Abstract
Despite advances in genetic mapping of quantitative traits and in phylogenetic comparative approaches, these two perspectives are rarely combined. The joint consideration of multiple crosses among related taxa (whether species or strains) not only allows more precise mapping of the genetic loci (called quantitative trait loci, QTL) that contribute to important quantitative traits, but also offers the opportunity to identify the origin of a QTL allele on the phylogenetic tree that relates the taxa. We describe a formal method for combining multiple crosses to infer the location of a QTL on a tree. We further discuss experimental design issues for such endeavors, such as how many crosses are required and which sets of crosses are best. Finally, we explore the method’s performance in computer simulations, and we illustrate its use through application to a set of four mouse intercrosses among five inbred strains, with data on HDL cholesterol.
Keywords: quantitative trait loci (QTL), phylogenetic tree, evolution, multiple crosses, combining crosses
THE analysis of experimental crosses to identify the genetic loci (called quantitative trait loci, QTL) that contribute to variation in quantitative traits has become a standard approach in evolutionary biology. The properties of the QTL responsible for phenotypic differences between populations or species—including the number of QTL, their effect sizes, and their modes of action—provide insights into the mechanisms of evolution. QTL data have been brought to bear on a wide range of evolutionary processes, including adaptation (Doebley and Stec 1991; Bradshaw et al. 1998; Orr 1998; Mauricio 2001; Peichel et al. 2001; Rieseberg et al. 2002; Mitchell-Olds et al. 2007; Steiner et al. 2007; Hall et al. 2010) and speciation (Bradshaw et al. 1995; Moehring et al. 2006; Oka et al. 2007; Shaw et al. 2007; Moyle and Nakazato 2008; McDermott and Noor 2011; White et al. 2011).
By modeling the distribution of trait values across a tree, phylogenetic comparative methods also help to reconstruct the dynamics of phenotypic evolution. These approaches address several key issues, including the values of traits in ancestors (Schluter et al. 1997; Pagel 1999; Garland and Ives 2000; Pagel et al. 2004), rates of phenotypic evolution (Garland 1992; Venditti et al. 2011), the connection between trait evolution and speciation/extinction (Maddison et al. 2007; Fitzjohn et al. 2009), and the role of natural selection vs. genetic drift (Hansen 1997; Freckleton and Harvey 2006).
Despite the successful application of QTL mapping and phylogenetic comparative methods to fundamental questions in evolutionary biology, the two frameworks are rarely integrated. Methods that combine the portraits of genetic architecture obtained from QTL mapping with the logic of phylogenetic comparisons would offer several benefits. First, QTL data would provide a mechanistic basis for the dynamics of phenotypic evolution uncovered by phylogenetic comparative approaches. Although trait shifts along trees are caused by mutations, the methods for reconstructing these shifts do not currently incorporate genetic information.
Second, situating QTL data within a phylogenetic framework would directly account for the statistical dependencies that accompany any mapping comparison among three or more taxa. The tree connecting the species used in genetic mapping constrains the configurations of shared and divergent QTL that are possible, but this information is currently ignored by most QTL mapping methods.
Most importantly, a combined method could reveal the history of genetic differences between species. The mutations that underlie QTL occur along a phylogeny. Assigning these mutations to branches of the tree would pinpoint their evolutionary origins and allow testable predictions regarding the temporal accumulation of mutations (Moyle and Payseur 2009).
The ability to assign QTL to branches of phylogenetic trees would benefit genetic research beyond evolutionary biology. Collectively or individually, researchers often map QTL for the same phenotype in multiple sets of strains, especially in agricultural and biomedical model organisms. In addition to refining QTL position (Li et al. 2005), joint analysis of these crosses can pinpoint the genetic backgrounds (strains) on which QTL arose, providing further insights into the genetic architecture of traits involved in food quality or disease.
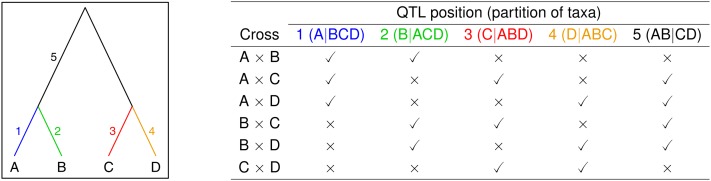
To envision the problem, consider the tree in Figure 1, and imagine the presence of a single diallelic QTL. The mutant allele at the QTL could have arisen in one of five possible locations on the tree, and each location is associated with a particular partition of the four taxa into two groups (those with the “high” allele and those with the “low” allele). For each such partition, the QTL will segregate in a different subset of the possible crosses between pairs of taxa. Throughout, we focus solely on unrooted trees. The two edges on either side of the root in Figure 1, labeled 5, cannot be distinguished. Also, a mutation arising above the root cannot be distinguished from the null model of no QTL.
Figure 1 .
Illustration of the basic concepts behind the mapping of a QTL to a phylogenetic tree. On the left is an example tree relating four taxa. The locations of possible origins of a diallelic QTL are indicated by the numbers 1–5. In the table on the right, we indicate the presence or absence of a QTL in each of the six possible crosses among pairs of taxa, according to the location of the QTL on the tree. Each possible QTL location on the tree corresponds to a partition of the taxa into two groups.
With data on multiple crosses, the simplest approach to identifying the location on the tree at which a QTL arose is to compare the pattern of presence and absence of the QTL in the individual crosses and match that to the ideal (see the table in Figure 1). We describe a more formal approach, combining ideas from Li et al. (2005), regarding the joint analysis of multiple crosses, with ideas from MacDonald and Long (2007), regarding partitioning multiple QTL alleles into two groups.
We discuss experimental design issues for such endeavors, such as how many crosses are required and which sets of crosses are best, explore the method’s performance in computer simulations, and illustrate its use through application to a set of four mouse intercrosses among five inbred strains, with data on HDL cholesterol.
Methods
To develop methods for mapping a QTL to a phylogenetic tree, we begin with several simplifying assumptions: The taxa are represented by inbred lines, the tree relating the taxa is known without error, the quantitative trait of interest is affected by a single diallelic QTL, and there are no background effects (i.e., the effect of the QTL is the same in the different crosses in which it is segregating). We consider the case of intercrosses among pairs of taxa, consider only autosomal loci, and assume a common genetic map.
The basic idea, illustrated in Figure 1, is that each possible location for the origin of a diallelic QTL on the tree corresponds to a different partition of the taxa into two groups, with the two groups corresponding to the two QTL alleles. For different partitions, the QTL will segregate in different sets of crosses. In the case of very large crosses, with each having high power to detect the QTL, if present, we could simply consider the crosses individually and use the pattern of presence/absence of QTL to identify the correct partition of the taxa. Note that one does not need data on all possible crosses. For the case illustrated in Figure 1, with four taxa, it would be sufficient to consider the crosses A × B, A × C, and B × D, as with just these three crosses, the five possible partitions have distinct patterns of presence/absence of the QTL. In the following, we focus on partitions of the taxa into two groups, in place of locations of the QTL on the tree.
Given limited resources and crosses of limited size, there will be incomplete power to detect the QTL in a given cross, and so the naive approach based on the presence or absence of the QTL in the different crosses will likely be misleading. A more formal approach, in which the likelihoods for the different possible partitions are evaluated and compared, will provide a clear assessment of the evidence for the different locations for the QTL on the tree.
