Abstract
RIG-I (Retinoic acid Inducible Gene - I) is a cytoplasmic pathogen recognition receptor that recognizes pathogen-associated molecular pattern (PAMP) motifs to differentiate between viral and cellular RNAs. RIG-I is activated by blunt-ended double-stranded (ds) RNA with or without a 5′-triphosphate (ppp), single-stranded (ss) RNA marked by 5′-ppp1 and poly-uridine sequence2,3. Upon binding to such PAMP motifs, RIG-I initiates a signaling cascade that induces innate immune defenses and inflammatory cytokines to establish an antiviral state. The RIG-I pathway is highly regulated and aberrant signaling leads to apoptosis, altered cell differentiation, inflammation, autoimmune diseases, and cancer4,5. The helicase and repressor domain (RD) of RIG-I recognize dsRNA and 5′-ppp RNA to activate the amino-terminal two CAspase Recruitment Domains (CARDs) for signaling. To understand the synergy between helicase and RD for RNA binding and the contribution of ATP hydrolysis to RIG-I activation, we determined the structure of human RIG-I helicase-RD in complex with dsRNA and an ATP-analog. The helicase-RD organizes into a ring around dsRNA, capping one end, while contacting both strands utilizing previously uncharacterized motifs to recognize dsRNA. Small angle X-ray scattering (SAXS), limited proteolysis, and differential scanning fluorimetry (DSF) suggest that RIG-I is in an extended and flexible conformation that compacts upon binding RNA. These results provide a detailed view of the helicase role in dsRNA recognition, the synergy between RD and the helicase for RNA binding, organization of full-length RIG-I bound to dsRNA, and evidence of a conformational change upon RNA binding. The RIG-I helicase-RD structure is consistent with dsRNA translocation without unwinding and cooperative binding to RNA. The structure yields unprecedented insight into innate immunity and has broader impact into other areas of biology, including RNA interference and DNA repair, which utilize homologous helicase domains within Dicer and FANCM.
To investigate the contributions of the individual domains of RIG-I to RNA binding, we employed fluorescence anisotropy and determined the equilibrium dissociation constant (Kd) of the protein-RNA complexes. The tightest RNA affinity was observed with the helicase-RD, while the full-length RIG-I, helicase domain, and RD bind dsRNA with a 24-fold, 8600-fold, and 50-fold weaker affinity, respectively, and all proteins bind RNA with a 1:1 stoichiometry (Table 1 and Supplemental Fig. 1). Consistent with the affinities, full-length RIG-I and helicase-RD demonstrated robust dsRNA stimulated ATPase activity, whereas the helicase domain showed a weak ATPase (Table 1). Interestingly, the presence of a 5′-ppp in dsRNA does not alter the stoichiometry of RNA binding, although the 5′-ppp dsRNA binds more tightly than the 5′-OH dsRNA to full-length RIG-I.
Table 1.
Summary of ATPase rates and affinity measurements of dsRNA binding by RD, Helicase, and Helicase-RD and Full length RIG-I in the presence and absence of ADP•BeF3
| ATPase | Affinity measurements* | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| RNA | ATPase Activity M/(M×s) | RNA | Nucleotide analog | Dissociation constant (Kd, nM) | Final Anisotropy | |
| RD | - | - | 14bp dsRNA-F | No nucleotide | 2.6 ± 1.20 | 0.109 |
|
| ||||||
| Helicase | 14bp dsRNA | 0.89±0.04 | 14bp dsRNA-F | No nucleotide | 436 ± 121 | 0.138 |
| -RNA | undetectable | ADP•BeF3 | 1300 ± 300** | 0.138 | ||
|
| ||||||
| Helicase- RD | 14bp dsRNA | 21.8±0.70 | 14bp dsRNA-F | No nucleotide | 0.05 ± 0.02 | 0.213 |
| 14bp pal dsRNA | 24.4±0.21 | ADP•BeF3 | 0.02 ± 0.07 | 0.280 | ||
|
|
||||||
| -RNA | undetectable | 5′-ppp14bp dsRNA-F | No nucleotide | 0.03 ± 0.03 | 0.190 | |
|
| ||||||
| Full- length RIG-I | 14bp dsRNA | 28.2±1.42 | 14bp dsRNA-F | No nucleotide | 1.2±0.6** | 0.187 |
| -RNA | undetectable | 5′-ppp 14bp dsRNA | No nucleotide | 0.44±0.27 | 0.183 | |
Mean ATPase rate constant from two independent measurements and range is shown.
Affinity measurements and range from two or more independent experiments are shown.
Initial anisotropy of the free fluorescein-labeled dsRNA was 0.10 for all experiments.
Data was fitted to a 1:1 (protein:RNA) binding model
Value from single measurement and fitting error is shown.
