Abstract
Iron overload has been associated with carcinogenesis in humans. Intraperitoneal administration of ferric nitrilotriacetate initiates a Fenton reaction in renal proximal tubules of rodents that ultimately leads to a high incidence of renal cell carcinoma (RCC) after repeated treatments. We performed high-resolution microarray comparative genomic hybridization to identify characteristics in the genomic profiles of this oxidative stress-induced rat RCCs. The results revealed extensive large-scale genomic alterations with a preference for deletions. Deletions and amplifications were numerous and sometimes fragmented, demonstrating that a Fenton reaction is a cause of such genomic alterations in vivo. Frequency plotting indicated that two of the most commonly altered loci corresponded to a Cdkn2a/2b deletion and a Met amplification. Tumor sizes were proportionally associated with Met expression and/or amplification, and clustering analysis confirmed our results. Furthermore, we developed a procedure to compare whole genomic patterns of the copy number alterations among different species based on chromosomal syntenic relationship. Patterns of the rat RCCs showed the strongest similarity to the human RCCs among five types of human cancers, followed by human malignant mesothelioma, an iron overload-associated cancer. Therefore, an iron-dependent Fenton chemical reaction causes large-scale genomic alterations during carcinogenesis, which may result in distinct genomic profiles. Based on the characteristics of extensive genome alterations in human cancer, our results suggest that this chemical reaction may play a major role during human carcinogenesis.
Introduction
Cancer is a disease of accumulated genomic alterations, presumably caused by a systematic process during cellular injury and repair. Causative agents for carcinogenesis are numerous including γ-radiation, ultraviolet radiation, inflammation, chemicals and iron overload [1]. Genomic data of a variety of human cancers is currently analyzed either with array-based comparative genomic hybridization (CGH) [2] or next-generation sequencing [3], [4]. These projects are performed to find causative gene mutations that will lead to identifying novel chemicals or antibodies directed for the interactions of responsible signaling molecules. These efforts are expected to result in developments of effective drugs. However, cancer prevention in daily life is as important as its therapy.
In the present study, we sought to resolve roles of iron-mediated oxidative stress during carcinogenesis using array-based CGH. Oxidative stress is constitutively caused by the metabolism of molecular oxygen [5], but is mainly regulated by various antioxidant systems. However, in a variety of pathological conditions, oxidative stress loads exceed the antioxidant capacity [6]. Iron is the most abundant heavy metal in mammals, such as rodents and humans. Whereas iron is essential for oxygen transport as a component of hemoglobin, excess iron has been associated with carcinogenesis [7], [8], presumably through a Fenton reaction [9]. Ionic forms of iron are barely soluble at a neutral pH, but ferric nitrilotriacetate (Fe-NTA), an iron chelate, is soluble at pH 7.4 and is an efficient catalytic agent for the Fenton reaction [10]. In the 1980s, our group established that repeated intraperitoneal administrations of Fe-NTA induce a high incidence of renal cell carcinoma (RCC) in rodents [11], [12]. Later, we showed that the renal injury occurs through a Fenton reaction with a variety of hydroxyl radical-mediated chemical products, such as 8-hydroxy-2′-deoxyguanosine [13], [14] and 4-hydroxy-nonenal [15], [16]. It is established that an iron overload in many pathological conditions is associated with the presence of catalytic iron [17], [18].
Accordingly, by evaluating whole genome of RCCs, we could find a general principle for the genomic alterations under oxidatively-stressed conditions. We reported a Cdkn2a/2b deletion using microsatellite analysis in this model [19]. In this study, we evaluated the whole genome of Fe-NTA-induced rat RCCs and their cell lines using array-based CGHs. Furthermore, we transformed the data into a human genome through chromosomal syntenic relationship and analyzed the association.
Results
Genome-wide Views of DNA Copy Number Alterations in Fe-NTA-induced Rat RCCs
Fifteen rat RCC DNA samples, which included 13 primary tumor samples and 2 cell line samples, were hybridized on Agilent oligonucleotide microarrays for CGH with 181,978 genomic loci (GEO accession: GSE36101). Comparing different array-based CGH profiles in a quantitative manner is difficult. A shift in the mean copy number is caused by polyploidy and the contamination of normal cells. Therefore, we have developed a statistical method that considers these factors to estimate the chromosomal copy number (Methods S1). In this paper, array-based CGH profile data analyses are based on the estimated copy numbers using this method.
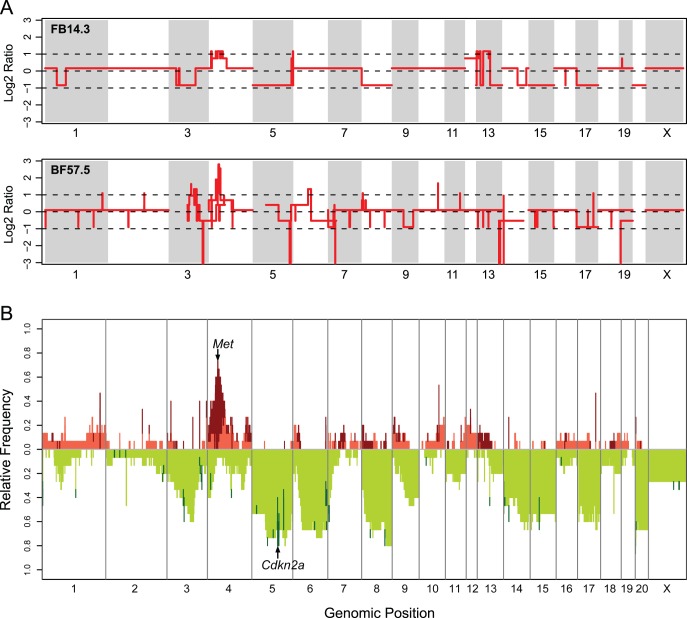
Array-based CGH profiling revealed that genomes of the Fe-NTA-induced rat RCCs are often complex and have many extensive chromosomal alterations ( Figs. 1A and S 1). A whole genome frequency analysis with 15 samples identified recurrent regions of a copy number aberration in the Fe-NTA-induced RCCs ( Fig. 1B ). Copy number aberrations were determined based on the distribution of the log2 ratio values that were recalculated with the estimated copy number for a set of 13 primary tumors and 2 cell lines (Fig. S2). In this distribution, the thresholds that represented gain and loss were chosen at ±0.377. A threshold representing amplification was chosen at +0.811 whereas a homozygous deletion (complete loss) was assigned to the position at which the copy number was estimated as 0. The most characteristic global feature uncovered by the frequency analysis was a predisposition to lose an extensively wide region of chromosomes, especially for chromosomes 3, 5, 6, 8, 9, 14, 15, 17 and 20. The second feature was a frequent amplification over a long pericentromeric region in chromosome 4.
