Abstract
Purpose
Human embryonic stem cells (hESCs) are self-renewing, pluripotent cells that are valuable research tools and hold promise for use in regenerative medicine. The need for new hESC lines motivated our attempts to find a new resource for the derivation of hESC lines. The aim of this work was to establish more hESC lines from abnormal fertilized zygotes and to meet the emerging requirements for their use in cell replacement therapies, disease modeling, and basic research.
Methods
A total of 130 tripronuclear human zygotes was collected 18–20 h post-insemination and cultured in a modified culture medium. The inner cell mass of 12 blastocysts were isolated by a mechanical method in order to establish embryonic stem cell lines.
Results
We established four hESC lines derived from 130 trinuclear zygotes, one of which was triploid and the others were diploid. The efficiency of deriving hESC lines is 3.08 %. The ratio of deriving triploid and diploid hESC lines is 1:3. All of these hESC lines exhibited similar markers of undifferentiated hESCs and had the typical morphology of hESCs, a capacity for long-term proliferation, and pluripotent differentiation potential both in vivo and in vitro.
Conclusions
These abnormal zygotes, which otherwise would have been discarded, can serve as an alternative source for normal euploid hESC lines.
Keywords: Human embryonic stem cells, Pronucleus (PN), Human zygotes, Triploid
Introduction
Human embryonic stem cell (hESC) lines are typically derived from the inner cell mass (ICM) of blastocyst stage embryos [1]. hESCs have indefinite proliferation potential, and they have been shown to differentiate into cells of the three embryonic germ layers. These characteristics provide hESCs with great potential for use in modern regenerative medicine and cell-based drug discovery.
HESCs are isolated from surplus cryopreserved embryos [1–3] or poor-quality embryos [4, 5] of in vitro fertilization (IVF) treatments. These embryos are developed to the blastocyst stage using in vitro culture prior to being plated for derivation purposes. However, the usage of embryos and derivation of hESCs for research and eventual medical applications has resulted in polarized ethical debates because the process involves the destruction of viable developing human embryos [6]. Therefore, the fertilized zygotes that are abnormal, which are usually disposed of, may be good resources for the derivation of hESCs because of less ethical debate.
Normal human zygotes consist of two pronuclei representing each parent. However, IVF often results in abnormal zygotes with one or three pronuclei (PN), and it is reported that approximately 7 % of fertilized eggs are polypronuclear in conventional IVF [7]. These zygotes cannot develop into normal human beings and are usually disposed of by IVF centers. However, reports have shown that the transfer of single-embryos with excessive polypronuclear zygote formation [8] or embryos developed from single-nucleated zygotes has resulted in healthy births[9]. Therefore, it is possible that a normal hESC line can be established from fertilized zygotes that are abnormal.
Recently, several groups have reported that diploid hESC lines were derived from a mononuclear zygote [10] and tripronuclear zygotes [11, 12]. However, no earlier deriving abnormal and normal hESC line at the same time has been reported from tripronuclear zygotes. The efficiency and ratio of derivation remain unknown. In the present study, we established four hESC lines derived from 130 trinuclear zygotes, one of which was triploid, and the others were diploid. The efficiency of deriving hESC lines is 3.08 %. The ratio of triploid and diploid hESC lines is 1:3. All of these hESC lines exhibited similar markers of undifferentiated hESCs and had the typical morphology of hESCs, a capacity for long-term proliferation, and pluripotent differentiation potential in vivo and in vitro.
Materials and methods
Source of tripronuclear human zygotes and dipronuclear human embryos
From February 2009 to November 2009, a total of 130 tripronuclear human zygotes with a visible second polar body were obtained 18–20 h post-insemination. As a control, 139 Day-3 poor-quality embryos were also collected. Embryos were morphologically graded, as described by Edwards[13] as follows grade 1, equal-sized symmetrical blastomeres; grade 2, uneven blastomeres with <10 % fragmentation; grade 3 (10–50 %), blastomeric fragmentation; and grade 4 (>50 %), blastomeric fragmentation embryos. Only grades 3 and grades 4 embryos that were not used for transfer or freezing on day 3 were used after obtaining cumulative embryo score (CES). The use of abnormally fertilized zygotes and poor-quality embryos that were donated by patients undergoing conventional in vitro fertilization (IVF) after obtaining informed consent was approved by the ethics committee of Guangzhou Medical College, China. The average ages of the zygote donors and the embryo donors were 32 ± 4 years.
