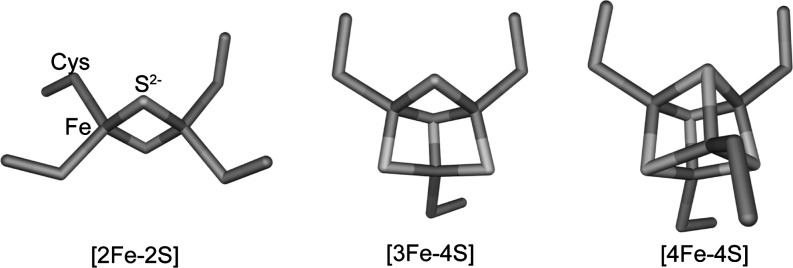
Iron–sulfur regulatory proteins contain clusters with either [2Fe-2S] or [4Fe-4S] cores (see Fig. 1). The [2Fe-2S] cluster consists of an [Fe2-(μ2-S)2] rhomb, with the protein amino acid residues that coordinate it constrained to lie in a plane perpendicular to the plane of the rhomb. The [4Fe-4S] cluster consists of two interpenetrating tetrahedra of iron and sulfide ions, forming a cube that is linked to the protein framework by four amino acid residues lying at the vertices of a tetrahedron. The [4Fe-4S] cluster may also be viewed as consisting of two Fe2-(μ2-S)2 rhombs: one on top of the other and at a 90° angle to each other. [3Fe-4S] clusters, derived from a [4Fe-4S] cluster by removal of one iron atom from one of the cube vertices, are found in biology, but they are not yet recognized in a stable form in a DNA regulatory protein. They have been found transiently, however, during cluster interconversion (see below). In all iron–sulfur clusters, each iron atom, present in either the +2 or +3 oxidation state, is tetrahedrally coordinated. Coordinating ligands may be provided by bridging sulfides, and by amino acid side-chains. These are usually cysteine thiolates (RS−), but other residues, such as histidine (N=), serine (R-O−), and aspartate (RCO2−), are sometimes found (see below). Since tetrahedrally coordinated iron(II) and (III) always adopt a high spin configuration, they have several unpaired electrons, and are therefore paramagnetic. However, paramagnetic exchange coupling within clusters can lead, in some cases, to a complete canceling of magnetic moments (known as antiferromagnetic coupling), resulting in an overall diamagnetic cluster (12). In cases where an odd number of total unpaired electrons are found, paramagnetism invariably arises, which is usually detectable using electron paramagnetic resonance (EPR), magnetic circular dichroism, or Mössbauer spectroscopy.
FIG. 1.
Iron–sulfur clusters common in nature. Structures of [2Fe-2S], [3Fe-4S], and [4Fe-4S] iron–sulfur clusters. Iron, sulfide, and cysteine residues are indicated.
Iron–sulfur clusters are coordination complexes containing ions that are redox active in physiological potential ranges and therefore can undergo redox reactions. Precisely which redox couple is involved (e.g., for [4Fe-4S]n+ clusters, redox couples where n=3+/2+ and 2+/1+ are commonly known) and at what potential is highly dependent on the protein environment of the cluster (12). The capacity to undergo redox chemistry underpins the functional importance of many clusters, but it also means that iron–sulfur clusters are susceptible to damage from reactions with redox-active species such as molecular oxygen (O2), superoxide ions, and hydrogen peroxide, leading, usually, to cluster conversion or even complete loss (70). Clusters may also be susceptible to damage resulting from reaction with strongly coordinating species. For example, it has been shown that a major cause of copper- and cobalt-mediated toxicity stems from the ability of these metals to disrupt iron–sulfur clusters of key metabolic enzymes, presumably as a result of their high affinities for sulfur ligands (94, 116). Nitric oxide (NO) also reacts with iron–sulfur clusters, forming various iron nitrosyl species. In such species, the iron is typically in a low oxidation state (formally +1), and so, in this case, reaction is coupled to redox chemistry (156).
This inherent reactivity makes iron–sulfur clusters very well suited to roles in sensing stress caused by reactive oxygen species (ROS) and reactive nitrogen species (RNS; see Table 1). While the cell usually strives to protect its complement of iron–sulfur clusters from this type of destructive reactant, the clusters of transcriptional regulators have evolved specifically to exploit such reactions, with sufficient sensitivity to and specificity for a particular analyte (e.g., O2 or NO). Although high-resolution structural information is generally lacking, regulatory cluster reactions, in most cases, must result in major structural changes, which have significant effects on interactions with DNA and thus provide a mechanism for transcriptional control.
Table 1.
Bacterial Iron–Sulfur Cluster Containing Regulatory Proteins That Are Currently Known
| Regulator | Cluster type | Function | References |
|---|---|---|---|
| FNR | [4Fe-4S] | Global regulator O2 sensor in a wide range of bacteria. Secondary function in NO sensing. | (26, 82) |
| NreB | [4Fe-4S] | O2 sensor in Staphylococci | (105) |
| ArnR | Unknown | O2 sensor in Corynebacterium | (107) |
| SoxR | [2Fe-2S] | Redox/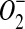 stress sensor stress sensor |
(59, 160) |
| NsrR | [2Fe-2S]/[4Fe-4S] | Global regulator of NO stress response | (85, 154) |
| Wbl | [4Fe-4S] | Key roles in cell developmental processes in Actinomycetes | (27, 139) |
| IscR | [2Fe-2S] | Sensor of cellular iron–sulfur cluster levels | (106, 128) |
| SufR | [4Fe-4S] | Sensor of cellular iron–sulfur cluster levels in Cyanobacteria | (132) |
| Bacterial aconitases | [4Fe-4S] | Autoregulation of aconitase and response to oxidative stress | (145) |
| RirA | Unknown | Iron regulator in alpha-proteobacteria | (151) |
| RsmA | [2Fe-2S] | SigM antisigma factor | (45) |
| ThnY | [2Fe-2S] | Regulation of pathway for utilization of tetralin as a carbon source | (41) |
NO, nitric oxide.
Although such regulators are still relatively few, they exhibit complex and fascinating chemistries, and are providing new insights into iron–sulfur cofactors. We will review these recent advances, paying particular attention to the nature of the cluster, and the molecular mechanisms by which they act as sensors.
Sensors of O2 and ROS
The remarkable ability of bacteria to thrive under a wide range of environmental conditions is a direct result of their metabolic flexibility, allowing them to adapt to the prevailing conditions. A prime example of this is the ability to respire both in the presence and in the absence of O2. While O2 is the preferred terminal electron acceptor for respiration (owing to the energetic advantages this conveys) in facultative anaerobic bacteria, when O2 is low, other compounds, such as nitrate and fumarate, can be used (155). This metabolic flexibility is underpinned by the ability to sense O2 and to reconfigure the cellular proteome accordingly. However, along with the energetic advantages of aerobic respiration come the dangers associated with the incomplete reduction of O2, which result in the generation of ROS, such as superoxide ion, hydrogen peroxide, and the hydroxyl radical. It is essential that cells can respond to and minimize the toxicity associated with ROS.
Fumarate and nitrate reduction regulator
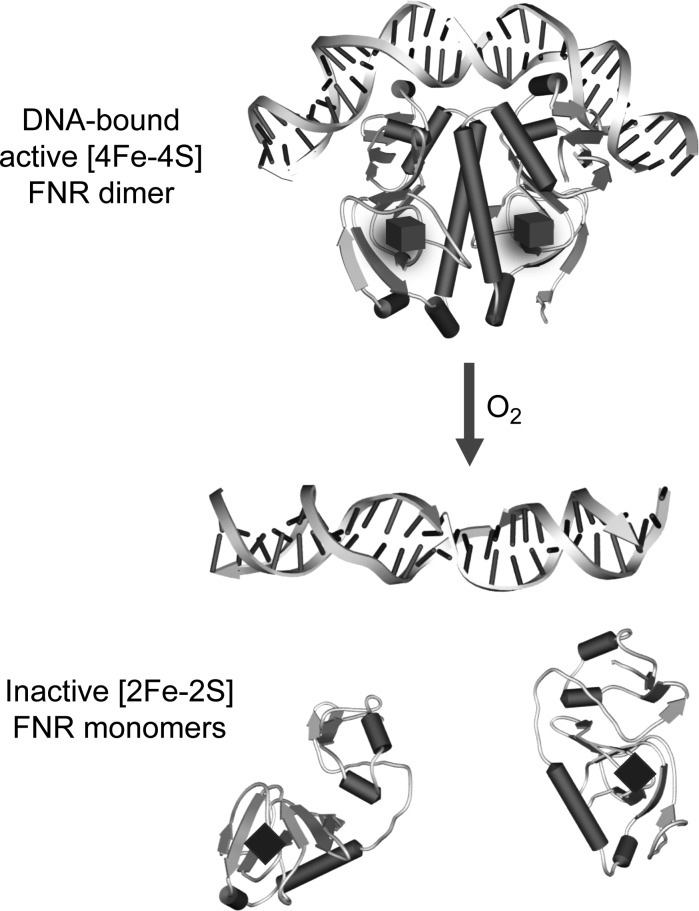
In Escherichia coli and many other bacteria the ability to sense environmental levels of O2 is achieved through the activity of the global O2-sensing fumarate and nitrate reduction (FNR) regulator (53, 82). The expression patterns of FNR-regulated genes indicate that the regulator responds to dissolved O2 tensions in the range >0–20 μM (11, 95). FNR belongs to the catabolite repressor protein (CRP)-FNR superfamily of regulators (87), characterized by an N-terminal sensory domain and a C-terminal DNA-binding domain containing a helix-turn-helix motif (54, 131). The N-terminal sensory unit varies between members of this super-family; some contain a cofactor (e.g., an iron–sulfur cluster or a heme group) (53, 80, 88), while others do not (162). By far the best characterized FNR protein is that from E. coli, considered to be the paradigm system, although there are clearly variations in properties between FNR proteins (see below). The E. coli protein becomes activated under anaerobic conditions by insertion of an O2-labile [4Fe-4S] cluster into the N-terminal sensory domain (53, 89, 114) by the iron–sulfur cluster (Isc) biosynthetic machinery (99, 127). This causes conformational changes that diminish inter-subunit electrostatic repulsion (102), resulting in the dimerization of the ∼30-kDa monomer. This enables the C-terminal DNA-binding domain to recognize specific binding sites within FNR-controlled promoters (53, 129). Structural information is not yet available for FNR, but a model, based on the structure of CRP (126), has been generated (see Fig. 2). Upon exposure to O2, the FNR dimer dissociates into monomers and is thus converted to a transcriptionally inactive state, in which favorable interactions with target DNA and the transcription machinery cannot take place (11, 53, 81, 89, 114). In this way, E. coli FNR regulates >300 genes (20, 52, 79, 125), most of which are associated with anaerobic metabolism.
FIG. 2.
A model of Escherichia coli fumarate and nitrate reduction (FNR) regulator and its reaction with O2. The FNR model is based on the structure of E. coli catabolite repressor protein (CRP) [pdb file 1CGP (126)]. The [4Fe-4S] cluster is represented by a cube. After the increase in O2 concentration, the DNA-bound dimer FNR protein undergoes reaction at its cluster, resulting in a [2Fe-2S] form (represented by a rhomb) that dissociates into monomers and no longer binds DNA.
In E. coli FNR, the iron–sulfur cluster is bound by the thiols of four cysteine residues (Cys20, 23, 29, and 122) in the N-terminal domain. Each of these is essential for function (55). E. coli FNR contains one other cysteine residue (Cys16), which can be removed without apparent effect (55), although it is conserved in a large subset of FNR proteins. In vitro biochemical studies of FNR were for a long time hampered by its extreme O2 sensitivity, until the development of robust anaerobic purification and manipulation protocols allowed progress in determining mechanistic details. Spectroscopic studies, including whole-cell Mössbauer, showed that the reaction of FNR with O2 results in conversion of the [4Fe-4S]2+ cluster into a [2Fe-2S]2+ form (77, 81, 114). This, together with considerations of the intracellular O2 concentration, indicated clearly that FNR senses O2 directly (77, 89), and not via another, intermediate signaling species (6, 155).