Consider a particular location in the genome as the site of a putative QTL, and consider a particular partition of the taxa into two QTL alleles. We assume a linear model with normally distributed errors
| , |
where yij is the phenotype for individual j in cross i, μi the average phenotype in cross i, α and δ are the additive and dominance effects of the QTL, respectively, and the εij are independent and identically distributed normal (0, σ2). The aij and dij denote encodings of the QTL genotypes, with aij = dij = 0 if the QTL is not segregating in cross i. For convenience, we call the two QTL alleles defined by the partition as the high allele (H) and the low allele (L), although we won’t actually constrain the high allele to increase the phenotype. If the QTL is segregating in cross i, then we take aij = −1, 0, or + 1, if individual j has QTL genotype gij = LL, HL, or HH, respectively, and dij = 1 if individual j has QTL genotype HL and dij = 0 otherwise.
For most putative QTL locations, the QTL genotypes are not be observed, but we may calculate (e.g., by a hidden Markov model) the conditional probabilities of the QTL genotypes given the available multipoint marker genotype data, . It is critical that we have a common map for the set of crosses, so that a putative QTL location is clearly defined in all crosses. It is not necessary, however, that the same markers be used in all crosses or that they be informative in all crosses. We may then use standard interval mapping (Lander and Botstein 1989) or an approximation such as Haley–Knott regression (Haley and Knott 1992) to fit the model, estimate the parameters μi, α, δ, and σ2, and calculate a LOD score, , where π denotes the partition of the taxa and λ denotes the location of the putative QTL. The LOD score is the log10 likelihood comparing the hypothesis of a single QTL at that location to the null hypothesis of no QTL but with the multiple crosses allowed to have separate phenotypic means, that is, yij ∼ normal (μi, σ2).
This analysis is just as in Li et al. (2005), in that one recodes the genotypes in the crosses in which the QTL is segregating, stacks them on top of one another, as if they were a single intercross, and performs interval mapping with cross indicators as additive covariates. The only difference is that we are considering all possible partitions of the taxa, while Li et al. (2005) assumed a particular one. There is one technicality: The crosses in which the QTL does not segregate also need to be included in the likelihood, and they contribute to the estimate of the residual variance.
We thus consider each possible partition, π, one at a time, and scan the genome to obtain a set of LOD curves, . We summarize these at the chromosome level, calculating the maximum LOD score for partition π on chromosome i, . The maximum on chromosome i, , indicates the evidence for a QTL on chromosome i.
To evaluate the relative support of the different partitions, we use an approximate Bayes procedure. Assuming the presence of a single diallelic QTL on chromosome i, we assign equal prior probabilities to the different possible partitions, π, treat the profile log likelihoods (in which we have maximized over all nuisance parameters, including the location of the QTL on the chromosome) as if they were true log likelihoods, and obtain posterior probabilities by taking and rescaling so that they sum to 1. That is,
| . |
We further use these approximate posterior probabilities to form a 95% Bayesian credible set of partitions. One could assign unequal prior probabilities to the partitions, for example, based on the branch lengths in the assumed phylogenetic tree, giving more weight to longer branches. One might also use a prior on partitions that assigns greater weight to partitions induced by the tree and lesser (but nonzero) weight to the other (possibly more numerous) partitions.
The 95% credible set of partitions is relevant only if there is sufficient evidence for a QTL on that chromosome. To evaluate the evidence for a QTL, we consider the maximum of the on chromosome i and derive a significance threshold, adjusting for the genome scan, by a stratified permutation test (Churchill and Doerge 1994). The permutation test is stratified in that we permute the phenotype data, relative to the genotype data, separately in each cross. For each permutation replicate, we calculate the LOD curve for each possible partition and then take the maximum LOD score across the genome and across partitions. The 95th percentile of these permutation results may be used as a significance threshold, or we may calculate a P-value that accounts for the search across partitions and across the genome.
One may restrict the analyses to the set of partitions induced by the assumed phylogenetic tree, or one may consider all possible partitions of the taxa into two groups. For example, for the four-taxon tree in Figure 1, one may consider only the five partitions that correspond to QTL locations on the tree, as in the accompanying table, or one may also consider the two additional partitions, and . The consideration of all possible partitions will be accompanied by some loss of power, particularly if there is a large number of taxa. However, the correct phylogenetic tree will seldom be known with certainty and will likely vary along the genome, particularly if the taxa are closely related. Moreover, if there is strong support for one of the partitions that is not associated with a QTL location on the assumed phylogenetic tree, one would certainly want to know this. Thus, we are inclined to always consider all possible partitions and not focus on those induced by an assumed phylogenetic tree.
Theory
In this section, we address a theoretical question of considerable interest: Which subsets of crosses are sufficient to identify the location of a QTL on the phylogenetic tree? With very large crosses, we can exactly determine which crosses are segregating a QTL and which are not. As discussed in the Introduction, one need not perform all possible crosses. For example, for the case in Figure 1, if one performs only the crosses A × B, A × C, and A × D, the ideal results perfectly discriminate among the possible locations of the QTL on the tree. However, if one performs only the crosses A × B, A × C, and B × C, several of the possible partitions of strains exhibit the same pattern of presence/absence of QTL and so are confounded. Clearly, all taxa must be involved in the chosen crosses.
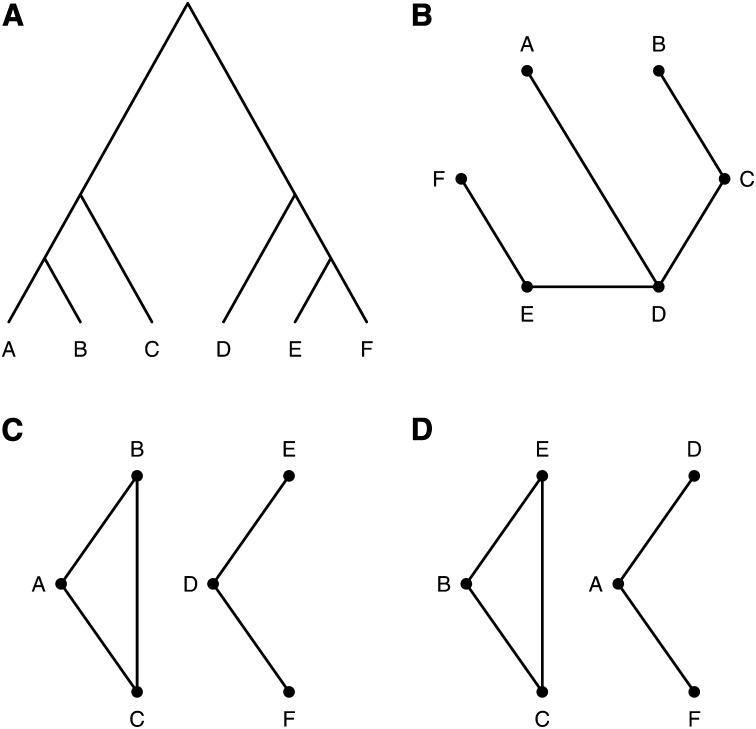
It is useful, in considering this problem, to represent a set of crosses by a graph, with nodes corresponding to taxa and edges indicating a cross between two taxa. For example, consider Figure 2. A phylogenetic tree relating six taxa is shown in Figure 2A. Three possible choices of a subset of five crosses among the six taxa are displayed in Figure 2, B–D.
Figure 2 .
A phylogenetic tree with six taxa (A) and three possible choices of five crosses among the six taxa, with nodes denoting taxa and edges denoting crosses (B–D).