Embryo culture
The zygotes were cultured according to our IVF laboratory standard protocol. The zygotes were grown in G1.5 medium (Vitrolife, Gothenburg, Sweden) until Day 3, and the embryos were then transferred into a modified culture medium [14] composed of G2.5 medium supplemented with 2000 U/ml of human recombinant leukemia inhibitory factor (LIF, Chemicon, Temecula, CA, USA), 10 ng/ml of human basic fibroblast growth factor (bFGF, Invitrogen, Carlsbad, CA, USA) and 10 % human serum albumin. The embryos were cultured for an additional 2–4 days at 37 C in 5 % CO2.
Blastocyst formation
The blastocyst quality was defined according to the criteria presented by Gardner et al. [15]. Briefly, quality 1 describes an early blastocyst with the blastocoel occupying <50 % of the volume, quality 2 describes an early blastocyst with the blastocoel expanded to 50–80 % of the volume, quality 3 describes a non-expanded blastocyst with a large blastocoel, quality 4 describes an expanded blastocyst, quality 5 indicates that the blastocyst has started to hatch [14], and quality 6 indicates that the blastocyst is completely hatched. The first letter describes the ICM: ‘A’ means a high number of tightly packed cells, ‘B’ means a few cells, loosely packed, and ‘C’ indicates almost no cells. The second letter reflects the cell number in the trophectoderm: ‘A’ indicates many cells, ‘B’ means few cells, and ‘C’ means almost no cells. For example, 6AA describes a hatched blastocyst possessing normal-looking cells in both the ICM and trophectoderm
Derivation and culture of human embryonic stem cells
On Day 5–7, all of the blastocysts were used to derive the hESC lines. The zona pellucida of the expanded blastocyst was mechanically removed. The ICMs of the blastocysts were isolated by a mechanical method[16], and the isolated ICMs were then placed on mitomycin C-treated MEF feeder layer, which was prepared as previous described [17], for further culture.
After the ICMs were seeded on the feeder layer with hESC medium [14], the formation of the dome structure was examined every other day. After 8–14 days of culture, the ICMs were then mechanically divided into 2–3 small clumps using a small pipette, and the ICM clumps were transferred to a freshly prepared feeder layer. These cells were again mechanically dissociated during the initial five passages. After five passages, the cells were incubated in 1 mg/ml collagenase IV (Invitrogen) for 20–25 min at 37 C before further culture on freshly prepared feeders. The cells were routinely passaged every 4–5 days, and the medium was changed every day.
Karyotype analysis
For the karyotype analysis, ESCs at passages 10, 20 and 30 were incubated in the culture medium with 0.25 mg/ml colcemid for 4 h, with 0.4 % sodium citrate and 0.4 % chloratum Kaliumat (1:1, v/v) at 37 C for 5 min, and were then fixed in methanol: acetic acid (3:1, v/v) solution. After Giemsa staining, at least 20 cells were examined in each group for the karyotype analysis.
Staining for ESC markers
Human ESC marker staining was performed after 20 passages as described previously [18]. To detect the alkaline phosphatase (AP) activity, the ESC colonies were fixed with PFA for 20 min, washed three times with PBS, and then stained with BCIP/NBT (AP substrate solution, Maxim Biotech Inc., USA) for 30 min. To detect the hESC stage-specific embryonic markers, the ESCs were fixed with 4 % PFA for 30 min and then incubated with 4 % goat serum for 1 h before ESC marker staining. The primary antibodies were the stage-specific embryonic antigens, (SSEA)-4, SSEA-1, tumor-rejection (TRA)-1-81 antigen and TRA-1-60 (Chemicon, Temecula, CA, USA). All of the antibodies were diluted 1:50 with PBS, and the cells were incubated with antibody solution at room temperature for 1 h. The cells were washed three times with PBS for 10 min and then incubated with the secondary antibody (goat anti-rat IgM, goat anti-mouse IgG and goat anti-mouse IgM, both 1:100 dilution, Santa Cruz) conjugated to fluorescein isothiocyanate for 30 min. Negative controls were carried out without the addition of the primary antibodies. The nuclei were stained with DAPI, and the cells were then washed again and examined under a confocal microscope.
