Abstract
Inversion polymorphisms have occupied a privileged place in Drosophila genetic research since their discovery in the 1920s. Indeed, inversions seem to be nearly ubiquitous, and the majority of species that have been thoroughly surveyed have been found to be polymorphic for one or more chromosomal inversions. Despite enduring interest, however, inversions remain difficult to study because their effects are often cryptic, and few efficient assays have been developed. Even in Drosophila melanogaster, in which inversions can be reliably detected and have received considerable attention, the breakpoints of only three inversions have been characterized molecularly. Hence, inversion detection and assay design remain important unsolved problems. Here, we present a method for identification and local de novo assembly of inversion breakpoints using next-generation paired-end reads derived from D. melanogaster isofemale lines. PCR and cytological confirmations demonstrate that our method can reliably assemble inversion breakpoints, providing tools for future research on D. melanogaster inversions as well as a framework for detection and assay design of inversions and other chromosome aberrations in diverse taxa.
Keywords: structural variation, long-range LD, In(3R)Mo, In(2R)NS, In(3R)K, In(1)Be, In(1)A
STURTEVANT (1917) discovered the first chromosomal inversion as a suppressor of recombination in Drosophila melanogaster. Shortly after his initial finding, Sturtevant produced evidence that inversions, structurally reversed segments of the linear map order of chromosome arms, account for this observation (Sturtevant 1926). A vast body of empirical work has followed this original discovery, yielding several key results that inform our understanding of the genetic, and potential evolutionary, implications of polymorphic inversions. First, single crossovers within the inverted regions of inversion heterokaryotypes are expected to yield aneuploid gametes, effectively suppressing exchange between arrangements (Sturtevant and Beadle 1936). Inversion heterokaryotypy redistributes chiasma elsewhere in the genome, both intrachromosomally in colinear segments and via the interchromosomal effect (Lucchesi and Suzuki 1968).
This primary effect—suppression of recombination in the inverted regions of heterokaryotypes, especially near the breakpoints—is the subject of much of the theoretical population genetic research focused on inversions. Generally, interpretations in the literature favor a model in which inversions achieve high frequencies by suppressing recombination between coadapted alleles located near the inversion breakpoints (Dobzhansky 1951), although there are many other possible mechanisms (Kirkpatrick and Barton 2006; Hoffmann and Reiseberg 2008). Empirical research on polymorphic inversions has been extensive as well, the central result being that chromosomal inversions are pervasive. Indeed, segregating inversions are found in abundance in the majority of organisms that have been examined in depth, including plants, insects, mammals, and humans (Hoffmann and Reiseberg 2008). However, the selective forces that govern the evolution of inversion polymorphisms remain largely obscure, with important exceptions being inversions associated with novel sex chromosomes (Charlesworth et al. 2005), sex ratio distortion (Jaenike 2001), and autosomal segregation distortion (Kusano et al. 2003; Lyon 2003).
D. melanogaster is highly polymorphic for chromosomal inversions. In fact, since the pioneering work of Sturtevant (1931) and Dubinin et al. (1937), >500 segregating inversions have been found in natural populations of D. melanogaster, encompassing a broad-frequency spectrum ranging from present at low frequency in single populations to present in virtually all populations worldwide (Ashburner and Lemeunier 1976; Aulard et al. 2004). This distribution is conventionally subdivided into four classes that correspond to the inversions’ prevalence in natural populations: unique endemic, recurrent endemic, rare cosmopolitan, and common cosmopolitan (Ashburner and Lemeunier 1976; Krimbas and Powell 1992).
The latter class has received by far the most attention. Common cosmopolitan inversions exhibit frequency clines, diminishing from high frequency in equatorial regions, to nearly absent in higher lattitudes. This pattern is replicated independently on several continents, suggesting that strong selective forces govern the distributions of these inversions (Knibb et al. 1981). This observation has prompted numerous attempts to identify the traits that are experiencing selection, and several ecologically relevant traits have been associated with common cosmopolitan inversions (Van Delden and Kamping 1991; Frydenberg et al. 2003; Kennington et al. 2007). However, it remains unknown if the genetic elements that confer these traits are the targets of selection or hitchhiking as a result of reduced recombination and the relatively young age of these inversions (Andolfatto et al. 1999; Matzkin et al. 2005).