Figure 1. Genome-wide views of DNA copy number alterations in Fe-NTA induced rat renal cell carcinomas (RCCs).
(A) Representative array-based CGH profiles from two RCC tumors. The red lines show log2 ratios of the estimated copy number over the inferred cancer ploidy versus the genomic position for all of the CGH microarray probes. (B) Frequency distribution of copy number aberrations across the whole rat genome. The relative frequencies of amplification (dark red), gain (tomato), loss (green yellow) and homozygous deletion (dark green) within 13 RCC tumors and two RCC cell lines are plotted at each genomic position. Two cancer-related loci, Met and Cdkn2a, which were most frequently affected by copy number aberration are indicated by the arrows.
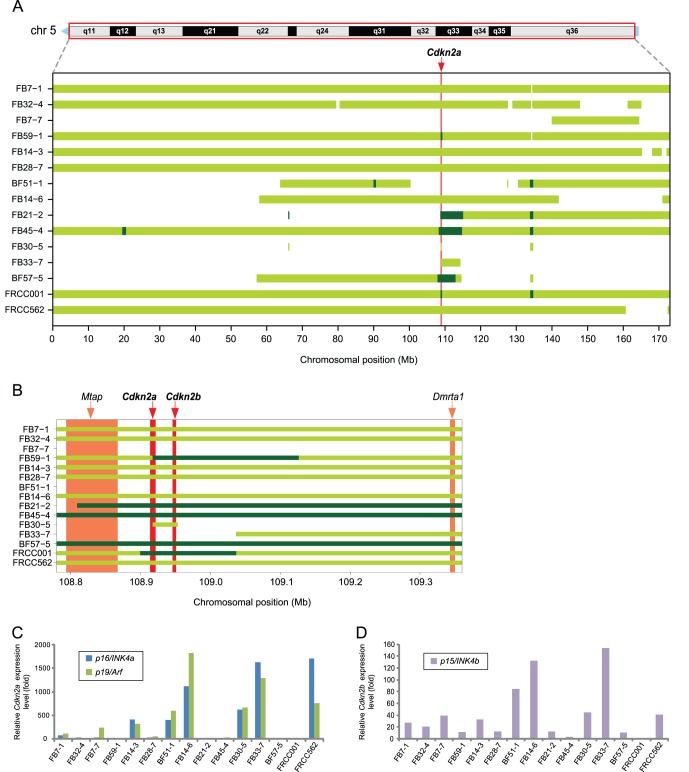
Frequent Chromosomal Loss in Rat Chromosome 5 and Homozygous Deletion at the Cdkn2a/2b Locus
Chromosome 5 underwent an extensive loss in copy number, not less than that in other chromosomes (e.g., chromosomes 6, 8 and 20) ( Fig. 1B ). As it relates to extensive loss, homozygous deletions were most frequently observed at the Cdkn2a/2b locus on chromosome 5 ( Fig. 2A ). This commonly deleted region included two loci (Cdkn2a and Cdkn2b) for three distinct tumor suppressor genes (p16 and p19 in Cdkn2a; p15 in Cdkn2b) ( Fig. 2B ). Shutdown of p16/p19 and p15 mRNA expression was confirmed in the samples that contained a homozygous deletion at the Cdkn2a/2b locus ( Figs. 2C and 2D ). In samples with a hemizygous deletion at the Cdkn2a locus (i.e., FB32-4, FB28-7), p16 and p19 expressions were downregulated, presumably because the promoter regions of the remaining alleles were methylated. However, some of the samples with either a hemizygous or no deletion (e.g., FB14-3; BF51-1; FB14-6; FB30-5 and FB33-7) showed a marked overexpression of p16 and p19.
Figure 2. Frequent extensive chromosomal losses in rat chromosome 5, and homozygous deletions at the region including the Cdkn2a and Cdkn2b loci.
(A) The bar chart represents the regions of chromosomal loss (green yellow) and homozygous deletion (dark green) along chromosome 5 for 13 RCC tumors and two RCC cell lines. The vertical red line on the background indicates the position of the Cdkn2a locus. (B) Magnified view of the bar chart centered on the Cdkn2a/2b loci. The genomic regions of all of the RefSeq genes included in the displayed range of the chromosome are depicted as vertical bars on the background. (C) Expression analysis of Cdkn2a (p16Ink4a and p19Arf) for 13 RCC tumors and two RCC cell lines, using real-time PCR with specific primer pairs for each different transcript. The values on the y-axis indicate relative mRNA expression level compared to an average of those in normal kidneys of three control rats. (D) Expression analysis of Cdkn2b (p15Ink4b) for 13 RCC tumors and two RCC cell lines by real-time PCR. The values on the y-axis indicate relative mRNA expression level compared to an average of those in normal kidneys of three control rats.
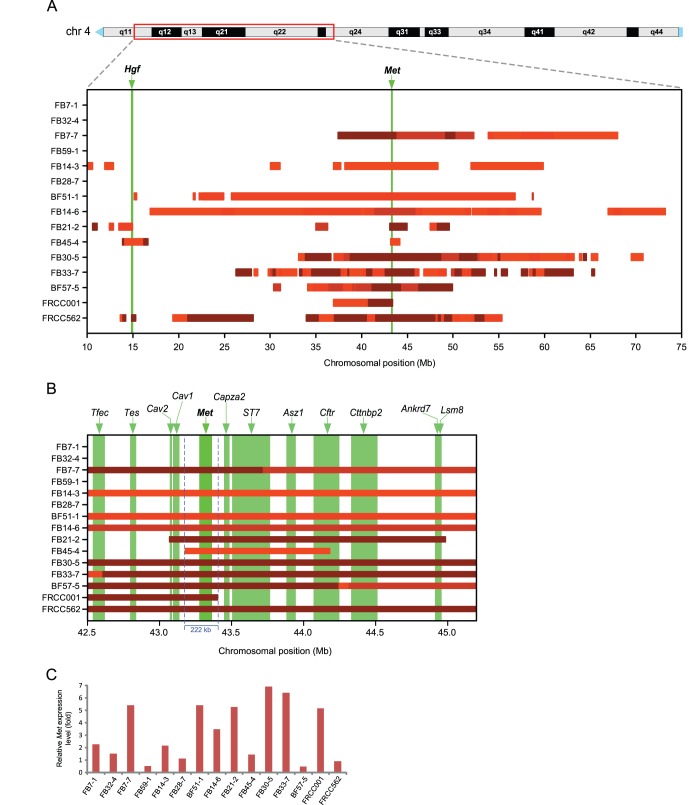
Frequent Amplifications Over the Pericentromeric Region of Chromosome 4 and Amplification at the Met Locus
Over a long portion of the pericentromeric region in chromosome 4, frequent copy number gain and amplification were observed ( Fig. 1B ). Genomic amplification and gene expression in the corresponding areas of chromosome 4 were mostly proportional (Fig. S3). A bar plot of the amplified region along the pericentromeric region in chromosome 4 revealed that Met oncogene resides in the most common overlapping genomic section ( Fig. 3A ). The most overlapping section with a length of approximately 222 kb consisted of an amplified region in 11/15 samples and contained only one RefSeq-curated gene (Met) ( Fig. 3B ). A greater than 5-fold increase in Met mRNA expression was observed in 6 samples among the 9 tumors that contained a genomic amplification of Met ( Fig. 3C ).