Expression of molecular markers in hESC lines
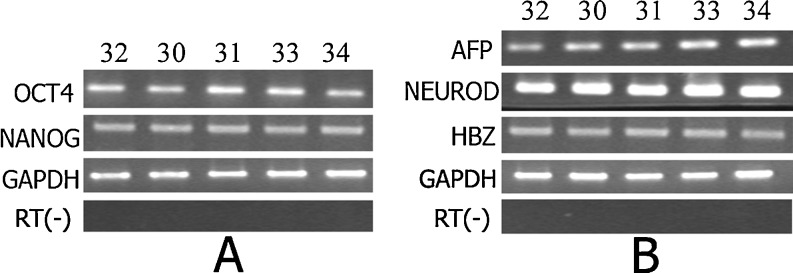
Total RNA was extracted from cells using the Trizol Kit (Invitrogen), and was transcribed into cDNA using oligo (dT) and ReverTra Ace reverse transcriptase (Toyobo). PCR reactions were carried out by mixing 1 μl of cDNA template, 250 nM of each primer, 200 M dNTP mixture and 1U of Taq DNA polymerase in a volume of 20 μl. Samples were amplified in a thermocycler. Pluripotency molecular marker genes, including OCT4 and NANOG, and marker genes for differentiated HESC, such as AFP (endoderm), NEUROD1 (ectoderm) and HBZ (mesoderm), were analyzed (Table 1). The relative expression levels of the detected genes from these cells were estimated visually by comparing relative band intensities with the expression level of housekeeping gene, GAPDH.
Table 1.
The derivation efficiency of hESC lines from 3PN zygotes
| No. of embryos received from IVF lab | No.of blastocysts formation(% total) | No. of cell lines derived(derivation efficiency % blastocysts) | No. of cell lines derived(derivation efficiency % total) | |
|---|---|---|---|---|
| 2PN group | 139 | 30(21.58 %)a | 5(16.67 %) | 5(3.58 %) |
| 3PN group | 130 | 12(9.23 %)b | 4(33.33 %) | 4(3.08 %) |
abThe different superscripts in the same column indicate significantly difference (P < 0.05)
DNA fingerprinting
The total DNA was extracted using the Qiagen DNeasy Tissue Kit (Qiagen) according to the manufacturer’s instructions. The extracted DNA was amplified for 16 different genetic loci using the AmpFLSTR Identifiler kit (Applied Biosystems, USA), and capillary electrophoresis was performed using an automated ABI 3100 Genetic Analyzer (Applied Biosystems). The 16 short tandem repeat (STR) loci were D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818, amelogenin and FGA.
In vitro and in vivo differentiation of hESC
Embryoid bodies (EBs) were created as described [14]. For teratoma formation, 5 × 106 hESCs were injected into the inguinal groove of 6-week-old male severe combined immunodeficiency (SCID) mice. Twelve weeks later, the mice were euthanized, and the teratomas were removed, fixed in 4 % PFA and embedded in paraffin. The sections were prepared, stained with hematoxylin and eosin, and examined for the presence of tissues derived from the three germ layers.
Statistical analysis
The results were compared using the SPSS 11.0 software (SPSS, Chicago, IL, USA). Percentages which presented in Tables 1 and 2 were compared by contingency table analysis followed by chi-square. P values of <0.05 were considered to be significant.
Table 2.
Compare the derivation efficiency of hESC lines from 1PN or 3PN zygotes
| No. of embryos received from IVF lab | No.of blastocysts formation(% total) | No. of cell lines derived(derivation efficiency % blastocysts) | P VALUE | |
|---|---|---|---|---|
| 3PN group | 130 | 12(9.23 %) | 4(33.33 %)a | |
| Ref.10 | 1.000 | |||
| 1PN | 2 | 1(50 %)a | ||
| Ref.11 | ||||
| 3PN | 5 | 1(20 %)a | 1.000 | |
| 1PN | 9 | 1(11.11 %)a | 0.506 | |
| Ref.12 | ||||
| 3PN | 2 | 1(50 %)a | 1.000 |
aSuperscripts in the same line indicate no difference (P > 0.05)
Results
Blastocyst formation and derivation of the hESC lines
Using our modified medium for the blastocyst culture, 12 of the 130 tripronuclear zygotes developed into blastocysts. In the control group, 30 of 139 poor-quality embryos developed into blastocysts. These zygotes showed either typical blastocyst morphology or irregular blastocyst morphology, that is, large peripheral cells and an unclear ICM. The blastocyst formation in the 3PN group was notably lower than that in the control group (9.23 %: 21.58 %, P = 0.005).
In total, 12 blastocysts from the 3PN zygotes were used to isolate the ICMs. Of the 12 ICMs attached to the feeder layer, 7 started to grow out. Four permanent hESC lines were obtained (FY-hES-30, 31, 33, and 34). As controls, five hESC lines were produced (FY-hES-27 to 29, 32, and 35) from Day-3 poor-quality embryos. There was no difference in the derivation efficiency using the 3PN zygotes and the Day-3 poor-quality embryos (3.08 %: 3.58 %, P = 0.813) (Table 1).
According to the data in those publications[10–12], there is no difference in the derivation efficiency of hESCs from abnormal fertilized zygotes between our lab and other labs (Table 2).
