Abstract
Background
Parainfluenza virus type 3 (PIV3) was isolated from dairy buffaloes (Bubalus bubalis) naturally affected with respiratory and reproductive clinical conditions.
Results
Examination of nasal and vaginal swabs collected from 12 diseased buffaloes led to the isolation of three paramyxovirus isolates from two animals. Antigenic, morphological and biological characteristics of these three isolates were essentially similar to those of members of the Paramyxoviridae family. Antigenic analysis by direct immunofluorescence and cross neutralization test placed these isolates together with bovine parainfluenza virus type 3 (BPIV3). Nucleotide and amino acid phylogenetic analysis of partial matrix gene sequences of the buffalo isolates and six field BPIV3 isolates from bovines in Argentina were studied. Buffalo isolates were similar to genotype B (BPIV3b) while the six BPIV3 isolates were similar to genotypes A (BPIV3a) and C (BPIV3c).
Conclusions
This is the first characterization of BPIV3 in water buffalo.
According to the samples analyzed, in Argentina, the genotype B was found in buffalo and the genotypes A and C were found in cattle.
Background
Paramyxoviruses are well-known pathogens of the central nervous and respiratory systems of many host species. In the last few decades, many novel paramyxoviruses have emerged causing devastating illnesses in different aquatic and terrestrial animals, including in some cases a species jump to humans [1]. Parainfluenza virus type 3 (PIV3) is an enveloped, single-stranded negative sense RNA virus within the Respirovirus genus of the Paramyxoviridae family [2,3]. The Respirovirus genus includes human parainfluenza virus types 1 and 3 (HPIV1 and HPIV3, respectively), Sendai virus (murine PIV1) and bovine parainfluenza virus type 3 (BPIV3) [2]. In some instances when animals are subjected to high stressful conditions, infection with BPIV3 can contribute to tissue damage and immunosuppression, resulting in severe bronchopneumonia from secondary bacterial infections [4]. The resulting disease is part of the bovine respiratory disease complex (BRDC) and is considered as the most significant illness associated with feedlot cattle in the USA [5], and possibly worldwide. Other respiratory viruses such as bovine herpesvirus 1, bovine viral diarrhea virus (BVDV) and bovine respiratory syncytial virus (BRSV) have also been associated with BRDC development in feedlot cattle [6]. The clinical presentation of bovine infections with BPIV3 can vary considerably, ranging from asymptomatic infections to severe respiratory illness. In most cases where BPIV3 is implicated in disease, usual clinical signs include coughing, fever and nasal discharge [6].
Three genotypes, A (BPIV3a), B (BPIV3b) and C (BPIV3c) have been described, based on genetic and phylogenetic analysis [7]. The BPIV3b genotype could hypothetically be a lineage from a strain that recently crossed from another host species into cattle [8]. PIV3 infections were found in a wide variety of mammals including cattle, humans, sheep [9], goats [10], bison [11], guinea pigs [12], black and white rhinoceros [13], moose [14], bighorn sheep [15] and camels [16]. Cross-species infections have been reported in numerous instances, including HPIV-3 in guinea pigs [12], BPIV3 in a human, BPIV3 in sheep, and ovine PIV3 in cattle [17]. In Argentina, serological studies conducted in 1980 and 1984 showed a high incidence of antibodies against BPIV3 (77%) in cattle from the main livestock breeding regions [18,19]. In addition, positive serology was reported in domestic and wild South American camelids [20-22] and the virus was isolated from cattle and sheep [23]. However, little is known about the circulation of BPIV3 in cattle and other ruminants in Argentina, as well as about the genotypes that are present in this Southern American country.
Production systems have evolved to mixed managements, where alternative production species, such as the water buffalo (Bubalus bubalis), coexist in the same habitat as with cattle. Water buffalo breeding represents an important economic alternative in Argentina, which allows access to national and international markets. This species is susceptible to several viral infections [20,24,25] including BPIV3 as reported in a very early in 1966 in Egypt [26]. In addition to its potential relevance with respect to water buffalo health, this finding bears epidemiological significance due to the risk of transmission of the virus to cattle. The aim of this study was to characterize antigenically and genetically BPIV3 isolates from a respiratory and reproductive syndrome outbreak in dairy buffaloes and compare the buffalo isolate with contemporaneous BPIV3 circulating in cattle.
Results
Morphological and biological properties of buffalo viral isolates
PIV3 was successfully isolated from two out of twelve water buffaloes studied, in one case from both nasal (7 N) and vaginal (7 V) swabs, and in the other only from the vaginal swab (2 V).