A sufficient condition for identifying the true partition of the strains is the use of a set of crosses that connect all of the taxa, as in Figure 2B. Choose an arbitrary taxon (e.g., A) and assign it an arbitrary QTL allele. With sufficient numbers of individuals in each cross, we may determine whether the QTL is segregating in a cross, which indicates that the two taxa have different QTL alleles, or is not segregating, which indicates that the two strains have the same QTL allele. Thus, one may move between taxa connected by a cross and assign QTL alleles, and so if the set of crosses connect all of the taxa, one can assign QTL alleles to all taxa and so identify the correct partition of taxa.
If the set of crosses are not connected (as in Figure 2, C and D), then some partitions of taxa will be confounded. For example, for the crosses in Figure 2C, the partition will give the same set of QTL results as under the null hypothesis of no QTL. Other pairs of partitions are confounded in this example, such as and .
If one is considering all possible partitions of the taxa (and not just those induced by the tree), then graph connectivity is also a necessary condition for identifying the true partition: If the crosses do not connect all taxa there will always be some partitions that are confounded.
However, if one focuses solely on those partitions induced by the tree (that is, partitions that result from a split on an edge in the tree), then it is not necessary that the crosses connect all taxa. An example is shown in Figure 2D. For the pairs of partitions that are confounded with this choice of crosses, no more than one of each pair corresponds to a split on the tree in Figure 2A; each possible partition induced by the tree gives a distinct set of QTL results for these crosses. Moreover, in this case one may omit any one of the three crosses, B × C, B × E, C × E: Only four crosses are necessary to distinguish among the nine partitions induced by the tree in Figure 2A.
That the crosses connect all taxa is a necessary and sufficient criterion to distinguish among all possible partitions, but it is not a necessary condition to distinguish among the partitions induced by the tree. Note that a cross between two taxa corresponds to a path along the tree from one leaf to another. Further, the QTL will be segregating in crosses whose paths go through the edge with the QTL, but it will not be segregating in crosses whose paths do not go through that edge. A necessary and sufficient criterion for a set of crosses to distinguish the partitions induced by the tree (i.e., to distinguish the possible locations of the QTL on the tree) is that each edge is covered by at least one cross and that no two edges appear only together.
If an edge was not covered by a cross, then a QTL on that edge could not be distinguished from the null model, of no QTL. If two edges only appear together in crosses, then those two QTL locations cannot be distinguished. Thus, the criterion is necessary. For sufficiency, note that a cross in which the QTL is segregating will limit the possible QTL locations to the edges on the corresponding path through the tree. As every pair of edges along such a path will appear separately in different crosses, we see that the specific edge containing the QTL may be identified.
For n taxa (with ), the minimal number of crosses to distinguish among all possible partitions is . To distinguish among the partitions induced by the tree, the minimal number of crosses is (the smallest integer that is greater than ; a proof appears in the Appendix). For , these are the same; for , fewer crosses are needed to distinguish among the tree partitions.
As discussed in the previous section, we recommend that one not restrict oneself to the partitions induced by the tree but rather always consider all possible partitions, possibly with different prior weights. As a result, we recommend that one use, at a minimum, a set of crosses that connect all taxa. However, this is based on the assumption of a small number of taxa. If the number of taxa, n, is large, the total number of non-null partitions (2n−1 − 1) will vastly exceed the number of partitions induced by the tree (2n − 3), and so there is great potential advantage in focusing on the tree partitions.
Of course, in practice crosses are of finite size and so one cannot identify the true partition of the taxa without some degree of uncertainty. In the next section we explore, via computer simulation, the relative performance of the proposed method with different possible choices of crosses.
Simulations
In this section, we investigate the performance of our approach via computer simulation. We begin by comparing our proposed method to the naive approach of considering the crosses individually and comparing the pattern of presence/absence of a QTL in the crosses to what is expected for different possible partitions. We then compare the performance of our approach with all possible crosses to different choices of a minimal set of crosses.
Comparison to naive approach
We consider the case of four taxa and use of all six possible intercrosses among pairs of taxa, with 75 individuals per cross (a total sample size of 450). We consider a single autosome of length 127 cM, with markers at an approximately 10-cM spacing, and with a single diallelic QTL placed in the center of an interval between two markers, near the middle of the chromosome. The QTL alleles were assumed to act additively (that is, no dominance), and the percentage phenotypic variance explained by the QTL, in the crosses in which it was segregating, was 10%. We assumed either the partition or ; other possible partitions are equivalent to one of these. To reduce computation time, we used Haley–Knott regression (Haley and Knott 1992) for all simulation studies, with LOD score calculations performed on a 1-cM grid. Recombination was simulated assuming no crossover interference.
For the naive approach, we applied a given significance threshold and inferred the presence or absence of a QTL in a cross if the maximum LOD score on the chromosome was above or below the threshold, respectively. If the presence/absence pattern matched that for a possible partition, that partition was inferred.
For the proposed approach, we applied a given significance threshold on and then formed a 95% Bayesian credible set of partitions, using equal prior probabilities on all seven possible partitions. If was greater than the threshold but the 95% credible set did not contain the truth, the result was considered a false positive.
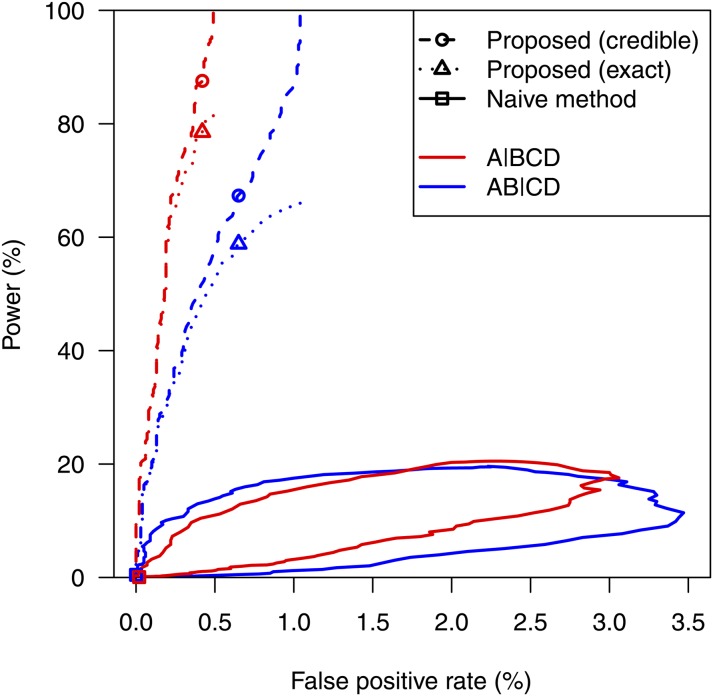
The results, based on 10,000 simulations, are displayed in Figure 3 as receiver operating characteristic (ROC) curves: the power (the rate of true positives) vs. the false positive rate, for varying significance thresholds. We display two sets of curves for the proposed method: For the dashed curves, the power indicates that exceeded the threshold and the true partition was contained within the 95% credible set; the dotted curves are more stringent and require that the credible set contained only the true partition. Points are plotted at the results with a nominal 5% significance threshold, adjusting for an autosomal genome scan, with the genome modeled after the mouse and the thresholds estimated by 10,000 simulations under the null hypothesis of no QTL. (The estimated thresholds are displayed in Supporting Information, Table S1 and Table S2.)
Figure 3 .
Estimated receiver operating characteristic (ROC) curves for the naive method (solid curves), the proposed method, with power indicating that the true partition is contained within the 95% credible set (dashed curves), and the proposed method, with power indicating that the 95% credible set contains only the true partition (dotted curves), in the case of four taxa, with each of the six possible intercrosses having a sample size of 75, and a QTL responsible for 10% of the phenotypic variance in the crosses in which it is segregating. The red and blue curves correspond to the case that the true partition is and , respectively. Points indicate the power and false positive rates for a 5% significance threshold. The results are based on 10,000 simulation replicates.