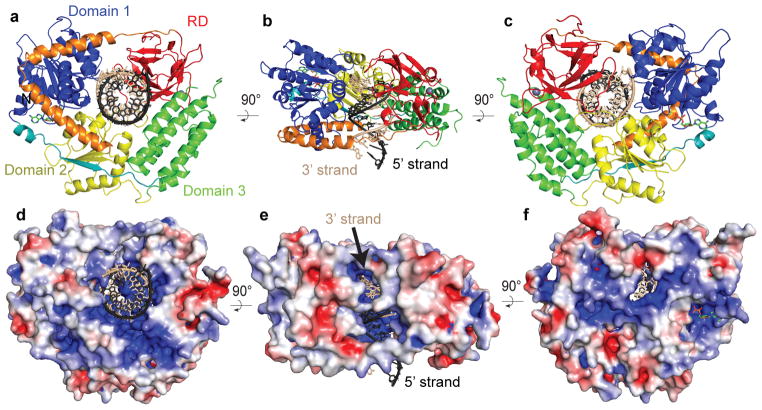
Crystals of RIG-I helicase-RD in complex with ADP•BeF3 and 14 base pair palindromic (pal) dsRNA diffracted to 2.9 Å resolution (Supplementary Table 1). The dsRNA maintains an A-form helical conformation and interacts with all four domains of helicase-RD, which are arranged into a ring around dsRNA (Fig. 1a–c). The RecA-like helicase domain (domain 1) progresses to an alpha helical domain (domain 3) and into the second RecA-like helicase domain (domain 2) that ends with the RD. The striking features of the RIG-I structure are the linkers that connect the domains. Domains 1 and 3 are connected by a beta strand that is part of the parallel beta sheet in domain 2. The RD is connected to domain 2 via a prominent V-shaped linker, consisting of two alpha helices that interact extensively with domains 1 and 2 with a buried surface over 1,500 Å2. The V-shaped linker extends into a proline-rich (796KPKPVPD) loop that makes the final connection to the RD. Interestingly, T770 at the vertex of the V-shaped linker is one of the phosphorylation sites that regulates RIG-I signaling6.
Figure 1. Structural overview of RIG-I helicase-RD.
(a, b, and c) Schematic representation of the RIG-I helicase-RD, highlighting the RecA-like domain 1 (blue), the alpha-helical domain 3 (green), RecA-like helicase domain 2 (yellow), and RD (red). The linker connecting Domain 1 with Domain 3 is colored teal, while the V-shape linker between Domain 2 and RD is colored orange. The ADP•BeF3 and dsRNA are shown in stick representation with the 5′ and 3′ strands of the RNA colored black and beige, respectively. A grey sphere denotes the position of the zinc ion in RD. The 3′ and 5′ strands are colored beige and black, respectively. (d, e, f) Surface of RIG-I helicase-RD colored for electrostatic potential at ±5 kT/e; blue (basic), white (neutral), and red (acidic). The views in panels a, b, and c are identical to d, e and f, respectively.
Molecular surface analysis shows that one end of the dsRNA is capped by the entire helicase-RD molecule while the opposite end of the RNA is exposed (Fig. 1d–f). At the capped end, the 5′ terminus of the dsRNA abuts the RD domain but the 3′ terminus is somewhat exposed through a highly basic channel (Figs. 1b,e). The 3′ terminus RNA strand will be referred to as the 3′ strand, while the opposite strand as the 5′ strand. This basic channel may allow RIG-I to recognize dsRNA with 3′ nucleotide overhangs or 3′ monophosphate that are products of RNase L digestion7. Additionally there are visible channels along the long axis of the dsRNA, which may allow the domains to flex and accommodate bulges or noncanonical base pairs (Fig. 1e–1f).
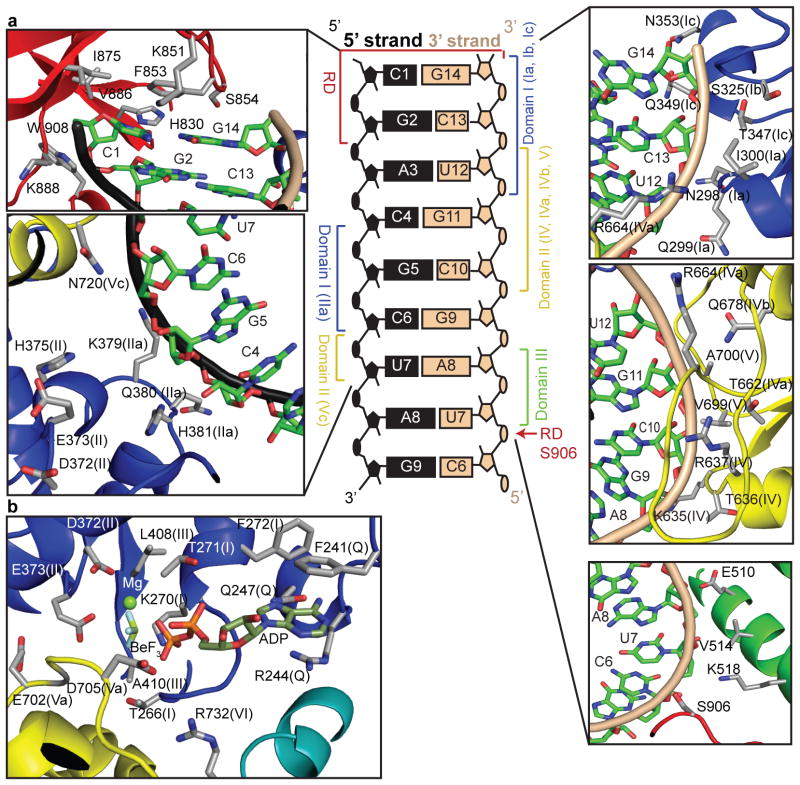
All four domains of the helicase-RD participate in dsRNA binding, burying a total of 1,500 Å2 of surface area to encircle about eight base pairs (Fig. 2a). Most contacts are with the sugar phosphate backbone of both strands but there are few base specific interactions. The F853 in the RD stacks over the terminal C1-G14 base pair, and H830 and S854 form hydrogen bonds with the ribose 2′-OH of C1 (5′-strand) and G14 (3′-strand), respectively. Phosphorylation of S854 and S855, shown to negatively regulate RIG-I signaling6, would be predicted to adversely impact RNA binding. The RD-RNA contacts in the helicase-RD structure are identical to those identified in the structures of the isolated RD bound to dsRNA8–10. Residues H847, K858, and K861 of RD, reported to interact with the 5′-ppp, are not making any new interactions in the helicase-RD structure. The only RD contact with the 3′ strand, other than at the blunt end, is between S906 and the U7 phosphate backbone.