Isolates produced characteristic cytopathic effects (CPE) on MDBK cells, similar to those of the BPIV3 reference strain, with many scattered, rounded, refractory cells and small syncytia. CPE were first visualized on the second passage, after three days post inoculation (DPI), and CPE were observed at day 4 or 5 DPI. Both bovine polyclonal anti-BPIV3 serum and FITC-anti-BPIV3 IgG reacted with the cells infected by these isolates (Figure 1 A).
Figure 1.
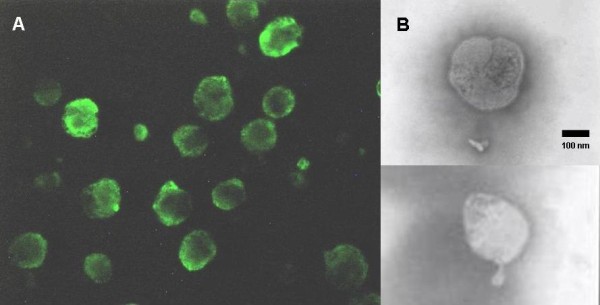
Microscopic examination of PIV3 isolated from buffaloes. A: Positive MDBK cells by IF, as observed by epifluorescence in an Olympus BX 40 + H hal microscope (400X magnification). B: Electron micrograph of two purified virions of the 7 N isolate, morphologically indistinguishable from paramixoviruses.
Transmission electron microscopy revealed spherical to pleomorphic virions, approximately 100–300 nm in diameter, which were morphologically indistinguishable from paramyxoviruses. Intact virions were enveloped and densely packed, as has been described for BPIV3. Nucleocapsids were visible and exhibited a typical “herringbone” pattern (Figure 1 B).
The three isolates (7 N, 7 V and 2 V) were able to agglutinate red blood cells (RBCs), with UHA titers of 16, 8 and 4 respectively. The agglutinated RBCs eluted after 1 h suggesting neuraminidase function/activity. The HI test showed 58% of positive reactions, with titers ranging between 1.6 and 2.5 (data not shown).
Molecular characterization RT-PCR and sequencing
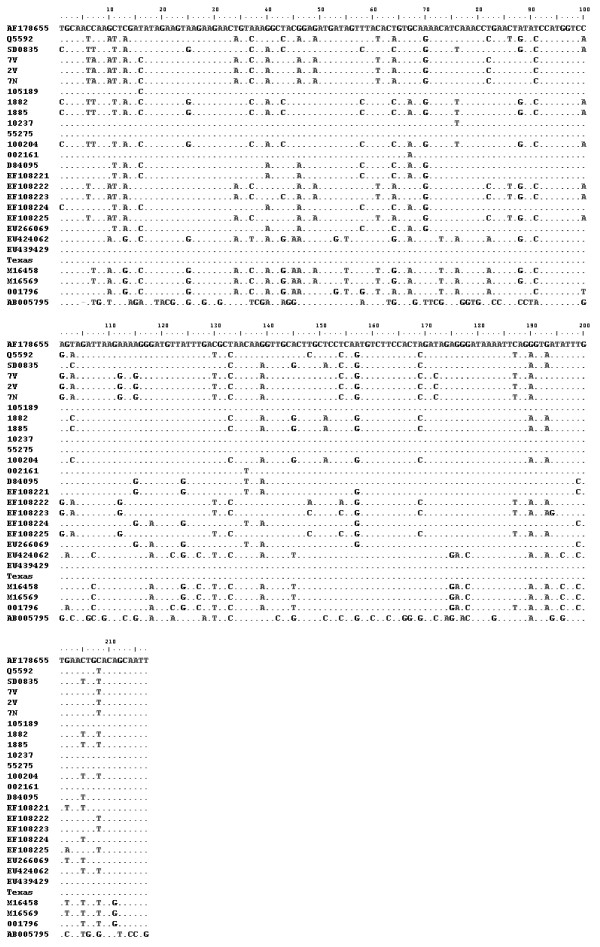
A fragment of the M gene, consistent with the expected size of 328 bp was amplified from the 3 virus isolates (7 N, 7 V and 2 V) by RT-PCR. As expected, the primer pairs did not amplify using genomic material of BRSV as template. Analysis of the alignments of 271 pb of the corresponding sequences revealed that they are related to BPIV3 strain Q5592 from Australia with 94% nucleotide identity (Figure 2) and 100% predicted amino acid identity (Table 1).
Figure 2.