Figure 3. Frequent wide-ranging amplifications over a long pericentromeric region of chromosome 4 with the Met oncogene residing in the most overlapping section.
(A) The bar chart represents the amplification regions along a 65 Mb pericentrometic region of chromosome 4 for 13 RCC tumors and two RCC cell lines. Four grades of amplification are indicated by bar color gradation; the darker the red, the larger the amplitude. (B) A magnified view of the bar chart above shows the vicinity of the most overlapping region. The genomic regions of all of the RefSeq genes included in the displayed range of the chromosome are depicted as vertical bars in the background. (C) Expression analysis of Met for 13 RCC tumors and two RCC cell lines by real-time PCR. The values on the y-axis indicate relative mRNA expression level compared to an average of those in normal kidneys of three control rats.
Collectively, regarding chromosomal aberrations at these two cancer-related loci, every examined carcinoma including the two cell lines contained either the Met amplification or the Cdkn2a/2b deletion ( Table 1 , Figs. 2B and 3B ). Other common genetic alterations are summarized in Table S1 (20 deleted genes, common in 2–4 RCC tumors) and Table S2 (340 amplified genes, common in 2–9 RCC tumors). Among those Zbtb38 amplification was confirmed for overexpression (Fig. S4).
Table 1. Features of 13 cases of Fe-NTA-induced renal cell carcinomas.
| Copy number aberration at | |||||||
| Tumor case | Size (mm) | Metastasis | Invasion | Nuclear atypia grade | Growth pattern | Met locus | Cdkn2a/2b locus |
| FB7-1 | 20 | None | None | Low | Intermediate | None | Loss |
| FB32-4 | 15 | Lung | None | Intermediate | Intermediate | None | Loss |
| FB7-7 | 60 | Lung | None | Intermediate | Expansive | Amplification | None |
| FB59-1 | 15 | Lung | Peritoneal | Intermediate | Infiltrating | None | HD |
| FB14-3 | 15 | None | None | High | Expansive | Amplification | Loss |
| FB28-7 | 30 | None | None | High | Intermediate | None | Loss |
| BF51-1 | 28 | None | None | High | Intermediate | Amplification | None |
| FB14-6 | 30 | Lung | Peritoneal | High | Intermediate | Amplification | Loss |
| FB21-2 | 40 | None | None | High | Infiltrating | Amplification | HD |
| FB45-4 | 40 | Lung | None | High | Infiltrating | Amplification | HD |
| FB30-5 | 60 | Lung | Peritoneal | High | Infiltrating | Amplification | Loss |
| FB33-7 | 70 | Lung | Peritoneal | High | Infiltrating | Amplification | None |
| BF57-5 | 25 | Lung | Peritoneal | High | Infiltrating | Amplification | HD |
Fe-NTA: ferric nitrilotriacetate; HD: homozygous deletion.
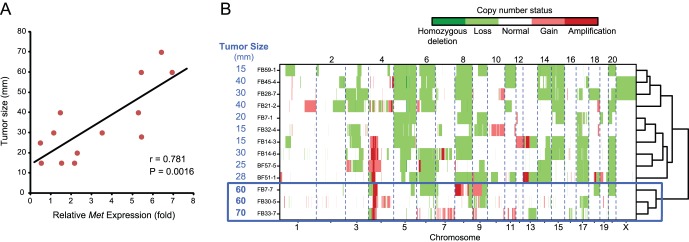
Tumor Size of Fe-NTA-induced RCCs is Regulated by Genetic Features
We examined the associations between genetic alterations and various RCC traits including those summarized in Table 1 . Among all of the relationships examined, Met expression and tumor size were proportionally associated ( Fig. 4A ). Furthermore, a hierarchical clustering of tumors based on whole patterns of chromosomal changes revealed that a group of large tumors (i.e., FB7-7, FB30-5 and FB33-7) corresponded to a distinct cluster ( Fig. 4B ). Therefore, a tumor trait, size at the time the tumor was clinically overt, was associated with the entire array-based CGH profiles.
Figure 4. Tumor sizes of Fe-NTA induced RCCs are controlled by the genetic features.
(A) Met expression is significantly correlated with tumor size. Pearson’s correlation coefficient (r) and the corresponding P value are written on the plot area. (B) Hierarchical clustering of the RCC tumors based on the whole genome patterns of the copy number changes. The large-size tumors form a distinct cluster.
Comparison of Copy Number Alteration Profiles in Cancer Genomes Between Rats and Humans
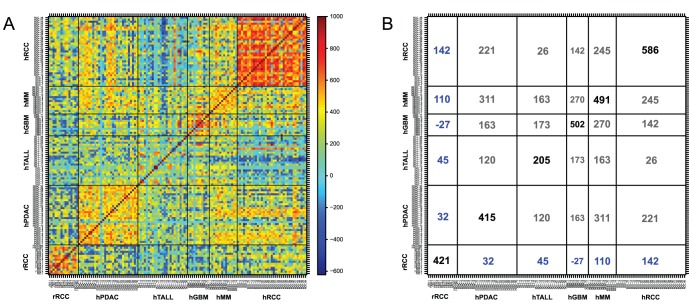
To determine the general principle of large-scale genomic changes in cancer across mammalian species, we compared cancer genomes of rats and humans as a whole spectrum of chromosomal alterations. First, we transformed the rat array-based CGH profiles onto human chromosomes according to a synteny between the two species. Thereafter, we compared the whole patterns based on estimated copy numbers using multidimensional data analysis methods. We found that Fe-NTA-induced rat RCC was most similar to human RCCs, followed by human malignant mesothelioma ( Figs. 5A and 5B ).