Even less is known about the rare cosmopolitan and recurrent endemic inversions, which are comparatively understudied and sometimes not recorded in published frequency assays (Knibb et al. 1981; Krimbas and Powell 1992). The rare cosmopolitans are distributed worldwide, but often entirely absent from populations, while the recurrent endemic inversions may be at high frequency in one geographic region, but have rarely or never been identified elsewhere (Krimbas and Powell 1992). A detailed understanding of their limited distributions and selective potentials is essential both as a comparison to the more “successful” common cosmopolitan inversions and to a nuanced and complete conception of polymorphic inversions in D. melanogaster and the broader topic of genome evolution.
Despite continuing interest in the inversion polymorphisms of D. melanogaster, only three inversions, all common cosmopolitans, can be assayed directly using molecular means (Wesley and Eanes 1994; Andolfatto et al. 1999; Matzkin et al. 2005). Others must be identified via the laborious original method: crossing to a stock with known chromosomal arrangements and examining the banding patterns of the giant salivary gland polytene chromsomes in the larval progeny. In fact, all inversion breakpoints that have been characterized molecularly in D. melanogaster were identified using this convenient cytogenetic feature in combination with fluorescent in situ hybridization techniques (Wesley and Eanes 1994; Andolfatto et al. 1999; Matzkin et al. 2005). By hybridizing larval polytene chromosomes with DNA fragments of known mapping positions, it is possible to narrow down the breakpoint regions through successively closer hybridizations (Wesley and Eanes 1994). This method is both time-consuming and, perhaps most problematic, completely impractical for organisms that lack visible polytene chromosomes. Considering the largely quantitative goals of population genetic research, a more general and efficient means of inversion detection and assay design is essential to furthering our understanding of inversion polymorphisms in natural populations.
Genomic techniques have presented two appealing alternatives for identifying and characterizing structural polymorphisms segregating within populations. One method, originally developed by Tuzun et al. (2005), is based on sequencing both ends of short DNA fragments with an approximate known distance and orientation with respect to each other. By first mapping paired reads to a reference sequence, and subsequently identifying clusters of read pairs that do not map in the expected orientation or distance relative to one another, it is possible to reliably identify the breakpoints of structural polymorphisms (Tuzun et al. 2005; Kidd et al. 2008; Medvedev et al. 2009). This approach is appealing because it can be used to fine-map structural breakpoint and it has previously been validated as a tool for interrogation of structural polymorphisms in D. melanogaster (Cridland and Thornton 2010). While these methods have high sensitivity for breakpoint detection, it is often not possible based solely on the breakpoints to distinguish inversions from other structural rearrangements, such as duplications that reinsert in inverted orientation (Cridland and Thornton 2010). The relatively short length of inserts that are currently used in the majority of resequencing projects, as opposed to the long inserts used in the previous landmark studies (Tuzun et al. 2005; Kidd et al. 2008), may exacerbate this issue, as short inserts provide little resolution beyond detected breakpoints.
Alternative approaches, which rely on data from many densely genotyped individuals, use an expected signature of nucleotide variation to detect polymorphic inversions (Bansal et al. 2007; Sindi and Raphael 2010). While these methods have provided valuable insights and many candidate inversions, the inherent genomic limitations of SNP genotype data have proved to be a major impediment. To accurately predict inversions, these methods require substantial minor allele frequencies of inversion and large sample sizes (Bansal et al. 2007; Sindi and Raphael 2010). Additionally, genotyping approaches offer poor resolution of inversion breakpoints, which may be of interest for population genetic analyses, designing PCR assays, and studying the mechanisms of inversion formation and DNA repair.
Here we present a hybrid method of inversion detection that incorporates features of breakpoint and genotype-based methods outlined above. Briefly, our method infers putative breakpoints using map positions of paired-end reads across many samples. We subsequently identify breakpoints that are shared between samples on the basis of overlap of detected breakpoints. We then filter potential breakpoints by scanning the surrounding regions for a signature of nucleotide variation, heightened FST, which is expected to be associated with inversion breakpoints. We apply this method to >100 D. melanogaster haploid genomes, in which we identify breakpoints and design molecular assays for five previously uncharacterized polymorphic inversions. We also improve on existing assays for two previously characterized inversions. Cytology as well as previously developed PCR assays confirm that our method is highly accurate. We expect that these primer pairs will be immediately useful as tools for researchers interested in assaying inversions directly in individual D. melanogaster or existing stocks. Additionally, this general framework could be implemented to detect and design molecular assays for structural polymorphisms in other species.