Characterization of hESC lines
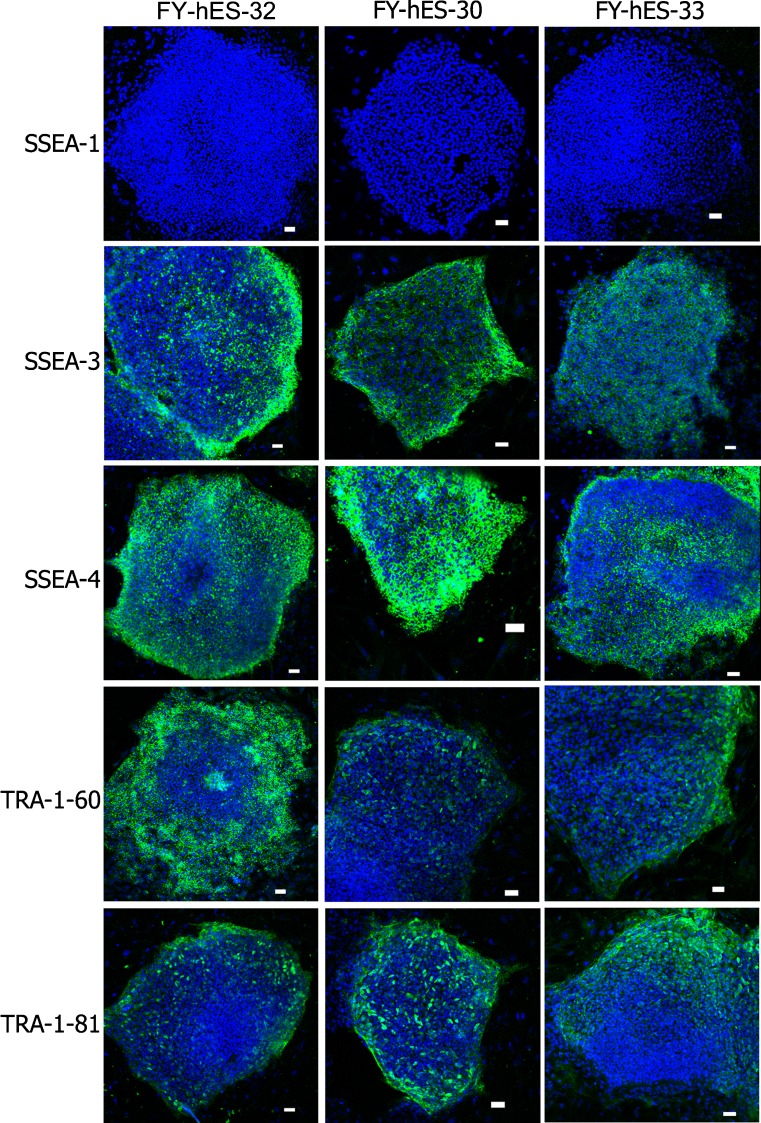
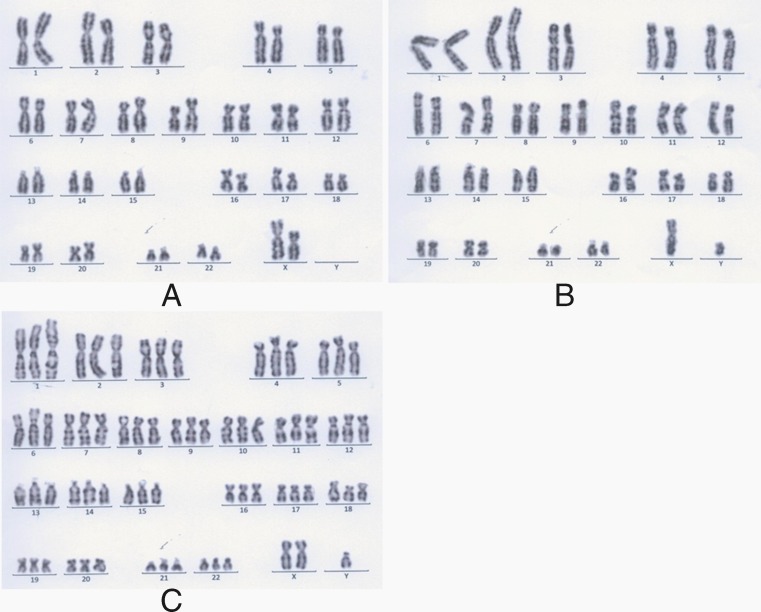
All nine of the hESC lines exhibited AKP (Fig. 1) activity, and were positive for the SSEA-3,SSEA-4, TRA-1-60, TRA-1-81 marker, but not SSEA-1 (Fig. 2). All of the karyotypes of the five hESC lines from the Day-3 poor-quality embryos were normal 46, XX (Figs. 3A and 4A). The karyotypes of three of the four hESC lines from 3PN zygotes were normal 46, XY (FY-hES-30, 31, and 34) (Figs. 3B and 4B). The karyotype of FY-hES-33 was 69, XXY (Fig. 3C and 4C). OCT4 and Nanog, which are specific marker genes for undifferentiated hESCs, were expressed in the cell lines (Fig. 5A).
