Before 1982, the Centers for Disease Control and Prevention (CDC) had identified only one Escherichia coli isolate of the O157:H7 serotype; it had been isolated from a patient with bloody diarrhea (1). Then, in 1982, two outbreaks of severe bloody diarrhea occurred among people who had eaten hamburgers at a fast food chain. The CDC isolated E. coli O157:H7 from people who had become ill as well as from a hamburger patty (1). In 1983, Karmali et al. discovered an association between infection with E. coli that produce Shiga toxin (then called Vero toxin), including O157:H7 strains, and another severe and sometimes fatal condition, the hemolytic uremic syndrome (HUS) (2). Last year, the CDC estimated that strains of E. coli O157:H7 cause approximately 73,000 illnesses and 60 deaths per year in the United States, and non-O157:H7 Shiga toxin-producing E. coli (STEC) add an additional 37,000 estimated cases (3).
Human infection with E. coli O157:H7 has been associated with a variety of contaminated foods, water, and person-to-person transmission (4–6). Cattle are considered the primary reservoir of E. coli O157:H7 that infect humans (Fig. 1). Adult cattle and weaned calves that carry E. coli O157:H7 generally remain asymptomatic but shed the bacteria into the environment in their feces (7, 8). Many of the foods implicated in human disease are of bovine origin, and epidemiological studies have associated the contamination of crops and water with the use of manure as fertilizer or with the close proximity of the vegetable fields or water supplies to cattle (Fig. 1). In this issue of PNAS, Elder et al. report the results of their investigation of the prevalence of E. coli O157 contamination in beef cattle and carcasses at four different meat processing plants (9). Their data reveal that the prevalence of E. coli O157:H7 in cattle and on carcasses is much higher than previously estimated, and the level of carcass contamination correlates with the level of E. coli O157:H7 in the cattle before processing. They also found evidence that current processing practices are reducing contamination levels. However, their study overall underscores the need to reduce the level of E. coli O157:H7 in cattle on the farm.
Figure 1.
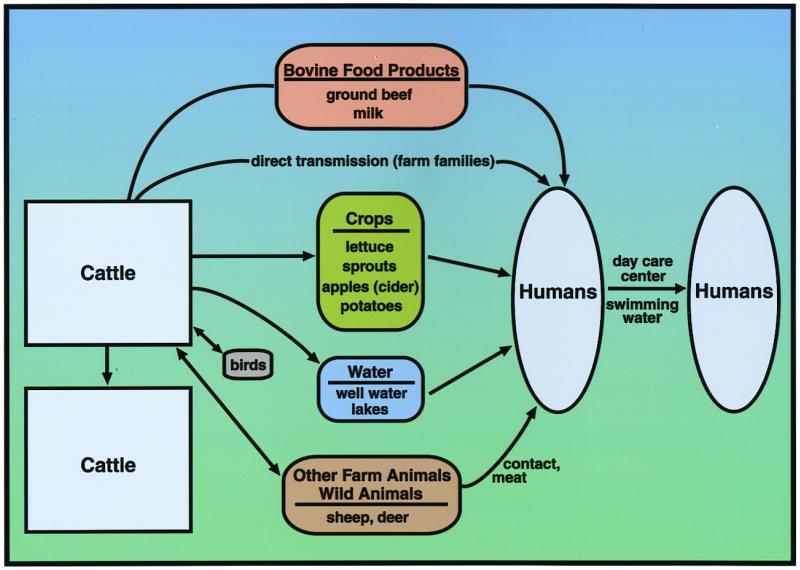
Model for transmission of E. coli O157:H7 from cattle to humans. The figure represents data from numerous studies and depicts examples of the major classes of foods and other sources of E. coli O157:H7 infection that have been reported. The contamination of crops and water sources is associated with the use of manure in fertilizer or with potential fecal contamination from nearby cattle. The sources of human infection with E. coli O157:H7 were identified first by epidemiological methods. In some cases, E. coli O157:H7 was isolated from the suspected food or other source; in many of these cases, including outbreaks associated with ground meat, PFGE or phage typing provided additional confirmation that the bacteria isolated from the patient and the suspected food or other source were the same strain. PFGE typing has also been useful in linking geographically separated outbreaks to a common source of contaminated meat. The finding of E. coli O157:H7 in birds, deer, and other animals has led to speculation that these organisms may also be vehicles for O157:H7 transmission. For reviews, see refs. 4–6, 8, and 11.