Materials and Methods
Details for all fly stocks used in this study can be found at http://www.dpgp.org. In short, all short-read sequences are derived from 76-bp Illumina paired-end reads, separated by ∼300-bp inserts, and represent numerous African populations and one French population. All genomic regions analyzed are haploid through chromosomal extractions, inbreeding, or haploid embryo extractions (Langley et al. 2011). The average coverage depth is ∼31× (range: 8–47).
We aligned short reads for each line to the D. melanogaster reference sequence v5.22 (Adams et al. 2000) using ELAND v2 as a standard part of the Illumina Casava pipeline. Alignments were performed over the course of more than a year; as such, several versions of the Casava pipeline were used (see http://www.dpgp.org for details). Eland alignments were ported to MAQ using the “export2maq” utility in the MAQ v0.7.3 software package (Li et al. 2008). We called consensus sequences for each line, requiring that each site called have a minimum read depth of 3 and a minimum quality of 30. All other sites were excluded from the resulting assemblies. We identified seven lines showing excess identity by descent as those with regions of little pairwise divergence on more than one chromosome arm; these are obvious in cursory inspections, so this was done by hand and primarily excluded genomes that are drawn from the same isofemale line. From each pair showing identity by descent, we discarded the line that was sequenced to lower depth.
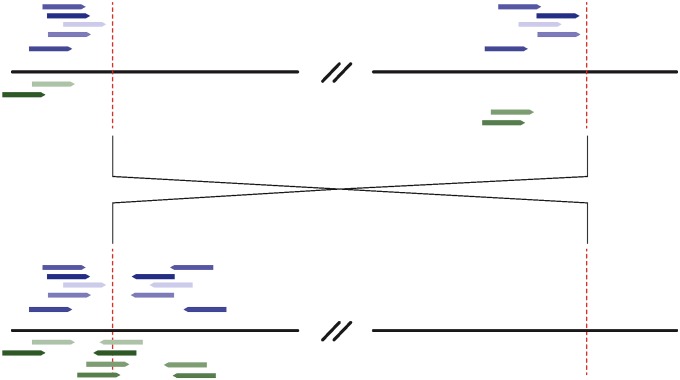
From the alignment files, we discarded read pairs that share identical mapping coordinates with another pair, as well as those for which one read maps to an annotated transposable element. Next, we parsed read pairs for which both reads mapped uniquely to the reference sequence but mapped in parallel orientation (≫ or ≪). We restricted this set of reads to those that mapped to the same chromosome arm. For each line individually, we assigned aberrant read pairs to clusters, requiring that both reads in a pair mapped within 500 bp of another read included in that cluster. Thus, each cluster contains sets of read pairs for which one of the pair maps to one 500-bp region of the genome, and the other read maps to another 500-bp region of the same chromosome arm. We further required that all reads in one cluster map in the same orientation and that genomic positions within each cluster be >1 megabase from each other. Clusters supported by fewer than five clones were discarded. Finally, we parsed from the “export.txt” file read pairs for which one read maps to the same genomic location and in the same orientation as the clusters identified previously, and the other read is unmapped. The unmapped reads are expected to cross the breakpoint. We then folded these reads into the original cluster (Figure 1).
Figure 1 .
Mapping positions of reads used for de novo assembly of the breakpoint sequences. (Top) Reads mapping positions on the reference sequence. (Bottom) Their inferred positions on the inverted haplotype. Reads pairs that span the breakpoint are shown in shades of blue, while reads for which one end maps and the pair crosses the junction on the reference sequence are shown in shades of green. All reads shown are used to produce de novo assemblies. For simplicity, reads corresponding to this inversion’s other breakpoint are not shown.
We compared each line’s set of read clusters to all other lines’ sets of read clusters. Overlapping clusters in the same genomic position and orientation were identified as potentially confirming the same inversion breakpoint. Due to their unique origins, which results in zero polymorphisms in the inverted population at formation, inversion breakpoints will immediately attain high FST relative to the standard arrangement population. As genetic exchange is almost completely suppressed near inversion breakpoints (Novitski and Braver 1954; Wesley and Eanes 1994; Andolfatto et al. 1999), genetic differentiation will be maintained between arrangements and is an expected signature of all inversion breakpoints. Provided that an inversion is present in more than two individuals, this expectation suggests an ideal way to sift through identified breakpoints for inversion false positives. For lines sharing identical breakpoints, we compared the consensus sequences in 20-kb windows centered on each potential breakpoint and calculated FST between the lines that share the putative breakpoint and the lines that do not. We retained candidate inversions for which both breakpoints’ FST was >0.25. We calculated FST as described in Hudson et al. (1992), using only sites that were called in all lines. Importantly, we did not weight FST by sample size, which enables the detection of low-frequency inversions. Because of this, even immediately after formation, we expect to observe strong genetic differentiation, and inversions’ FST’s will initially be ∼0.5.