The ROC curves for the naive method form interesting shapes, with the lower part of each corresponding to low thresholds and the upper part corresponding to high thresholds, and indicate terrible performance: The false positive rate is well controlled, but power is low. The problem is that, with only moderate power to detect the QTL in a given cross, one has low power to detect the QTL in all of the crosses in which it is segregating, which is necessary to identify the correct partition of the taxa. Lowering the significance threshold below the 5% level helps somewhat, but the power to detect the true partition is no higher than 21%. The naive approach might actually perform better if one considered a smaller set of crosses, but we have not explored this further.
The proposed method performs reasonably well, and the false positive rate is well controlled at the nominal 5% significance threshold (the points in Figure 3). Lowering the threshold could give some improvement in power while maintaining the false-positive rate below the target level, at least in the simulated situations.
All crosses vs. minimal crosses
In the previous section, we noted that it is not necessary to use all possible crosses among taxa. To distinguish among all possible partitions, one need only choose a set of crosses that connect all taxa. Sets of crosses that connect all taxa and are of minimal size (i.e., crosses for n taxa) are called minimal sets. We now turn to the question of whether it is better to use all crosses, with a smaller number of individuals per cross, or a minimal set of crosses, with a larger number of individuals per cross. We use the same general settings as for the simulations comparing the proposed method to the naive approach, with four taxa and the true partition being either or , but here we vary the total sample size among 300, 450, and 600 individuals, and we vary the percentage phenotype variance explained by the QTL from 2.5 to 15%. We consider either all six crosses or a minimal set of three crosses, and we consider all 16 choices of three crosses that include all four taxa. We also compared the consideration of all seven possible partitions, or just the five partitions induced by the tree in Figure 1. We estimated 5% genome-wide significance thresholds by simulations under the null hypothesis of no QTL (see Table S2).
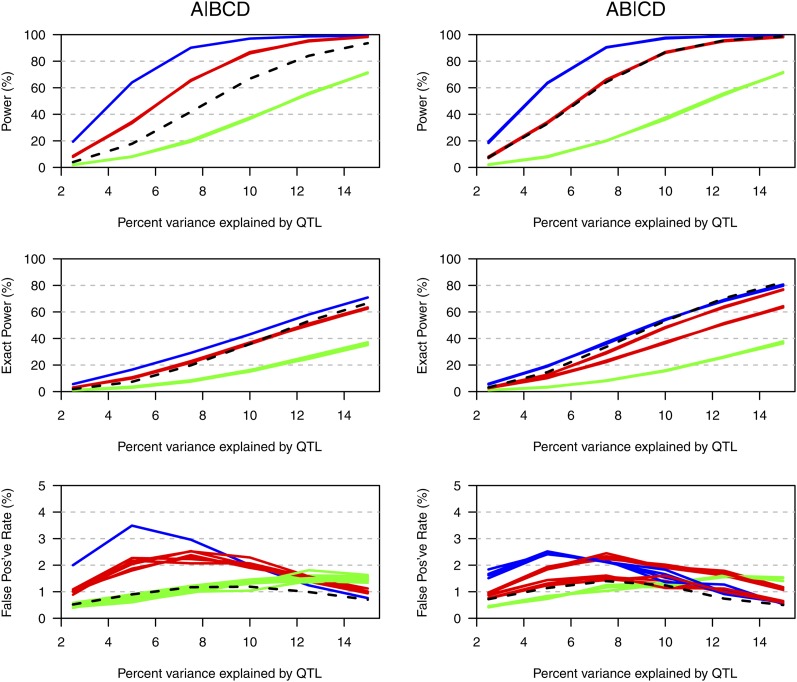
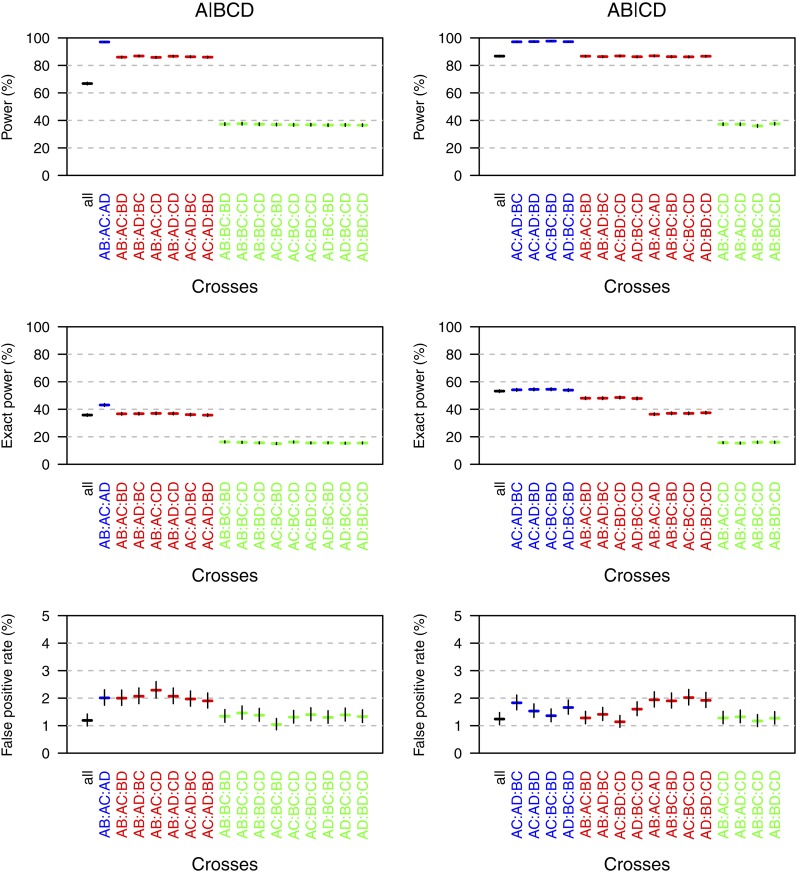
Figure 4 displays the simulation results, as a function of the effect of the QTL, for the case that the total sample size was 450 (i.e., 75 individuals per cross when considering all crosses and 150 individuals per cross when considering a minimal set of three crosses) and when all possible partitions were considered. The results with other sample sizes and with analysis restricted to the five partitions induced by the tree in Figure 1 are shown in Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, and Figure S6. The top of each figure indicates the power (the chance that exceeded its threshold and the true partition was contained in the 95% credible set); the middle indicates the “exact” power (the chance that exceeded its threshold and that the credible set contained only the true partition); the bottom indicates the false positive rate. The left and right correspond to the true partition being or , respectively. The black dashed curves correspond to the use of all six possible crosses; the solid curves correspond to the different choices of a minimal set of three crosses, with blue, red, and green corresponding to cases in which 3, 2, or 1 of the crosses are segregating a QTL.
Figure 4 .
Estimated power (top), “exact” power (middle), and false-positive rates (bottom) in the case of four taxa with a total sample size of 450, as a function of the percentage phenotypic variance explained by the QTL. The black dashed curves correspond to the use of all six possible crosses. The other curves are for the various choices of a minimal set of three crosses, with the curves in blue, red, and green corresponding to cases in which three, two, and one of the crosses are segregating the QTL, respectively. The results are based on 10,000 simulation replicates, with analyses considering all possible partitions of the taxa.