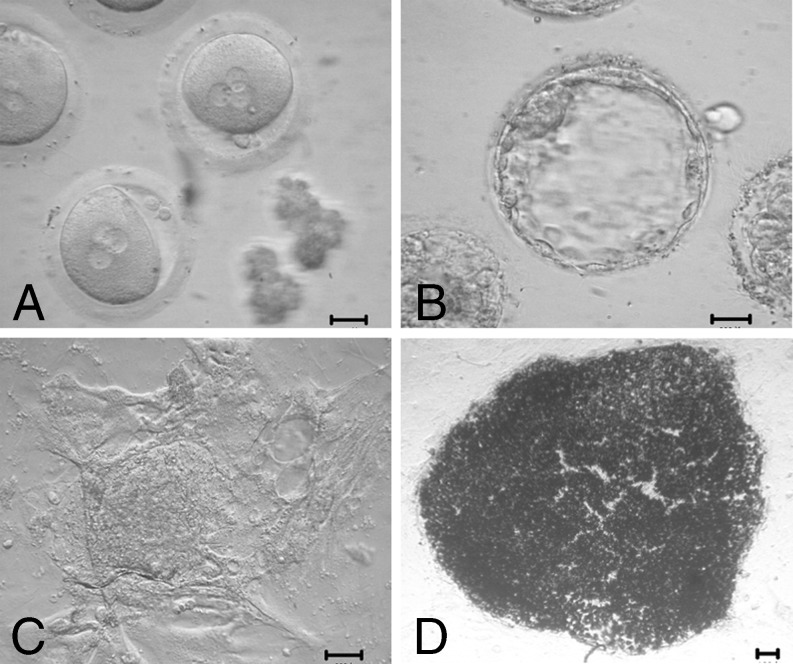
Fig. 1.
Derivation of an hESC line from 3PN zygotes. a 3PN zygotes. b Blastocysts developed from 3PN zygotes. c Outgrowth of FY-hES-30. d AKP staining of FY-hES-30. Scale bar = 50 μm
Fig. 2.
Immunostaining analysis of hESC lines. FY-hES-32, FY-hES-30 and FY-hES-33 with anti-SSEA-1 (green), anti-SSEA-3 (green), anti-SSEA-4 (green), anti-TRA-1-60 (green), anti-TRA-1-81 (green). Nuclei are stained blue with DAPI. Scale bar = 50 μm
Fig. 3.
Karyotype analysis of hESC lines by G-banding. a Normal female karyotype of FY-hES-32, 46 XX. b Normal male karyotype of FY-hES-30, 46 XY. c Triploid male karyotype of FY-hES-33, 69 XXY
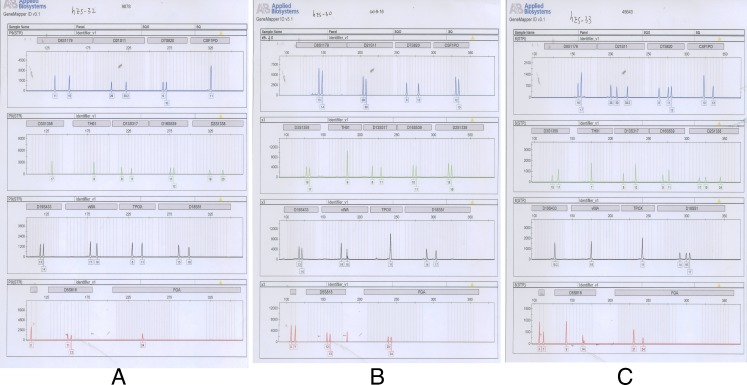
Fig. 4.