Figure 2. Interactions of RIG-I helicase-RD with dsRNA and ADP•BeF3.
A schematic representation showing the interactions between RIG-I domains and helicase motifs (given in parentheses) with dsRNA is located in the center. Detailed contacts are shown in the surrounding panels. Stick representation detailing the RIG-I helicase motifs interactions with ADP•BeF3 and Mg2+ is shown in the lower left panel.
The core helicase (domains 1 and 2) contains characteristic motifs critical for RNA binding and ATP hydrolysis11 (Supplementary Fig. 3). The two helicase domains together contact five nucleotides from the 3′-end of the 3′-strand (G14-C10) and four nucleotides in the middle of the 5′-strand (C4-U7) (Fig. 2a). Motif Ib (S325 and G326) and motif Ic (T347, Q349 and N353) of domain 1 contact the ribose-phosphate backbone of the terminal base of the 3′-strand (G14), while motif Ia (N298, Q299, and I300) interacts with C13 and U12. Motifs IV, IVa, IVb, and V in domain 2 interact with the 3′-strand from C13 to C10. Residues in motif IV (K635, T636, and R637) interact with the phosphate backbone of G11-C10. Similarly motif IVa (T662, G663, and R664) and motif IVb (Q678) contact the backbone of C13-G11, and motif V (V699) interacts with the RNA backbone at C10.
All RIG-I-like helicases contain a large insertion (domain 3) between the core helicase domains12. The structure of the RIG-I like helicase, Hef, showed a similar alpha-helical domain 3, except it is rotated in RIG-I due to dsRNA interactions (Supplementary Fig. 4). An alpha helix within domain 3 (residue 506–522) of RIG-I runs almost perpendicular to the minor groove of the dsRNA and interacts with the 3′ strand without contacting the 5′-strand (Fig. 2a). The interactions of the domain 3 with the dsRNA via residues E510, V514 and K518 extend the helicase contacts to U7 and A8 of the 3′-strand.
Although most interactions of the RIG-I are with the 3′ strand, all three helicase domains are in close proximity to the 5′-strand and two new motifs contact the 5′ strand. Motif IIa (379KQHPY) immediately follows motif II (372DECH) and interacts with C4–C6 of the 5′ strand. Similarly, N720 in motif Vc in domain 2 contacts the 5′ strand backbone at C6 and U7. These new motifs may represent a general feature of helicases binding to dsRNA or dsDNA since a region similar to motif IIa was identified in the Swi/Snf2 family helicases and a Rad54 homolog dsDNA complex structure13.
RIG-I helicase-RD contains one ADP•BeF3 molecule bound at the interface of domains 1 and 2 via the conserved helicase motifs Q, I, II, III, V, and VI, which are generally involved in ATP binding/hydrolysis (Fig. 2b)12. The Q motif makes adenine specific contacts, while the other motifs are involved in binding the triphosphate moiety and the Mg2+. Motif I (267GCGKT) contacts the phosphates and the BeF3, which mimics the γ-phosphate of ATP as observed in other SF2 helicases12. The D372 and E373 of motif II (372DECH) coordinate the Mg2+ to stabilize the ATPanalog. The helicase motifs Va and VI, which contact the ribose and phosphates of the ATP in other helicases, are close to ADP•BeF3.