E. coli O157:H7 possess a potent combination of virulence factors that undoubtedly contributes to its low infectious dose; additionally, these bacteria survive well under adverse conditions, such as low pH (4, 5, 10). The capacity of E. coli O157:H7 to cause bloody diarrhea and HUS derives from the activity of the Shiga toxins (reviewed in refs. 4 and 6). There are two major types of Shiga toxins expressed by E. coli associated with human disease, Stx1 and Stx2. These toxins are structurally similar, and both are cytotoxins that block eukaryotic translation. Other virulence factors include genes in the locus of enterocyte effacement (LEE); these factors allow the bacteria to attach tightly to mammalian epithelial cells, disrupt the cytoskeletal structure and signaling pathways, and efface the intestinal brush border to form the characteristic attaching and effacing lesion (reviewed in ref. 10). Among the genes on the LEE locus is eae (originally called eaeA), which encodes intimin, an outer membrane protein required for E. coli O157:H7 to adhere to mammalian cells (10).
In addition to Shiga toxins, eae, and the O157 serogroup of the O-antigen (the extracellular branched polysaccharide that is part of the lipopolysaccharide coat), another hallmark of this pathogen is the presence of a large plasmid. Probes for the hemolysin gene encoded on this plasmid have been useful for identifying O157:H7 strains (11). Most E. coli O157:H7 lack the capacity to ferment sorbitol; this trait has also been useful to microbiologists because it distinguishes this pathogen from the majority of other E. coli strains (4, 11). Some O157 strains do not express functional flagella (the H antigen); these bacteria are designated O157:H− or O157:NM (nonmotile).
A large number of O157:H7 strains have been associated with human disease. Nearly all carry one or more stx genes, eae, and the large plasmid, but they are distinguishable by pulse-field gel electrophoresis (PFGE) patterns after restriction enzyme digestion and by plaque phenotypes of a specific set of phage isolated for typing purposes (11). PFGE and phage typing have been valuable epidemiological tools (see Fig. 1) (5, 11).
In earlier studies of the prevalence of E. coli O157:H7 in dairy and beef cattle, researchers generally estimated that fewer than 10% of cattle carry this pathogen; many estimates were lower than 2% (12). More sensitive culture techniques have been developed to detect these bacteria, and several studies have reported increased prevalence in cattle during warmer months of the year (11–15). These data on cattle carriage of O157:H7 correlate with the seasonal variation in the incidence of human disease (4). Thus, estimates of the prevalence of O157:H7 in cattle that average isolation rates over warm and cold seasons or that only include samplings from cooler months dilute the impact of high shedding in the warmer months. For example, during a year-long study in England in 1997, E. coli O157:H7 were isolated from the feces from as many as 38% of cattle presented for slaughter in the spring, but only 4.8% during the winter (15). Similar high prevalence rates and seasonal variation were obtained in recent studies in Canada and the Netherlands (13, 14).
To determine the prevalence of E. coli O157:H7 in beef cattle and to assess whether levels of E. coli O157:H7 in the cattle presented for slaughter affect the level of carcass contamination during processing, Elder and colleagues assayed cattle and carcasses from four meat processing plants in the United States (9). They visited each plant twice during July and August, followed three or four lots of cattle per plant at each visit, and sampled at least 20% of each lot. Samples were taken from hide and feces before processing, and from carcasses at three points during processing: preevisceration, postevisceration, and after anti-microbial intervention steps. Fecal samples from 327 cattle and hide swabs from 357 cattle were taken, and each of approximately 330 carcasses was sampled at different stages of the processing procedure.
The enormous number of commensal microorganisms in the bovine intestinal tract (16) and the absence of observable disease caused by E. coli O157:H7 in cattle present a challenge for detection of this pathogen. The culture methods used by Elder et al. incorporate several strategies that were found to be effective in previous studies. These include growth of the samples in broth followed by the use of immunomagnetic beads coated with antibodies to the O157 antigen, rather than direct plating of samples, and the addition of cefixime and tellurite to the sorbitol-MacConkey agar (SMACct) used to screen for sorbitol nonfermenters (11, 13–15, 17). The broth chosen by Elder et al. for the initial growth of the fecal samples was different from the broth used for the hide and carcass samples. Although they do not specify their rationale for this difference in the paper, perhaps these media were chosen for selectivity against different background flora that might colonize these sites.