Distinct short tandem repeat (STR) profiles of hESC lines. The isogenic analysis of four blue peaks (from left to right): loci D8S1179(8), D21S11(21q11.2–q21), D7S820(7q11.21–22), CSF1PO(5q33.3–34); five green peaks: D3S1358(3p), TH01(11p15.5), D13S317 (13q22–31), D16S539(16q24-qter), D2S1338 (2q35–37.1); four black peaks: D19S433(19q12–13.1), vWA(12p12-pter), TPOX(2p23–2per), D18S51(18q21.3); three red peaks: Amelogenin (Xp22.1–22.3 Yp11.2), D5S818(5q21–23), FGA (4q28). These peaks represent locations of polymorphisms for each STR marker. Each single peak, double peaks or triple peaks of one locus shows how the STR is expressed at this locus. According to the different distances from the same locus to the markers, we can distinguish one cell line from another easily by its own genetic labels. a Normal female STR profile of FY-hES-32. b Normal male STR profile of FY-hES-30. c Triploid male STR profile of FY-hES-33
Fig. 5.
The expression of pluripotency gene and Differentiated marker gene. a Pluripotency gene expression of FY-hES-30, 31, 33, and 34 based on RT-PCR using FY-hES-32 expression as a control. b RT-PCR analysis of the expression of genes representative of the three germ layers of FY-hES-30, 31, 33, and 34 differentiated cells in vitro
In vitro and in vivo differentiation potential of the hESC line
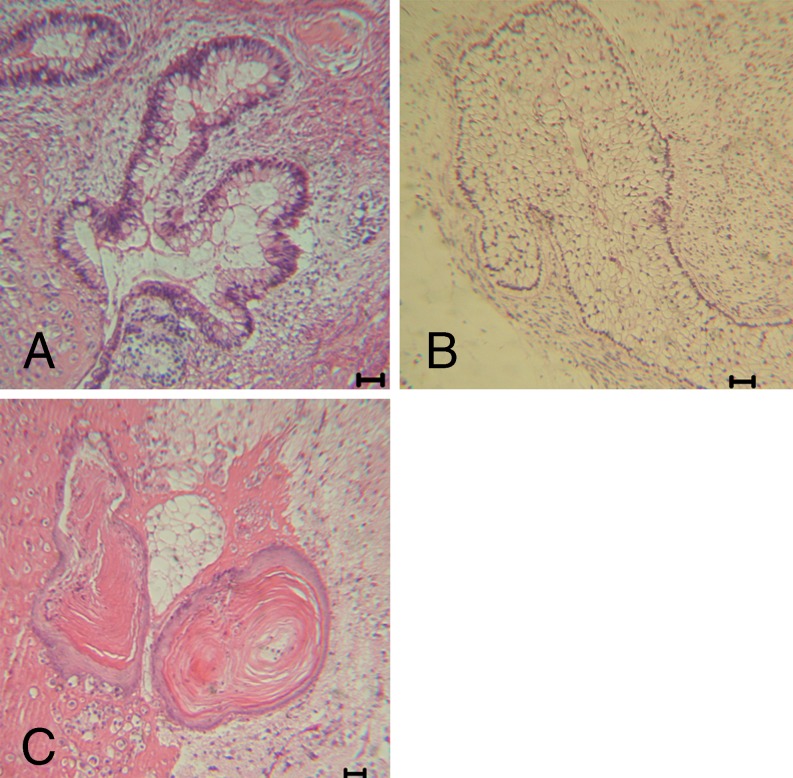
EBs were formed from each hESC line. Differentiated hESC marker genes, such as AFP (endoderm), NEUROD1 (ectoderm) and HBZ (mesoderm), were expressed in the hESC cell lines (Fig. 5B). The results demonstrate that these cells can differentiate into three germ layers in vitro. Teratomas were first observed 4–5 weeks after the hESCs were injected into the SCID mice. A histological examination revealed that the teratomas contained the tissues of three embryonic germ layers, including the endoderm (glandular tissue), mesoderm (fat cells) and ectoderm (squamous cells)(Fig. 6).
Fig. 6.
Immunohistological analysis of teratomas resulting from in vivo differentiation assay using hESC lines derived from 3PN zygotes. a Glandular tissue (endoderm). b Fat cells (mesoderm). c Squamous cells (ectoderm). Scale bar = 50 μm
Discussion
hESCs possess a huge potential for modern regenerative medicine. To use hESCs in cell-based therapies, it is necessary to establish many cell lines, as each line may have its own characteristics and advantages for different applications. Furthermore, to overcome the immune rejection of hESCs after transplantation, it was proposed to establish a stem cell bank that contains hundreds of thousands of immunophenotyped stem cell lines to cover the vast spectrum of transplant antigens and to avoid rejection [19, 20]. Although recent reports suggest that hESCs are derived more efficiently from high-quality embryos, it is difficult to obtain good-quality human embryos for hESC derivation [21, 22]. However, the usage of embryos and derivation of hESCs for research has resulted in polarized ethical debates with most of the controversy centered on embryo destruction[6, 23]. Therefore, these abnormal fertilized zygotes, which are destined to be discarded from IVF cycles, are alternative resources for hESC production. This option could be of particular interst in countries where the use of normal embryos or the creation of human embryos for stem cell research is not allow (i.e., Germany,Italy). In our IVF lab, ~50 % of the couples having abnormal fertilized zygotes agreed to donate them for derivation of hESC lines, a percentage that is much higher than the percentage (~20 %) of couples agreeing to donate euploid spare embryos. A few groups have derived diploid hESC lines from both a mononuclear zygote [10] and tripronuclear zygotes [11, 12]. However, it is necessary to investigate the efficiency and the ratio of deriving abnormal and normal hESC lines from tripronuclear zygotes to know more about the developmental potential of abnormal zygotes. In the present study, we collected 130 tripronuclear zygotes that were donated by patients undergoing conventional in vitro fertilization (IVF); four hESC lines were established, one of which was triploid, and the others were diploid.