In choosing among the possible minimal sets of crosses, power is highest when a larger number of crosses are segregating the QTL. For a fixed total sample size, the use of all possible crosses (with fewer individuals per cross) has better performance than the worst of the possible minimal sets of crosses, but is not as good as the best of the possible minimal sets of crosses. The use of all possible crosses has greater power when the true partition is (in which case four of the six crosses are segregating the QTL) than when the true partition is (in which case three of the six crosses are segregating the QTL). The false-positive rate (Figure 4, bottom) is well controlled throughout.
The use of a total sample size of 300 or 600 gives qualitatively similar results (see Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, and Figure S6; Figure S7 and Figure S8 contain the false negative rates), although we note that while a larger sample size results in a great improvement in power, it gives only a slight improvement in the chance that the credible set includes only the true partition.
Restricting the analysis to the five partitions induced by the tree has little effect on power (compare Figure S1 and Figure S2), but improves the chance that the credible set includes only the true partition (compare Figure S3 and Figure S4), and results in a somewhat lower false-positive rate (compare Figure S5 and Figure S6).
The performance of the proposed method with different possible choices of minimal crosses is largely predicted by the number of crosses that are segregating a QTL: The solid curves of a given color (which indicates the number of crosses segregating a QTL) are largely coincident, but there are some differences (red curves in Figure 4, middle right). To explore this further, the results for the individual choices of crosses, when the percentage phenotypic variance explained by the QTL is 10% and the total sample size is 450, are displayed in Figure 5. (For other sample sizes and for the analyses restricted to the partitions induced by the tree in Figure 1, see Figure S9, Figure S10, Figure S11, Figure S12, Figure S13, Figure S14, Figure S15, and Figure S16.)
Figure 5 .
Detailed results on the estimated power (top), “exact” power (middle), and false positive rates (bottom), for individual choices of crosses, in the case of four taxa with a total sample size of 450, and with the QTL being responsible for 10% of the phenotypic variance in crosses in which it is segregating. Blue, red, and green correspond to cases in which three, two, and one of the crosses are segregating the QTL, respectively. The results are based on 10,000 simulation replicates, with analyses considering all possible partitions of the taxa. The black vertical line segments indicate 95% confidence intervals.
In the case that the true partition is , there are some differences among the choices of three crosses when two of the three are segregating the QTL, in terms of the chance that the 95% credible set contains only the true partition (Figure 5, middle). For example, the use of the crosses A × B, A × C, and B × D gives “exact” power of ∼50%, while the use of A × B, A × C, and A × D gives “exact” power of ∼40%.
To understand the difference, we need to consider the sign of the QTL effect in different crosses for the true partition and the best alternative partition; these are shown in Table 1. If the true partition is , with C and D having an allele that results in an increase in the phenotype, the A × B cross does not segregate a QTL, while each of A × C, B × D, and A × D have a segregating QTL with the latter taxon in each cross increasing the phenotype. With the crosses A × B, A × C, and A × D, the best alternative partition after would be , in which A × C and A × D are also segregating the QTL, but A × B should also be segregating the QTL, and note that for both partitions and , the QTL has effect in the same direction in the A × C and A × D crosses. On the other hand, with the crosses A × B, A × C, and B × D (which was seen to have better performance), in the only alternative partition with two crosses segregating a QTL, , the two crosses should have QTL effects in opposite directions (the A and D alleles both result in a decrease in phenotype), and so this should be easy to distinguish from the partition. For this choice of three crosses, all other partitions have a QTL segregating in just one of A × C or B × D but not both. As a result, the chance that the credible set contains only the true partition is slightly higher.
Table 1. Signs of the QTL effects in the case of four taxa, for each possible cross and each possible partition.
| Partition of taxa |
|||||||
|---|---|---|---|---|---|---|---|
| Cross | |||||||
| A × B | + | − | 0 | 0 | 0 | + | + |
| A × C | + | 0 | − | 0 | + | 0 | + |
| A × D | + | 0 | 0 | − | + | + | 0 |
| B × C | 0 | + | − | 0 | + | − | 0 |
| B × D | 0 | + | 0 | − | + | 0 | − |
| C × D | 0 | 0 | + | − | 0 | + | − |
For each partition, the taxa to the right of the vertical bar have the high allele. For each cross, the sign of the effect is for the right vs. the left taxon.
While no such differences among the choices of minimal crosses are seen when the true partition is and all possible partitions are considered in the analysis, these sorts of differences do arise when the analysis is restricted to the five partitions induced by the tree in Figure 1. (See Figure S12.)
Application
To illustrate our approach, we consider the data from Li et al. (2005), originally reported in Lyons et al. (2003a,c,b) and Wittenburg et al. (2003, 2005) and available at the QTL Archive (http://www.qtlarchive.org). These data concern four intercrosses among five inbred mouse strains, CAST/Ei (C), DBA/2 (D), I/LnJ (I), PERA/Ei (P), and 129S1/SvImJ (S). The four intercrosses performed were C × D, C × S, D × P, and I × P. The C × D and C × S crosses were all males and had 277 and 275 mice, respectively. The D × P and I × P crosses had approximately equal numbers of males and females and had a total of 282 and 322 mice, respectively. As in Li et al. (2005), we focus on a single phenotype, the square root of plasma HDL cholesterol. Note that the four intercrosses form a daisy chain, S × C × D × P × I, and so satisfy the connectedness condition necessary for inference of the correct partition of the strains at a diallelic QTL.
We used the genetic map from Cox et al. (2009), with marker locations obtained using the Mouse Map Converter at the Jackson Laboratory (http://cgd.jax.org/mousemapconverter). We used standard interval mapping (Lander and Botstein 1989) and considered all 15 possible partitions of the five strains, without attempting to infer a phylogenetic tree relating the strains. To handle the two sexes, we included sex as an additive covariate (that is, we allowed for a shift in the average phenotype between the sexes and assumed no QTL × sex interaction). We used permutation tests with 10,000 replicates to obtain 5% significance thresholds for the individual crosses and for . The estimated significance thresholds for the individual crosses were approximately 3.44 for all four crosses; the estimated threshold on was 5.39.
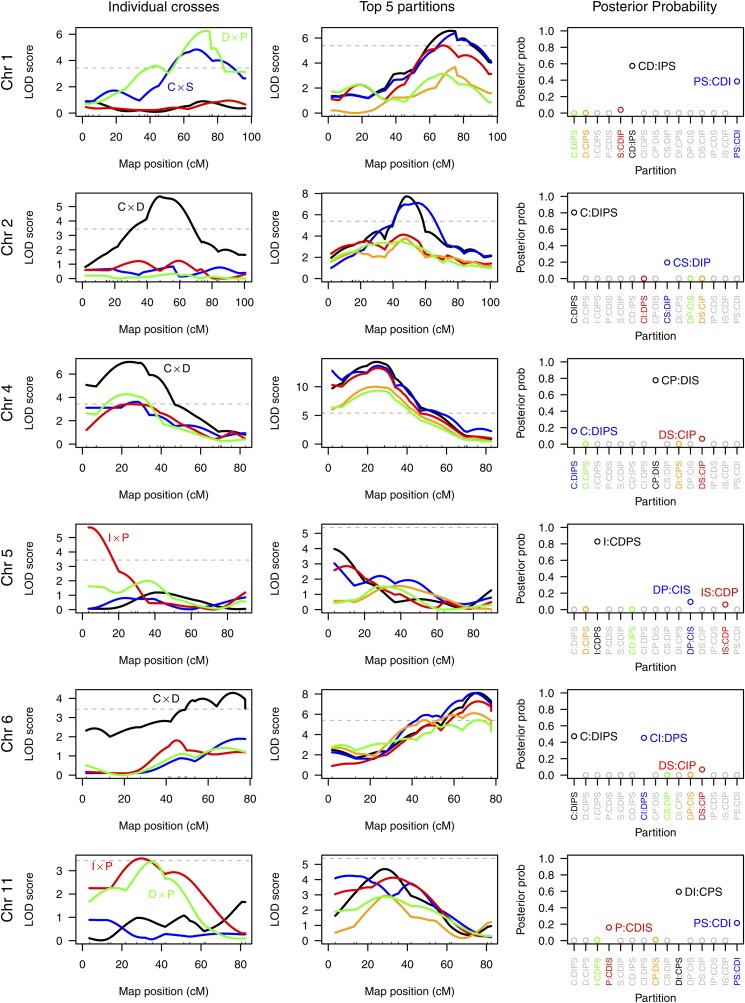
Following Li et al. (2005), we focused on chromosomes 1, 2, 4, 5, 6, and 11. The LOD curves for the individual crosses are displayed in Figure 6, left. The LOD curves for the top five partitions on each chromosome are in Figure 6, middle. The posterior probabilities of the different partitions, assuming the presence of a single diallelic QTL, are on the right. In all cases, the 95% credible set of partitions contains either two or three partitions.