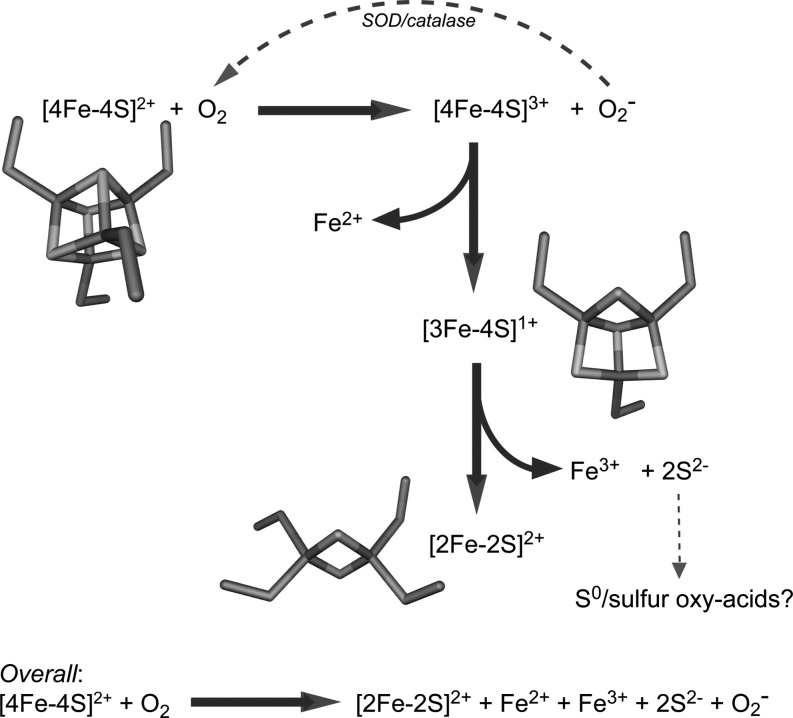
Two proposals of cluster conversion mechanisms have been made. First, the reaction proceeds as a concerted single-step reaction involving the release of two Fe(II) ions and partial oxidation of sulfide (81, 142). Second, a two-step reaction takes place involving a [3Fe-4S]1+ intermediate and sequential release of Fe(II) and Fe(III) ions (21, 23, 24). Evidence for the latter came through the detection of a transient EPR species with characteristics of a [3Fe-4S]1+ cluster, whose rate of formation and loss was similar to those obtained from optical observations of a two-step conversion reaction. Fe(II) is released from the [4Fe-4S]2+ cluster to generate the [3Fe-4S]1+ cluster containing three Fe(III) ions (24). Since the initial [4Fe-4S]2+ cluster contained two Fe(II) and two Fe(III) ions, the reaction with O2 results in oxidation to a superoxidized state, [4Fe-4S]3+, that is unstable and immediately ejects an Fe(II) ion, forming a [3Fe-4S]1+ species. This chemistry has previously been characterized in the [4Fe-4S]2+ clusters of Clostridium pasteurianum 8Fe ferredoxin after one electron oxidation (17, 40). Consistent with this, the one electron reduction product of O2, superoxide ion, could be detected during this process (24), although this undergoes disproportionation to O2 and H2O2 (see Fig. 3). Once formed, the [3Fe-4S]1+ cluster is only transiently stable, ejecting one Fe(III) ion and two sulfide ions (S2−) (25) to generate the product, [2Fe-2S]2+.
FIG. 3.
The chemistry of FNR cluster conversion. Scheme summarizing the mechanism of the reaction of [4Fe-4S] FNR with O2. See section Fumarate and nitrate reduction regulator for details.
Hence, oxidation of the metal ions rather than of sulfide ions occurs. However, it has also been shown that the final oxidation state of the iron released is dependent on the chelating power of the environment in which the reaction takes place (23). Thus, in the presence of reductant, two Fe(II) ions can be detected. It is possible, therefore, that the sulfide ions released from the [4Fe-4S] cluster are also susceptible to redox reactions. Oxidation, for example, could generate S0 (carried as polysulfides) or oxy-acids of sulfur (142). These studies also revealed that the chelating power of the medium can also influence the relative rates of formation and loss of the [3Fe-4S]1+ intermediate; for example, in the presence of a strong Fe(III) chelator (e.g., phosphate), the rate of intermediate decomposition, relative to its formation, was enhanced so that the intermediate could not be detected (23).
Dismutation of superoxide to O2 and H2O2, followed by disproportionation of the latter to O2 and H2O, catalyzed by superoxide dismutase and catalase enzymes, respectively, could result in recycling of the signal molecule, which would maximize the sensitivity of FNR to O2 (24). Further insight into the site of O2 attack was gained from mutagenesis studies of E. coli FNR (73). Site-directed replacement of residues lying next to cluster coordinating cysteine residues revealed that replacement of Ser24, particularly by bulky hydrophobic residues, had the greatest effect on in vivo activity. In vitro studies of an S24F variant showed that although the mechanism was conserved, this variant reacted significantly more slowly with O2 than the wild-type protein (73). It was proposed that the bulkier Phe side chain shields O2 from binding at the iron coordinated to Cys23, suggesting that this is a primary site of O2 reaction. A decrease in O2 sensitivity was also observed for L28H FNR, which, like S24F FNR, contains a substitution of a residue lying next to a cluster-coordinating cysteine (10). These data imply that the O2 sensitivity of FNR proteins can be finely tuned, through residue substitutions (leading to structural changes), to ensure that the protein senses O2 over the physiologically relevant concentration ranges (73).
The [2Fe-2S]2+ cluster of FNR is also not very stable in the presence of O2 and slowly degrades to form cluster-free (apo-) protein. This occurs in vitro and is enhanced by, for example, superoxide as well as in vivo (1, 119, 143). However, the extended time period over which this occurs (at least in vitro) offers the possibility that the [4Fe-4S] to [2Fe-2S] conversion could reverse. Indeed, some [4Fe-4S]2+ cluster was observed after addition of dithionite to air-oxidized [2Fe-2S]2+ FNR in vitro (81). The apo-protein can also incorporate a fresh cluster after oxidative disassembly of the original one, as demonstrated by in vitro reconstitution experiments and, more importantly, in vivo through reactivation of FNR in cells where protein synthesis was inhibited (31, 36, 98).
Thus, FNR appears to function by a continual process of cycling between active [4Fe-4S], inactive [2Fe-2S], and apo forms, with the level of O2 determining which form predominates and, therefore, the extent to which FNR is transcriptionally active. Such a mechanism requires that the levels of FNR in the cell are tightly controlled (73). It has been shown that this is the case in E. coli, where FNR concentrations vary little under different growth conditions (98, 142). Progress toward achieving a detailed system level understanding of FNR-mediated regulation has been made recently, based on experimentally determined kinetic models and parameter values that enabled the large number of unknowns to be minimized (73, 152). Using a relatively simple model, it was shown that small changes in the rate at which the [4Fe-4S] cluster reacts with O2 have a significant effect on the balance in vivo between active and inactive forms (73). A more sophisticated modeling study was able to predict closely the in vivo behavior of cells containing wild-type and variant forms of FNR, and to enable distinction between different forms of FNR (e.g., active and inactive) that cannot be easily measured experimentally (152).
FnrP from Paracoccus denitrificans is an FNR ortholog that binds a [4Fe-4S] cluster, but has a different arrangement of cysteine residues at its N-terminus (68), while FNR from Bacillus subtilis exhibits significant differences to the E. coli protein. In the B. subtilis protein the [4Fe-4S] cluster is located in the C-terminal domain, and, unusually, is coordinated by three cysteine residues (Cys227, 230, and 235) plus an aspartate (Asp141) (58, 118). The cluster is not required for dimerization, but is essential for the activation of FNR-regulated genes, and, therefore, functions as a sensory unit. In contrast to E. coli, levels of B. subtilis FNR are variable depending on the O2 concentration (up-regulated at low O2 and down-regulated at high O2). This, together with the major differences in the cluster binding and location, suggests that the B. subtilis protein may function differently from E. coli FNR. Mechanistic details of the O2-reaction are now needed.
The role of FNR as a master regulator of the anaerobic to aerobic switch means that it is likely to be important for virulence of pathogens that encounter fluctuations in O2 concentration as they seek to establish infection. In Salmonella enterica serovar Typhimurium, FNR was shown to be essential for infection in a mouse model and for survival in the macrophage (38), and this was associated with the inability of the fnr mutant to respond to the cytotoxic oxidative burst associated with the NADPH phagocyte oxidase. Furthermore, the type III secretion system critical to Shigella flexneri virulence is controlled by FNR-mediated regulation of Ipa secretion (96). This regulation ensures that type III secretion is functional only at its precise site of action, that is, in the partially oxygenated environment in the vicinity of the gastrointestinal mucosa.
NreB
Staphylococci apparently lack an FNR protein and the regulation of genes associated with anaerobic respiration of nitrate and nitrite is controlled by the products of the nreABC operon (37). NreA is believed to be involved in sensing nitrate, while NreBC constitutes a two-component O2-sensor. NreC is activated upon phosphorylation to bind to specific sequences upstream of anaerobic respiratory nar and nir operons. NreB is a cytoplasmic histidine kinase that phosphorylates and, therefore, activates NreC (37, 78).
NreB contains four cysteine residues at its N-terminus (Cys59, 62, 74, and 77) and can bind a [4Fe-4S] cluster, which is degraded in the presence of O2 (78, 105) to a [2Fe-2S] form, and subsequently to the apo-form. NreB kinase activity was found to be dependent on the presence of the [4Fe-4S] cluster. Thus, like FNR, NreB is a direct O2 sensor, but, unlike FNR, it does not itself act as a transcriptional regulator, but rather activates NreC (78). How NreA influences this signal transduction system is not yet clear.
ArnR
Anaerobic respiration in the presence of nitrate in Corynebacterium glutamicum is regulated by the novel transcriptional regulator ArnR, which is distinct from any previously identified regulator of nitrate reduction (107, 108). It contains a predicted N-terminal DNA-binding domain and three conserved cysteine residues (Cys179, 193, and 223) at the C-terminus. Anaerobically purified ArnR was colored and had absorbance properties characteristic of an iron–sulfur cluster (107). Remarkably, AnrR functions as a repressor of the narKGHJI operon under aerobic conditions by directly binding to a specific site in the promoter region, thus contrasting with the anaerobic activation of the equivalent genes by FNR in E. coli and B. subtilis. Currently, little is known about the nature of the ArnR iron–sulfur cluster or its role in the protein's regulatory function.
SoxR
The SoxRS system constitutes an unusual two-part regulatory system in enteric bacteria, in which the two proteins act sequentially to activate transcription of more than 100 genes (112) in response to redox stress. SoxR is activated in response to redox stress and, in enteric bacteria, switches on transcription of soxS. The SoxS protein is also a transcriptional activator and in turn switches on expression of the SoxRS regulon. SoxS is subject to proteolytic degradation, ensuring that the system's response remains dependent on the signal (56). From early studies of this system in E. coli, it was known that a range of redox active molecules (e.g., paraquat and menadione) could lead to activation of SoxR (4). For a long time it was believed that this was an indirect activation resulting from the capacity of such molecules to generate superoxide (O2−) in vivo (4). However, it was concluded very recently that SoxR responds not to superoxide, but instead to bacteria- and plant-derived redox-cycling molecules themselves, independent of superoxide (59). SoxR activation through oxidation by DNA-guanine radicals was also reported, with the potential for long-range electron transfer mediated by DNA (92). Although SoxR is widely distributed in bacteria, SoxS is absent in nonenterics, and so SoxR acts directly in regulating all of the regulon members (33). Interestingly, this type of SoxR protein (e.g., SoxR from Pseudomonas aeruginosa and Streptomyces coelicolor) is known to be activated by small redox active molecules (32, 134). Precisely how SoxR becomes activated currently remains controversial (69, 93).
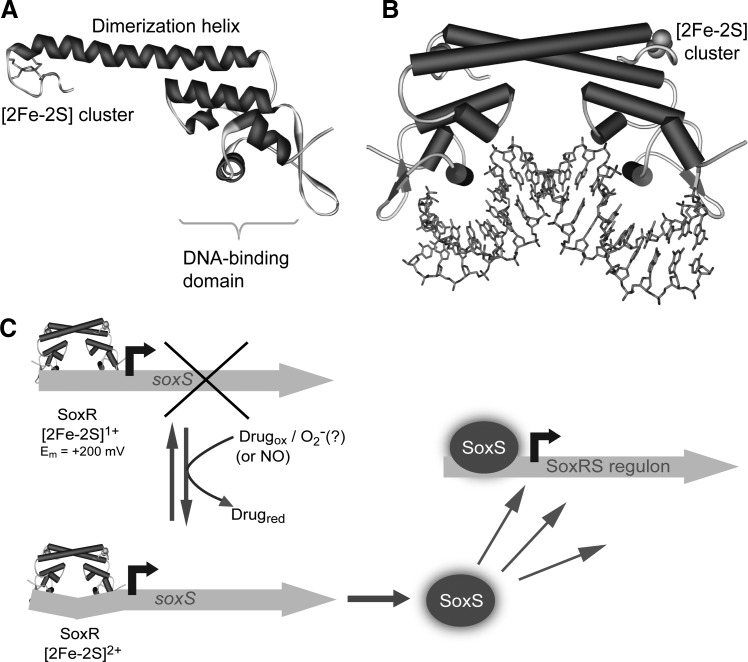
SoxR is an ∼17-kDa protein belonging to the MerR family that forms a homodimer in solution, with each monomer containing a [2Fe-2S] cluster, essential for transcriptional activation (66), bound by four cysteine residues organized as a Cys-X2-Cys-X-Cys-X5-Cys motif (see Fig. 4A). High-resolution structural information for iron–sulfur cluster regulatory proteins is not abundant, owing to the sensitivity/fragility of the cluster. However, the X-ray structure of [2Fe-2S] SoxR was recently solved, in the free form and in a complex with soxS promoter DNA. The structure revealed overall similarity to other MerR family members, with significant differences in the alignment of the dimerization helix with the DNA-binding domain (160). Also, in comparison to other structurally characterized [2Fe-2S] clusters, the rhomb plane of the cluster (in the sensory domain) is tilted (by 20°) relative to the plane of coordinating cysteine residues (160). Importantly, the cluster iron atoms were found to be solvent-exposed, in an asymmetric electrostatic environment, where one of the two sulfides interacts (via H-bonds and van der Waals contacts) with backbone amides, while the other is solvent exposed and only makes van der Waals contacts with oxygen and carbon atoms. Furthermore, the cluster is partially stabilized through interactions with residues belonging to the other monomer of the dimer, including a number in the DNA-binding domain (160).