An important step in the establishment of a hESC line is to culture the human embryo until it forms a good-quality blastocyst. Therefore, securing high-quality embryos and improving embryo culture conditions are important for improving the efficiency of the establishment of the hESCs. Most human dispermic preimplantation embryos show an apparent normal development in vitro until the first 2 or 3 cell cycles. Thereafter, they exhibit retarded, arrested or abnormal development, as revealed by blastomeres of unequal size, cellular fragmentation, multinucleation, and spindle defects [24, 25]. This may be the reason that only approximately 10 % of the 3PN zygotes reached the blastocyst stage [24, 25]. In our previous study, we had utilized a modified culture medium to increase the blastocyst formation and the efficiency of the hESC derivation [14]. Therefore, we cultured those human 3PN zygotes in sequential G1.5/2.5 media in combination with bFGF and LIF to obtain high-quality blastocysts. In the present study, we cultured 130 human 3PN zygotes using our modified culture medium, which resulted in 12 blastocysts (9.23 %), from which four cell lines were derived (33.33 %). The blastocyst formation in the 3PN group was notably lower than that in the control group (9.23 %: 21.58 %, P = 0.005). However, there was no difference in the derivation efficiency using the 3PN zygotes and day-3 poor-quality embryos (3.08 %: 3.58 %, P = 0.813). Our results suggested that in the modified culture medium the developmental potential of the 3PN zygotes was lower than that of control group, but the efficiency of the hESC derivation was similar in the two group. According to the data in those publications[10–12], there is also no difference in the derivation efficiency of hESCs from mononuclear and tripronuclear zygotes between our lab and other labs (Table 2).
Because the 3PN human zygotes used in this study were obtained through conventional IVF and second polar bodies were identified, the additional pronucleus observed in these 3PN zygotes arose from supernumerary sperm that penetrated the oocyte [26]. Kola and Trounson have found that during the first mitotic division of human dispermic zygotes, 62 %, 24 %, and 14 % of 3PN human zygotes cleave directly into 3-cells, 2-cells, and 2-cells plus an extrusion, respectively [27]. The karyotype of 3-cells produced directly from zygotic cleavage is severely abnormal because of randomly distribution of the chromosomes. Zygotes that cleave into 2-cells seem to form a bipolar spindle, and the karyotype of the resulting 2-cell embryo is triploid. As for the third pattern of division, the 2 cells have a diploid number of chromosomes whereas the extrusion is haploid. Further evidence showing that preimplantation embryos derived from dispermic 3PN zygotes have variable chromosome patterns has been provided by Rosenbush et al. [25, 28].
In our study, 3 out of the 4 hESC lines (75 %) were karyotypically normal. The data suggested the following: (1) the diploid hESC lines originated from the tripronucear zygotes that arose from the third pattern of division; and (2) the in vitro selection favored diploid cells. This is in accordance with previously reported results [11, 12, 29]. The present report is, we believe, the first record of deriving normal and abnormal hESC lines from 3PN zygotes and comparing the characteristics of these two kinds of hESC lines. Because the embryos were from multiple donors and were subjected to hESC derivation at various time points, the influence of any differences in the genetic background and variations in the culture conditions may play a role.
In summary, the embryos that we used for the derivation of new hESC lines were identified as tripronuclear and had been destined to be discarded. Surprisingly, three out of the four hESC lines were found to carry a normal euploid karyotype (Fig. 3). These results led us to conclude that these abnormal zygotes, which otherwise would have been discarded, can serve as an alternative source for normal euploid hESC lines. Most couples, who had abnormal fertilized zygotes agreed to donate them for the derivation of hESCs. This new source of zygotes, together with both arrested and poor-quality embryos, expands the pool of embryos that can be used for the derivation of euploid hESC lines [15, 16].