Figure 5. Comparison of copy number alteration profiles in cancer genomes between Fe-NTA-induced rat RCCs and human tumors.
(A) The color plot represents a similarity matrix across the rat RCCs and various human cancers. rRCC, rat renal cell carcinoma; hPDAC, human pancreatic ductal adenocarcinoma; hTALL, human T-cell acute lymphoblastic leukemia; hGBM, human glioblastoma multiforme; hRCC, human renal cell carcinoma. (B) Numerical summary of the similarity matrix. The number in each square indicates an average value of similarity index (defined between −1000 and 1,000). Refer to the Materials and Methods section for details.
Discussion
In this study, we report for the first time analyses of the entire data from array-based CGH applied to a Fenton chemistry-induced carcinogenesis in a rat kidney model [20]. We found that oxidative stress causes extensive genomic alterations in the induced cancer genome at chromosomal level ( Fig. 1A ). It is well known that a majority of human malignant solid tumors possess gains or losses in numerous chromosomes [21], [22], and amplification can be a suitable target for cancer chemotherapy. During the carcinogenic process of such tumors, chromosomal instability is thought to contribute as a driving factor [23]. Among wild-type rodent carcinogenesis models, however, few models report using primary tumor samples extensive genetic alterations because of chromosomal instability [24], [25]. Radiation-induced murine malignant lymphoma [26] and murine lung adenocarcinoma induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a carcinogen present in tobacco smoke [27], revealed slightly more gross chromosomal aberrations than the corresponding spontaneous tumors, albeit the low resolution in the report (bacterial artificial chromosome [BAC] array of ∼6,500). The facts that control rats exhibit no RCCs [11], [12] and Fe-NTA-induced rat RCC model exhibits an equivalence to human cancers in genomic alterations at chromosomal level strongly support the idea that this carcinogenesis model mimics an actual carcinogenic process in those humans who lack strong cancer susceptibility traits.
Conversely, mice with multiple genetically-engineered cancer-associated genes show genetic alteration of this kind [28]. We think that those experiments correspond to an established mutator phenotype [29] and, thus, to the carcinogenic process in humans who have strong cancer susceptibility traits such as Li-Fraumeni syndrome (p53) [30] or melanoma kindreds (p16) [31]. As a hereditary rat RCC model, we also analyzed the RCCs of Eker rats, which do not show aggressive characteristics, such as metastasis [32], [33], [34]. We observed that these RCCs showed null or subtle genetic alterations (Fig. S5). Accordingly, oxidative stress, including that induced due to excess iron, could be one of the causes of human renal carcinogenesis. Indeed, numerous epidemiological studies have associated iron and steel industry workers with an increased RCC risk [35].
A frequency plot analysis revealed two remarkable features. First, the chromosomal aberrations showed a preference for loss against the ploidy of each cancer genome. Mostly, the aberration was represented by a deletion at either a whole chromosomal level (monosomy) or at a segmental level ( Fig. 1B ). The most common target for loss was the Cdkn2a/2b locus. The predominance of loss in the profile of chromosomal alterations may be attributed to the early stage of carcinogenesis. We previously demonstrated that cells with a hemizygous deletion at Cdkn2a appear as early as a few weeks after initiating a Fe-NTA administration [36].
Our present analysis revealed that the monoallelic loss of chromosome 5 in its entirety or of an equivalently wide region is the major first event. Indeed, we found only one case (6.7%) of a monoallelic loss of an extremely narrow region (∼350 kilobases; Figs. 2A and 2B ). Fe-NTA catalyzes the generation of hydroxyl radicals through a Fenton reaction specifically in the lumina of renal proximal tubules, which leads to degeneration and necrosis/apoptosis of those cells [37], [38]. Because kidney is a vital organ that performs urea excretion, reabsorption of valuable molecules as well as ionic homeostasis maintenance, regeneration from the remaining tubular cells is intensely promoted. Under chronic oxidative stress by repeated Fe-NTA administrations, this degeneration and regeneration process would continue for months to years, increasing the risk of mitotic events simultaneously with the repair of oxidative DNA damage. We believe that this oxidative stress causes abnormal DNA replication and chromosomal missegregation, which leads to the emergence of aneuploid cells. Surprisingly, this series of events appear to occur in months, leading to a high incidence (∼90%) of RCC in rats within two years. Aneuploid cells usually exhibit phenotypes consistent with increased energy need and proteotoxic stress. However, aneuploidy can promote tumorigenesis under the following two hypothetical mechanisms: 1) aneuploidy may cause a proliferative advantage through loss of G1/S transition control under conditions in which normal euploid cells do not divide [39] and 2) aneuploidy can advance tumorigenesis by promoting genomic instability, hence increasing the evolvability of tumors [40]. The frequent deletion of the Cdkn2a/2b locus is observed in the rat peritoneal mesothelioma, an iron overload-associated tumor [41], induced either by another iron compound (ferric saccharate) [42] or by asbestos [43]. These common traits of animal models strongly suggest that Cdkn2a/2b is a principal target in iron-mediated carcinogenesis. The same genetic alteration is observed in rat mesothelioma induced by multi-walled carbon nanotubes [44], in which iron involvement is not yet established. We would like to add here for the biological significance of our results that homozygous deletion of CDKN2A/2B is frequently observed in human mesothelioma associated with asbestos exposure [45], [46].
Some of the tumors with a remaining Cdkn2a/2b allele showed extremely high expression levels of two products from this locus, p16/Ink4a and p19/Arf. This is a currently debated issue in human cancer [47]. There is considerable evidence that several neoplasms exhibit significant p16 levels in cytoplasm [48]. This can be an unsuccessful attempt to stop cell proliferation in the case of downstream Rb dysregulation [49] or may represent an alternative mechanism for modulating unidentified pathways. Our data exhibit ∼50% hemizygous deletion of Rb. This requires further clarification with epigenetic analysis.