FIG. 4.
Structure of [2Fe-2S] SoxR and its regulatory mechanism. (A) Structure of the SoxR monomer, with the dimerisation helix, DNA-binding domain, and [2Fe-2S] cluster indicated in sticks representation [pdb file 2ZHH (160)]. (B) Structure of SoxR bound to DNA (pdb file 2ZHG). Note that the cluster is represented in space-filling mode to ensure contrast with DNA, which is in sticks representation. (C) Scheme summarizing the SoxRS system in E. coli. Nonenteric bacteria do not contain SoxS and so SoxR itself directly regulates the Sox regulon. See section SoxR for details.
The SoxR [2Fe-2S] cluster can exist in the +1 and +2 oxidation states connected by a reduction potential of −285 mV (vs. standard hydrogen electrode at pH 7.6) (35, 46). Both forms bind DNA with similar affinities (as does the cluster-free [apo] form), but it is only the oxidized [2Fe-2S]2+ form that can activate transcription of the soxS gene. Electrochemical measurements on DNA-modified electrodes revealed a dramatic shift in the reduction potential to +200 mV for SoxR bound to its cognate DNA (51), indicating that only strongly oxidizing species should be able to oxidize the cluster to the +2 state. This implies that SoxR only becomes activated under conditions of oxidative stress and does not respond to small fluctuations in the cellular redox balance. Activation occurs through a remodeling of the −35 and −10 promoter elements such that they become optimally positioned for interaction with RNA polymerase (160). Thus, SoxR is distinct from other iron–sulfur cluster regulators (see below) in that the switch which controls transcriptional activity does not involve major structural changes at the cluster, but instead simply involves a single-electron oxidation process. The solvent exposure of the cluster likely facilitates the oxidation reaction, and suggests a rather broad specificity, consistent with the finding that a number of different redox active molecules can activate expression (59). Cluster oxidation is a reversible process and a specific reductase system, encoded by rseC and rsxABCDGE, was identified in E. coli (86), in which inactivation of either rseC or any of the genes in the rsx operon resulted in constitutive expression of soxS.
It is still not fully understood how a simple one electron redox process can generate the structural changes (in SoxR and in the promoter DNA) that are required for transcriptional activation. The sensory domain interacts directly with the DNA-binding domain, providing a direct route for transmission of the signal. The asymmetric charge environment of the cluster may also be important, and it was proposed that oxidation state change is likely to result in structural rearrangements that maximize favorable electrostatic interactions (160). The current understanding of the SoxRS system is summarized in Figure 4B.
Sensors of NO
NO is a reactive, lipophilic radical that can freely diffuse into cells. In eukaryotes, NO functions [at nanomolar concentrations (100)] as a signal via the reversible coordination of NO to the heme group in guanylate cyclase to facilitate vasodilation, and also as a defense within mammalian macrophages [at micromolar concentrations (50)] in response to infection by pathogenic bacteria (16). Soil bacteria are also exposed to this gas (161), through anaerobic respiration with nitrate and some contain an NO synthase that generates NO at micromolar concentrations (61), which may be involved in protection against a broad spectrum of antibiotics (60, 97). Thus, for some bacteria at least, NO is a natural metabolite. The ability of NO to react readily with a variety of targets means that, at elevated concentrations, it is cytotoxic through processes such as thiol S-nitrosation, recently discovered in bacteria (67, 120); N-nitrosation of certain amino acids (e.g., tryptophan); and oxidative DNA damage. Metal cofactors are also a major target for NO reactions. Thus, proteins containing iron–sulfur clusters are highly susceptible to NO-induced damage (75).
Bacteria must be able to counter the deleterious effects of NO (at high concentration), but also to respond to environmental stress conditions that may be signaled at lower NO concentrations. For example, the presence of NO is associated with hypoxia (62). As a result, many bacteria have evolved a suite of specific proteins to sense NO (141), leading to its detoxification and, in some cases, to dramatic metabolic reorganization (such as occurs in sporulation) in order to adapt to stress conditions.
NsrR
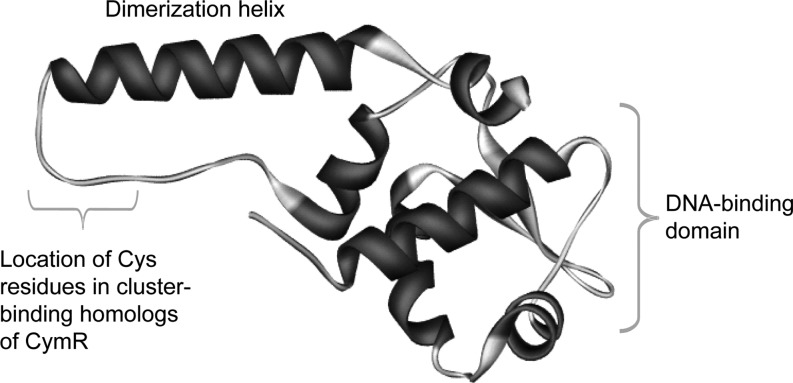
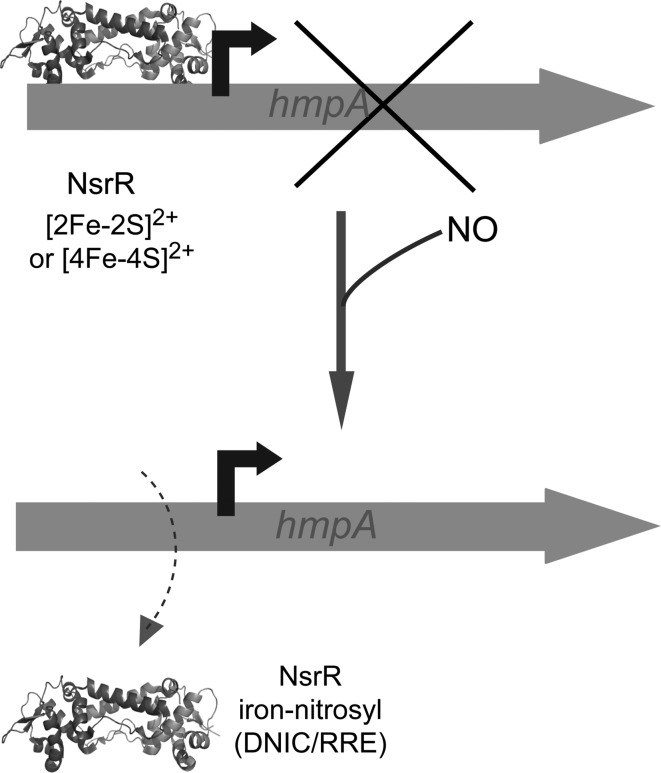
NsrR has been shown to sense NO in E. coli (15) and to switch on a regulon of at least 60 genes (111), including hmp, which encodes an NO detoxifying flavohaemoglobin (113). Thus, NsrR is a global regulator of the response to NO-induced stress and has been shown to be important for virulence in the pathogen S. Typhimurium (9, 49), and for establishing symbiosis between Vibrio fischeri and its squid host (159). NsrR belongs to the largely understudied Rrf2 family (122) that includes the [2Fe-2S] cluster-containing regulator IscR (see below). Purified NsrR from S. coelicolor (153), Neisseria gonorrhoeae (71), and B. subtilis (169) have all been shown to be iron–sulfur cluster binding proteins with three C-terminal conserved cysteine residues [Cys93, 99, and 105 in S. coelicolor NsrR; note that the spacing between cysteine residues varies in different NsrR proteins (154)] that likely act as cluster ligands. Consistent with this, Cys to Ala substitutions in N. gonorrhoeae NsrR relieved repression of a target promoter and inhibited DNA-binding activity in vitro (71). The identity of the fourth coordinating residue is not yet known, but, unless cluster coordination occurs between NsrR monomers, it cannot be a cysteine residue. The first structure of an Rrf2 family regulator was recently published (133). CymR, which regulates cysteine uptake and biosynthesis, was shown to be a dimer, with each monomer consisting of a DNA-binding domain (containing a winged helix-turn-helix motif) and a dimerization domain (see Fig. 5). CymR is unusual in that it performs its regulatory function through reversible complex formation with CysK, a key enzyme of cysteine biosynthesis, and is not a member of the iron–sulfur cluster containing Rrf2 family regulators. Nevertheless, modeling studies of CymR indicated the likely location of the cluster in such proteins (133).
FIG. 5.
Structure of the Rrf2 family regulator CymR. Structure of the Bacillus subtilis CymR monomer, showing the dimerization helix and DNA-binding domain [pdb file 2Y75 (133)]. The likely location of cluster-binding cysteine residues found in homologous iron–sulfur cluster-binding Rrf2 proteins (e.g., IscR, NsrR, and RirA) is indicated.
In addition to the uncertainty about ligation, the nature of the cluster and the mechanism by which the NsrR protein functions to coordinate the response to NO stress are not clear. Purification of S. coelicolor NsrR from E. coli yielded a [2Fe-2S] cluster form that was found to bind specifically to the S. coelicolor hmpA1 and hmpA2 promoter regions in bandshift assays (153). Binding was abolished by addition of NO and this reaction resulted in the formation of iron nitrosyl species, including a tetrahedral dinitrosyl iron complex (DNIC), [FeI(NO)2(RS)2]−, species in which RS− are cysteinate residues (see Fig. 6A) (156). Interestingly, this DNIC only accounted for ∼9% of the original iron, indicating that the major product was EPR silent (153). Although mass spectrometry suggested that purified N. gonorrhoeae NsrR also contained a [2Fe-2S] cluster (71), B. subtilis NsrR was found to contain a [4Fe-4S] cluster (169). This form was very recently shown to bind the B. subtilis nasD (nitrite reductase) promoter in an NO-sensitive manner (85). Furthermore, it was shown that B. subtilis NsrR also bound DNA at other sites, and this was not dependent on the cluster, suggesting that apo-NsrR plays a role in regulation (85).
FIG. 6.
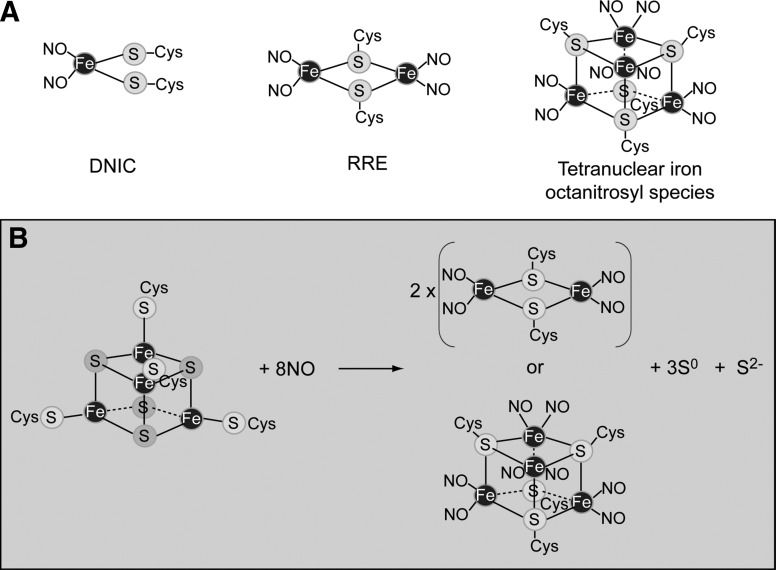
Iron-nitrosyl species. (A) The reaction of iron–sulfur clusters in regulatory proteins has been reported to result in the formation of at least two different iron-nitrosyl species: dinitrosyl iron complex (DNIC) and Roussin's red ester (RRE). The formation of a novel tetranuclear iron octanitrosyl cluster species was recently discussed (27). (B) A scheme illustrating the reaction of WhiB-like (Wbl) [4Fe-4S]2+ clusters with nitric oxide (NO) (27, 139).
The current literature on NsrR does not, therefore, provide a consistent view of the type of cluster: both [2Fe-2S] and [4Fe-4S] forms have been reported for NsrR proteins from different organisms that each binds DNA specifically. Given the quite different spatial arrangement of ligating residues required to coordinate cubane [4Fe-4S] and planar [2Fe-2S] clusters, it is unlikely that NsrR containing each cluster type could adopt the same conformation, and therefore each form might be expected to recognize different operator sequences and regulate distinct sets of genes. Clearly, further information is required, but a general scheme describing NsrR regulatory function is shown in Figure 7.
FIG. 7.