Figure 6 .
Analysis results for selected chromosomes for the data from Li et al. (2005): LOD curves for individual crosses (left), LOD curves for the top five partitions (middle), and approximate posterior probabilities for each partition (right). The partitions corresponding to the five LOD curves in the middle are indicated on the right. The labeled points on the right indicate the partitions included in the 95% Bayesian credible sets. On the left and in the middle, dashed horizontal lines are plotted at the 5% significance thresholds.
For chromosome 1, significant evidence for a QTL is seen in the crosses C × S and D × P but not in C × D or I × P. By the naive approach, we would infer the partition , and this is the partition that Li et al. (2005) assumed. Our proposed method does give this partition the highest posterior probability (57%), but also gives reasonable weight to the alternative (posterior probability 39%), in which case the QTL would also be segregating in the I × P cross.
For chromosome 2, we see a QTL just in cross C × D. By the naive approach (given the set of crosses performed), we would infer the partition , which is the partition that Li et al. (2005) assumed. However, by the proposed method, has a posterior probability of only 20%, while the partition (in which the QTL would also be present in the C × S cross) has a posterior probability of 80%.
For chromosome 4, we have evidence for a QTL in all four crosses (although in the cross I × P, the maximum LOD score was 3.42, just missing the threshold of 3.44). If we assume that there is no QTL segregating in I × P, we would infer the partition , while if we take the evidence for a QTL in I × P as sufficient, we would infer the partition , and this is the partition that Li et al. (2005) assumed. The latter is the partition with the highest posterior probability (78%), while the former has posterior probability 7%, and a third partition, , in which case the QTL is segregating in neither I × P nor D × P, has posterior probability 16%.
For chromosome 5, we see a QTL only in cross I × P, and so by the naive approach we would infer the partition ; this partition does have the highest posterior probability (83%) and was the partition that Li et al. (2005) assumed. But the maximum LOD score for this partition was 3.98, which doesn’t meet the 5% significance threshold. (The genome-scan-adjusted P-value was 0.37.) Thus, by our proposed approach, we would not infer the presence of a QTL. But if we do allow that there is a QTL, two other partitions are contained within the 95% credible set: , with posterior probability 9%, in which case the QTL is also segregating in the cross C × D, and , with posterior probability 6%, in which case the QTL is also segregating in the cross C × S.
For chromosome 6, we have significant evidence for a QTL only in cross C × D (the other three crosses have maximum LOD scores of 1.5–1.9 on chromosome 6), and so the naive method would give the partition CS|DIP, which has posterior probability <0.01% and is not contained in the 95% credible set. The partitions with highest posterior are (47%), with the QTL also segregating in C × S, and (45%), with the QTL also segregating in C × S and I × P. The 95% credible set also contains a third partition, , with posterior probability 7%. Li et al. (2005) had assumed the partition , which is the partition with highest posterior probability.
For chromosome 11, there was significant evidence for a QTL only in the cross I × P, although the cross D × P has a maximum LOD score of 3.16 (corresponding to a genome-scan-adjusted P-value of 0.093). The naive approach would give the partition , which has posterior probability 0.9% and is not contained in the 95% credible set. If we consider the evidence for a QTL in D × P to be sufficient, we would infer the partition , which has posterior 16% and was the one that Li et al. (2005) assumed. The partition with highest posterior probability is (posterior probability 60%), in which case the QTL is also segregating in C × D. The 95% credible set also contains the partition (posterior probability 21%), in which case the QTL is segregating in C × S but not C × D. As with chromosome 5, the maximum LOD score across partitions (4.70) does not meet our 5% significance threshold, and so by our proposed method we would not infer the presence of a QTL. (The genome-scan-adjusted P-value was 0.14.)
Discussion
We have described a formal approach for the joint analysis of multiple crosses to map the origin of QTL alleles to a position on a phylogenetic tree. Our approach unites QTL mapping with phylogenetic comparative methods to provide a view of the genetic mechanism underlying phenotypic evolution. Further, our approach partitions taxa according to their QTL allele, facilitating haplotype analyses for the fine mapping of QTL. In addition, as part of this work, we have begun to evaluate a variety of experimental design issues for such research, which provides some guidance to researchers seeking to take advantage of this approach.
The goal of the work in Li et al. (2005) was to combine multiple related crosses to more precisely map QTL. The key difficulty in applying this idea is that one must define a unique partition of the strains into the two QTL alleles, a priori. In the presence of multiple QTL, the phenotypes of the strains cannot be trusted for inferring the QTL alleles, and in the current application, the six QTL partition the five strains in diverse ways. Li et al. (2005) used the pattern of QTL in the different crosses to infer the appropriate partition, which we have (perhaps overly harshly) characterized as the naive approach. We have proposed a formal method for comparing the different possible partitions. For two of the six loci, we find that the partition with strongest support is different from that assumed by Li et al. (2005), and for all six loci there are multiple partitions with reasonable support.
Our approach thus provides an important improvement on the method of Li et al. (2005). As seen in Figure 6, middle, the different partitions can have quite different LOD curves and so provide different information on the likely location of the QTL. Thus, our formal approach to identifying the well-supported partitions can improve localization of a QTL. Moreover, one could combine the information from the multiple partitions to better define the location of the QTL, taking account of the uncertainty in the partition.
Furthermore, while the application of these ideas to evolutionary studies remains our primary interest, the more straightforward application is in biomedical or agricultural research, as in Li et al. (2005), for the combined use of multiple crosses to more precisely map a QTL and, subsequently, with an inferred partition (or partitions) of strains in hand, to inform the analysis of the haplotypes of the strains (see, for example, Burgess-Herbert et al. 2008) in the search for the underlying causal polymorphism. The results are also valuable for the design of future experiments, if additional crosses are to be performed.
Our approach has some similarities to the use of local phylogenetic trees to define possible partitions of multiple alleles (Pan et al. 2009; Zhang et al. 2012) and to coalescent-based approaches (Zöllner and Pritchard 2005) for genome-wide association studies. The key distinction of our method is that we seek not just to establish association but also to identify the appropriate partition and so define the origin of the mutant QTL allele on the local phylogenetic tree. In our approach, the QTL location on the tree is not a nuisance parameter but rather is the target of inference.