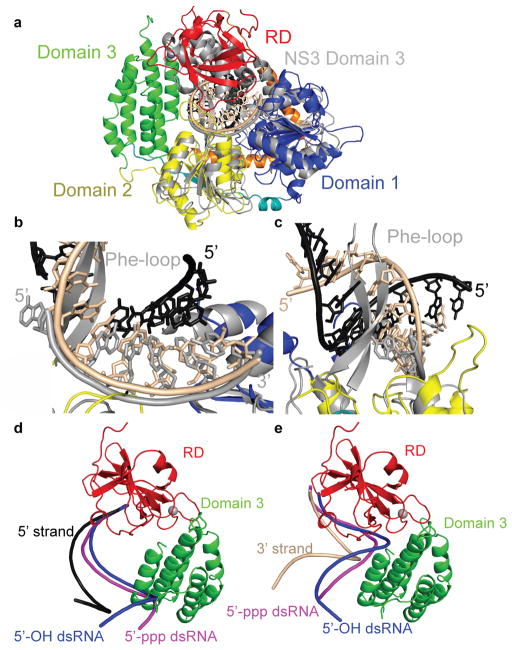
RIG-I has been shown to have translocation activity on dsRNA14 and reported to unwind short dsRNA15, although we (data not shown) and others14 have failed to detect unwinding activity. Superposition of RIG-I with hepatitis C virus (HCV) NS3 helicase (NS3h) bound to ssDNA16 (Fig. 3a) makes predictions about translocation and lack of helicase activity of RIG-I. First, ssDNA in NS3h overlays with the 3′-strand of the dsRNA bound to RIG-I, suggesting that the RIG-I helicase makes principal motor contacts with the 3′-strand (Fig. 3b) when translocating along dsRNA. Second, a conserved Phe-loop in NS3h bisects the dsRNA, consistent with its implicated role in unwinding17 (Fig. 3b–c). The absence of such a motif in RIG-I could explain the lack of RNA unwinding activity. Interestingly, the RIG-I RD positions over domain 3 of NS3h, implicating that this domain of NS3h may interact with the 5′ strand of dsRNA (Fig. 3a).
Figure 3. Comparison of RIG-I helicase RD with HCV NS3h and RD bound to 5′-OH and 5′-ppp dsRNA.
(a, b, and c) Ribbons diagram showing the superposition of RIG-I helicase-RD•dsRNA•ADP•BeF3 structure and NS3h bound to ssDNA (PDB code 3KQH) (grey). The helicase core domains 1 and 2 from RIG-I helicase-RD superimpose well, while domain 3 of NS3h is positioned over the RD. (b) Superposition of RIG-I helicase-RD with NS3h demonstrates that the ssDNA bound to NS3h overlays with the 3′-strand (beige) of the dsRNA bound to the helicase-RD. (c) The location of the Phe-loop of NS3h relative to the dsRNA of the RIG-I helicase-RD•dsRNA•ADP•BeF3 structure. (d and e) Superposition of the 5′-OH (blue; PDB code 3OG8) and 5′-ppp dsRNA (magenta; PDB code 3LRR) based on the location of RD. For clarity the 5′ strands (d) and 3′ strands (e) are shown separately.
The RD superimposes well with a root mean square deviation of ~0.6 Å with the structures of isolated RD bound to dsRNA with and without 5′-ppp8–10. However, the trajectory of the RNA helix in the helicase-RD is different. Superposition of the three RD structures demonstrates that the dsRNA bound to the isolated RD clashes with domain 3 (Fig 3d–e). Thus, the dsRNA in helicase-RD is rotated and the previously reported contacts (R811, K849, K851, and H871) between RD and RNA are no longer observed in the helicase-RD structure.
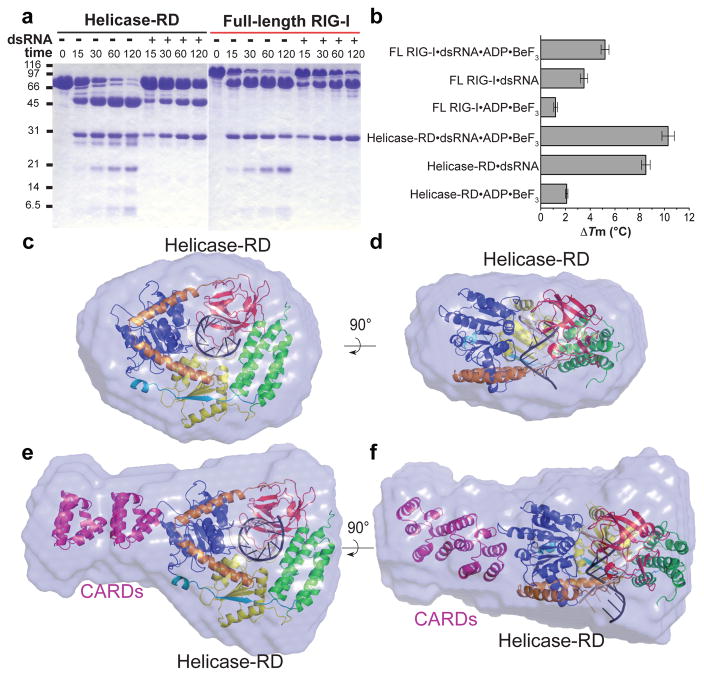
It is proposed that RIG-I exists in an autoinhibited state18 in the absence of RNA with undetectable ATPase activity (Table 1) and activates upon binding viral RNAs. SAXS, limited proteolysis, and DSF were performed to gain insights into how RNA binding activates RIG-I. Full-length RIG-I and helicase-RD undergo conformational changes and exhibit greater stability upon RNA binding (Fig. 4). The radius of gyration (Rg) of helicase-RD and full-length RIG-I decreases upon dsRNA binding by 10 and 2.3 Å, respectively (Supplementary Table 2), consistent with the results from size exclusion chromatography (Supplemental Fig. 1b). The SAXS Kratky plots of helicase-RD and full-length RIG-I with RNA are symmetrical parabolic curves (Supplemental Fig. 5), consistent with folded and globular complexes. The parabolic shape of the Kratky plot is lost and the peak amplitude decreases in the absence of RNA, indicative of greater flexibility19. Limited trypsin digestion and DSF shows a greater stabilization with dsRNA and a small increase in stability upon addition of the ATP analog (Fig. 4a–4b). These data suggest that RIG-I is composed of globular domains connected with flexible linkers that become ordered upon RNA binding. Such conformational stabilization upon ATP and RNA binding has been documented in other helicases 20–22.