After broth enrichment and immunomagnetic separation, the organisms were screened on SMACct. Potential O157 colonies were assayed with commercial diagnostic products and were confirmed to be either O157:H7 or O157:NM by monoclonal antibody assays and by direct examination of motility. Elder et al. note as unpublished data that their fecal culture method was more sensitive than two other diagnostic veterinary procedures when tested on the same samples, although they did not specify the steps of these other methods.
Among the O157:H7 or O157:NM strains isolated from the cattle and carcasses, all were positive by PCR for eaeA, the hemolysin gene, and rfbO157 [a gene associated with production of the O157 O-antigen (18)]. Additionally, most isolates carried stx2 or both stx2 and stx1; only 1.4% carried stx1 alone, and only one isolate was stx−. Strains that produce Stx2 or both toxins are more commonly associated with human illness than strains that produce only Stx1 (11). Thus, the strains isolated from this random sampling of cattle displayed characteristics similar to strains associated with human disease.
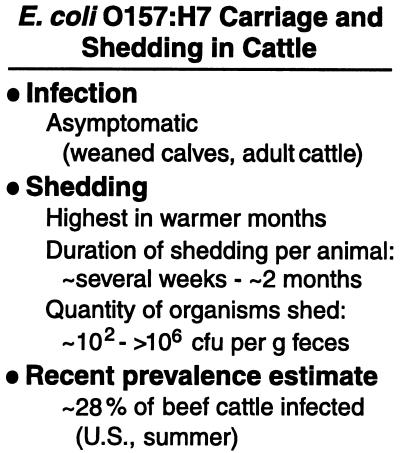
All but two of the 30 lots tested by Elder and colleagues contained cattle or carcasses that were positive for E. coli O157:H7; there was no clustering of fecal and hide or carcass prevalence by processing plant. Among individual cattle, the prevalence of E. coli O157:H7 in fecal samples was 28% overall. This number is similar to the recent high estimates obtained in other countries and much higher than earlier estimates in the U.S. This result reinforces the concept that intervention strategies on the farm are warranted to prevent contamination of food and water supplies (Fig. 2). The level of contamination on hides was relatively low; Elder et al. noted that this was surprising.
Figure 2.
Data for infection and shedding are reviewed in refs. 5 and 8; the prevalence value (from fecal samples) is reported by Elder et al. (9) in this issue of PNAS.
When they followed the animals through processing, they found that 43% of the carcasses sampled preevisceration contained E. coli O157:H7 contamination. They note that this value is much higher than previous estimates of carcass contamination in the U.S. (19). Among the postprocessing samples, however, many fewer (1.8%) were positive. This suggests that current antimicrobial intervention strategies in the plants are working.
The total preharvest (fecal and hide) prevalence correlated with the prevalence of contamination of carcasses within a given lot. The correlation suggests that preharvest infection may influence contamination during processing. The prevalence levels detected in the fecal samples were also generally lower than those detected in the preevisceration carcass samples from the same lot. This result may simply indicate that their culture methods were more sensitive for detecting E. coli O157:H7 in carcass swabs than in fecal samples. However, Elder et al. noted that this finding may reflect some level of cross-contamination of the carcasses, and they suggest that preevisceration carcass contamination may be a critical control point for further intervention strategies. The Food Safety and Inspection Service of the U.S. Department of Agriculture proposed in 1996 that all meat (and poultry) establishments implement Hazard Analysis and Critical Control Points (HACCP), a system of controls to reduce the level of microbial pathogens in food products (20).
Reducing the amount of E. coli O157:H7 in live cattle will likely lower contamination not only of meat but also of other foods and water supplies that come into contact with bovine fecal matter. A variety of strategies have been proposed to reach this goal. These include modification of farm practices and bovine diet (12, 21), vaccination (22, 23), and administration of lytic phage or probiotic bacteria (24, 25).
As a final note, although O157:H7 E. coli are the most important Shiga-toxin producing E. coli with respect to human disease in the United States, other STEC strains are emerging as important pathogens in the U.S. and throughout the world. These include E. coli of non-O157 serotypes as well as O157 strains that can ferment sorbitol (3, 11). The increased prevalence of E. coli O157:H7 found by Elder et al. with improved detection methods raises the question of the actual prevalence of these other pathogens.
Acknowledgments
Work in this area in our laboratory is supported by grants from the National Institutes of Health (Grant AI20148) and the U.S. Department of Agriculture (Grant 97-35201-4578) to A.D.O.
Footnotes
See companion article on page 2999.