The second feature determined using frequency plot analysis was a high incidence of amplification along a limited chromosomal region toward the centromere of chromosome 4, pointing to the Met locus. A region spanning 80 Mb from the centromere of rat chromosome 4 is syntenic to human chromosome 7. Various human cancers, such as glioblastoma [50] and non-small cell lung cancer [51], are reported to harbor amplifications in chromosome 7. Tyrosine kinase MET is a receptor for a hepatocyte growth factor and is situated upstream of ras in the signal transduction pathway, thus serving as an advantage for cell proliferation [52]. Therefore, it is conceivable that tumor size was proportionally associated with the Met expression level ( Figs. 3C and 4A ). Because we dissect the animal as soon as we recognize the tumor, we believe that large-sized tumors are more aggressive in nature. It is of note that tumor size was also related to the genome alteration pattern ( Fig. 4B ), and was associated with amplification and overexpression of Zbtb38 located on chromosome 8 (Fig. S4). ZBTB38 (CIBZ) represses the transcription of methylated templates [53], thus presumably regulating epigenetic mechanisms. Down-regulation of ZBTB38, a novel substrate of caspase-3, induces apoptosis [54] and this gene is localized in a prostate cancer susceptibility locus [54]. These results confirm the possibility of tumor classification using array-based CGH. Met arose evolutionally late and is unique to mammals [52]; it could thereby be associated with the unique amplification in the whole genome.
Genomic amplification is hypothesized to occur via the breakage-fusion-bridge cycle [55], [56], [57]. A Fenton reaction causes double-stranded DNA breakage [10]. Our results revealed that these amplifications consisted of a mixture of wide-range low-level amplifications and fragmented, narrow high-level amplifications ( Fig. 3A ). This suggests a mechanism of positive feedback for amplification, starting from wide-range low-level amplification. We suspect an involvement of double-minutes, and a presence of susceptible genomic loci. This hypothesis requires further study. It was interesting that two tumor suppressive genes, Cav1 [58], [59] and ST7 [60], surrounded the Met locus ( Fig. 3B ). This may be the reason why the Met locus was a denominator for the rat RCCs. Whole exome or genome sequencing may further reveal new findings regarding point mutation and chromosomal translocation.
Finally, we compared the present rat results with corresponding human tumors by transforming data based on chromosomal synteny ( Figs. 5A and 5B ). It was expected that the genomic alteration of Fe-NTA-induced rat RCC was most similar to human RCC presumably because target cells are the same. However, surprisingly, human mesothelioma was the second most similar. It is now established that most human mesothelioma results from exposure to asbestos, and the primary pathogenic process involved is iron overload [8] , [61]. The same mesodermal origin of renal tubular cells and mesothelial cells may cause the similarity of the array-based CGH profiles. Endodermal tumor, such as pancreatic ductal adenocarcinoma (PDAC), and ectodermal tumor, such as glioblastoma multiforme (GBM), exhibited a significant difference in genomic profiles.
In conclusion, we showed that repeated Fenton reactions in wild-type rats induced cancer that recapitulated genomic alterations similar to those in human cancers, suggesting the involvement of oxidative stress as a major factor in human carcinogenesis. In this renal carcinogenesis model, preferred alterations were deletion; Cdkn2A/2B deletion and Met amplification were the major target gene modifications. A comparative interspecies analysis would contribute to identifying candidate carcinogenic agents.
Materials and Methods
Fe-NTA-induced Renal Cell Carcinoma Model
Fe-NTA-induced carcinogenesis experiments were performed using male F1 hybrid rats that were a cross between Fischer 344 and Brown-Norway inbred strains (Charles River, Yokohama, Japan) as previously described [62]. Thirteen RCC cases were used in this study, and the histological grade of the tumor was determined according to the modified World Health Organization classification as we previously described [62]. The details are summarized in Table 1 . The animal experiment committees of the Graduate School of Medicine, Kyoto University Graduate School of Medicine and Nagoya University Graduate School of Medicine approved this study. FRCC001 and FRCC562 cell lines were established from primary Fe-NTA-induced RCCs as previously described [63].
Array-based Comparative Genomic Hybridization
Genomic DNA from the tumors and the cell lines was isolated with DNeasy (Qiagen, Valencia, CA). Array-based CGH was performed with an Agilent 185 K rat genome CGH microarray (Agilent Technologies, Santa Clara, CA) as previously described [64]. Thirteen primary tumors and two cell lines of Fe-NTA induced RCCs were analyzed using reference DNA extracted from a normal kidney of a rat from Brown-Norway imbred strain. One RCC sample of a female Eker rat [32] was analyzed using reference DNA extracted from a normal kidney of a male Eker rat. Two additional RCC samples of male Eker rats were analyzed with Rat Genome CGH Microarray 105A (G4436A; Agilent Technologies), using reference DNA extracted from a normal liver of another male Eker rat. The normalized array-based CGH data were processed to generate a segmented profile by circular binary segmentation (CBS) [65] with an altered significance level (alpha = 0.0001). The procedure of data processing for copy number estimation is detailed in Methods S1.
Quantitative RT-PCR Analysis
Total RNA was isolated using Isogen reagents (Nippon Gene Co. Ltd., Tokyo, Japan) according to the manufacturer’s protocol. cDNA was synthesized using an RNA PCR kit ver. 3.0 (Takara Bio, Shiga, Japan) with random primers. A Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen, Carlsbad, CA) and a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA) were used for quantitative real-time PCR analysis. Rat β-actin was used as an internal control. The primers used were as follows: p16Ink4a, 5′-aaacgccccgaacactttc-3′ and 5′-gttcgaatctgcaccatagga-3′; p19Arf, 5′-accccaagtgagggttttct-3′ and 5′-agagctgccactttgacgtt-3′; p15Ink4b, 5′-tccacaggctagagggaaaa-3′ and 5′-gtgcaggtgactccttggtt-3′; Met, 5′-ttaagcgagagcacgacaaat-3′ and 5′-ccacataggaaaacgcactgt-3′; Zbtb38, 5′-gtagctgctgctccaaatcc-3′ and 5′-cctgttgagggtggtgaact-3′; β-actin, 5′-tgtgttgtccctgtatgcctctg-3′ and 5′-atagatgggcacagtgtgggtg-3′.
Human Data
We used human array-based CGH data of pancreatic ductal adenocarcinoma (hPDAC) [28], T-cell acute lymphoblastic leukemia (hTALL) [28], glioblastoma (hGBM) [66], mesothelioma cell line (hMM) [67] and renal cell carcinoma (hRCC) [56]. The Agilent CGH-array data of the former four types of human cancer were obtained from NCBI’s Gene Expression Omnibus (GEO) website (http://www.ncbi.nlm.nih.gov/geo/). The GEO accession numbers for the data sets are GSE7599 (hPDAC), GSE7603 (hTALL), GSE9177 (hGBM) and GSE22237 (hMM). The human RCC data was obtained through analyses with BAC microarrays (4,361 clones) [68]. We defined similarity index between the two array-based CGH profiles as follows. First, we calculated the correlation coefficient with log2 ratios of the estimated copy number over the inferred cancer ploidy for the genomic positions corresponding to all of the Agilent 44 K human CGH microarray probes. Then, we multiplied the value by 1×103 after changing the absolute value into its square root.