Figure 4. Limited trypsin digestion, DSF and SAXS analyses of helicase-RD and full-length RIG-I in the presence and absence of dsRNA.
(a) SDS-PAGE analysis of a time course (minutes) of limited trypsin digestion of helicase-RD or full-length RIG-I in the absence or presence of 14 base pair pal-dsRNA. (b) DSF of RIG-I helicase-RD or full-length RIG-I in the presence of 14 base pair pal-dsRNA and/or ADP•BeF3 with respect to protein alone. The bar graph displays the mean melting temperature difference (ΔTm) and the error bars represent the standard deviation from three independent measurements. (c and d) Ab initio envelope of helicase-RD and dsRNA overlaid with the crystal structure of helicase-RD•dsRNA (dsRNA truncated to 10 base pairs). The view in d is rotated 90° about a horizontal axis from panel c. (e and f) Ab initio envelope of full-length RIG-I and dsRNA overlaid with the crystal structure of helicase-RD•dsRNA with two copies of CARDs added (PDB code 2VGQ). The view in f is rotated 90° about a horizontal axis from panel e.
A model of full-length RIG-I bound to dsRNA was established from ab initio SAXS envelope followed by rigid body refinement of helicase-RD•dsRNA complex and homologous CARD structures (Supplementary Table 2, and Fig. 4c–4f). The helicase-RD•dsRNA complex and two copies of the CARD are accommodated in full-length RIG-I SAXS envelope. The two CARDs project from domain 1. Such an orientation of the CARDs would allow for interaction with downstream signaling factors23–28. Interestingly, the second CARD is positioned adjacent to the V-shaped linker and close to the T770 phosphorylation site. The close proximity of the linker to the second CARD suggests a possible mechanism where the two alpha helices may serve as a hinge for RD movement upon RNA binding leading to RIG-I activation. The model suggests that the dsRNA extends along a perpendicular axis relative to the CARDs and hence can accommodate multiple RIG-I molecules29.
Method Summary
Recombinant full-length RIG-I (1–925), helicase-RD (232–925), helicase (232–794), RD (795–925), and selenomethione derivatized helicase-RD were expressed in Escherichia coli and purified to homogeneity using immobilized metal ion affinity, hydroxyapatite and heparin affinity chromatography. Fluorescence anisotropy titrations were performed at 25°C30 (λexcitation 494 nm and λemission 516 nm) using a fluorescein-labeled 14 base pair dsRNA prepared by annealing 5′GGAGAGAACCGCCU and 3′CCUCUCUUGGCGGA-F RNA, where F is fluorescein. Crystals of the native and selenomethione helicase-RD with pal-dsRNA (5′CGACGCUAGCGUCG) and ADP•BeF3•Mg2+ were obtained in 25% (w/v) PEG 3350, 0.25 M NaSCN, 100 mM MOPS (pH 7.8), 3% (v/v) 2,2,2-Trifluoroethanol at 20°C by hanging drop. The crystals belong to space group P6522 with cell parameters a=b=174.9 Å and c=110.9 Å. The structure was determined by SAD to 3.2 Å resolution and refined against a 2.9 Å resolution native data set. The final model has an Rwork and Rfree of 0.199 and 0.287, respectively. SAXS measurements were preformed on full-length RIG-I and helicase-RD in the absence and presence of dsRNA (5′GCGCGCGCGC). Buffer subtraction and Rg were calculated from Guinier plots. Dmax was determined by scanning a range of values and comparing experimental I(s) values to P(r) transforms. Ten ab initio models were averaged and normalized spatial discrepancy (NSD) values were calculated. The helicase-RD•dsRNA and a homologous CARD structure were positioned into the ab initio model and χ2 were determined.