We de novo assembled the set of reads corresponding to each remaining potential breakpoint using Phrap v1.090518 (Green 1996) All contigs were aligned to the D. melanogaster reference sequence v5.22 (Adams et al. 2000), using Blast v2.2.25 (Altschul et al. 1990), and contigs with significant alignments to both sides of the expected breakpoint were retained. Blast alignments with corresponding e-values <10−10 and alignment lengths >30 bp were considered significant.
To assist in assigning identities for novel inversions, we compared the cytogenetic positions of the inversions’ breakpoints with reported breakpoint coordinates. To do this, we downloaded the cytologically predicted positions of inversion breakpoints and the map conversion table for cytological coordinates from FlyBase (http://www.flybase.org).
Inversion breakpoints may take two forms: cut-and-paste and inverted duplication (see Ranz et al. 2007 for a description of breakpoint structure). After aligning breakpoint-spanning contigs to the reference genome, we inferred the structure on the basis of the following criteria. If both breakpoint-spanning contigs appear to map in convergent orientations to within 50 bp of each other at both ends of the inversion, they are assumed to be cut-and-paste breakpoints. Otherwise, we assume that the sequence between mapping positions is present as a duplication at the other breakpoint. We confirmed these structural predictions via comparisons with the three inversions that have previously been examined (Wesley and Eanes 1994; Andolfatto et al. 1999; Matzkin et al. 2005) and by comparison with the copy-number variation analysis performed by C. H. Langley et al. (2012), whose stocks are known to bear many of these inversions.
To confirm breakpoints, we developed a PCR-based inversion assay. We designed primers using Primer3 (http://frodo.wi.mit.edu/primer3/) that would produce an amplicon unique to the standard or inverted chromosomal arrangement on the basis of these putative breakpoints. We extracted genomic DNA from flies using the Quick Fly Genomic DNA Prep provided by the Berkeley Drosophila Genome Project (http://www.fruitfly.org/about/methods/inverse.pcr.html). Briefly, we ground 30 flies in Buffer A (100 mM Tris–HCl, 100 mM EDTA, 100 mM NaCl, 0.5% SDS) and incubated the flies at 65° for 30 min. We added a 1:2.5 solution (1 part 5 M KAc to 2.5 parts 6 M LiCl) to the samples and incubated them on ice for at least 10 min. The DNA was precipitated with isopropanol, washed, and resuspended in ddH2O.
All of the PCR inversion assays [except for the standard chromosomal arrangement of In(3R)P] used standard PCR reaction conditions: 2.0 mM MgCl2, 0.2 mM each of dNTPs, 0.5 uM each of forward and reverse primers, 1 unit of Taq, and 50 ng of DNA. Specific PCR conditions for each reaction are described in Supporting Information, Table S1. Appropriately sized amplicons were identified with agarose gels. We used long PCR to assay the In(3R)P standard chromosomal arrangement. This was necessary because the inverted duplications present at each breakpoint are too long for standard PCR. We followed the manufacturer’s PCR general reaction mixture and conditions (TaKaRa LA Taq) with a few exceptions: a final MgCl2 concentration of 1.75 mM (Table S1) and an annealing/elongation time of 5 min. We included the reference strain y; cn bw; sp as well as several other standard orientation lines as negative controls for inversion-specific primers and positive controls for standard specific primers. For inversion-positive controls, we obtained several lines known by cytology to harbor the putative inversion from the Bloomington Stock Center and C. H. Langley et al. (2012; Table S1).
For each inversion that had not been detected previously, we sequenced via Sanger PCR at least one breakpoint to further validate Phrap assemblies. Sequences were assembled from forward and reverse chromatograms using phredPhrap, which is distributed as a part of the Consed package. We inspected all assembled PCR fragments by hand in Consed v1.090518 (Gordon et al. 1998). These sequences are available in File S1.