NsrR regulatory systems. Scheme summarizing the mechanism of NsrR NO-sensing and regulation. Specific DNA binding has been reported for NsrR proteins from different organisms in which the form of the cluster ([4Fe-4S] or [2Fe-2S]) is different (85, 153). The gene hmpA encodes an NO-detoxifying enzyme. Note that the high-resolution structure of NsrR is not yet available, and the schematic representation included here is based on the recently published structure of the Rrf2 family protein CymR (133). Structural information on the likely iron–nitrosyl species formed following reaction of NsrR with NO is given in Figure 6. Although not illustrated, apo-NsrR has also been reported to bind DNA (at a different sequence) and, therefore, may also fulfill a regulatory function (85).
Wbl proteins
The WhiB-like (Wbl) family of regulators (named after the first discovered WhiB protein from S. coelicolor) is, remarkably, found only in the Actinobacteria, a phylum of Gram-positive bacteria that includes Streptomyces, the most abundant source of clinically important antibiotics, and important pathogens such as Mycobacterium tuberculosis and Corynebacterium diphtheriae (140). Wbl proteins are generally small (∼10–15 kDa) and contain a highly conserved pattern of cysteine residues Cys-Xn-Cys-X2-Cys-X5-Cys that bind an iron–sulfur cluster (22, 72).
S. coelicolor encodes 11 Wbl proteins of which five, WblA, WhiB, WblC, WhiD, and WhiE, are well conserved in actinobacteria (39). In S. coelicolor, WblA and WhiB are required for the initiation of sporulation and septation, whereas WhiD is required for the late stages of sporulation (39, 101). A S. coelicolor wblC mutant is hyper-sensitive to antibiotics, as is the equivalent whiB7 mutant in M. tuberculosis, suggesting an important role for WblC/WhiB7 in antibiotic tolerance (39, 103). The function of WblE is not known, but the wblE gene could not be disrupted in S. coelicolor, suggesting that its function may be essential (39). In M. tuberculosis, Wbl proteins bestow the ability on the pathogen to persist within its host for long periods, as well as its remarkable tolerance to a wide range of antibiotics (103). The function of Wbl proteins has been the subject of some controversy. While initially believed to be regulatory proteins, in vitro studies revealed that cluster-free Wbl proteins exhibit disulfide reductase activity (2). This led to the proposal that loss of the iron–sulfur cluster through oxidative stress leads to activation of Wbl proteins as disulfide bond reductases. It seems unlikely that this is a general property of Wbl proteins, however, as investigations of other Wbl proteins failed to detect any general reductase activity (22, 139). It is possible that Wbl proteins may act as reductases of specific cellular targets, and in the case of M. tuberculosis WhiB1, a specific disulfide-mediated interaction with the alpha (1,4)-glucan branching enzyme GlgB was detected (44). However, the majority of evidence now indicates that Wbl proteins are indeed DNA regulatory proteins.
Recently, M. tuberculosis WhiB3, which contributes to virulence and is induced in mouse lungs and macrophages, was shown to regulate lipid and polyketide biosynthesis, including tri-acylglycerol accumulation, in vivo, in response to activated macrophages (135). The [4Fe-4S] form of WhiB3 is O2 sensitive and reacts with NO (136), leading to the suggestion that WhiB3 acts as a sensor of O2 and NO to control expression of genes involved in intermediary metabolism. This provides a key connection to the well-documented accumulation in M. tuberculosis of tri-acylglycerol (which is also present in the sputum of tuberculosis patients) in response to hypoxia and NO exposure (29, 30). Recently, M. tuberculosis whiB1, which encodes a homolog of S. coelicolor WblE, was shown to be an essential gene. The [4Fe-4S] form of WhiB1 did not bind to whiB1 promoter DNA, but the reduced and oxidized (disulfide containing) forms of apo-WhiB1 did. Binding was also observed after cluster nitrosylation (139). It is becoming clear that the reaction of NO with Wbl proteins is a central signaling process and that the iron–sulfur cluster plays a key functional role in controlling DNA binding.
Many literature reports of reactions of NO with protein-bound iron–sulfur clusters ([2Fe-2S], [3Fe-4S], or [4Fe-4S]) have identified the product as a DNIC (Fig. 6A). This species has invariably been detected, both in vivo and in vitro, by means of its distinctive S=½ EPR signal, at g=2.03. However, when quantified by spin integration, the amount of DNIC accounts for only a few percent of the iron in the original cluster [see (27) and references therein]. Thus, the fate of most of the cluster iron and, indeed, the sulfide ion is unclear. Insight into this was recently provided by studies of the reaction of the Wbl proteins WhiD from S. coelicolor and WhiB1 from M. tuberculosis with NO (27), which revealed several novel aspects of cluster nitrosylation and a common mechanism (27, 139). The addition of NO resulted in an extremely rapid, multi-phasic reaction, with a t½ of ∼1 s, which is about 104-fold faster than the reaction of these clusters with O2 and by far the most rapid iron–sulfur cluster nitrosylation reaction reported to date. Remarkably, the reaction was stoichiometric, giving a product(s) with a total of eight NO molecules per [4Fe-4S] cluster that was EPR silent. The product is, therefore, not a DNIC. Instead, it was proposed that a pair of EPR-silent, dinuclear iron tetra-nitrosyl species, [FeI2(NO)4(SR)2], known as Roussin's red ester (RRE) complexes were formed (see Fig. 6A, B) (18). If two such RRE species were to lie close to one another, the product could be described as a novel, tetranuclear octanitrosyl cluster (see Fig. 6A, B).
Iron nitrosyl complexes typically contain iron in the +1 state. Six electrons are therefore required to reduce all the iron ions of the [4Fe-4S]2+ cluster to this oxidation state (since the cluster contains 2×Fe(III) and 2×Fe(II) ions). The reductant is likely to be the cluster sulfide (S2−) ions. Analysis revealed that three of them were indeed oxidized to S0 during the reaction, yielding the six electrons required (27).
EPR-silent RRE species have also been obtained after the reaction of NO with model iron–sulfur clusters (63), and recent studies of the Rieske protein using the novel technique of nuclear resonance vibrational spectroscopy showed that, upon addition of NO, the [2Fe-2S] cluster forms, in high yield, an RRE rather than a DNIC species (149). It is not known currently how commonly RRE species are formed when iron–sulfur clusters react with NO, but it is apparent that, at least in some cases, they are the major iron–nitrosyl species formed.
Sensors of Iron or Iron–Sulfur Clusters
As well as exploiting the reactivity of iron–sulfur clusters toward small gaseous molecules, iron–sulfur regulatory proteins are also involved in sensing cellular concentrations of iron–sulfur clusters and iron themselves, where the binding of the cluster, rather than its reaction with an analyte, is the signal to which the regulator responds.
IscR
Iron–sulfur cluster regulator (IscR) is a member of the Rrf2 family of transcriptional regulators that also includes NsrR (see above) and was first discovered as a regulator of the Isc iron–sulfur biogenesis pathway in bacteria (128). When purified anaerobically, the 17 kDa E. coli protein was found to contain a [2Fe-2S] cluster, with a characteristic EPR spectrum indicative of a reduced cluster (in the +1 state) with a total spin, S,=½ (128). The protein contains three conserved cysteine residues (Cys92, 98, and 104), which are all functionally essential (167), and, in Acidithiobacillus ferrooxidans IscR, it was shown that a conserved Glu residue located toward the N-terminus is required for cluster binding, implying that this may be the fourth cluster ligand in this organism (170).
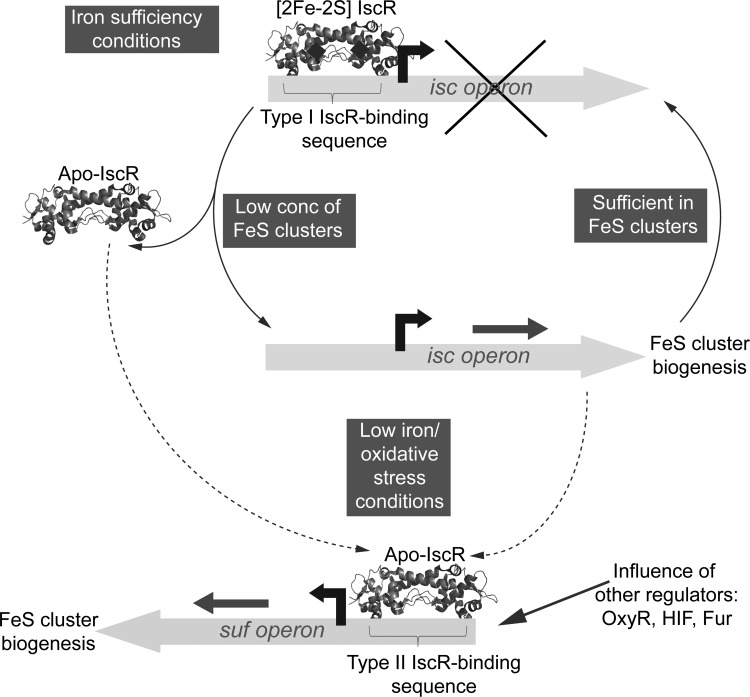
In its cluster-bound state, IscR represses expression of the iscSUA-hscBA-fdx genes that encode the housekeeping Isc iron–sulfur biogenesis system in E. coli. Under conditions of low iron or oxidative stress, E. coli utilizes a different iron–sulfur biogenesis system encoded by the sufABCDSE operon (110, 144). Regulation of the Suf system is complex, involving OxyR, IHF, Fur, and IscR (91, 110, 167). Under inducing conditions, suf operon expression is activated by IscR and it is the cluster-free form that regulates the suf promoter (106, 167). DNAse I footprinting studies revealed that IscR exhibits different DNA-binding properties in its apo and holo states: type I sites in the isc promoter are recognized by holo-IscR, while the type II site in the suf promoter is recognized by both apo- and holo-IscR (48, 106). Subsequently, it was demonstrated that a number of other IscR-regulated genes, including those encoding anaerobically produced iron–sulfur enzymes, contained type II sites and that these, too, did not require the [2Fe-2S] cluster and are regulated by apo-IscR (106). That type II sites can bind both apo- and holo-IscR raises the question of how regulation is achieved in vivo. It is proposed that under low iron and/or stress, iscR expression is activated, resulting in a significant increase in apo-IscR, leading to activation of suf and other type II-regulated genes (106). Thus, through the control of its own expression, IscR regulates different subsets of genes in cluster-bound and cluster-free forms and, in doing so, integrates regulation of iron–sulfur cluster supply by sensing the cluster assembly status of cells. Figure 8 summarizes the mechanism of IscR-mediated regulation.
FIG. 8.
The IscR regulatory system. Scheme summarizing the mechanism of IscR regulation of isc and suf operons. Under iron replete conditions (upper part of the figure), and when the cellular supply of iron–sulfur clusters is sufficient, cluster-bound IscR represses the housekeeping Isc iron–sulfur biogenesis system in E. coli. When iron–sulfur cluster demand increases, the apo-form of IscR accumulates, leading to depression of the cluster biosynthesis system. Under conditions of low iron or oxidative stress (lower part of the figure), E. coli utilizes a different iron–sulfur biogenesis system encoded by the Suf system. Under inducing conditions, suf operon expression is activated by apo-IscR, which accumulates under conditions of low iron–sulfur cluster supply through both the loss of cluster from cluster-bound IscR and depression of the isc operon, which includes iscR. See section IscR for details. Note that the high-resolution structure of IscR is not yet available, and the schematic representation included here is based on the recently published structure of the Rrf2 family protein CymR (133).
The importance of IscR is becoming increasingly apparent. Disruption of the regulation of iron–sulfur cluster supply was found to dramatically attenuate the virulence of the pathogens S. flexneri, Erwinia chrysanthemi, Burkholderia mallei, and P. aeruginosa (75, 84, 121, 124), and IscR was also reported to control biofilm formation by E. coli through regulation of type I fimbria expression (166).
SufR
Although both Isc and Suf systems are present in cyanobacteria, their relative importance appears to be different compared to many other bacteria (158). Many of the suf genes are essential, while isc genes are not, indicating that the Suf system is the primary iron–sulfur biogenesis system in cyanobacteria. This is consistent with the observation that a Suf-like system is present in plant chloroplasts, whereas an Isc system is not (7). Thus, rather than IscR functioning as a master regulator of iron–sulfur cluster biogenesis, cyanobacteria contain an iron–sulfur cluster-binding protein, SufR, which regulates the suf operon.
SufR is a homodimeric protein with an N-terminal helix-loop-helix DNA-binding motif and a C-terminal domain containing four conserved cysteine residues (Cys164, 177, 191, and 206, unusually arranged as Cys-X12-Cys-X13-Cys-X14-Cys), which can accommodate a [4Fe-4S] cluster (132). However, only three of these cysteine residues (Cys164, 177, and 206) are actually involved in coordinating the cluster, indicating that the fourth coordinating residue is not a cysteine (132). This points to some similarities with B. subtilis FNR (see above), which also includes a dimeric state that is independent of the type of cluster. Reduction of the cluster from the +2 to the +1 state gave a mixture of spin S=3/2 and 1/2 ground states, something occasionally observed in ferredoxins with noncysteine coordination (47).