In our simulation studies, we compared the use, for a fixed total sample size, of all possible crosses to different choices of a minimal set of crosses. Depending on the underlying true partition of taxa at a QTL, one can choose a minimal set of crosses with considerably higher power. However, given the prior uncertainty in the true partition, and the possibility of multiple QTL that each partition the taxa differently, it is prudent to consider all or at least a larger number of possible crosses. An even more important experimental design question, which we have not considered here, is how to choose which taxa, out of a large number of related taxa, to consider, in the effort to characterize the genetic architecture of a quantitative trait.
We have focused on a set of intercrosses. The approach could be adapted for the analysis of a set of backcrosses, although these would likely need to be of a special form, with the F1 hybrids all crossed to a common parent.
There are a number of additional ways in which our analytical framework could be extended. Most quantitative traits are affected by multiple QTL, rather than single QTL as assumed here. The restriction that a QTL has a common effect in all crosses in which it segregates might be relaxed, particularly for traits that are heavily shaped by epistasis, such as hybrid sterility and hybrid inviability (Coyne and Orr 2004). Prior distributions of QTL partitions could incorporate phylogenetic branch lengths (taxa separated by shorter evolutionary distances are more likely to share QTL alleles) as well as topologies. Finally, future developments might account for variation in the tree. This variation includes both statistical uncertainty associated with phylogenetic inference and real phylogenetic discordance across the genome, which results from incomplete lineage sorting and introgression in recently diverged taxa (Pamilo and Nei 1988; Maddison 1997; Pollard et al. 2006; White et al. 2009). The power of reconstructing QTL evolution as well as the increasing capacity for genetic mapping of complex traits and phylogenetic reconstruction should provide motivation for these extensions in the evolutionary, biomedical, and agricultural communities.
Software incorporating the proposed methods are available as part of R/qtl (Broman et al. 2003, http://www.rqtl.org), an add-on package to the general statistical software R (R Development Core Team 2010).
Supplementary Material
Acknowledgments
The authors thank Beverly Paigen for making her data publicly available, Gary Churchill for developing the QTL Archive, from which the data were drawn, and an anonymous reviewer for identifying a gap in the Theory section and encouraging its closure. This work was supported in part by National Institutes of Health grant GM074244 (to K.W.B.) and by National Science Foundation grant DEB 0918000 (to B.A.P.).
Appendix
In this section, we demonstrate that the minimal number of crosses to distinguish among the partitions induced by a tree with n taxa is , the smallest integer ≥2n/3. Recall, from the Theory section, that such a set of crosses must cover each edge in the tree, and no two edges can appear only together in crosses.
First note that such a minimal set of crosses defines a graph on the set of taxa. The connected components in this graph must each contain three or more taxa. If a component has just one taxon, then it is not covered by any cross. If a component has just two taxa, say A and B, then the edges in the tree leading to A and B will only appear together (in the cross A × B) and so cannot be distinguished. Thus, a minimal set of crosses must divide the taxa into groups of at least three, and so the minimal number of crosses is ≥2n/3, since every three taxa will need two crosses to connect them.
We now describe an algorithm to construct a minimal set of crosses, through which we show that the tree partitions can be distinguished with crosses. If the tree contains n < 6 taxa, form any set of n − 1 crosses that connect all taxa. Otherwise, we pull out three taxa at a time, to form nonoverlapping subtrees, , that together cover the entire tree, and for which the minimal crosses within each subtree, combined together, are sufficient to distinguish the tree partitions.
Define a cherry to be a pair of taxa that form a clade, and let T denote the full tree. Any unrooted tree contains at least two cherries. If there are exactly two cherries, then pick one taxon from each cherry and a third arbitrarily from among taxa not part of a cherry. If there are more than two cherries, then choose one taxon each from three different cherries. The three taxa chosen form T1. Let denote the subtree formed from all other taxa. If contains fewer than six taxa, we stop; otherwise, we apply the same technique recursively to until we reach a set of fewer than six taxa.
As an illustration, consider Figure S17; the top depicts a tree with nine taxa, and with four cherries. We first pick three taxa, one each from three different cherries: A, C, and F. We then consider the subtree of the remaining six taxa, displayed in Figure S17, bottom. We now pick one taxon from each of the two cherries and one of the two remaining taxa: B, E, and H. There are three taxa remaining (D, G, and I), and so we stop. We may then pick any two crosses among each group of three taxa, say A × C, C × F, B × E, E × H, D × G, and G × I. These six crosses are sufficient to distinguish all 15 partitions induced by the tree of nine taxa.
We turn to the proof. Again, let T denote the full tree, let T1 denote the subtree formed by the first three taxa chosen by the algorithm above, and let denote the subtree with the remaining n − 3 taxa. Note that any internal edge in T is covered by ; otherwise, there would be two taxa in T1 that form a cherry. Further, any internal edge in T is either represented in both T1 and or it is maintained as a distinct internal edge in , or both. (In Figure S17, top, internal edge f is covered by but not T1. The other five internal edges are each covered by both T1 and .) In other words, any internal branch in T that is collapsed with another branch in forming must be covered by T1: Any internal edge that gets collapsed with another edge in forming is either leading to a cherry and so being collapsed with an external edge or there are two internal edges that are being collapsed together, in which case there must be cherries on either side, each with one taxon becoming part of T1.
Consider any pair of crosses that cover T1 and any set of crosses that are sufficient to distinguish the edges in . These crosses clearly cover all edges, as all edges are covered by either T1 or or both. What remains is to show that no two edges in T will appear only together in these crosses. There are four classes of edges to consider: the external edges that become part of T1, the external edges that become part of , the internal edges that remain as distinct internal edges in , and the other internal edges, covered by both T1 and .
Now consider two distinct edges from T. If one is an external edge in T1, then the other is either another external edge in T1, in which case they are distinguished by the crosses within T1, or it is represented in , in which case there is a cross in that contains the latter but not the former edge. If one edge is an external edge in , then the other is either represented in T1, in which case there is a cross in T1 containing the latter but not the former, or it is a distinct external or internal edge in , in which case they are distinguished by the crosses within . A similar argument applies in the case of an internal edge that remains a distinct internal edge in . The final case to consider is of two internal edges that are each collapsed with another edge in forming . These are both represented in both T1 and , and they must be part of distinct edges in either T1 or or both, and so they can be distinguished by crosses in either T1 or or both.
Finally, we consider the recursion: If all edges in T are distinguished by T1 and , and if the edges in are similarly identified by , then all edges in T are identified by . This follows immediately by noting that our arguments above applied to any set of crosses that distinguished the edges within .
At the risk of sounding pedantic, note that our algorithm partitions T into subtrees , with for i < p and |Tp| = 3, 4 or 5. Since subtrees with three, four, and five taxa will be covered by two, three, and four crosses, respectively, we see that for any tree with n taxa, there is a minimal set of crosses that distinguish all edges.