Results
Across all genomes, we recovered >15,000 breakpoints that map in parallel orientation to a single chromosome arm. After pooling across all samples, we found >200 breakpoints that were present in more than one line. Finally, after applying the FST filter, we found 12 aberrant read clusters whose corresponding consensus sequences showed increased FST around both breakpoints. Heightened FST is an expected signature of nucleotide variation between inverted and standard arrangements owing to the unique origin of inversion and suppressed exchange between arrangements immediately surrounding each breakpoint (Novitski and Braver 1954; Wesley and Eanes 1994; Andolfatto et al. 1999). Importantly, heightened FST at both breakpoints is not expected for breakpoints associated with other rearrangements that may occur at higher frequencies. This is because only a single breakpoint actually harbors the novel insertion event; the other “breakpoint” reflects reads that map uniquely to the single copy present in the reference sequence, but are not actually linked to this genomic location. We also surveyed all clusters present in more than one line for breakpoints consistent with cytologically known inversions that have been identified in populations from sub-Saharan Africa (Aulard et al. 2002). We did not find any additional breakpoints that were consistent with these inversions; thus, FST appears to be a successful filter for inversion true positives.
We successfully assembled contigs that spanned all breakpoints identified. Since five pairs of breakpoints were in perfect linkage disequilibrium and the breakpoint coordinates are very similar, we surmised that these breakpoints corresponded to the same inversion. Thus, we were able to recover both breakpoints for five inversions, and only one breakpoint for two others (Table S1). That we recovered only a single breakpoint for In(2L)t and In(3R)P is likely due to the presence of fixed repetitive elements immediately adjacent to the proximal breakpoints of each inversion (Andolfatto et al. 1999; Matzkin et al. 2005).
In a recent study, C. H. Langley et al. (2012) found a pattern of excess long-distance linkage disequilibrium and significantly decreased nucleotide diversity associated with a large paracentric inversion, In(3R)Mo, in a Raleigh, North Carolina, population of D. melanogaster. Because the sequence data used in this study were derived from single-end reads, they are not suitable for direct comparison via our method, which relies on independently mapped paired-end reads. We did identify one line, FR310, which shares this pattern of long-distance linkage disequilibrium and that had been sequenced using paired-end reads. Because of this inversion’s unexpected prevalence in this Raleigh, North Carolina, population, we surveyed this line specifically for potential breakpoints, without requiring that the identified read clusters be corroborated by clones derived from another line. We found two breakpoints whose positions are consistent with our expectations for this inversion on the basis of the observed pattern of nucleotide diversity and confirmed these breakpoints via PCR in the eight lines identified by C. H. Langley et al. (2012).
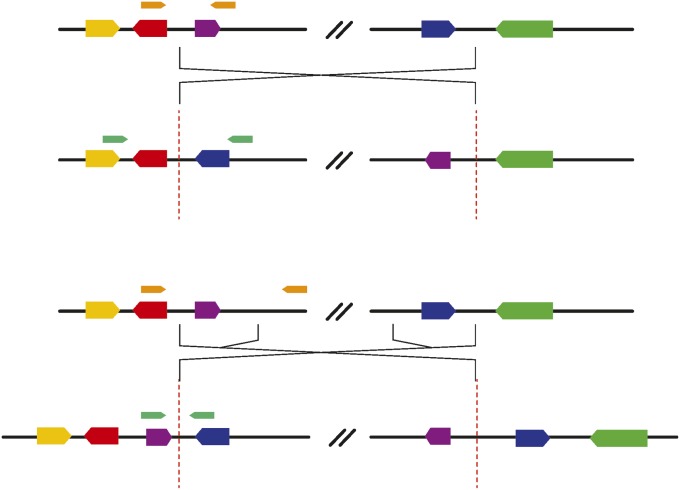
We recovered two classes of breakpoints: simple cut-and-paste breakpoints and staggered break-plus-inverted-duplication breakpoints (Figure 2). We were able to confirm structural predictions for three inversions whose breakpoints were previously characterized (Wesley and Eanes 1994; Andolfatto et al. 1999; Matzkin et al. 2005). In(3R)K, In(3R)Mo, In(1)A, and In(2R)NS are all present in the sample analyzed by C. H. Langley et al. (2012), and we were able to support breakpoint structure predictions for each on the basis of comparisons to the copy-number variation analysis included in that work. Five of the eight inversions contain inverted duplications at their breakpoints (Table S2), which is similar to the proportions that were found by a study that focused on inversions that fix between species in the melanogaster subgroup (17 of 29) (Ranz et al. 2007).
Figure 2 .
We found two types of inversion breakpoints: cut-and-paste breakpoints (top), and staggered breakpoints (bottom), which create inverted duplications at the breakpoints. The duplicated regions are shown as purple and blue “genes.” PCR primers for the standard (in orange) and inverted (in teal) arrangements were designed on the basis of assembled contigs that span the breakpoints and amplify a unique product for either arrangement.