Deletion of the sufR gene led to constitutive expression of the sufBCDS operon in Synechocystis, indicating that the protein acts as a repressor (158). The region between juxtaposed but divergently transcribed sufR and sufBCDS contains two SufR inverted repeat binding sites that are highly conserved in cyanobacteria. Although apo-SufR and SufR, containing reduced [4Fe-4S]1+, were able to bind the Suf operator sequence, as judged from DNA mobility shifts assays, a much larger effect was observed with the [4Fe-4S]2+ form, implying that this is the repressing form of the regulator. Thus, when iron–sulfur cluster levels drop, SufR can no longer bind, thus alleviating the repression of the Suf system (132). Further investigation is required to understand the effect of the redox state of the SufR cluster on regulation.
Bacterial aconitases
Aconitase is an enzyme of the citric acid cycle that is localized in the mitochondrion of eukaryotes. Its [4Fe-4S] cluster is key to its dehydratase activity in converting citrate to isocitrate. A close homolog of aconitase is found in the cytoplasm of vertebrates and is known as cytoplasmic aconitase or iron regulatory protein 1, IRP1. It is a key regulator of cellular iron levels [reviewed in refs. (117, 157)]. Under conditions of iron starvation or oxidative/nitrosative stress, the protein loses its cluster and becomes activated for specific binding to iron regulatory elements (IREs) contained within the mRNA resulting from transcription of genes related to iron metabolism. The specific effect of binding is dependent on whether the IRE is at the 5′ or 3′ end of the mRNA: binding to the 5′ end results in stabilization of transferrin receptor mRNA, whereas binding to the 3′ end inhibits translation of ferritin mRNA.
This remarkable dual enzyme/regulator behavior of aconitase is also found in some bacteria (3, 8, 42, 43, 130, 145, 164). Two types of bacterial aconitases have been identified: one is similar to the eukaryotic aconitases and is expressed under stress conditions (AcnA), while the other, which serves as the principal citric acid cycle enzyme, is unique to bacteria in that it contains an additional N-terminal domain and a different domain arrangement (AcnB) (57, 163). In E. coli the two aconitases are expressed under different conditions and exhibit different stabilities [AcnA is more stable than AcnB (76)], but both types have been shown to bind to specific sequences in the 3′ untranslated regions of acnA and acnB mRNA in their cluster-free forms (145), thereby promoting the production of the aconitase proteins under conditions of stress (e.g., iron starvation, or oxidative/nitrosative stress) that depletes the cluster-bound active form. Thus, it is suggested that these enzymatically inactive aconitases mediate a post-transcriptional positive autoregulatory switch.
In aconitase, the cluster is coordinated by only three cysteine residues, such that one iron has a vacant coordination site for substrate binding. Thus, the cluster is susceptible to losing that iron, generating an enzymatically inactive [3Fe-4S] cluster that can degrade further to the apo-protein, which in turn binds mRNA. Iron levels, or stresses that lead to degradation of iron–sulfur clusters, could be sensed through the [4Fe-4S] to apo-protein transition. Alternatively, because the conversion [4Fe-4S]2+ to give a [3Fe-4S]0 cluster plus Fe2+ is an equilibrium reaction dependent on the concentration of Fe(II), it could serve as a means of sensing iron. There is evidence that the mechanism of cluster degradation is different in the two E. coli aconitases: AcnB is a homo-dimer under iron-sufficient conditions. The dimerization interface was traced to the N-terminal region, where it is proposed a second iron-binding site is located, and is important in the switch of AcnB from a holo-dimer (catalytic) to an apo-monomeric (regulatory) form (146). It is possible that the two aconitases have difference sensitivities to ROS/RNS and that this might be functionally important.
In addition to autoregulation, aconitases in E. coli are involved in regulating (positively and negatively) superoxide dismutase synthesis (148). In Salmonella aconitase is involved in a regulatory cascade linking central metabolism, oxidative stress, and motility (147). The aconitases of B. subtilis and M. tuberculosis were found to bind to IRE-like sequences in mRNA, suggesting that the mode of binding to mRNA is similar to that found in eukaroytes (3, 8). The B. subtilis protein is involved in the regulation of a major respiratory oxidase and an iron uptake system and was found to be important for sporulation (3, 130), while roles in iron homeostasis have been proposed for the aconitases of M. tuberculosis (8) and Xanthomonas campestris (164). There is still much to learn about the importance of the regulatory roles of aconitases in bacteria, and of the mechanisms associated with their regulatory functions.
RirA
Rhizobial iron regulator (RirA), another member of the Rrf2 family of transcriptional regulators, was first discovered in Rhizobium leguminosarum (151) and is found in α-proteobacteria that are closely related to Rhizobium, Agrobacterium, Brucella, and Bartonella (123). Mutations in the rirA gene led to constitutive expression of genes encoding a variety of iron-uptake systems, and it was shown that RirA-regulated genes contain an iron-responsive operator (RirA-box) within, or close to, their promoter sequences (168). Proteomic studies revealed that RirA is a global (>100 genes) regulator of iron homeostasis, and also plays a role in regulation of the suf gene homologs in R. leguminosarum and Sinorhizobium meliloti (19, 150). Although biochemical studies of RirA have not yet been reported, it contains three conserved cysteine residues (Cys91, 100, and 106) and is predicted to bind an iron–sulfur cluster. How it senses iron is not yet clear, but RirA functions in concert with a second global iron regulator, Irr, that is more widely distributed among α-proteobacteria, and which represses a wide range of genes under low iron availability by binding to operator sequences (ICE-boxes) that are upstream of the target genes. Repression by Irr is relieved by the binding of heme (137, 138) and so it has been proposed that these two regulators, together, sense the physiologically relevant status of iron in the form of iron–sulfur clusters and heme, respectively, rather than as the concentration of Fe2+, as seems to be the case in organisms that employ Fur or DtxR (74). Alternatively, RirA could sense Fe2+ directly through a cluster conversion reaction (for example, [3Fe-4S] ↔ [4Fe-4S]). Biochemical data are now required to address these questions.
Other Iron–Sulfur Regulatory Proteins
While iron–sulfur cluster regulator proteins are mainly involved in the cellular response to O2, ROS/RNS, and iron/iron–sulfur cluster concentrations, they have also been found to play other roles, but even in these cases, the cluster is likely to be responding to a form of stress.
The group 3 sigma factor SigM of S. coelicolor becomes activated in response to osmotic stress (90). The activity of SigM is controlled by the antisigma factor RsmA, which has homology with the HATPase_c family of antisigma factors but is unusual in that it contains seven cysteine residues (45). Isolation and characterization of the protein revealed that it binds a [2Fe-2S] cluster and that the interaction between RsmA and SigM is dependent on the presence of the cluster. Thus, it is proposed that RsmA regulates the activity of SigM in response to an as yet unknown signal that leads to the loss of its iron–sulfur cluster (45).
The thn genes encode a system for the utilization of tetralin, an aromatic compound found at low concentrations in different crude oils, as a carbon and energy source by Sphingomonas macrogolitabida strain TFA (64). Regulation of expression is complex, but involves the LysR-type activator ThnR and also ThnY, which has homology to bacterial oxygenase-coupled NAD(P)H-dependent ferredoxin reductases (41). Purification and characterization of ThnY revealed that it is an iron–sulfur flavoprotein containing an FAD and a plant ferredoxin-like [2Fe-2S] cluster. Electrophoretic mobility shift experiments showed that ThnY enhanced specific DNA-binding by ThnR, indicating that ThnY functions by promoting thn transcription activation by ThnR, most likely via the formation of a complex with ThnR, and its capacity to do this may be modulated by the redox state of the ThnY [2Fe-2S] cluster (41).
Hierarchy of Regulatory Responses
In this review we have categorized the various known iron–sulfur regulators according to the primary signal to which they respond. However, in many cases, a regulator will respond to other signals, and this raises questions about the specificity of regulatory responses and also about the hierarchy of regulators that are involved in the response to a particular signal. Thus, although not part of the primary response to NO (104), SoxR and thus the SoxRS regulon are activated by NO (34, 65, 109) and in E. coli, it plays an important role in virulence (109). Activation results from the direct reaction of NO with the [2Fe-2S] cluster, generating DNICs (34). Whereas nitrosylation of iron–sulfur clusters normally results in inactivation, in this case, SoxR becomes transcriptionally active. How the nitrosylated form of SoxR is able to elicit the same response as the one-electron oxidation of the [2Fe-2S] cluster is unclear (see section SoxR). Although stable in vitro, the DNIC forms of SoxR are rapidly lost in vivo, pointing to the existence of a system for the clearance of iron-nitrosyl species (34).
The O2 sensor FNR also appears to be involved in sensing NO. Studies of FNR (or FnrP) regulation of gene expression in Azotobacter vinelandii (165), P. denitrificans (68), and E. coli (28) showed that NO significantly diminishes FNR activation. Reaction of the FNR [4Fe-4S] cluster with NO resulted in the formation of a mixture of DNIC- and RRE-type iron-dinitrosyl species (28) (see section Wbl proteins). Anaerobic exposure of E. coli to NO led to up-regulation of multiple FNR-repressed genes and down-regulation of FNR-activated genes, including nrfA, which encodes cytochrome c nitrite reductase, providing strong evidence that there is NO inactivation of FNR (115). Apart from NsrR and NorR, other E. coli global regulators apparently affected by NO were IscR, Fur, and SoxR (115).
Wbl proteins, which are discussed above in terms of their extreme sensitivity to NO, may also respond in vivo to O2 and ROS (22, 136). In this case, the O2 reaction occurs much more slowly, but it cannot be discounted that such a response could be physiologically important.
Concluding Remarks
Iron–sulfur cluster-containing regulators, upon reaction with their analyte molecules, exhibit rich and variable chemistries, including simple cluster oxidation state change, and more complex cluster reactions, involving conversion/loss, or formation of iron nitrosyl adducts. Most act as transcriptional repressors or activators, and may bind to specific operator sequences in their cluster-bound (e.g., FNR, IscR, and NsrR) or cluster-free (e.g., IscR, NsrR, and Wbl) forms, and perhaps even in nitrosylated forms (Wbl proteins). In some cases, the regulator binds mRNA rather than DNA (aconitases), and in others, they do not bind nucleic acids at all, but rather regulate a target protein that does (e.g., NreB and RsmA). In each case, it is predicted/assumed that the cluster reaction drives a conformational change in the protein that alters its interaction with DNA (or phosphorylation activity or protein affinity) with a resulting significant effect on transcription/translation and, consequently, the cellular proteome, thus effecting an appropriate response to the changing environment.
With one notable exception, there is a lack of structural information on iron–sulfur regulators, undoubtedly reflecting the difficulties associated with working with such sensitive proteins. However, more structural information will be needed to complement the growing mechanistic understanding of how these proteins function. With recent advances it should now be possible to begin to address key questions of specificity, such as why apparently similar clusters in two different regulators exhibit widely different reactivities with the same analyte molecule, which will reveal the general principles governing the specificity of reaction of a protein-bound cluster. This has clear physiological importance, as regulators have evolved such that their cluster reactivity is tuned to enable them to sense concentrations of their analyte in a particular physiological range. Modeling studies have shown that changes in reactivity can significantly shift this range. An important related question concerns the factors that govern the specificity of any one cluster for one particular analyte over others (e.g., O2 vs. NO). Answers to these questions will enable researchers to develop more sophisticated integrated models of how cells respond to particular stresses, and what the contributions from the various regulators are. For example, S. coelicolor contains 11 Wbl proteins and NsrR, and so it is not clear whether one regulator dominates the response under particular conditions, and what the key determinants of this are. Future research will require data from in vivo studies of regulatory responses and from in vitro studies, which can provide kinetic and mechanistic information on the reaction and its products, facilitating ever-more complex (and comprehensive) modeling and leading to a greater understanding of these complex regulatory responses.
Abbreviations Used
- CRP
catabolite repressor protein
- DNIC
dinitrosyl iron complex
- EPR
electron paramagnetic resonance
- IREs
iron regulatory elements
- Isc
iron–sulfur cluster
- IscR
iron–sulfur cluster regulator
- NO
nitric oxide
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- RRE
Roussin's red ester
- Wbl
WhiB-like
Acknowledgments
Our work on iron–sulfur regulatory proteins has been generously supported by the UK's BBSRC, most recently through grants BB/G019347/1, BB/G018960/1, and BB/D00926X/1 and The Wellcome Trust, through a Joint Infrastructure Award and Grant 078731/Z/05/Z.