Supporting Information
Array-CGH profiles from all the RCCs examined. Red lines show log2 ratios of estimated copy number over inferred cancer ploidy versus genomic position for all the CGH microarray probes.
(PDF)
Distribution of log2 ratio values of estimated copy number for all the probes in all the microarrays performed.
(PDF)
Example of global expression changes in line with genomic alteration. Differences in genome and transcriptome are analyzed between two RCCs, FB7-7 having wide-range amplification on chromosome 4 versus FB28-7 having no substantial genomic alteration on chromosome 4. (A) Sky blue circle plot indicates ratio of estimated copy numbers based on array-based CGH (FB7-7 vs FB28-7). Red circle plot indicates ratio of normalized signals on Affymetrix expression microarray (FB7-7 vs FB28-7). (B) Expression ratio values are averaged along the chromosome. Here, red circle plot indicates average value in 2-Mb windows.
(PDF)
Zbtb38 mRNA expression is demonstrably associated with its chromosomal copy number. (A) Array-CGH profiles of two RCC tumors harboring amplification over Zbtb38 locus on chromosome 8. (B) Expression analysis of Zbtb38 on 13 RCC tumors by real-time PCR. The values of the y-axis indicate the relative mRNA expression level compared to an average of those in normal kidneys of three control rats.
(PDF)
Array-CGH profiles of three hereditary (Eker rat) renal tumors. Red lines show log2 ratios of estimated copy number over inferred cancer ploidy versus genomic position for all the CGH microarray probes.
(PDF)
List of genes completely deleted in more than two RCC tumors.
(XLS)
List of genes amplified in more than two RCC tumors.
(XLS)
Supplementary methods.
(DOC)
Funding Statement
This study was supported by Princess Takamatsu Cancer Research Fund (10-24213); a Grant-in-Aid for Cancer Research from the Ministry of Health, Labour and Welfare of Japan; and a Grant-in Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Weinberg RA (2007) The biology of cancer. New York: Garland Science, Tailor & Francis Group, LLC.
- 2. Klijn C, Holstege H, de Ridder J, Liu X, Reinders M, et al. (2008) Identification of cancer genes using a statistical framework for multiexperiment analysis of nondiscretized array CGH data. Nucleic Acids Res 36: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pfeifer GP, Hainaut P (2011) Next-generation sequencing: emerging lessons on the origins of human cancer. Curr Opin Oncol 23: 62–68. [DOI] [PubMed] [Google Scholar]
- 5. Toyokuni S (1999) Reactive oxygen species-induced molecular damage and its applicaton in pathology. PatholInt 49: 91–102. [DOI] [PubMed] [Google Scholar]
- 6.Halliwell B, Gutteridge JMC (2007) Free radicals in biology and medicine. New York: Oxford University Press.
- 7. Toyokuni S (1996) Iron-induced carcinogenesis: the role of redox regulation. Free Radic Biol Med 20: 553–566. [DOI] [PubMed] [Google Scholar]
- 8. Toyokuni S (2009) Role of iron in carcinogenesis: Cancer as a ferrotoxic disease. Cancer Sci 100: 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fenton HJH (1894) Oxidation of tartaric acid in presence of iron. J Chem Soc 65: 899–910. [Google Scholar]
- 10. Toyokuni S, Sagripanti JL (1992) Iron-mediated DNA damage: sensitive detection of DNA strand breakage catalyzed by iron. J Inorg Biochem 47: 241–248. [DOI] [PubMed] [Google Scholar]
- 11. Ebina Y, Okada S, Hamazaki S, Ogino F, Li JL, et al. (1986) Nephrotoxicity and renal cell carcinoma after use of iron- and aluminum- nitrilotriacetate complexes in rats. J Natl Cancer Inst 76: 107–113. [PubMed] [Google Scholar]
- 12. Li JL, Okada S, Hamazaki S, Ebina Y, Midorikawa O (1987) Subacute nephrotoxicity and induction of renal cell carcinoma in mice treated with ferric nitrilotriacetate. Cancer Res 47: 1867–1869. [PubMed] [Google Scholar]
- 13. Toyokuni S, Mori T, Dizdaroglu M (1994) DNA base modifications in renal chromatin of Wistar rats treated with a renal carcinogen, ferric nitrilotriacetate. Int J Cancer 57: 123–128. [DOI] [PubMed] [Google Scholar]
- 14. Toyokuni S, Tanaka T, Hattori Y, Nishiyama Y, Ochi H, et al. (1997) Quantitative immunohistochemical determination of 8-hydroxy-2′-deoxyguanosine by a monoclonal antibody N45.1: its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab Invest 76: 365–374. [PubMed] [Google Scholar]
- 15. Toyokuni S, Uchida K, Okamoto K, Hattori-Nakakuki Y, Hiai H, et al. (1994) Formation of 4-hydroxy-2-nonenal-modified proteins in the renal proximal tubules of rats treated with a renal carcinogen, ferric nitrilotriacetate. Proc Natl Acad Sci USA 91: 2616–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Toyokuni S, Luo XP, Tanaka T, Uchida K, Hiai H, et al. (1997) Induction of a wide range of C2–12 aldehydes and C7–12 acyloins in the kidney of Wistar rats after treatment with a renal carcinogen, ferric nitrilotriacetate. Free Radic Biol Med 22: 1019–1027. [DOI] [PubMed] [Google Scholar]
- 17. Gutteridge J, Rowley D, Halliwell B (1981) Superoxide-dependent formation of hydroxyl radicals in the presence of iron salts. Detection of ‘free’ iron in biological systems by using bleomycin-dependent degradation of DNA. Biochem J 199: p263–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sasaki K, Ikuta K, Tanaka H, Ohtake T, Torimoto Y, et al. (2011) Improved quantification for non-transferrin-bound iron measurement using high-performance liquid chromatography by reducing iron contamination. Mol Med Report 4: 913–918. [DOI] [PubMed] [Google Scholar]
- 19. Tanaka T, Iwasa Y, Kondo S, Hiai H, Toyokuni S (1999) High incidence of allelic loss on chromosome 5 and inactivation of p15INK4B and p16INK4A tumor suppressor genes in oxystress-induced renal cell carcinoma of rats. Oncogene 18: 3793–3797. [DOI] [PubMed] [Google Scholar]
- 20. Toyokuni S, Akatsuka S (2006) What has been learned from the studies of oxidative stress-induced carcinogenesis: Proposal of the concept of oxygenomics. J Clin Biochem Nutr 39: 3–10. [Google Scholar]