We designed assays for each class that amplify a product of unique length for either the standard or the inverted haplotype. Cut-and-paste breakpoints can be assayed easily as described in Andolfatto et al. (1999). Because in staggered-break inversion structures there is no single breakpoint that is unique to the standard arrangement, primers that span a single breakpoint cannot be used to distinguish between inversion heterozygotes and inversion homozygotes. Our solution is to design primers that span the duplicated regions at either end of the inversion (Figure 2). This produces a PCR product that is unique to the standard arrangement and may be more robust than an allele-specific PCR approach (e.g., Anderson et al. 2005).
To make use of this advantage, we designed new primers for the standard arrangement of In(3R)P (Table S1), for which only ambiguous or allele-specific primers were previously available. Likely due to the presence of a repetitive element immediately adjacent to the proximal breakpoints of In(3R)P (Matzkin et al. 2005), we recovered only a single breakpoint for this inversion. Fortunately, Matzkin et al. (2005) have previously sequenced both breakpoints via long-range PCR for In(3R)P. We downloaded these sequences from GenBank (accession nos. AY886890–AY886892) and used them to design a novel set of primers that yield a unique amplicon for the standard arrangement. All PCR fragments that we sequenced are identical to the Phrap assemblies, except in the low-quality bases toward the ends of the traces (File S1).
For the three inversions that were previously characterized at the molecular level, breakpoint coordinates are known, and we identified these breakpoints directly in our sequence data. We also confirmed our results for these inversions using published primers (Table S3). For In(3L)P, the existing primers did not work reliably. We elected to design new primers (Table S1) and have found these to be more reliable. For four other inversions, all putative inversion identities were confirmed by positive controls (Table S1).
For all previously uncharacterized inversions, the cytologically derived mapping positions based on previous surveys were within 100 kb of the breakpoint that we identified. In most previously uncharacterized inversions, it was also possible to test our primers on stocks known via cytology to bear the inversion. This was not possible for one inversion on the X chromosome for which no independent positive controls are available. However, the breakpoint coordinates, geographic distribution, and frequency of this inversion are all consistent with In(1)Be and do not suggest any other known inversions. Hence, although we cannot be certain of the identity, we refer to this inversion as In(1)Be (Table S1).
Discussion
While our method was quite successful and has immediate applications to many short-read sequencing projects, it should be noted that there are two important drawbacks, both of which will be ameliorated by imminent advances in sequencing technologies. First, our method requires accurate mapping information and a well-characterized reference genome. Already, several species’ genomes have been fully assembled using next-generation sequencing technologies (e.g., Li et al. 2009); hence, the anticipated availability of many additional reference sequences may make the proposed method widely serviceable. Second, the extent to which transposable elements contribute to the formation of chromosomal inversions remains an open question (Mathiopoulos et al. 1998; Caceres et al. 1999). Although this does not appear to be a common mechanism of inversion formation in the D. melanogaster subgroup (Ranz et al. 2007), it is possible that transposable elements contribute more to structural polymorphisms in other species. Because of the modest insert lengths used in sequencing, our method has little power to detect inversions that form via ectopic recombination between repetitive elements. However, this limitation will also diminish in importance with the increasing availability and quality of larger insert sizes in library preparation, which will be able to span individual repetitive elements. Hence, if anything, the applicability and usefulness of this approach will increase as sequencing technologies continue to progress.
Despite these potential drawbacks, our method has performed well. A recent survey of African D. melanogaster inversion polymorphisms (Aulard et al. 2002) reported generally the same set of polymorphic inversions at moderate frequencies. So, while we cannot estimate a true false-negative rate, this suggests that we have recovered the majority of inversions that are likely to be segregating at frequencies >2 in this sample. Requiring FST calculations means that our method will miss inversions present in only one individual. This drawback is unavoidable, since there are thousands of aberrant read clusters in individual genomes, and it is not always possible to distinguish between inversions and other structural variants solely on the basis of breakpoint coordinates. Regardless of the error rates, our method is a vast improvement over conventional methods, and it allows us to rapidly characterize and develop novel molecular assays for five chromosomal inversions and to improve on two existing assays. This more than doubles the available assays, providing a substantial improvement in the tools available for studies of the polymorphic inversions of D. melanogaster.