References
- 1.Achebach S. Selmer T. Unden G. Properties and significance of apo FNR as a second form of air-inactivated [4Fe-4S] FNR of Escherichia coli. FEBS J. 2005;272:4260–4269. doi: 10.1111/j.1742-4658.2005.04840.x. [DOI] [PubMed] [Google Scholar]
- 2.Alam MS. Garg SK. Agrawal P. Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: a [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol Microbiol. 2007;63:1414–1431. doi: 10.1111/j.1365-2958.2007.05589.x. [DOI] [PubMed] [Google Scholar]
- 3.Alen C. Sonenshein AL. Bacillus subtilis aconitase is an RNA-binding protein. Proc Natl Acad Sci U S A. 1999;96:10412–10417. doi: 10.1073/pnas.96.18.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amabilecuevas CF. Demple B. Molecular characterization of the SoxRS genes of Escherichia coli −2 genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnon DI. Whatley FR. Allen MB. Triphosphopyridine nucleotide as a catalyst of photosynthetic phosphorylation. Nature. 1957;180:182–185. doi: 10.1038/180182a0. [DOI] [PubMed] [Google Scholar]
- 6.Arras T. Schirawski J. Unden G. Availability of O2 as a substrate in the cytoplasm of bacteria under aerobic and microaerobic conditions. J Bacteriol. 1998;180:2133–2136. doi: 10.1128/jb.180.8.2133-2136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balk J. Pilon M. Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends Plant Sci. 2011;16:218–226. doi: 10.1016/j.tplants.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee S. Nandyala AK. Raviprasad P. Ahmed N. Hasnain SE. Iron-dependent RNA-binding activity of Mycobacterium tuberculosis aconitase. J Bacteriol. 2007;189:4046–4052. doi: 10.1128/JB.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bang IS. Liu L. Vazquez-Torres A. Crouch ML. Stamler JS. Fang FC. Maintenance of nitric oxide and redox homeostasis by the salmonella flavohemoglobin hmp. J Biol Chem. 2006;281:28039–28047. doi: 10.1074/jbc.M605174200. [DOI] [PubMed] [Google Scholar]
- 10.Bates DM. Popescu CV. Khoroshilova N. Vogt K. Beinert H. Munck E. Kiley PJ. Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S](2+) cluster to oxygen. J Biol Chem. 2000;275:6234–6240. doi: 10.1074/jbc.275.9.6234. [DOI] [PubMed] [Google Scholar]
- 11.Becker S. Holighaus G. Gabrielczyk T. Unden G. O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J Bacteriol. 1996;178:4515–4521. doi: 10.1128/jb.178.15.4515-4521.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beinert H. Holm RH. Munck E. Iron-sulfur clusters: nature's modular, multipurpose structures. Science. 1997;277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 13.Beinert H. Lee W. Evidence for a new type of iron containing electron carrier in mitochondria. Biochem Biophys Res Commun. 1961;5:40–45. doi: 10.1016/0006-291x(61)90077-8. [DOI] [PubMed] [Google Scholar]
- 14.Bian SM. Cowan JA. Protein-bound iron-sulfur centers. Form, function, and assembly. Coord Chem Rev. 1999;192:1049–1066. [Google Scholar]
- 15.Bodenmiller DM. Spiro S. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol. 2006;188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruckdorfer R. The basics about nitric oxide. Mol Aspects Med. 2005;26:3–31. doi: 10.1016/j.mam.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Busch JL. Breton JL. Bartlett BM. Armstrong FA. James R. Thomson AJ. [3Fe-4S] ↔ [4Fe-4S] cluster interconversion in Desulfovibrio africanus ferredoxin III: properties of an Asp14 → Cys mutant. Biochem J. 1997;323:95–102. doi: 10.1042/bj3230095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler AR. Glidewell C. Li M. Nitrosyl complexes of iron-sulfur clusters. Adv Inorg Chem. 1988;32:335–393. [Google Scholar]
- 19.Chao TC. Buhrmester J. Hansmeier N. Puhler A. Weidner S. Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation. App Environ Microbiol. 2005;71:5969–5982. doi: 10.1128/AEM.71.10.5969-5982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Constantinidou C. Hobman JL. Griffiths L. Patel MD. Penn CW. Cole JA. Overton TW. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem. 2006;281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- 21.Crack J. Green J. Thomson AJ. Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR) J Biol Chem. 2004;279:9278–9286. doi: 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- 22.Crack JC. den Hengst CD. Jakimowicz P. Subramanian S. Johnson MK. Buttner MJ. Thomson AJ. Le Brun NE. Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD. Biochemistry. 2009;48:12252–12264. doi: 10.1021/bi901498v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crack JC. Gaskell AA. Green J. Cheesmant MR. Le Brun NE. Thomson AJ. Influence of the environment on the [4Fe-4S]2+ to [2Fe-2S]2+ cluster switch in the transcriptional regulator FNR. J Am Chem Soc. 2008;130:1749–1758. doi: 10.1021/ja077455+. [DOI] [PubMed] [Google Scholar]
- 24.Crack JC. Green J. Cheesman MR. Le Brun NE. Thomson AJ. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc Natl Acad Sci U S A. 2007;104:2092–2097. doi: 10.1073/pnas.0609514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crack JC. Green J. Le Brun NE. Thomson AJ. Detection of sulfide release from the oxygen-sensing [4Fe-4S] cluster of FNR. J Biol Chem. 2006;281:18909–18913. doi: 10.1074/jbc.C600042200. [DOI] [PubMed] [Google Scholar]
- 26.Crack JC. Jervis AJ. Gaskell AA. White GF. Green J. Thomson AJ. Le Brun NE. Signal perception by FNR: the role of the iron-sulfur cluster. Biochem Soc Trans. 2008;36:1144–1148. doi: 10.1042/BST0361144. [DOI] [PubMed] [Google Scholar]
- 27.Crack JC. Smith LJ. Stapleton MR. Peck J. Watmough NJ. Buttner MJ. Buxton RS. Green J. Oganesyan VS. Thomson AJ. Le Brun NE. Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins. J Am Chem Soc. 2011;133:1112–1121. doi: 10.1021/ja109581t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruz-Ramos H. Crack J. Wu GG. Hughes MN. Scott C. Thomson AJ. Green J. Poole RK. NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daniel J. Deb C. Dubey VS. Sirakova TD. Abomoelak B. Morbidoni HR. Kolattukudy PE. Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in Mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J Bacteriol. 2004;186:5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daniel J. Maamar H. Deb C. Sirakova TD. Kolattukudy PE. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 2011;7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dibden DP. Green J. In vivo cycling of the Escherichia coli transcription factor FNR between active and inactive states. Microbiology. 2005;151:4063–4070. doi: 10.1099/mic.0.28253-0. [DOI] [PubMed] [Google Scholar]
- 32.Dietrich LEP. Price-Whelan A. Petersen A. Whiteley M. Newman DK. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol. 2006;61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 33.Dietrich LEP. Teal TK. Price-Whelan A. Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding HG. Demple B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc Natl Acad Sci U S A. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding HG. Hidalgo E. Demple B. The redox state of the [2Fe-2S] clusters in SoxR protein regulates its activity as a transcription factor. J Biol Chem. 1996;271:33173–33175. doi: 10.1074/jbc.271.52.33173. [DOI] [PubMed] [Google Scholar]
- 36.Engel P. Trageser M. Unden G. Reversible interconversion of the functional state of the gene regulator FNR from Escherichia coli in vivo by O2 and iron availability. Arch Microbiol. 1991;156:463–470. doi: 10.1007/BF00245393. [DOI] [PubMed] [Google Scholar]
- 37.Fedtke I. Kamps A. Krismer B. Gotz F. The nitrate reductase and nitrite reductase operons and the narT gene of Staphylococcus carnosus are positively controlled by the novel two-component system NreBC. J Bacteriol. 2002;184:6624–6634. doi: 10.1128/JB.184.23.6624-6634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fink RC. Evans MR. Porwollik S. Vazquez-Torres A. Jones-Carson J. Troxell B. Libby SJ. McClelland M. Hassan HM. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s) J Bacteriol. 2007;189:2262–2273. doi: 10.1128/JB.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fowler-Goldsworthy K. Gust B. Mouz S. Chandra G. Findlay KC. Chater KF. The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2) Microbiology. 2011;157:1312–1328. doi: 10.1099/mic.0.047555-0. [DOI] [PubMed] [Google Scholar]
- 40.Gao-Sheridan HS. Kemper MA. Khayat R. Tilley GJ. Armstrong FA. Sridhar V. Prasad GS. Stout CD. Burgess BK. A T14C variant of Azotobacter vinelandii ferredoxin I undergoes facile [3Fe-4S]0 to [4Fe-4S]2+ conversion in vitro but not in vivo. J Biol Chem. 1998;273:33692–33701. doi: 10.1074/jbc.273.50.33692. [DOI] [PubMed] [Google Scholar]
- 41.Garcia LL. Rivas-Marin E. Floriano B. Bernhardt R. Ewen KM. Reyes-Ramirez F. Santero E. ThnY is a ferredoxin reductase-like iron-sulfur flavoprotein that has evolved to function as a regulator of tetralin biodegradation gene expression. J Biol Chem. 2011;286:1709–1718. doi: 10.1074/jbc.M110.184648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner PR. Superoxide-driven aconitase Fe-S center cycling. Biosci Rep. 1997;17:33–42. doi: 10.1023/a:1027383100936. [DOI] [PubMed] [Google Scholar]
- 43.Gardner PR. Costantino G. Szabo C. Salzman AL. Nitric oxide sensitivity of the aconitases. J Biol Chem. 1997;272:25071–25076. doi: 10.1074/jbc.272.40.25071. [DOI] [PubMed] [Google Scholar]
- 44.Garg S. Alam MS. Bajpai R. Kishan KVR. Agrawal P. Redox biology of Mycobacterium tuberculosis H37Rv: protein-protein interaction between GlgB and WhiB1 involves exchange of thiol-disulfide. BMC Biochem. 2009;10:1. doi: 10.1186/1471-2091-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaskell AA. Crack JC. Kelemen GH. Hutchings MI. Le Brun NE. RsmA is an anti-sigma factor that modulates its activity through a [2Fe-2S] cluster cofactor. J Biol Chem. 2007;282:31812–31820. doi: 10.1074/jbc.M705160200. [DOI] [PubMed] [Google Scholar]
- 46.Gaudu P. Weiss B. SoxR, a [2Fe-2S] transcription factor, is active only in its oxidized form. Proc Natl Acad Sci U S A. 1996;93:10094–10098. doi: 10.1073/pnas.93.19.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.George SJ. Armstrong FA. Hatchikian EC. Thomson AJ. Electrochemical and spectroscopic characterization of the conversion of the 7Fe into the 8Fe form of ferredoxin III from Desulfovibrio africanus. Identification of a [4Fe-4S] cluster with one non-cysteine ligand. Biochem J. 1989;264:275–284. doi: 10.1042/bj2640275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giel JL. Rodionov D. Liu MZ. Blattner FR. Kiley PJ. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 49.Gilberthorpe NJ. Lee ME. Stevanin TM. Read RC. Poole RK. NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFN-gamma-stimulated J774.2 macrophages. Microbiology. 2007;153:1756–1771. doi: 10.1099/mic.0.2006/003731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gobert AP. McGee DJ. Akhtar M. Mendz GL. Newton JC. Cheng Y. Mobley HL. Wilson KT. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc Natl Acad Sci U S A. 2001;98:13844–13849. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorodetsky AA. Dietrich LEP. Lee PE. Demple B. Newman DK. Barton JK. DNA binding shifts the redox potential of the transcription factor SoxR. Proc Natl Acad Sci U S A. 2008;105:3684–3689. doi: 10.1073/pnas.0800093105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grainger DC. Aiba H. Hurd D. Browning DF. Busby SJ. Transcription factor distribution in Escherichia coli: studies with FNR protein. Nucleic Acids Res. 2007;35:269–278. doi: 10.1093/nar/gkl1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green J. Bennett B. Jordan P. Ralph ET. Thomson AJ. Guest JR. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem J. 1996;316:887–892. doi: 10.1042/bj3160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green J. Scott C. Guest JR. Functional versatility in the CRP-FNR superfamily of transcription factors: FNR and FLP. Adv Microb Physiol. 2001;44:1–34. doi: 10.1016/s0065-2911(01)44010-0. [DOI] [PubMed] [Google Scholar]
- 55.Green J. Sharrocks AD. Green B. Geisow M. Guest JR. Properties of FNR proteins substituted at each of the 5 cysteine residues. Mol Microbiol. 1993;8:61–68. doi: 10.1111/j.1365-2958.1993.tb01203.x. [DOI] [PubMed] [Google Scholar]
- 56.Griffith KL. Shah IM. Wolf RE., Jr. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol Microbiol. 2004;51:1801–1816. doi: 10.1046/j.1365-2958.2003.03952.x. [DOI] [PubMed] [Google Scholar]
- 57.Gruer MJ. Artymiuk PJ. Guest JR. The aconitase family: three structural variations on a common theme. Trends Biochem Sci. 1997;22:3–6. doi: 10.1016/s0968-0004(96)10069-4. [DOI] [PubMed] [Google Scholar]