References
- 1.Riley L W, Remis R S, Helgerson S D, McGee H B, Wells J G, Davis B R, Hebert R J, Olcott E S, Johnson L M, Hargett N T, et al. N Eng J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 2.Karmali M A, Steele B T, Petric M, Lim C. Lancet. 1983;1:619–620. doi: 10.1016/s0140-6736(83)91795-6. [DOI] [PubMed] [Google Scholar]
- 3.Mead P S, Slutsker L, Dietz V, McCaig L F, Bresee J S, Shapiro C, Griffin P M, Tauxe R V. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin P M. In: Infections of the Gastrointestinal Tract. Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. New York: Raven; 1995. pp. 739–761. [Google Scholar]
- 5.Armstrong G L, Hollingsworth J, Morris J G., Jr Epidemiol Rev. 1996;18:29–51. doi: 10.1093/oxfordjournals.epirev.a017914. [DOI] [PubMed] [Google Scholar]
- 6.O'Brien A D, Kaper J B. In: Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains. Kaper J B, O'Brien A D, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 1–11. [Google Scholar]
- 7.Cray W C, Jr, Moon H W. Appl Environ Microbiol. 1995;61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wallace J S. In: Escherichia coli O157 in Farm Animals. Stewart C S, Flint H J, editors. Wallingford, U.K.: CABI Publishing; 1999. pp. 195–223. [Google Scholar]
- 9.Elder R O, Keen J E, Siragusa G R, Barkocy-Gallagher G A, Koohmaraie M, Laegreid W W. Proc Natl Acad Sci USA. 2000;97:2999–3003. doi: 10.1073/pnas.060024897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaper J B, Elliott S, Sperandio V, Perna N T, Mayhew G F, Blattner F R. In: Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains. Kaper J B, O'Brien A D, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 163–182. [Google Scholar]
- 11.Strockbine N A, Wells J G, Bopp C A, Barrett T J. In: Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains. Kaper J B, O'Brien A D, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 331–356. [Google Scholar]
- 12.Hancock D D, Besser T E, Rice D H. In: Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains. Kaper J B, O'Brien A D, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 85–91. [Google Scholar]
- 13.Van Donkersgoed J, Graham T, Gannon V. Can Vet J. 1999;40:332–338. [PMC free article] [PubMed] [Google Scholar]
- 14.Heuvelink A E, van den Biggelaar F L A M, Zwartkruis-Nahuis J T M, Herbes R G, Huyben R, Nagelkerke N, Melchers W J G, Monnens L A H, de Boer E. J Clin Microbiol. 1998;36:3480–3487. doi: 10.1128/jcm.36.12.3480-3487.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapman P A, Siddons C A, Cerdan Malo A T, Harkin M A. Epidemiol Infect. 1997;119:245–250. doi: 10.1017/s0950268897007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan S H, Scott K P, Flint H J, Stewart C S. In: Escherichia coli O157:H7 in Farm Animals. Stewart C S, Flint H J, editors. Wallingford, U.K.: CABI Publishing; 1999. pp. 71–89. [Google Scholar]
- 17.Sanderson M W, Gay J M, Hancock D D, Gay C C, Fox L K, Besser T E. J Clin Microbiol. 1995;33:2616–2619. doi: 10.1128/jcm.33.10.2616-2619.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bilge S S, Vary J C, Jr, Dowell S F, Tarr P I. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.U.S. Department of Agriculture Food Safety and Inspection Service. Nationwide Beef Microbiological Baseline Data Collection Program: Steers and Heifers, October 1992–September 1993. 1994. [Google Scholar]
- 20.61 Federal Register 144 (1996), p. 38806.
- 21.Diez-Gonzalez F, Callaway T R, Kizoulis M G, Russell J B. Science. 1998;281:1666–1668. doi: 10.1126/science.281.5383.1666. [DOI] [PubMed] [Google Scholar]
- 22.Gyles C L. In: Escherichia coli O157:H7 and Other Shiga Toxin-Producing E. coli Strains. Kaper J B, O'Brien A D, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 434–444. [Google Scholar]
- 23.Dean-Nystrom E A, Bosworth B T, Moon H W, O'Brien A D. Infect Immun. 1998;66:4560–4563. doi: 10.1128/iai.66.9.4560-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudva I T, Jelacic S, Tarr P I, Youderian P, Hovde C J. Appl Environ Microbiol. 1999;65:3767–3773. doi: 10.1128/aem.65.9.3767-3773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao T, Doyle M P, Harmon B G, Brown C A, Mueller P O E, Parks A H. J Clin Microbiol. 1998;36:641–647. doi: 10.1128/jcm.36.3.641-647.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]