Another advantage of this approach is its broad applicability. Cytological methods, beyond being time-consuming, require visible polytene chromosomes, as well as an approximate idea of where inversion breakpoints might be expected to occur and in what strains. Our method circumvents these issues, and it allows us to examine numerous individuals simultaneously and to identify polymorphic inversions without requiring any prior knowledge of the lines or inversion content of the genome. Hence, we expect this will be a useful framework for researchers interested in characterizing and developing molecular assays for polymorphic inversions, especially in developing model systems. Although we analyzed haploid data, this method could feasibly be extended to accommodate diploid samples with sufficient sequencing or sampling depth.
It should also be emphasized that clustering putative breakpoints across samples would allow detection not only of inversions but also of other types of chromosome aberrations. These need not necessarily be aberrations transmitted through the germ line. For example, our approach may have applications in the study of rearrangements among somatic or cancer cells where relevant independent sampling can be conducted.
Chromosomal inversion polymorphisms are a ubiquitous evolutionary phenomenon. They are present in virtually all species and may have potent evolutionary effects ranging from resisting gene flow in hybrid zones, to maintaining co-adapted gene complexes, to the long-term maintenance of epistatically interacting segregation distortion systems (Hoffmann and Reiseberg 2008). However, a complete understanding of the selective effects of polymorphic inversions is elusive. Even—perhaps especially—in the species in which chromosomal inversions were originally discovered, D. melanogaster inversions remain an enigmatic and intriguing feature of virtually all populations. A central impediment to quantitative studies of these polymorphisms, especially rare cosmopolitan and recurrent endemic inversions, is a lack of low-cost efficient assays. Here, we provide these tools. Although there are certainly numerous interesting population genetics questions that could feasibly be addressed using these data (many of which are subjects of ongoing research), our goal with this work is to make these resources available to the community as soon as possible.
Supplementary Material
Acknowledgments
We thank John Pool, Daniel Hartl, and two anonymous reviewers for helpful comments on this manuscript, as well as Shu Fang for providing cytology information. We also acknowledge funding by National Institutes of Health grant HG02942 (to C.H.L.). R.B.C-D. is supported by a Harvard Prize Fellowship.
Footnotes
Communicating editor: B. A. Payseur
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 3: 403–410 [DOI] [PubMed] [Google Scholar]
- Anderson A. R., Hoffmann A. A., McKechnie S. W., Umina P. A., Weeks A. R., 2005. The latitudinal cline in the In(3R)Payne inversion polymorphism has shifted in the last 20 years in Australian Drosophila melanogaster populations. Mol. Ecol. 14: 851–858 [DOI] [PubMed] [Google Scholar]
- Andolfatto P., Wall J. D., Kreitman M., 1999. Unusual haplotype structure at the proximal breakpoint of In(2L)t in a natural population of Drosophila melanogaster. Genetics 153: 1297–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Lemeunier F., 1976. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). 1. Inversion polymorphisms in Drosophila melanogaster and Drosophila simulans. Proc. R. Soc. Lond. 193: 137–157 [DOI] [PubMed] [Google Scholar]
- Aulard S., David J. R., Lemeunier F., 2002. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genet. Res. 79: 49–63 [DOI] [PubMed] [Google Scholar]
- Aulard S., Monti L., Chaminade N., Lemeunier F., 2004. Mitotic and polytene chromosomes: comparisons between Drosophila melanogaster and Drosophila simulans. Genetica 120: 137–150 [DOI] [PubMed] [Google Scholar]
- Bansal V., Bashir A., Bafna V., 2007. Evidence for large inversion polymorphisms in the human genome from HapMap data. Genome Res. 17: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres M., Ranz J. M., Barbadilla A., Long M., Ruiz A., 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 285: 415–418 [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128 [DOI] [PubMed] [Google Scholar]
- Cridland J., Thornton K., 2010. Validation of rearrangement breakpoints identified by paired-end sequencing in natural populations of Drosophila melanogaster. Genome Biol. Evol. 2: 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1951. Genetics and the Origin of Species, Ed. 3 Columbia University Press, New York [Google Scholar]
- Dubinin N. P., Sokolov N. N., Tiniakov G. G., 1937. Intraspecific chromosomal variability. Biol Zh. 6: 1007 [Google Scholar]
- Frydenberg J., Hoffmann A. A., Loeschcke V., 2003. DNA sequence variation and latitudinal associations in hsp23, hsp26 and hsp27 from natural populations of Drosophila melanogaster. Mol. Ecol. 12: 2025–2032 [DOI] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P., 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8: 195–202 [DOI] [PubMed] [Google Scholar]