- 58.Gruner I. Fradrich C. Bottger LH. Trautwein AX. Jahn D. Hartig E. Aspartate 141 Is the fourth ligand of the oxygen-sensing [4Fe-4S]2+ cluster of Bacillus subtilis transcriptional regulator Fnr. J Biol Chem. 2011;286:2017–2021. doi: 10.1074/jbc.M110.191940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu MZ. Imlay JA. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol Microbiol. 2011;79:1136–1150. doi: 10.1111/j.1365-2958.2010.07520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gusarov I. Shatalin K. Starodubtseva M. Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gusarov I. Starodubtseva M. Wang ZQ. McQuade L. Lippard SJ. Stuehr DJ. Nudler E. Bacterial nitric-oxide synthases operate without a dedicated redox partner. J Biol Chem. 2008;283:13140–13147. doi: 10.1074/jbc.M710178200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hagen T. Taylor CT. Lam F. Moncada S. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1alpha. Science. 2003;302:1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 63.Harrop TC. Tonzetich ZJ. Reisner E. Lippard SJ. Reactions of synthetic [2Fe-2S] and [4Fe-4S] clusters with nitric oxide and nitrosothiols. J Am Chem Soc. 2008;130:15602–15610. doi: 10.1021/ja8054996. [DOI] [PubMed] [Google Scholar]
- 64.Hernaez MJ. Andujar E. Rios JL. Kaschabek SR. Reineke W. Santero E. Identification of a serine hydrolase which cleaves the alicyclic ring of tetralin. J Bacteriol. 2000;182:5448–5453. doi: 10.1128/jb.182.19.5448-5453.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hidalgo E. Demple B. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 1994;13:138–146. doi: 10.1002/j.1460-2075.1994.tb06243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hidalgo E. Ding HG. Demple B. Redox signal transduction via iron-sulfur clusters in the SoxR transcription activator. Trends Biochem Sci. 1997;22:207–210. doi: 10.1016/s0968-0004(97)01068-2. [DOI] [PubMed] [Google Scholar]
- 67.Husain M. Jones-Carson J. Song M. McCollister BD. Bourret TJ. Vazquez-Torres A. Redox sensor SsrB Cys203 enhances Salmonella fitness against nitric oxide generated in the host immune response to oral infection. Proc Natl Acad Sci U S A. 2010;107:14396–14401. doi: 10.1073/pnas.1005299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hutchings MI. Crack JC. Shearer N. Thompson BJ. Thomson AJ. Spiro S. Transcription factor FnrP from Paracoccus denitrificans contains an iron-sulfur cluster and is activated by anoxia: identification of essential cysteine residues. J Bacteriol. 2002;184:503–508. doi: 10.1128/JB.184.2.503-508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Imlay J. Gu M. Many plants and bacteria excrete redox-cycling compounds. Free Rad Biol Med. 2011;50:1814–1815. doi: 10.1016/j.freeradbiomed.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 70.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Ann Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Isabella VM. Lapek JD., Jr. Kennedy EM. Clark VL. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol Microbiol. 2009;71:227–239. doi: 10.1111/j.1365-2958.2008.06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jakimowicz P. Cheesman MR. Bishai WR. Chater KF. Thomson AJ. Buttner MJ. Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe-4S] cluster. J Biol Chem. 2005;280:8309–8315. doi: 10.1074/jbc.M412622200. [DOI] [PubMed] [Google Scholar]
- 73.Jervis AJ. Crack JC. White G. Artymiuk PJ. Cheesman MR. Thomson AJ. Le Brun NE. Green J. The O2 sensitivity of the transcription factor FNR is controlled by Ser24 modulating the kinetics of [4Fe-4S] to [2Fe-2S] conversion. Proc Natl Acad Sci U S A. 2009;106:4659–4664. doi: 10.1073/pnas.0804943106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnston AW. Todd JD. Curson AR. Lei S. Nikolaidou-Katsaridou N. Gelfand MS. Rodionov DA. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals. 2007;20:501–511. doi: 10.1007/s10534-007-9085-8. [DOI] [PubMed] [Google Scholar]
- 75.Jones-Carson J. Laughlin J. Hamad MA. Stewart AL. Voskuil MI. Vazquez-Torres A. Inactivation of [Fe-S] metalloproteins mediates nitric oxide-dependent killing of Burkholderia mallei. PLoS One. 2008;3:e1976. doi: 10.1371/journal.pone.0001976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jordan PA. Tang Y. Bradbury AJ. Thomson AJ. Guest JR. Biochemical and spectroscopic characterization of Escherichia coli aconitases (AcnA and AcnB) Biochem J. 1999;344:739–746. [PMC free article] [PubMed] [Google Scholar]
- 77.Jordan PA. Thomson AJ. Ralph ET. Guest JR. Green J. FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett. 1997;416:349–352. doi: 10.1016/s0014-5793(97)01219-2. [DOI] [PubMed] [Google Scholar]
- 78.Kamps A. Achebach S. Fedtke I. Unden G. Gotz F. Staphylococcal NreB: an O2-sensing histidine protein kinase with an O2-labile iron-sulphur cluster of the FNR type. Mol Microbiol. 2004;52:713–723. doi: 10.1111/j.1365-2958.2004.04024.x. [DOI] [PubMed] [Google Scholar]
- 79.Kang Y. Weber KD. Qiu Y. Kiley PJ. Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol. 2005;187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khoroshilova N. Beinert H. Kiley PJ. Associtation of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA-binding. Proc Natl Acad Sci U S A. 1995;92:2499–2503. doi: 10.1073/pnas.92.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khoroshilova N. Popescu C. Munck E. Beinert H. Kiley PJ. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci U S A. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kiley PJ. Beinert H. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol Rev. 1998;22:341–352. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 83.Kiley PJ. Beinert H. The role of Fe-S proteins in sensing and regulation in bacteria. Curr Opin Microbiol. 2003;6:181–185. doi: 10.1016/s1369-5274(03)00039-0. [DOI] [PubMed] [Google Scholar]
- 84.Kim SH. Lee BY. Lau GW. Cho YH. IscR modulates catalase A (KatA) activity, peroxide resistance, and full virulence of Pseudomonas aeruginosa PA14. J Microbiol Biotech. 2009;19:1520–1526. doi: 10.4014/jmb.0906.06028. [DOI] [PubMed] [Google Scholar]
- 85.Kommineni S. Yukl E. Hayashi T. Delepine J. Geng H. Moenne-Loccoz P. Nakano MM. Nitric oxide-sensitive and -insensitive interaction of Bacillus subtilis NsrR with a ResDE-controlled promoter. Mol Microbiol. 2010;78:1280–1293. doi: 10.1111/j.1365-2958.2010.07407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koo MS. Lee JH. Rah SY. Yeo WS. Lee JW. Lee KL. Koh YS. Kang SO. Roe JH. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Korner H. Sofia HJ. Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- 88.Lanzilotta WN. Schuller DJ. Thorsteinsson MV. Kerby RL. Roberts GP. Poulos TL. Structure of the CO sensing transcription activator CooA. Nat Struct Biol. 2000;7:876–880. doi: 10.1038/82820. [DOI] [PubMed] [Google Scholar]
- 89.Lazazzera BA. Beinert H. Khoroshilova N. Kennedy MC. Kiley PJ. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 90.Lee EJ. Karoonuthaisiri N. Kim HS. Park JH. Cha CJ. Kao CM. Roe JH. A master regulator sigmaB governs osmotic and oxidative response as well as differentiation via a network of sigma factors in Streptomyces coelicolor. Mol Microbiol. 2005;57:1252–1264. doi: 10.1111/j.1365-2958.2005.04761.x. [DOI] [PubMed] [Google Scholar]
- 91.Lee JH. Yeo WS. Roe JH. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol Microbiol. 2004;51:1745–1755. doi: 10.1111/j.1365-2958.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- 92.Lee PE. Demple B. Barton JK. DNA-mediated redox signaling for transcriptional activation of SoxR. Proc Natl Acad Sci U S A. 2009;106:13164–13168. doi: 10.1073/pnas.0906429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liochev SI. Fridovich I. Is superoxide able to induce SoxRS? Free Rad Biol Med. 2011;50:1813. doi: 10.1016/j.freeradbiomed.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 94.Macomber L. Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marshall FA. Messenger SL. Wyborn NR. Guest JR. Wing H. Busby SJ. Green J. A novel promoter architecture for microaerobic activation by the anaerobic transcription factor FNR. Mol Microbiol. 2001;39:747–753. doi: 10.1046/j.1365-2958.2001.02262.x. [DOI] [PubMed] [Google Scholar]
- 96.Marteyn B. West NP. Browning DF. Cole JA. Shaw JG. Palm F. Mounier J. Prevost MC. Sansonetti P. Tang CM. Modulation of Shigella virulence in response to available oxygen in vivo. Nature. 2010;465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCollister BD. Hoffman M. Husain M. Vazquez-Torres A. Nitric oxide protects bacteria from aminoglycosides by blocking the energy-dependent phases of drug uptake. Antimicrob Agents Chemother. 2011;55:2189–2196. doi: 10.1128/AAC.01203-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mettert EL. Kiley PJ. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J Mol Biol. 2005;354:220–232. doi: 10.1016/j.jmb.2005.09.066. [DOI] [PubMed] [Google Scholar]
- 99.Mettert EL. Outten FW. Wanta B. Kiley PJ. The impact of O2 on the Fe-S cluster biogenesis requirements of Escherichia coli FNR. J Mol Biol. 2008;384:798–811. doi: 10.1016/j.jmb.2008.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miranda KM. Espey MG. Jourd'heuil D. Grisham MB. Fukuto JM. Feelisch M. Wink DA. Chemical biology of nitric oxide. In: Ignarro LJ, editor. Nitric Oxide Biology and Pathobiology. 2nd. San Diego, CA: Academic Press; 2009. pp. 41–55. [Google Scholar]
- 101.Molle V. Palframan WJ. Findlay KC. Buttner MJ. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3(2) J Bacteriol. 2000;182:1286–1295. doi: 10.1128/jb.182.5.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moore LJ. Kiley PJ. Characterization of the dimerization domain in the FNR transcription factor. J Biol Chem. 2001;276:45744–45750. doi: 10.1074/jbc.M106569200. [DOI] [PubMed] [Google Scholar]
- 103.Morris RP. Nguyen L. Gatfield J. Visconti K. Nguyen K. Schnappinger D. Ehrt S. Liu Y. Heifets L. Pieters J. Schoolnik G. Thompson CJ. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2005;102:12200–12205. doi: 10.1073/pnas.0505446102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mukhopadhyay P. Zheng M. Bedzyk LA. LaRossa RA. Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci U S A. 2004;101:745–750. doi: 10.1073/pnas.0307741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mullner M. Hammel O. Mienert B. Schlag S. Bill E. Unden G. A PAS domain with an oxygen labile [4Fe-4S]2+ cluster in the oxygen sensor kinase NreB of Staphylococcus carnosus. Biochemistry. 2008;47:13921–13932. doi: 10.1021/bi8014086. [DOI] [PubMed] [Google Scholar]
- 106.Nesbit AD. Giel JL. Rose JC. Kiley PJ. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J Mol Biol. 2009;387:28–41. doi: 10.1016/j.jmb.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nishimura T. Teramoto H. Vertes AA. Inui M. Yukawa H. ArnR, a novel transcriptional regulator, represses expression of the narKGHJI operon in Corynebacterium glutamicum. J Bacteriol. 2008;190:3264–3273. doi: 10.1128/JB.01801-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nishimura T. Vertes AA. Shinoda Y. Inui M. Yukawa H. Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. App Microbiol Biotechnol. 2007;75:889–897. doi: 10.1007/s00253-007-0879-y. [DOI] [PubMed] [Google Scholar]
- 109.Nunoshiba T. deRojas-Walker T. Wishnok JS. Tannenbaum SR. Demple B. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc Natl Acad Sci U S A. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Outten FW. Djaman O. Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 111.Partridge JD. Bodenmiller DM. Humphrys MS. Spiro S. NsrR targets in the Escherichia coli genome: new insights into DNA sequence requirements for binding and a role for NsrR in the regulation of motility. Mol Microbiol. 2009;73:680–694. doi: 10.1111/j.1365-2958.2009.06799.x. [DOI] [PubMed] [Google Scholar]
- 112.Pomposiello PJ. Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19:109–114. doi: 10.1016/s0167-7799(00)01542-0. [DOI] [PubMed] [Google Scholar]
- 113.Poole RK. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem Soc Trans. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 114.Popescu CV. Bates DM. Beinert H. Munck E. Kiley PJ. Mössbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:13431–13435. doi: 10.1073/pnas.95.23.13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pullan ST. Gidley MD. Jones RA. Barrett J. Stevanin TA. Read RC. Green J. Poole RK. Nitric oxide in chemostat-cultured Escherichia coli is sensed by Fnr and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J Bacteriol. 2007;189:1845–1855. doi: 10.1128/JB.01354-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ranquet C. Ollagnier-de-Choudens S. Loiseau L. Barras F. Fontecave M. Cobalt stress in Escherichia coli. The effect on the iron-sulfur proteins. J Biol Chem. 2007;282:30442–30451. doi: 10.1074/jbc.M702519200. [DOI] [PubMed] [Google Scholar]