Nucleotide alignment of M fragment sequences of Argentine buffalo and bovine isolates, other bovine parainfluenza virus 3 and human parainfluenza virus 3 that circulate in the world. Dots indicate identity with respect to SF strain (AF178655).
Table 1.
Comparison of identity percentages in amino acid and nucleotide sequences of the 217-bp region of the M gene among buffalo and bovine isolates and reference strains of BPIV3
| BPIV3a ref strain | BPIV3b ref strain | BPIV3c ref strain |
Buffalo isolates |
Bovine isolates |
Sendai virus | Amino acid identity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BPIV3b | BPIV3c | BPIV3a | ||||||||||||
| |
AF178655 |
Q5592 |
SD0835 |
7 V |
2 V |
7 N |
1882 |
1885 |
100204 |
105189 |
55275 |
10237 |
AB005795 |
|
|
AF178655 |
ID |
97 |
99 |
97 |
97 |
97 |
99 |
99 |
99 |
100 |
100 |
100 |
65 |
|
| Q5592 |
87 |
ID |
99 |
100 |
100 |
100 |
99 |
99 |
99 |
97 |
97 |
97 |
65 |
|
| SD0835 |
87 |
89 |
ID |
99 |
99 |
99 |
100 |
100 |
100 |
99 |
99 |
99 |
64 |
|
| 7 V |
86 |
94 |
88 |
ID |
100 |
100 |
99 |
99 |
99 |
97 |
97 |
97 |
65 |
|
| 2 V |
86 |
94 |
88 |
100 |
ID |
100 |
99 |
99 |
99 |
97 |
97 |
97 |
65 |
|
| 7 N |
86 |
94 |
88 |
100 |
100 |
ID |
99 |
99 |
99 |
97 |
97 |
97 |
65 |
|
| 1882 |
87 |
88 |
99 |
87 |
87 |
87 |
ID |
100 |
100 |
99 |
99 |
99 |
64 |
|
| 1885 |
87 |
88 |
99 |
87 |
87 |
87 |
100 |
ID |
100 |
99 |
99 |
99 |
64 |
|
| 100204 |
87 |
88 |
99 |
87 |
87 |
87 |
100 |
100 |
ID |
99 |
99 |
99 |
64 |
|
| 105189 |
100 |
86 |
87 |
87 |
87 |
87 |
87 |
87 |
87 |
ID |
100 |
100 |
65 |
|
| 55275 |
100 |
87 |
87 |
86 |
86 |
86 |
87 |
87 |
87 |
100 |
ID |
100 |
65 |
|
| 10237 |
100 |
86 |
88 |
86 |
86 |
86 |
87 |
87 |
87 |
99 |
100 |
ID |
65 |
|
|
AB005795 |
64 |
61 |
65 |
63 |
63 |
63 |
64 |
64 |
64 |
64 |
64 |
63 |
ID |
|
| Nucleotide identity | ||||||||||||||
ID: identical; ref: reference.
Data and phylogenetic analysis
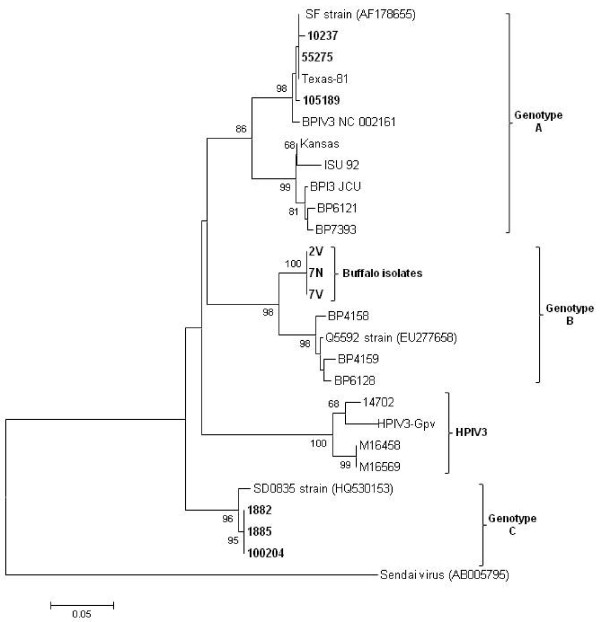
Phylogenetic analysis of the M gene fragment of the three Argentinean buffalo isolates (7 N, 7 V and 2 V) together with other paramyxoviruses, placed the buffalo isolates in the same clade as BPIV3 (Figure 3).
Figure 3.