- 21. Kallioniemi A, Kallioniemi OP, Sudar D, Rutovitz D, Gray JW, et al. (1992) Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 258: 818–821. [DOI] [PubMed] [Google Scholar]
- 22. Macconaill LE, Van Hummelen P, Meyerson M, Hahn WC (2011) Clinical implementation of comprehensive strategies to characterize cancer genomes: opportunities and challenges. Cancer Discov 1: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Negrini S, Gorgoulis VG, Halazonetis TD (2011) Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228. [DOI] [PubMed] [Google Scholar]
- 24. Adamovic T, McAllister D, Guryev V, Wang X, Andrae JW, et al. (2009) Microalterations of inherently unstable genomic regions in rat mammary carcinomas as revealed by long oligonucleotide array-based comparative genomic hybridization. Cancer Res 69: 5159–5167. [DOI] [PubMed] [Google Scholar]
- 25. Femia AP, Luceri C, Toti S, Giannini A, Dolara P, et al. (2010) Gene expression profile and genomic alterations in colonic tumours induced by 1,2-dimethylhydrazine (DMH) in rats. BMC Cancer 10: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takabatake T, Fujikawa K, Tanaka S, Hirouchi T, Nakamura M, et al. (2006) Array-CGH analyses of murine malignant lymphomas: genomic clues to understanding the effects of chronic exposure to low-dose-rate gamma rays on lymphomagenesis. Radiat Res 166: 61–72. [DOI] [PubMed] [Google Scholar]
- 27. Herzog CR, Desai D, Amin S (2006) Array CGH analysis reveals chromosomal aberrations in mouse lung adenocarcinomas induced by the human lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Biochem Biophys Res Commun 341: 856–863. [DOI] [PubMed] [Google Scholar]
- 28. Maser R, Choudhury B, Campbell P, Feng B, Wong K, et al. (2007) Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature 447: 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loeb LA, Bielas JH, Beckman RA (2008) Cancers exhibit a mutator phenotype: clinical implications. Cancer Res 68: 3551–3557. [DOI] [PubMed] [Google Scholar]
- 30. Hisada M, Garber JE, Fung CY, Fraumeni JF, Li FP (1998) Multiple primary cancers in families with Li-Fraumeni syndrome. J Natl Cancer Inst 90: 606–611. [DOI] [PubMed] [Google Scholar]
- 31. Gruis NA, van der Velden PA, Sandkuijl LA, Prins DE, Weaver-Feldhaus J, et al. (1995) Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nat Genet 10: 351–353. [DOI] [PubMed] [Google Scholar]
- 32. Kobayashi T, Hirayama Y, Kobayashi E, Kubo Y, Hino O (1995) A germline insertion in the tuberous sclerosis (Tsc2) gene give rise to the Eker rat model of dominantly inherited cancer. Nature Genet 9: 70–74. [DOI] [PubMed] [Google Scholar]
- 33. Hino O (2004) Multistep renal carcinogenesis in the Eker (Tsc 2 gene mutant) rat model. Curr Mol Med 4: 807–811. [DOI] [PubMed] [Google Scholar]
- 34. Yeung RS (2004) Lessons from the Eker rat model: from cage to bedside. Curr Mol Med 4: 799–806. [DOI] [PubMed] [Google Scholar]
- 35. Huang X (2003) Iron overload and its association with cancer risk in humans: evidence for iron as a carcinogenic metal. Mutat Res 533: 153–171. [DOI] [PubMed] [Google Scholar]
- 36. Hiroyasu M, Ozeki M, Kohda H, Echizenya M, Tanaka T, et al. (2002) Specific allelic loss of p16 (INK4A) tumor suppressor gene after weeks of iron-mediated oxidative damage during rat renal carcinogenesis. Am J Pathol 160: 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Toyokuni S, Okada S, Hamazaki S, Minamiyama Y, Yamada Y, et al. (1990) Combined histochemical and biochemical analysis of sex hormone dependence of ferric nitrilotriacetate-induced renal lipid peroxidation in ddY mice. Cancer Res 50: 5574–5580. [PubMed] [Google Scholar]
- 38. Zhang D, Okada S, Yu Y, Zheng P, Yamaguchi R, et al. (1997) Vitamin E inhibits apoptosis, DNA modification, and cancer incidence induced by iron-mediated peroxidation in Wistar rat kidney. Cancer Res 57: 2410–2414. [PubMed] [Google Scholar]
- 39. Torres EM, Williams BR, Tang YC, Amon A (2010) Thoughts on aneuploidy. Cold Spring Harb Symp Quant Biol 75: 445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torres EM, Williams BR, Amon A (2008) Aneuploidy: cells losing their balance. Genetics 179: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Toyokuni S (2009) Mechanisms of asbestos-induced carcinogenesis. Nagoya J Med Sci 71: 1–10. [PMC free article] [PubMed] [Google Scholar]
- 42. Hu Q, Akatsuka S, Yamashita Y, Ohara H, Nagai H, et al. (2010) Homozygous deletion of CDKN2A/2B is a hallmark of iron-induced high-grade rat mesothelioma. Lab Invest 90: 360–373. [DOI] [PubMed] [Google Scholar]
- 43. Jean D, Thomas E, Manie E, Renier A, de Reynies A, et al. (2011) Syntenic relationships between genomic profiles of fiber-induced murine and human malignant mesothelioma. Am J Pathol 178: 881–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagai H, Okazaki Y, Hwu C, Misawa N, Yamashita Y, et al.. (2011) Diameter of multi-walled carbon nanotubes is a critical factor in mesothelial injury and subsequent carcinogenesis. Proc Natl Acad Sci USA. [DOI] [PMC free article] [PubMed]
- 45. Cheng J, Jhanwar S, Klein W, Bell D, Lee W, et al. (1994) p16 alterations and deletion mapping of 9p21-p22 in malignant mesothelioma. Cancer Res 54: 5547–5551. [PubMed] [Google Scholar]
- 46. Xio S, Li D, Vijg J, Sugarbaker D, Corson J, et al. (1995) Codeletion of p15 and p16 in primary malignant mesothelioma. Oncogene 11: p511–515. [PubMed] [Google Scholar]
- 47. Romagosa C, Simonetti S, Lopez-Vicente L, Mazo A, Lleonart ME, et al. (2011) p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 30: 2087–2097. [DOI] [PubMed] [Google Scholar]
- 48. Evangelou K, Bramis J, Peros I, Zacharatos P, Dasiou-Plakida D, et al. (2004) Electron microscopy evidence that cytoplasmic localization of the p16(INK4A) “nuclear” cyclin-dependent kinase inhibitor (CKI) in tumor cells is specific and not an artifact. A study in non-small cell lung carcinomas. Biotech Histochem 79: 5–10. [DOI] [PubMed] [Google Scholar]