Full Methods
Protein expression and purification
All the protein constructs were expressed with an N-terminal 6xHis-SUMO fusion. Human full-length RIG-I (1–925), helicase-RD (232–925), and helicase (232–794) were expressed in Escherichia coli strain Rosetta (DE3) (Novagen) as soluble proteins. The soluble fraction of helicase-RD was purified from the cell lysate using a Ni2+-nitrilotriacetate (Qiagen) column. The recovered protein was then digested with Ulp1 protease to remove the 6xHis-SUMO tag and further purified by hydroxyapatite column (CHT-II, Bio-Rad) and heparin sepharose column (GE Healthcare). Finally, purified helicase-RD was dialyzed against 50 mM HEPES (pH 7.5), 50 mM NaCl, 5 mM DTT, 10% glycerol overnight at 4°C, snap frozen in liquid nitrogen, and stored at −80 °C. The RIG-I RD (795–925) was expressed in Escherichia coli BL21 Star (DE3) cells and the soluble fraction was purified to homogeneity using a Ni2+-nitrilotriacetate column, cation exchange (HiTrap SP, GE Healthcare) and gel filtration chromatography. Selenomethionine (SeMet)-labeled helicase-RD was produced in Rosetta (DE3) cells grown in M9 minimal medium supplemented with 50 mg ml−1 L-SeMet (Sigma) and specific amino acids to inhibit endogenous methionine synthesis. The SeMet protein was purified using the same procedure as the unmodified protein.
Fluorescence anisotropy titrations
Fluorescence anisotropy measurements31 were carried out on a Fluoro-Max-4 spectrofluorimeter (Horiba Jobin Yvon). Fluorescein-labeled 14 base pair dsRNA (Dharmacon) (10 nM) was titrated with full-length RIG-I, helicase domain, RD, or helicase-RD in 50 mM Tris-acetate (pH 7.5), 100 mM Na-acetate, 10 mM Mg-acetate, 5% glycerol, 5 mM DTT, 0.05% Tween 20 buffer. 5′-ppp dsRNA (GGAGAGAACCGCCU) was transcribed using T7 RNA polymerase, PAGE purified, and annealed to a complementary, fluorescein-labeled RNA. Fluorescein anisotropy was measured at 25°C with excitation at 494 nm and emission at 516 nm. The helicase-RD titrations were also performed at lower dsRNA concentrations (1 and 2 nM) to obtain an accurate Kd. The observed fluorescence anisotropy (robs) as a function of protein concentration (Pt) was fit to equations 1 and 2 to obtain the equilibrium dissociation constant, Kd.
| Eq. 1 |
Where, rf and rb are the anisotropy values of free RNA and of the complex, fb is the fraction of RNA bound in the protein-RNA complex and fb = [PR]/[Rt] (PR is the concentration of the protein-RNA complex and Rt is the total RNA concentration).
| Eq. 2 |
Initial anisotropy of the free fluorescein-labeled dsRNA was 0.10 for all experiments. The measurements and errors are from two independent experiments in Table 1.
ATPase activity
A time course (0–30 min) of the ATPase reaction was performed using RIG-I helicase-RD (5 nM), ATP (1 mM) spiked with [γ-32P]ATP with and without 14 base pair dsRNA (80 nM) in buffer containing 50mM MOPS (pH 7.4), 5mM MgCl2, 5mM DTT, 0.01% Tween 20 at 37°C. The ATPase activity of the RIG-I helicase domain was measured using a higher amount of protein (100 nM) and RNA (1 μM). The quenched reactions were analyzed by PEI-Cellulose-F TLC (Merck) developed in 0.4 M potassium phosphate buffer (pH 3.4). The TLC plates were exposed to a phosphorimager plate, analyzed on a Typhoon phosphor-imager, and quantified using ImageQuant software. The ATPase rate was determined from the plots of [Pi] produced versus time and the rate constant values were calculated by dividing the ATPase rate by the respective enzyme concentration. Mean rate constant from 2 independent measurements and range is shown in Table 1.
Analytical size-exclusion chromatography
Analytical size-exclusion chromatography was carried out on an AKTA FPLC system (GE Healthcare). Proteins were loaded on to Superdex 200 10/300 GL column (GE Healthcare) equilibrated with a buffer containing 50 mM HEPES (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 5 mM DTT. The eluate was monitored by ultraviolet absorbance at 280 nm. For the complex, helicase-RD was incubated with 14 base pair dsRNA at the molar ratio of 1:1.2 on ice for 15 min before the sample was applied onto the column.
Preparation of Helicase-RD•dsRNA
The palindrome RNA oligonucleotide (5′-CGACGCUAGCGUCG-3′) (Dharmacon) was deprotected and desalted into 20 mM potassium phosphate (pH 7.4) before use. The dsRNA was prepared by incubating the RNA at 95 °C for 1 min followed by gradual cooling to 4 °C. The resulting dsRNA were mixed with purified helicase-RD in a RNA:protein molar ratio of 1.2:1, incubated at room temperature for 15 min, and then purified by size-exclusion chromatography (Superdex200, GE Healthcare) with an elution buffer of 50 mM HEPES (pH 7.5), 50 mM NaCl, 5 mM DTT, 5 mM MgCl2.