Phylogenetic analysis based on M gene fragment. Phylogenetic analysis was performed using MEGA 4 software with 1,000 bootstrap replicates. Phylogenetic analysis based on M gene fragment nucleotide sequences compared to other Respirovirus genus members. The Argentinean isolates are indicated in bold. GenBank accession numbers are presented in Materials and Methods.
In particular, the isolates grouped with the BPIV3b genotype represented in the tree by an Australian isolate (Q5592). BPIV3b is distinct from the previously characterized members of BPIV3 represented by genotypes A and C.
In order to determine whether the PIV3 genotype found in buffalo is also circulating in cattle, the same gene fragment was amplified by RT-PCR using as template BPIV3 RNA isolated from six bovines of the Buenos Aires region. Three of the six isolates show 98% nucleotide identity and 100% amino acid identity with BPIV3c (SD0835 strain) reference isolate, and the remaining three, 99% nucleotide identity and 100% amino acid identity with BPIV3a (SF strain) reference isolate (Table 1). Phylogenetic characterization showed that the bovine isolates belong to the BPIV3a and BPIV3c genotypes, which have been reported so far in North America, Europe and China (Figure 3).
Serum neutralization test
Buffalo sera (n = 24) were analyzed by cross-neutralization, Three were negative for both viruses, BPIV3a and b, 9 showed higher titers against BPIV3b isolated from buffalo, 4 showed higher titers against BPIV3a and the remaining 11 had same titers for both viruses. (Table 2). In general, sera from buffaloes showed higher antibody titers against the isolated buffalo virus than against the reference virus.
Table 2.
Comparison of neutralizing titers against BPIV3b isolated from buffaloes and against a BPIV3a reference strain
| 24 buffaloes |
*Antibody titers against BPIV3b isolate from buffalo |
Subtotal | |||||
|---|---|---|---|---|---|---|---|
| Negative | 0.3 | 0.6 | 0.9 | 1.2 | |||
| *Antibody titers against BPIV3a SF strain |
Negative |
3 |
1 |
2 |
1 |
|
7 |
| 0.3 |
|
|
|
1 |
|
1 |
|
| 0.6 |
|
1 |
2 |
1 |
2 |
6 |
|
| 0.9 |
|
|
3 |
4 |
1 |
8 |
|
| 1.2 |
|
|
|
|
2 |
2 |
|
| Subtotal | 3 | 2 | 7 | 7 | 5 | 24 | |
* Neutralization titers were calculated as the decimal logarithm of the reciprocal of the last dilution with clear cytopathic effects.
Each cell contains the number of animals with the antibody titers against each virus.
Discussion
In the present study, we describe the isolation of BPIV3 from two water buffaloes with respiratory and reproductive signs. The virus was isolated from nasal and vaginal swabs from two out of twelve animals with reproductive and respiratory signs studied. To our knowledge little is known about the pathogenesis of this virus in buffalo. Should there be a syndrome similar to that present in cattle where the virus is transiently excreted [27], the unsuccessful virus isolation from the then remaining symptomatic animals could be related to the time point of sampling, which could have taken place at the end of the acute infection.
The IF assay showed strong and specific binding of anti-BPIV3 antibodies to cells infected with these three positive clinical isolates. Sera from buffaloes were able to neutralize the activity of the 7 N, 7 V and 2 V isolates and that of the reference bovine virus (BPIV3 strain SF), but the neutralization titers against the latter were lower than against buffalo isolates, suggesting antigenic differences.
The sequence of the M amplified fragment showed 94% nucleotide identity between buffalo isolates and BPIV3b strain Q5592 from Australia [8]. This is the first report of molecular characterization of a BPIV3 isolated from buffaloes. In order to determine homology between PIV3 isolates from Argentinean bovine and buffaloes, we characterized retrospectively six isolates collected from bovine respiratory outbreaks that occurred in 2009 and 2011. These isolates showed high percentages of nucleotide and amino acid identity with A and C genotypes (Table 1). There are only few reports about BPIV3 circulation in Argentina, Epstein et al described the isolation, virological and physicochemical characterization of PIV3 from Argentinean cattle and sheep in 1974 [23]. However, phylogenetic studies of these isolates were not carried out. Phylogenetic reconstructions based on the alignment of the M-gene nucleotide sequences demonstrated that the 7 N, 7 V and 2 V isolates clustered in the B genotype group. So far, this genotype has only been previously reported in Australia [8]. The phylogenetic position of the Argentinean buffalo isolates might be correlated with the antigenic differences observed by cross-neutralization studies, where the reference strain belonging to the BPIV3a group was lesser neutralized than the buffalo viral isolates by sera of buffaloes. Therefore the serological and molecular characterization of buffalo BPVI3 isolates show that these viruses are more similar to B than to A (SF reference strain) and C BPVI3 genotypes (SD0835 reference strain). Interestingly, the genotype B isolated from buffaloes was not found among the bovine samples, analyzed in this work, that clustered with genotypes A and C. Genotype B was until now observed only in Australia with the hypothesis of a recent crossing from another species into cattle [8]. This evidence of genotype B in buffaloes should need further investigation to know if this species could be one of the host species of this genotype. Therefore analysis of a larger number of isolates from cattle and buffaloes is needed to determine whether the three genotypes circulate in both species, or if the distribution observed is representative. Importantly, this is the first report of bovine B and C genotype circulation out of Australia and China, respectively. The circulation of genotype C only in China was attributed to a geographic restriction [7]. This hypothesis can be ruled out by our results and this observation was most likely due to the relative frequencies of these genotypes with respect to the samples analyzed.