Footnotes
Communicating editor: K. M. Nichols
Literature Cited
- Bradshaw H. D., Wilbert S. M., Otto K. G., Schemske D. W., 1995. Genetic mapping of floral traits associated with reproductive isolation in monkey flowers (Mimulus). Nature 376: 762–765 [Google Scholar]
- Bradshaw H. D. J., Otto K. G., Frewen B. E., McKay J. K., Schemske D. W., 1998. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 149: 367–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Wu H., Sen S., Churchill G. A., 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19: 889–890 [DOI] [PubMed] [Google Scholar]
- Burgess-Herbert S. L., Cox A., Tsaih S.-W., Paigen B., 2008. Practical applications of the bioinformatics toolbox for narrowing quantitative trait loci. Genetics 180: 2227–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A., Ackert-Bicknell C. L., Dumont B. L., Ding Y., Bell J. T., et al. , 2009. A new standard genetic map for the laboratory mouse. Genetics 182: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation. Sinauer Associates, Sunderland, MA [Google Scholar]
- Doebley J., Stec A., 1991. Genetic analysis of the morphological differences between maize and teosinte. Genetics 129: 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzjohn R. G., Maddison W. P., Otto S. P., 2009. Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58: 595–611 [DOI] [PubMed] [Google Scholar]
- Freckleton R. P., Harvey P. H., 2006. Detecting non-Brownian trait evolution in adaptive radiations. PLoS Biol. 4: e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T., Jr, 1992. Rate tests for phenotypic evolution using phylogenetically independent contrasts. Am. Nat. 140: 509–519 [DOI] [PubMed] [Google Scholar]
- Garland T., Jr, Ives A. R., 2000. Using the past to predict the present: confidence intervals for regression equations in phylogenetic comparative methods. Am. Nat. 155: 346–364 [DOI] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324 [DOI] [PubMed] [Google Scholar]
- Hall M. C., Lowry D. B., Willis J. H., 2010. Is local adaptation in Mimulus guttatus caused by trade-offs at individual loci? Mol. Ecol. 19: 2739–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T., 1997. Stabilizing selection and the comparative analysis of adaptation. Evolution 51: 1341–1351 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D., 1989. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Lyons M. A., Wittenburg H., Paigen B., Churchill G. A., 2005. Combining data from multiple inbred line crosses improves the power and resolution of quantitative trait loci mapping. Genetics 169: 1699–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons M. A., Wittenburg H., Li R., Walsh K. A., Churchill G. A., et al. , 2003a Quantitative trait loci that determine lipoprotein cholesterol levels in DBA/2J and CAST/Ei inbred mice. J. Lipid Res. 44: 953–967 [DOI] [PubMed] [Google Scholar]
- Lyons M. A., Wittenburg H., Li R., Walsh K. A., Leonard M. R., et al. , 2003b Lith6: a new QTL for cholesterol gallstones from an intercross of CAST/Ei and DBA/2J inbred mouse strains. J. Lipid Res. 44: 1763–1771 [DOI] [PubMed] [Google Scholar]
- Lyons M. A., Wittenburg H., Li R., Walsh K. A., Leonard M. R., et al. , 2003c New quantitative trait loci that contribute to cholesterol gallstone formation detected in an intercross of CAST/Ei and 129S1/SvImJ inbred mice. Physiol. Genomics 14: 225–239 [DOI] [PubMed] [Google Scholar]
- MacDonald S. J., Long A. D., 2007. Joint estimates of quantitative trait locus effect and frequency using synthetic recombinant populations of Drosophila melanogaster. Genetics 176: 1261–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W., 1997. Gene trees in species trees. Syst. Biol. 46: 523–536 [Google Scholar]
- Maddison W. P., Midford P. E., Otto S. P., 2007. Estimating a binary character’s effect on speciation and extinction. Syst. Biol. 56: 701–710 [DOI] [PubMed] [Google Scholar]
- Mauricio R., 2001. Mapping quantitative trait loci in plants: uses and caveats for evolutionary biology. Nat. Rev. Genet. 2: 370–381 [DOI] [PubMed] [Google Scholar]
- McDermott S. R., Noor M. A., 2011. Genetics of hybrid male sterility among strains and species in the Drosophila pseudoobscura species group. Evolution 65: 1969–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds T., Willis J. H., Goldstein D. B., 2007. Which evolutionary processes influence natural genetic variation for phenotypic traits. Nat. Rev. Genet. 8: 845–856 [DOI] [PubMed] [Google Scholar]
- Moehring A. J., Llopart A., Elwyn S., Coyne J. A., Mackay T. F., 2006. The genetic basis of prezygotic reproductive isolation between Drosophila santomea and D. yakuba due to mating preference. Genetics 173: 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle L. C., Nakazato T., 2008. Comparative genetics of hybrid incompatibility: sterility in two Solanum species crosses. Genetics 179: 1437–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle L. C., Payseur B. A., 2009. Reproductive isolation grows on trees. Trends Ecol. Evol. 24: 591–598 [DOI] [PubMed] [Google Scholar]
- Oka A., Aoto T., Totsuka Y., Takahashi R., Ueda M., et al. , 2007. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics 175: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1998. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149: 2099–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M., 1999. Inferring the historical patterns of biological evolution. Nature 401: 877–884 [DOI] [PubMed] [Google Scholar]
- Pagel M., Meade A., Barker D., 2004. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53: 673–684 [DOI] [PubMed] [Google Scholar]
- Pamilo P., Nei M., 1988. Relationships between gene trees and species trees. Mol. Biol. Evol. 5: 568–583 [DOI] [PubMed] [Google Scholar]
- Pan F., Yang L., McMillan L., Pardo-Manuel De Villena F., Threadgill D., et al. , 2009. TreeQA: quantitative genome-wide association mapping using local perfect phylogeny trees. Pac. Symp. Biocomput. 426: 415–426 [PMC free article] [PubMed] [Google Scholar]
- Peichel C. L., Nereng K. S., Ohgi K. A., Cole B. L., Colosimo P. F., et al. , 2001. The genetic architecture of divergence between threespine stickleback species. Nature 414: 901–905 [DOI] [PubMed] [Google Scholar]
- Pollard D. A., Iyer V. N., Moses A. M., Eisen M. B., 2006. Widespread discordance of gene trees with species tree in Drosophila: evidence for incomplete lineage sorting. PLoS Genet. 2: 1634–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2010. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Rieseberg L. H., Widmer A., Arntz A. M., Burke J. M., 2002. Directional selection is the primary cause of phenotypic diversification. Proc. Natl. Acad. Sci. USA 99: 12242–12245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D., Price T., Mooers A. O., Ludwig D., 1997. Likelihood of ancestor states in adaptive radiation. Evolution 51: 1699–1711 [DOI] [PubMed] [Google Scholar]
- Shaw K. L., Parsons Y. M., Lesnick S. C., 2007. QTL analysis of a rapidly evolving speciation phenotype in the Hawaiian cricket Laupala. Mol. Ecol. 16: 2879–2892 [DOI] [PubMed] [Google Scholar]
- Steiner C. C., Weber J. N., Hoekstra H. E., 2007. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 5: e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti C., Meade A., Pagel M., 2011. Multiple routes to mammalian diversity. Nature 479: 393–396 [DOI] [PubMed] [Google Scholar]
- White M. A., Ané C., Dewey C. N., Larget B. R., Payseur B. A., 2009. Fine-scale phylogenetic discordance across the house mouse genome. PLoS Genet. 5: e1000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Genetic dissection of a key reproductive barrier between nascent species of house mice. Genetics 189: 289–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenburg H., Lyons M. A., Li R., Churchill G. A., Carey M. C., et al. , 2003. FXR and ABCG5/ABCG8 as determinants of cholesterol gallstone formation from quantitative trait locus mapping in mice. Gastroenterology 125: 868–881 [DOI] [PubMed] [Google Scholar]
- Wittenburg H., Lyons M. A., Li R., Kurtz U., Mossner J., et al. , 2005. Association of a lithogenic Abcg5/Abcg8 allele on chromosome 17 (Lith9) with cholesterol gallstone formation in PERA/EiJ mice. Mamm. Genome 16: 495–504 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang X., Wang W., 2012. HTreeQA: using semi-perfect phylogeny trees in quantitative trait loci study on genotype data. G3: Genes, Genomes, Genetics 2: 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zöllner S., Pritchard J. K., 2005. Coalescent-based association mapping and fine mapping of complex trait loci. Genetics 169: 1071–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.