Crystallization and X-ray diffraction data collection
Helicase-RD•dsRNA•ADP•BeF3 ternary complex was reconstituted by incubating 0.17 mM helicase-RD with 0.17 mM dsRNA, 2 mM ADP, 2 mM BeCl2 and 10 mM NaF on ice for 30 min prior to crystallization. Crystals of native and SeMet-substituted complex were grown by hanging drop vapor diffusion method at 20°C. Aliquots (2.5 μl) of 15 mg ml−1 of helicase-RD•dsRNA•ADP•BeF3 complex in 50 mM HEPES (pH 7.5), 50 mM NaCl, 5 mM DTT, 5 mM MgCl2 were mixed with 2.5 μl of reservoir solution containing 25% (w/v) PEG 3350, 0.25 M NaSCN, 100 mM MOPS (pH 7.8), 3% (v/v) 2,2,2-Trifluoroethanol. Crystals appeared after 2–3 days, and they grew to a maximum size of 0.15 × 0.15 × 0.5 mm over the course of 8 days. 5-iodo-uridine derivative helicase-RD•dsRNA•ADP•BeF3 crystals were grown under identical conditions. For cryogenic data collection, crystals were transferred into crystallization solutions containing 5% (v/v) (2R,3R)-(−)-2,3-butanediol as cryoprotectant and then flash-cooled at 100 K. SeMet single-wavelength anomalous dispersion (SAD) dataset was collected at the X29A beam line of the National Synchrotron Light Source (NSLS). Native data set was recorded at the LRL-CAT 31-ID beam line of the Advanced Photon Source (APS). Data from iodouridine derivative crystals were collected at the Cornell High Energy Synchrotron Source (CHESS) F1 beam line. All diffraction data were integrated using iMosfilm and scaled in SCALA32.
Structure determination and refinement
Phases were determined using the SAD method. SHELXD/HKL2MAP33 detected a total of 12 out of 14 possible selenium sites in the asymmetric unit. Initial phases were calculated with Phaser32 and followed by density modification by DM32. Phase extension to 3.2 Å by SOLOMON32 produced an electron-density map into which most of the protein and RNA residues could be built unambiguously. The model was built using COOT34 and refined in PHENIX35. The final model, comprising ADP•BeF3, 12 base pair dsRNA, RIG-I residues 240–495, 504–522, 527–687, and 690–923 has Rwork and Rfree values of 0.199 and 0.287, respectively. Model validation demonstrated no outliers and 90% of the residues located the most favorable region of the Ramachandran plot35. Statistics of the data processing and structure refinement are summarized in Supplementary Table 1.
Small angle X-ray scattering and structural modeling
SAXS data were collected at the CHESS beamline G1 using a Finger Lakes CCD X-ray detector with a sample-to-detector distance 1450mm, covering the range of scattering vectors 0.01 < s < 0.233 Å −1, where s= 4πsinθ/λ (2θ is the scattering angle and λ= 1.296 Å). All samples were buffer exchanged into 50 mM HEPES (pH 7.5), 50 mM NaCl, 5 mM DTT, 5% glycerol by size exclusion chromatography (Hiload 16/26 Superdex200, GE Healthcare) to minimize the discrepancies in background subtraction. SAXS data was reduced using Data Squeeze. Various programs in ATSAS software package were used to process and evaluate the scattering data. Radius of gyration (Rg) was analyzed using the Guinier approximation with low angle data (s < 1.3/Rg) using PRIMUS36. The residuals from the Guinier plots did not show signs of protein aggregation. The probability distribution of distances between scattering atoms within the macromolecule, P(r), and the maximum atom pair distance, Dmax, were determined from the scattering data using the GNOM algorithm37. The program DAMMIF38 was used to calculate low-resolution ab initio reconstructions from experimental SAXS profiles. Ten to twenty independent models were aligned, filtered, and averaged based on the occupancy using SUPCOMB and DAMAVER39 to reconstruct the final ab initio envelope, as judged by averaged normalized spatial discrepancies (NSD) less than 1.0. The structure modeling was refined against the solution scattering data by rigid body docking using SASREF program40. The orientation and position of individual domains in the structure models were further manually adjusted to minimize the discrepancies (χ2) between the calculated scattering intensities and experimental scattering intensities computed using program CRYSOL41. Kratky plots were calculated using PRIMUS36.
Differential scanning fluorimetry
Thermal shift assay was conducted with 10 μM of RIG-I helicase-RD or full-length RIG-I with or without 12 μM of 14 base pair pal-dsRNA and/or 2 mM ADP•BeF3 in 50 mM HEPES (pH 7.5), 50 mM NaCl, 5 mM MgCl2, 5 mM DTT, and a 5x dilution of SYPRO Orange dye (Invitrogen) as described42. The fluorescence signal as a function of temperature was recorded using a Real Time PCR machine (Applied Biosystems). The temperature gradient is performed in the range of 25–80°C with a ramp of 0.2°C over the course of 60 minutes. Control assays were carried out with buffer in the presence or absence of RNA. Data were analyzed with the Excel-based worksheet DSF analysis, and Boltzmann model was used to fit the fluorescence data to obtain the midpoint temperature for the thermal protein unfolding transition (Tm) using the curve-fitting software Prism.