To our knowledge this is the first report of the isolation of a parainfluenza virus type 3 from vaginal secretions. However, it is important to highlight that the presence of the virus in the secretions could be due to oro-genital contact between animals. Evidence of viral replication in the genital mucosa needs new investigations.
Conclusion
The isolation and characterization of BPIV3 from water buffaloes were reported. We identified the isolates as bovine parainfluenza virus type 3 genotype B that differs from genotypes A and C circulating in Argentinean cattle herds reported so far. Further studies would be required to determine if A and C genotypes also circulate in buffalo herds, as well as to elucidate the role of buffalo as reservoirs, especially of the B genotype, and in the transmission of this virus to cattle.
Methods
Samples
Nasal and vaginal swabs were obtained from 12 water buffaloes of a dairy farm in the Argentinean province of Chaco, during an outbreak of respiratory and reproductive disease in July 2009. These animals had white stringy mucus in the nose and vulva. These samples were inoculated on Madin Darby bovine kidney (MDBK) cell cultures. The SF strain of BPIV3 was used as reference. Cells were daily examined under optical microscope to observe the appearance of cytopathic effects.
Bovine isolates were obtained from cattle with respiratory syndromes, 5 of them in the year 2009 and the remaining isolate in 2011. They were collected and amplified from Azul laboratories (Azul-Buenos Aires province) from outbreaks of Buenos Aires province.
Immunofluorescence test
Virus isolates were inoculated into confluent MDBK cells in 8-well chambers at a MOI of 1.0, 0.1, and 0.01. For direct IF test, Cells with cytopathic effects were fixed with 99.5% acetone and then incubated with FITC anti-BPVI3 IgG (Ames). For indirect IF, cells were incubated with antibovine PIV3 polyclonal antibody (NVSL) diluted 1:100 in PBS at 37 °C in a moist environment for 1 h. Then, cells were incubated with FITC antibovine IgG (KPL). In both cases, slides were layered with buffered glycerin and observed under epifluorescence in an Olympus BX 40 + H hal microscope.
Transmission electron microscopy
Twenty-five μl of viral suspension (16 HAU) were placed on a collodion membrane supported on a copper grid, negatively stained with phosphotungstic acid, and observed with a JEOL 1200 EX II transmission electron microscope [28].
Hemagglutination assay (HA) and Haemagglutination inhibition assay (HIA)
Serial two-fold dilutions of culture supernatants from infected MDBK (50 ul) were titrated against 50 ul Guinea pig erythrocytes in V-bottom microtiter plates (Nunc). After incubation at 4°C for 4 h, hemagglutination was evaluated by the appearance or absence of a red cell button. Results were expressed as hemagglutinating units/50 μl (HAU/50 μl). The last dilution at which hemagglutination was observed was taken as the endpoint of the hemagglutinating activity. The reciprocal of this dilution expresses the number of hemagglutinating units or virus titer in UHA.
For the HI assay serial two-fold dilutions of sera from buffaloes were allowed to react with a fixed dose of reference viral haemagglutinin (8 UHA), followed by the addition of guinea pig agglutinable erythrocytes. Titers were expressed as the log10 of the reciprocal of the highest dilution hemagglutination inhibitory multiplied by a constant factor of 8.