- 117.Recalcati S. Minotti G. Cairo G. Iron regulatory proteins: from molecular mechanisms to drug development. Antioxid Redox Signal. 2010;13:1593–1616. doi: 10.1089/ars.2009.2983. [DOI] [PubMed] [Google Scholar]
- 118.Reents H. Gruner I. Harmening U. Bottger LH. Layer G. Heathcote P. Trautwein AX. Jahn D. Hartig E. Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state. Mol Microbiol. 2006;60:1432–1445. doi: 10.1111/j.1365-2958.2006.05198.x. [DOI] [PubMed] [Google Scholar]
- 119.Reinhart F. Achebach S. Koch T. Unden G. Reduced apo-fumarate nitrate reductase regulator (ApoFNR) as the major form of FNR in aerobically growing Escherichia coli. J Bacteriol. 2008;190:879–886. doi: 10.1128/JB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rhee KY. Erdjument-Bromage H. Tempst P. Nathan CF. S-nitroso proteome of Mycobacterium tuberculosis: enzymes of intermediary metabolism and antioxidant defense. Proc Natl Acad Sci U S A. 2005;102:467–472. doi: 10.1073/pnas.0406133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rincon-Enriquez G. Crete P. Barras F. Py B. Biogenesis of Fe/S proteins and pathogenicity: IscR plays a key role in allowing Erwinia chrysanthemi to adapt to hostile conditions. Mol Microbiol. 2008;67:1257–1273. doi: 10.1111/j.1365-2958.2008.06118.x. [DOI] [PubMed] [Google Scholar]
- 122.Rodionov DA. Dubchak IL. Arkin AP. Alm EJ. Gelfand MS. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol. 2005;1:e55. doi: 10.1371/journal.pcbi.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rodionov DA. Gelfand MS. Todd JD. Curson ARJ. Johnston AWB. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-proteobacteria. PLoS Comp Biol. 2006;2:1568–1585. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Runyen-Janecky L. Daugherty A. Lloyd B. Wellington C. Eskandarian H. Sagransky M. Role and regulation of iron-sulfur cluster biosynthesis genes in Shigella flexneri virulence. Infect Immun. 2008;76:1083–1092. doi: 10.1128/IAI.01211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Salmon K. Hung SP. Mekjian K. Baldi P. Hatfield GW. Gunsalus RP. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J Biol Chem. 2003;278:29837–29855. doi: 10.1074/jbc.M213060200. [DOI] [PubMed] [Google Scholar]
- 126.Schultz SC. Shields GC. Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 127.Schwartz CJ. Djaman O. Imlay JA. Kiley PJ. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:9009–9014. doi: 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Schwartz CJ. Giel JL. Patschkowski T. Luther C. Ruzicka FJ. Beinert H. Kiley PJ. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci U S A. 2001;98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Scott C. Partridge JD. Stephenson JR. Green J. DNA target sequence and FNR-dependent gene expression. FEBS Lett. 2003;541:97–101. doi: 10.1016/s0014-5793(03)00312-0. [DOI] [PubMed] [Google Scholar]
- 130.Serio AW. Pechter KB. Sonenshein AL. Bacillus subtilis aconitase is required for efficient late-sporulation gene expression. J Bacteriol. 2006;188:6396–6405. doi: 10.1128/JB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shaw DJ. Rice DW. Guest JR. Homology between CAP and FNR, a regulator of anaerobic respiration in Escherichia coli. J Mol Biol. 1983;166:241–247. doi: 10.1016/s0022-2836(83)80011-4. [DOI] [PubMed] [Google Scholar]
- 132.Shen G. Balasubramanian R. Wang T. Wu Y. Hoffart LM. Krebs C. Bryant DA. Golbeck JH. SufR coordinates two [4Fe-4S](2+,1+) clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator of sufR in cyanobacteria. J Biol Chem. 2007;282:31909–31919. doi: 10.1074/jbc.M705554200. [DOI] [PubMed] [Google Scholar]
- 133.Shepard W. Soutourina O. Courtois E. England P. Haouz A. Martin-Verstraete I. Insights into the Rrf2 repressor family—the structure of CymR, the global cysteine regulator of Bacillus subtilis. FEBS J. 2011;278:2689–2701. doi: 10.1111/j.1742-4658.2011.08195.x. [DOI] [PubMed] [Google Scholar]
- 134.Shin JH. Singh AK. Cheon DJ. Roe JH. Activation of the SoxR regulon in Streptomyces coelicolor by the extracellular form of the pigmented antibiotic actinorhodin. J Bacteriol. 2011;193:75–81. doi: 10.1128/JB.00965-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Singh A. Crossman DK. Mai D. Guidry L. Voskuil MI. Renfrow MB. Steyn AJC. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Singh A. Guidry L. Narasimhulu KV. Mai D. Trombley J. Redding KE. Giles GI. Lancaster JR. Steyn AJC. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc Natl Acad Sci U S A. 2007;104:11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Singleton C. White GF. Todd JD. Marritt SJ. Cheesman MR. Johnston AW. Le Brun NE. Heme-responsive DNA binding by the global iron regulator Irr from Rhizobium leguminosarum. J Biol Chem. 2010;285:16023–16031. doi: 10.1074/jbc.M109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Small SK. Puri S. O'Brian MR. Heme-dependent metalloregulation by the iron response regulator (Irr) protein in Rhizobium and other Alpha-proteobacteria. Biometals. 2009;22:89–97. doi: 10.1007/s10534-008-9192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smith LJ. Stapleton MR. Fullstone GJM. Crack JC. Thomson AJ. Le Brun NE. Hunt DM. Harvey E. Adinolfi S. Buxton RS. Green J. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem J. 2010;432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Soliveri JA. Gomez J. Bishai WR. Chater KF. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology. 2000;146:333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- 141.Spiro S. Regulators of bacterial responses to nitric oxide. FEMS Microbiol Rev. 2007;31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- 142.Sutton VR. Mettert EL. Beinert H. Kiley PJ. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J Bacteriol. 2004;186:8018–8025. doi: 10.1128/JB.186.23.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sutton VR. Stubna A. Patschkowski T. Munck E. Beinert H. Kiley PJ. Superoxide destroys the [2Fe-2S]2+ cluster of FNR from Escherichia coli. Biochemistry. 2004;43:791–798. doi: 10.1021/bi0357053. [DOI] [PubMed] [Google Scholar]
- 144.Takahashi Y. Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 145.Tang Y. Guest JR. Direct evidence for mRNA binding and post-transcriptional regulation by Escherichia coli aconitases. Microbiology. 1999;145:3069–3079. doi: 10.1099/00221287-145-11-3069. [DOI] [PubMed] [Google Scholar]
- 146.Tang Y. Guest JR. Artymiuk PJ. Green J. Switching aconitase B between catalytic and regulatory modes involves iron-dependent dimer formation. Mol Microbiol. 2005;56:1149–1158. doi: 10.1111/j.1365-2958.2005.04610.x. [DOI] [PubMed] [Google Scholar]
- 147.Tang Y. Guest JR. Artymiuk PJ. Read RC. Green J. Post-transcriptional regulation of bacterial motility by aconitase proteins. Mol Microbiol. 2004;51:1817–1826. doi: 10.1111/j.1365-2958.2003.03954.x. [DOI] [PubMed] [Google Scholar]
- 148.Tang Y. Quail MA. Artymiuk PJ. Guest JR. Green J. Escherichia coli aconitases and oxidative stress: post-transcriptional regulation of sodA expression. Microbiology. 2002;148:1027–1037. doi: 10.1099/00221287-148-4-1027. [DOI] [PubMed] [Google Scholar]
- 149.Tinberg CE. Tonzetich ZJ. Wang H. Do LH. Yoda Y. Cramer SP. Lippard SJ. Characterization of iron dinitrosyl species formed in the reaction of nitric oxide with a biological Rieske center. J Am Chem Soc. 2010;132:18168–18176. doi: 10.1021/ja106290p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Todd JD. Sawers G. Johnston AWB. Proteomic analysis reveals the wide-ranging effects of the novel, iron-responsive regulator RirA in Rhizobium leguminosarum bv. viciae. Mol Gen Genom. 2005;273:197–206. doi: 10.1007/s00438-005-1127-8. [DOI] [PubMed] [Google Scholar]
- 151.Todd JD. Wexler M. Sawers G. Yeoman KH. Poole PS. Johnston AWB. RirA, an iron-responsive regulator in the symbiotic bacterium Rhizobium leguminosarum. Microbiology. 2002;148:4059–4071. doi: 10.1099/00221287-148-12-4059. [DOI] [PubMed] [Google Scholar]
- 152.Tolla DA. Savageau MA. Regulation of aerobic-to-anaerobic transitions by the FNR cycle in Escherichia coli. J Mol Biol. 2010;397:893–905. doi: 10.1016/j.jmb.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tucker NP. Hicks MG. Clarke TA. Crack JC. Chandra G. Le Brun NE. Dixon R. Hutchings MI. The transcriptional repressor protein NsrR senses nitric oxide directly via a [2Fe-2S] cluster. PLoS One. 2008;3:e3623. doi: 10.1371/journal.pone.0003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tucker NP. Le Brun NE. Dixon R. Hutchings MI. There's NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol. 2010;18:149–156. doi: 10.1016/j.tim.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 155.Unden G. Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta. 1997;1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- 156.Vanin AF. Dinitrosyl iron complexes with thiolate ligands: physico-chemistry, biochemistry and physiology. Nitric Oxide. 2009;21:1–13. doi: 10.1016/j.niox.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 157.Volz K. The functional duality of iron regulatory protein 1. Curr Opin Struct Biol. 2008;18:106–111. doi: 10.1016/j.sbi.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang T. Shen GZ. Balasubramanian R. McIntosh L. Bryant DA. Golbeck JH. The sufR gene (sll0088 in Synechocystis sp strain PCC 6803) functions as a repressor of the sufBCDS operon in iron-sulfur cluster biogenesis in cyanobacteria. J Bacteriol. 2004;186:956–967. doi: 10.1128/JB.186.4.956-967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang Y. Dunn AK. Wilneff J. McFall-Ngai MJ. Spiro S. Ruby EG. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol. 2010;78:903–915. doi: 10.1111/j.1365-2958.2010.07376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Watanabe S. Kita A. Kobayashi K. Miki K. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc Natl Acad Sci U S A. 2008;105:4121–4126. doi: 10.1073/pnas.0709188105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Watmough NJ. Butland G. Cheesman MR. Moir JW. Richardson DJ. Spiro S. Nitric oxide in bacteria: synthesis and consumption. Biochim Biophys Acta. 1999;1411:456–474. doi: 10.1016/s0005-2728(99)00032-8. [DOI] [PubMed] [Google Scholar]
- 162.Weber IT. Steitz TA. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 Å resolution. J Mol Biol. 1987;198:311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- 163.Williams CH. Stillman TJ. Barynin VV. Sedelnikova SE. Tang Y. Green J. Guest JR. Artymiuk PJ. E. coli aconitase B structure reveals a HEAT-like domain with implications for protein-protein recognition. Nat Struct Biol. 2002;9:447–452. doi: 10.1038/nsb801. [DOI] [PubMed] [Google Scholar]
- 164.Wilson TJ. Bertrand N. Tang JL. Feng JX. Pan MQ. Barber CE. Dow JM. Daniels MJ. The rpfA gene of Xanthomonas campestris pathovar campestris, which is involved in the regulation of pathogenicity factor production, encodes an aconitase. Mol Microbiol. 1998;28:961–970. doi: 10.1046/j.1365-2958.1998.00852.x. [DOI] [PubMed] [Google Scholar]
- 165.Wu G. Cruz-Ramos H. Hill S. Green J. Sawers G. Poole RK. Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr). Sensitivity to oxygen, reactive oxygen species, and nitric oxide. J Biol Chem. 2000;275:4679–4686. doi: 10.1074/jbc.275.7.4679. [DOI] [PubMed] [Google Scholar]
- 166.Wu Y. Outten FW. IscR controls iron-dependent biofilm formation in Escherichia coli by regulating type I fimbria expression. J Bacteriol. 2009;191:1248–1257. doi: 10.1128/JB.01086-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yeo WS. Lee JH. Lee KC. Roe JH. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol. 2006;61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 168.Yeoman KH. Curson ARJ. Todd JD. Sawers G. Johnston AWB. Evidence that the Rhizobium regulatory protein RirA binds to cis-acting iron-responsive operators (IROs) at promoters of some Fe-regulated genes. Microbiology. 2004;150:4065–4074. doi: 10.1099/mic.0.27419-0. [DOI] [PubMed] [Google Scholar]
- 169.Yukl ET. Elbaz MA. Nakano MM. Moenne-Loccoz P. Transcription factor NsrR from Bacillus subtilis senses nitric oxide with a 4Fe-4S cluster. Biochemistry. 2008;47:13084–13092. doi: 10.1021/bi801342x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zeng J. Zhang XJ. Wang YP. Ai CB. Liu Q. Qiu GZ. Glu43 is an essential residue for coordinating the [Fe2S2] cluster of IscR from Acidithiobacillus ferrooxidans. FEBS Lett. 2008;582:3889–3892. doi: 10.1016/j.febslet.2008.09.060. [DOI] [PubMed] [Google Scholar]