Limited trypsin proteolysis
Limited proteolysis with trypsin (Roche) was performed using 120 μg of purified full-length RIG-I or helicase-RD in the absence or presence of pal-dsRNA and incubated with trypsin at a protein:protease mass ratio of 300:1. The reaction mixtures were maintained at room temperature and aliquots were removed at 15, 30, 60, and 120 min. The reaction was stopped by the addition of SDS-PAGE loading buffer and analyzed by SDS-PAGE.
Supplementary Material
Acknowledgments
We acknowledge access to beamlines X29 at the NSLS, LRL-CAT at APS, and G1 and F1 at CHESS and thank the NSLS, APS and CHESS staff. NSLS and APS are supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886 and DE-AC02-06CH11357, respectively. CHESS is supported by the NSF & NIH/NIGMS via NSF award DMR-0936384, and the MacCHESS resource is supported by NIH/NCRR award RR-01646. Use of the LRL-CAT beam line facilities at Sector 31 was provided by Eli Lilly & Company. We would like to thank V. Rajagopal for initiating the biochemical experiments and guiding the project in the early stages. We thank E. Arnold, H. Berman, S.K. Burley, R. Gillilian, L. Morisco, W. Olson, T. Saito, A. Shatkin, A. Stock, H. Yang and M. Zhuravieva for providing helpful comments and assistance. This work was supported by NIH grants GM55310 to SSP and AI080659 to JM.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Authors’f contributions. The project was initiated by M.G., J.M. and S.S.P. J.M. and S.S.P. designed and supervised the project. M.G. provided reagents and consultation. F.J. designed protein constructs and established purification protocols. A.R. generated all RNA reagents. F.J. and A.R. purifed the complex and setup crystallization screens. F.J. optimized the crystal for data collection. J.M., M.M., F.J., and A.R. collected, processed, and analyzed the X-ray crystallographic data. M.M., F.J., and J.M. collected and analyzed the SAXS data. A.R., G.-Q.T., and S.S.P. collected and analyzed the RNA binding and ATPase assays. F.J. performed limited proteolysis and thermal melting assay. S.S.P. and J.M. wrote the paper and all authors contributed to editing.
Author information. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/natureCorrespondence and requests for materials should be addressed to J.M. (jmarco@cabm.rutgers.edu) and S.S.P. (patelss@umdnj.edu). The coordinates for this structure have been deposited in the Protein Data Bank under accession code (3TMI).
References
- 1.Schlee M, et al. Approaching the RNA ligand for RIG-I? Immunol Rev. 2009;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- 2.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsumiya T, Stafforini DM. Function and regulation of retinoic acid-inducible gene-I. Crit Rev Immunol. 2010;30:489–513. doi: 10.1615/critrevimmunol.v30.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chattopadhyay S, et al. Viral apoptosis is induced by IRF-3-mediated activation of Bax. Embo J. 2010;29:1762–1773. doi: 10.1038/emboj.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Z, Ren H, Liu Y, Teeling JL, Gu J. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J Virol. 2011;85:1036–1047. doi: 10.1128/JVI.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu C, Ranjith-Kumar CT, Hao L, Kao CC, Li P. Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5′ triphosphate. Nucleic Acids Res. 2011;39:1565–1575. doi: 10.1093/nar/gkq974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu C, et al. The Structural Basis of 5′ Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain. Structure. 2010 doi: 10.1016/j.str.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, et al. Structural and functional insights into 5′-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annual review of biochemistry. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 12.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 14.Myong S, et al. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science. 2009;323:1070–1074. doi: 10.1126/science.1168352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 16.Gu M, Rice CM. Inaugural Article: Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proceedings of the National Academy of Sciences. 2009 doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam AM, Keeney D, Frick DN. Two novel conserved motifs in the hepatitis C virus NS3 protein critical for helicase action. J Biol Chem. 2003;278:44514–44524. doi: 10.1074/jbc.M306444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 20.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 2. A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry. 1998;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- 21.Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 22.Theissen B, Karow AR, Kohler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci U S A. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loo YM, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 25.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 26.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 28.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 29.Binder M, et al. Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid inducible gene-I (RIG-I) J Biol Chem. 2011 doi: 10.1074/jbc.M111.256974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang GQ, Bandwar RP, Patel SS. Extended upstream A-T sequence increases T7 promoter strength. J Biol Chem. 2005;280:40707–40713. doi: 10.1074/jbc.M508013200. [DOI] [PubMed] [Google Scholar]
- 31.Tang GQ, Bandwar RP, Patel SS. Extended upstream A-T sequence increases T7 promoter strength. J Biol Chem. 2005;280:40707–40713. doi: 10.1074/jbc.M508013200. [DOI] [PubMed] [Google Scholar]
- 32.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. S0907444909052925 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konarev PV, Volkov VV, Sokolova AV, Koch MH, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- 37.Semenyuk AV, Svergun DI. GNOM - a program package for small-angle scattering data processing. Journal of Applied Crystallography. 1991;24:537–540. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Journal of Applied Crystallography. 36(203):860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 42.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.