RT-PCR and sequencing
Infected MDBK cells were scraped off the plates and homogenized by three cycles of freezing and thawing. After an initial centrifugation at 3,000 g for 15 min, polyethylene glycol 8000 (Sigma), 10% (w/v, final concentration) was added to the cell lysates, followed by incubation for 4 h at 4°C. The virus was pelleted at 12,000 g for 60 min at 4°C. Viral genomic RNA was extracted from the virus pellet using RNeasy Mini Kit (QIAgen). Oligonucleotide primers for BPIV3 detection and identification (Mfwd: 5´AGTGATCTAGATGATGATCCA 3´ nt - 3960 and Mrev: 5´GTTATTGATCCAATTGCTGT −3´ nt - 4288) were designed based on a 328 bp segment of the consensus BPIV3 Matrix (M) gene. cDNA was synthesized using the specific oligonucleotide primer (Mfw) and MMLV reverse transcriptase (Promega). Subsequently, Go Taq DNA Polymerase (Promega) was used to amplify the M gene fragment.
Mfw and Mrev primers were tested for specificity by RT-PCR on BRSV (bovine respiratory syncytial virus) RNA, giving negative results. PCR products were purified using a Qiaquick PCR purification kit (Qiagen). Sequencing reactions were performed twice in both directions with BigDye Terminator v3.0 kit (Applied Biosystems, Lennik, Belgium) and analyzed with ABI Prism 3730 DNA Analyzer (Applied Biosystems).
Phylogenetic analysis
M fragment nucleotide and predicted amino acid sequences of buffalo isolates were edited and analyzed with BioEdit version 7.0.5.3 [29]. Alignments of these sequences together with other from representative isolates of previously identified BPIV3 and human parainfluenza virus type 3 (HPIV3) were created by Clustal W. Nucleotide sequence alignments of the M gene fragment were used for phylogenetic analyses using MEGA software, version 4 [30]. Phylogenetic relationships were investigated by bootstrap analysis (1000 replicates) using the Neighbor joining method with the Kimura two-parameter model for nucleotide data analysis. GenBank accession numbers of the sequences used in the phylogenetic tree are as follows: BPIV3 reference strains: BPIV3 strain Kansas/15626/84, D84095; BPIV3 strain, NC 002161, AF178654; SF strain AF178655; Q5592, EU277658; SD0835, HQ530153; BPI3 JCU strain, EF 108221; BP4158 stain EF 108222; BP4159 strain, EF 108223; BP6121 strain, EF 108224; BP6128 strain, EF 108225; BP7393, EU266069; ISU92, EU439428; Texas-81, EU439429 and HPIV3 reference strains: HPIV strains, M16458, M16569; HPIV3-Gpv strain, NC 001796 and 14702 strain, EU424062. Sendai virus (AB005795) as outgroup.
Serum neutralization test
Cross-neutralization tests were performed in MDBK 96-well plates, by overnight incubation of 200 TCID50 on 50 ul of 7 N buffalo virus isolates or BPIV3 reference strain (SF) with 50 ul heat-inactivated two-fold diluted sera. The plate was incubated for 5 days at 37 °C, under 5% CO2, and then screened for the presence or absence of cytopathic effects under optical microscope (400x magnification) to determine the virus-neutralization titers. Neutralization titers were calculated as the decimal logarithm of the reciprocal of the last serum dilution with clear cytopathic effects.
Authors’ contributions
SM and SR designed the experiments, analyzed the data and drafted the manuscript together. SM performed the experiments. PL, GC, JK, and GC gently surrendered the field isolates. DR and VP kindly provided the reference strain. AM and ET participated in the molecular genetic studies, interpretation of data and contributed to the manuscript. MC helped to draft the manuscript. JD helped with electron microscopy. OZ helped with cell cultures. All authors read and approved the final manuscript.
Contributor Information
Silvina S Maidana, Email: smaidana@cnia.inta.gov.ar.
Patricia M Lomonaco, Email: mlomonaco@cnia.inta.gov.ar.
Gustavo Combessies, Email: mcraig@cnia.inta.gov.ar.
María I Craig, Email: mcraig@cnia.inta.gov.ar.
Julian Diodati, Email: jdiodati@cnia.inta.gov.ar.
Daniela Rodriguez, Email: drodriguez@cnia.inta.gov.ar.
Viviana Parreño, Email: vpareno@cnia.inta.gov.ar.
Osvaldo Zabal, Email: ozabal@cnia.inta.gov.ar.
José L Konrad, Email: konradjl@hotmail.com.
Gustavo Crudelli, Email: gacrudelli@hotmail.com.
Axel Mauroy, Email: amauroy@ulg.ac.be.
Etienne Thiry, Email: etienne.thiry@ulg.ac.be.
Sonia A Romera, Email: aromera@cnia.inta.gov.ar.