- Green P., 1996. Phrap documentation. Available at: http://www.phrap.org/phredphrap/phrap.html
- Hoffmann A. A., Rieseberg L. H., 2008. Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 39: 21–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., Slatkin M., Maddison W. P., 1992. Estimation of levels of gene flow from DNA-sequence data. Genetics 132: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike J., 2001. Sex chromosome meiotic drive. Annu. Rev. Ecol. Syst. 32: 25–49 [Google Scholar]
- Kennington W. J., Hoffmann A. A., Partridge L., 2007. Mapping regions within cosmopolitan inversion In(3R)Payne associated with natural variation in body size in Drosophila melanogaster. Genetics 177: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd J. M., Cooper G. M., Donahue W. F., Hayden H. S., Sampas N., et al. , 2008. Mapping and sequencing of structural variation from eight human genomes. Nature 453: 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Barton N., 2006. Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb W. R., Oakenshot J. G., Gibson J. B., 1981. Chromosome inversion polymorphism in Drosophila melanogaster.I. Latitudinal clines and associations between inversions in Australasian populations. Genetics 98: 833–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimbas C. B., Powell J. R., 1992. Drosophila Inversion Polymorphism. CRC Press, Boca Raton, FL [Google Scholar]
- Kusano A., Staber C., Chan H. Y. E., Ganetzky B., 2003. Closing the (Ran)GAP on segregation distortion in Drosophila. Bioessays 25: 108–115 [DOI] [PubMed] [Google Scholar]
- Langley C. H., Crepeau M., Cardeno C., Corbett-Detig R., Stevens K., 2011. Circumventing heterozygosity: sequencing the amplified genome of a single haploid Drosophila melanogster embryo. Genetics 188: 239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Stevens K., Cardeno C., Lee Y. C. G., Schrider D. R., et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192: 533–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ruan J., Durbin R., 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Fan W., Tian G., Zhu H., He L., et al. , 2009. The sequence and de novo assembly of the giant panda genome. Nature 463: 311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J. C., Suzuki D. T., 1968. The interchromosomal control of recombination. Annu. Rev. Genet. 2: 53–86 [Google Scholar]
- Lyon M. F., 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37: 393–408 [DOI] [PubMed] [Google Scholar]
- Mathiopoulos K. D., della Torre A., Predazzi V., Petrarca V., Coluzzi M., 1998. Cloning of inversion breakpoints in the Anopheles gambiae complex traces a transposable element at the inversion junction. Proc. Natl. Acad. Sci. USA 95: 12444–12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzkin L. M., Merritt T. J. S., Zhu C. T., Eanes W. F., 2005. The structure and population genetics of the breakpoints associated with the cosmopolitan chromosomal inversion In(3R)Payne in Drosophila melanogaster. Genetics 170: 1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev P., Stanciu M., Brudno M., 2009. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods 6: S13–S20 [DOI] [PubMed] [Google Scholar]
- Novitski E., Braver G., 1954. An analysis of crossing-over within a heterozygous inversion in Drosophila melanogaster. Genetics 39: 197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz J. M., Maurin D., Chan Y. S., Grotthuss M. V., Hillier L. W., et al. , 2007. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 5: 1366–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindi S. S., Raphael B. J., 2010. Identification and frequency estimation of inversion polymorphisms from haplotype data. J. Comput. Biol. 17(3): 517–531 [DOI] [PubMed] [Google Scholar]
- Sturtevant A. H., 1917. Genetic factors affecting the strength of linkage in Drosophila. Proc. Natl. Acad. Sci. USA 3: 555–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1926. A crossover reducer in Drosophila melanogaster due to inversion of a section of the third chromosome. Biol. Zentralbl. 46: 697–702 [Google Scholar]
- Sturtevant A. H., 1931. Known and probable inverted sections of the autosomes of Drosophila melanogaster. Carnegie Inst.Washington Publ. 421: 1–27 [Google Scholar]
- Sturtevant A. H., Beadle G. W., 1936. The relations of inversions in the X chromosome of Drosophila melanogaster to crossing over and disjunction. Genetics 21: 544–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzun E., Sharp A. J., Bailey J. A., Kaul R., Morrison V. A., et al. , 2005. Fine-scale structural variation of the human genome. Nat. Genet. 37: 727–732 [DOI] [PubMed] [Google Scholar]
- Van Delden W., Kamping A., 1991. Changes in relative fitness with temperature among second chromosome arrangements in Drosophila melanogaster. Genetics 127: 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley C. S., Eanes W. F., 1994. Isolation and analysis of the breakpoint sequences of chromosome inversion In(3L)Payne in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 91: 3132–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.