- 49. Reuschenbach M, Waterboer T, Wallin KL, Einenkel J, Dillner J, et al. (2008) Characterization of humoral immune responses against p16, p53, HPV16 E6 and HPV16 E7 in patients with HPV-associated cancers. Int J Cancer 123: 2626–2631. [DOI] [PubMed] [Google Scholar]
- 50. Wullich B, Sattler HP, Fischer U, Meese E (1994) Two independent amplification events on chromosome 7 in glioma: amplification of the epidermal growth factor receptor gene and amplification of the oncogene MET. Anticancer Res 14: 577–579. [PubMed] [Google Scholar]
- 51. Campbell JM, Lockwood WW, Buys TP, Chari R, Coe BP, et al. (2008) Integrative genomic and gene expression analysis of chromosome 7 identified novel oncogene loci in non-small cell lung cancer. Genome 51: 1032–1039. [DOI] [PubMed] [Google Scholar]
- 52. Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF (2003) Met, metastasis, motility and more. Nat Rev Mol Cell Biol 4: 915–925. [DOI] [PubMed] [Google Scholar]
- 53. Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, et al. (2006) A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol 26: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oikawa Y, Matsuda E, Nishii T, Ishida Y, Kawaichi M (2008) Down-regulation of CIBZ, a novel substrate of caspase-3, induces apoptosis. J Biol Chem 283: 14242–14247. [DOI] [PubMed] [Google Scholar]
- 55. Hellman A, Zlotorynski E, Scherer SW, Cheung J, Vincent JB, et al. (2002) A role for common fragile site induction in amplification of human oncogenes. Cancer Cell 1: 89–97. [DOI] [PubMed] [Google Scholar]
- 56. Albertson DG (2006) Gene amplification in cancer. Trends Genet 22: 447–455. [DOI] [PubMed] [Google Scholar]
- 57. Martinez AC, van Wely KH (2010) Are aneuploidy and chromosome breakage caused by a CINgle mechanism? Cell Cycle 9: 2275–2280. [DOI] [PubMed] [Google Scholar]
- 58. Wiechen K, Diatchenko L, Agoulnik A, Scharff KM, Schober H, et al. (2001) Caveolin-1 is down-regulated in human ovarian carcinoma and acts as a candidate tumor suppressor gene. Am J Pathol 159: 1635–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wiechen K, Sers C, Agoulnik A, Arlt K, Dietel M, et al. (2001) Down-regulation of caveolin-1, a candidate tumor suppressor gene, in sarcomas. Am J Pathol 158: 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol 24: 9630–9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nagai H, Ishihara T, Lee WH, Ohara H, Okazaki Y, et al. (2011) Asbestos surface provides a niche for oxidative modification. Cancer Sci 102: 2118–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nishiyama Y, Suwa H, Okamoto K, Fukumoto M, Hiai H, et al. (1995) Low incidence of point mutations in H-, K- and N-ras oncogenes and p53 tumor suppressor gene in renal cell carcinoma and peritoneal mesothelioma of Wistar rats induced by ferric nitrilotriacetate. Jpn J Cancer Res 86: 1150–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tanaka T, Akatsuka S, Ozeki M, Shirase T, Hiai H, et al. (2004) Redox regulation of annexin 2 and its implications for oxidative stess-induced renal carcinogenesis and metastasis. Oncogene 23: 3980–3989. [DOI] [PubMed] [Google Scholar]
- 64. Liu Y-T, Shang D-G, Akatsuka S, Ohara H, Dutta KK, et al. (2007) Chronic oxidative stress causes amplification and overexpresson of ptprz1 protein tyrosine phosphatase to activate β-catenin pathway. Am J Pathol 171: 1978–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Olshen AB, Venkatraman ES, Lucito R, Wigler M (2004) Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5: 557–572. [DOI] [PubMed] [Google Scholar]
- 66. Wiedemeyer R, Brennan C, Heffernan TP, Xiao Y, Mahoney J, et al. (2008) Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell 13: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Murakami H, Mizuno T, Taniguchi T, Fujii M, Ishiguro F, et al. (2011) LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res 71: 873–883. [DOI] [PubMed] [Google Scholar]
- 68. Arai E, Ushijima S, Tsuda H, Fujimoto H, Hosoda F, et al. (2008) Genetic clustering of clear cell renal cell carcinoma based on array-comparative genomic hybridization: its association with DNA methylation alteration and patient outcome. Clin Cancer Res 14: 5531–5539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Array-CGH profiles from all the RCCs examined. Red lines show log2 ratios of estimated copy number over inferred cancer ploidy versus genomic position for all the CGH microarray probes.
(PDF)
Distribution of log2 ratio values of estimated copy number for all the probes in all the microarrays performed.
(PDF)
Example of global expression changes in line with genomic alteration. Differences in genome and transcriptome are analyzed between two RCCs, FB7-7 having wide-range amplification on chromosome 4 versus FB28-7 having no substantial genomic alteration on chromosome 4. (A) Sky blue circle plot indicates ratio of estimated copy numbers based on array-based CGH (FB7-7 vs FB28-7). Red circle plot indicates ratio of normalized signals on Affymetrix expression microarray (FB7-7 vs FB28-7). (B) Expression ratio values are averaged along the chromosome. Here, red circle plot indicates average value in 2-Mb windows.
(PDF)
Zbtb38 mRNA expression is demonstrably associated with its chromosomal copy number. (A) Array-CGH profiles of two RCC tumors harboring amplification over Zbtb38 locus on chromosome 8. (B) Expression analysis of Zbtb38 on 13 RCC tumors by real-time PCR. The values of the y-axis indicate the relative mRNA expression level compared to an average of those in normal kidneys of three control rats.
(PDF)
Array-CGH profiles of three hereditary (Eker rat) renal tumors. Red lines show log2 ratios of estimated copy number over inferred cancer ploidy versus genomic position for all the CGH microarray probes.
(PDF)
List of genes completely deleted in more than two RCC tumors.
(XLS)
List of genes amplified in more than two RCC tumors.
(XLS)
Supplementary methods.
(DOC)