Acknowledgements
The authors wish to thank Mónica Florin-Christensen (INTA) for helping to draft the manuscript and Domnique Ziant for their excellent technical assistance (University of Liege, Belgium). This work was supported by Fonds de la Recherche Scientifique (FRS-FNRS) and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET). SM is a CONICET Fellow (Argentina).
References
- Wang LF, Collins PL, Fouchier RAM, Kurath G, Lamb RA, Randall RE, Rima BK. In: Virus taxonomy: Ninth report of the international committee on taxonomy of viruses. King AMQ, Lefkowitz E, Adams MJ, editor. Elsevier Academic Press, San Diego, California; 2011. family Paramyxoviridae; pp. 672–685. [Google Scholar]
- Turner RB, Couch RB. In: Fields Virology. 5th edition. Knipe DM, Howley PM, editor. Lippincott, Williams & Wilkins, Philadelphia; 2007. Rhinoviruses : Parainfluenza Viruses : the viruses and their replication; pp. 1449–1496. [Google Scholar]
- Pringle CR, Filipiuk AH, Robinson BS, Watt PJ, Higgins P, Tyrrell DA. Immunogenicity and pathogenicity of a triple temperature-sensitive modified respiratory syncytial virus in adult volunteers. Vaccine. 1993;11(4):473–478. doi: 10.1016/0264-410X(93)90290-E. [DOI] [PubMed] [Google Scholar]
- Haanes EJ, Guimond P, Wardley R. The bovine parainfluenza virus type-3 (BPIV-3) hemagglutinin/neuraminidase glycoprotein expressed in baculovirus protects calves against experimental BPIV-3 challenge. Vaccine. 1997;15(6–7):730–738. doi: 10.1016/s0264-410x(96)00231-9. [DOI] [PubMed] [Google Scholar]
- Snowder GD, Van Vleck LD, Cundiff LV, Bennett GL. Bovine respiratory disease in feedlot cattle: environmental, genetic, and economic factors. J Anim Sci. 2006;84(8):1999–2008. doi: 10.2527/jas.2006-046. [DOI] [PubMed] [Google Scholar]
- Bryson DG. In: Virus infections in Ruminants. Dinter Z, Morein B, editor. Elsevier publishers, Amsterdam; 1990. Parainfluenza-3 Virus in Cattle; pp. 319–333. [Google Scholar]
- Zhu YM, Shi HF, Gao YR, Xin JQ, Liu NH, Xiang WH, Ren XG, Feng JK, Zhao LP, Xue F. Isolation and genetic characterization of bovine parainfluenza virus type 3 from cattle in China. Vet Microbiol. 2010;149(3–4):446–451. doi: 10.1016/j.vetmic.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Horwood PF, Gravel JL, Mahony TJ. Identification of two distinct bovine parainfluenza virus type 3 genotypes. J Gen Virol. 2008;89(Pt 7):1643–1648. doi: 10.1099/vir.0.2008/000026-0. [DOI] [PubMed] [Google Scholar]
- Lyon M, Leroux C, Greenland T, Chastang J, Patet J, Mornex JF. Presence of a unique parainfluenza virus 3 strain identified by RT-PCR in visna-maedi virus infected sheep. Vet Microbiol. 1997;57(2–3):95–104. doi: 10.1016/s0378-1135(97)00104-1. [DOI] [PubMed] [Google Scholar]
- Yener Z, Saglam YS, Timurkaan N, Ilhan F. Immunohistochemical detection of parainfluenza type 3 virus antigens in paraffin sections of pneumonic caprine lungs. J Vet Med. 2005;52(6):268–271. doi: 10.1111/j.1439-0442.2005.00724.x. [DOI] [PubMed] [Google Scholar]
- Zarnke RL, Erickson GA. Serum antibody prevalence of parainfluenza 3 virus in a free-ranging bison (Bison bison) herd from Alaska. J Wildl Dis. 1990;26(3):416–419. doi: 10.7589/0090-3558-26.3.416. [DOI] [PubMed] [Google Scholar]
- Ohsawa K, Yamada A, Takeuchi K, Watanabe Y, Miyata H, Sato H. Genetic characterization of parainfluenza virus 3 derived from guinea pigs. J Vet Med Sci / the Japanese Soc Vet Sci. 1998;60(8):919–922. doi: 10.1292/jvms.60.919. [DOI] [PubMed] [Google Scholar]
- Fischer-Tenhagen C, Hamblin C, Quandt S, Frolich K. Serosurvey for selected infectious disease agents in free-ranging black and white rhinoceros in Africa. J Wildl Dis. 2000;36(2):316–323. doi: 10.7589/0090-3558-36.2.316. [DOI] [PubMed] [Google Scholar]
- Thorsen J, Henderson JP. Survey for antibody to infectious bovine rhinotracheitis (IBR), bovine virus diarrhea (BVD) and parainfluenza 3 (PI3) in moose sera. J Wildl Dis. 1971;7(2):93–95. doi: 10.7589/0090-3558-7.2.93. [DOI] [PubMed] [Google Scholar]
- Parks JB, Post G, Thorne T, Nash P. Parainfluenza-3 virus infection in Rocky Mountain bighorn sheep. J Am Vet Med Assoc. 1972;161(6):669–672. [PubMed] [Google Scholar]
- Eisa M, Karrar AE, Abdel Rahim AH. The occurrence of antibodies to parainfluenza3 virus in sera of some domestic animals of the Sudan. Br Vet J. 1979;135(2):192–197. doi: 10.1016/s0007-1935(17)32941-x. [DOI] [PubMed] [Google Scholar]
- Stevenson RG, Hore DE. Comparative pathology of lambs and calves infected with parainfluenza virus type 3. J Comp Pathol. 1970;80(4):613–618. doi: 10.1016/0021-9975(70)90060-5. [DOI] [PubMed] [Google Scholar]
- Lager L, Sadir A, Schudel A. Enfermedades respiratorias virales de bovinos. J Información y Desarrollo en Investigación Agropecuaria. 1983;1(2):55–58. [Google Scholar]
- Ruiz ML, Miquet J, Vena MM, Borca MV, Sadir AM, Estevez Mader J, León E, Marcovecchino FE, Schudel AA Parraud JR y. Enfermedades respiratorias de los bovinos: Neumonía de los terneros. Revista Veterinaria Argentina. 1984;10(1):948–954. [Google Scholar]
- Marcoppido G, Parreno V, Vila B. Antibodies to pathogenic livestock viruses in a wild vicuna (Vicugna vicugna) population in the Argentinean Andean altiplano. J Wildl Dis. 2010;46(2):608–614. doi: 10.7589/0090-3558-46.2.608. [DOI] [PubMed] [Google Scholar]
- Craig MI, Venzano A, Konig G, Morris WE, Jimenez L, Julia S, Capellino F, Blanco Viera J, Weber EL. Detection of bovine viral diarrhoea virus (BVDV) nucleic acid and antigen in different organs of water buffaloes (Bubalus bubalis) Res Vet Sci. 2008;85(1):194–196. doi: 10.1016/j.rvsc.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Puntel M. Seroprevalencia de infecciones virales en llamas (Lama Galma) en la república Argentina. Revista Argentina de microbiología veterinaria. 1997;29:38–46. [PubMed] [Google Scholar]
- Epstein B. Parainfluenza tipo III en bovinos y ovinos de Argentina. Patología. Aislamiento y caracterización. Revista Medicina Veterinaria. 1974;55(5):397–406. [Google Scholar]
- De Carlo E, Re GN, Letteriello R, Del Vecchio V, Giordanelli MP, Magnino S, Fabbi M, Bazzocchi C, Bandi C, Galiero G. Molecular characterisation of a field strain of bubaline herpesvirus isolated from buffaloes (Bubalus bubalis) after pharmacological reactivation. Vet Rec. 2004;154(6):171–174. doi: 10.1136/vr.154.6.171. [DOI] [PubMed] [Google Scholar]
- Martucciello A, De Mia GM, Giammarioli M, De Donato I, Iovane G, Galiero G. Detection of Bovine viral diarrhea virus from three water buffalo fetuses (Bubalus bubalis) in southern Italy. J Vet Diagn Invest. 2009;21(1):137–140. doi: 10.1177/104063870902100123. [DOI] [PubMed] [Google Scholar]
- Singh KV, Baz TI. Isolation of parainfluenza-3 virus from water buffaloes in Egypt. Nature. 1966;210(5036):656–657. doi: 10.1038/210656a0. [DOI] [PubMed] [Google Scholar]
- Clinical virology of ruminants. Collection Clinical Virology, France; 2007. Viral diseases of the respiratory tract: Bovine parainfluenza type 3 virus infection; pp. 32–35. [Google Scholar]
- Qiao D, Janke BH, Elankumaran S. Molecular characterization of glycoprotein genes and phylogenetic analysis of two swine paramyxoviruses isolated from United States. Virus genes. 2009;39(1):53–65. doi: 10.1007/s11262-009-0353-2. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biology Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]