Footnotes
Xinjie Chen, Yumei Luo, Yong Fan contributed equally to the work.
Capsule
Our results of this study show that these tripronuclear zygotes, which otherwise would have been discarded, can serve as an alternative source for normal euploid hESC lines.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Cowan CA, Klimanskaya I, McMahon J, Atienza J, Witmyer J, Zucker JP, Wang S, Morton CC, McMahon AP, Powers D, Melton DA. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–6. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 3.Chen AE, Egli D, Niakan K, Deng J, Akutsu H, Yamaki M, Cowan C, Fitz-Gerald C, Zhang K, Melton DA, Eggan K. Optimal timing of inner cell mass isolation increases the efficiency of human embryonic stem cell derivation and allows generation of sibling cell lines. Cell Stem Cell. 2009;4:103–6. doi: 10.1016/j.stem.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inamdar MS, Venu P, Srinivas MS, Rao K, VijayRaghavan K. Derivation and characterization of two sibling human embryonic stem cell lines from discarded grade III embryos. Stem Cells Dev. 2009;18:423–33. doi: 10.1089/scd.2008.0131. [DOI] [PubMed] [Google Scholar]
- 5.Lerou PH, Yabuuchi A, Huo H, Takeuchi A, Shea J, Cimini T, Ince TA, Ginsburg E, Racowsky C, Daley GQ. Human embryonic stem cell derivation from poor-quality embryos. Nat Biotechnol. 2008;26:212–4. doi: 10.1038/nbt1378. [DOI] [PubMed] [Google Scholar]
- 6.Landry DW, Zucker HA. Embryonic death and the creation of human embryonic stem cells. J Clin Invest. 2004;114:1184–6. doi: 10.1172/JCI23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho PC, Yeung WS, Chan YF, So WW, Chan ST. Factors affecting the incidence of polyploidy in a human in vitro fertilization program. Int J Fertil Menopausal Stud. 1994;39:14–9. [PubMed] [Google Scholar]
- 8.Matt DW, Ingram AR, Graff DP, Edelstein MC. Normal birth after single-embryo transfer in a patient with excessive polypronuclear zygote formation following in vitro fertilization and intracytoplasmic sperm injection. Fertil Steril. 2004;82:1662–5. doi: 10.1016/j.fertnstert.2004.04.069. [DOI] [PubMed] [Google Scholar]
- 9.Feenan K, Herbert M. Can ‘abnormally’ fertilized zygotes give rise to viable embryos? Hum Fertil (Camb). 2006;9:157–69. doi: 10.1080/14647270600636269. [DOI] [PubMed] [Google Scholar]
- 10.Suss-Toby E, Gerecht-Nir S, Amit M, Manor D, Itskovitz-Eldor J. Derivation of a diploid human embryonic stem cell line from a mononuclear zygote. Hum Reprod. 2004;19:670–5. doi: 10.1093/humrep/deh135. [DOI] [PubMed] [Google Scholar]
- 11.Huan Q, Gao X, Wang Y, Shen Y, Ma W, Chen ZJ. Comparative evaluation of human embryonic stem cell lines derived from zygotes with normal and abnormal pronuclei. Dev Dyn. 2010;239:425–38. doi: 10.1002/dvdy.22175. [DOI] [PubMed] [Google Scholar]
- 12.Strom S, Rodriguez-Wallberg K, Holm F, Bergstrom R, Eklund L, Stromberg AM, Hovatta O. No relationship between embryo morphology and successful derivation of human embryonic stem cell lines. PLoS One. 2010;5:e15329. doi: 10.1371/journal.pone.0015329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steer CV, Mills CL, Tan SL, Campbell S, Edwards RG. The cumulative embryo score: a predictive embryo scoring technique to select the optimal number of embryos to transfer in an in-vitro fertilization and embryo transfer programme. Hum Reprod. 1992;7:117–9. doi: 10.1093/oxfordjournals.humrep.a137542. [DOI] [PubMed] [Google Scholar]
- 14.Fan Y, Luo Y, Chen X, Sun X. A modified culture medium increases blastocyst formation and the efficiency of human embryonic stem cell derivation from poor-quality embryos. J Reprod Dev. 2010;56:533–9. doi: 10.1262/jrd.09-225M. [DOI] [PubMed] [Google Scholar]
- 15.Gardner DK, Lane M, Schoolcraft WB. Culture and transfer of viable blastocysts: a feasible proposition for human IVF. Hum Reprod. 2000;15(Suppl 6):9–23. [PubMed] [Google Scholar]
- 16.Liu W, Yin Y, Long X, Luo Y, Jiang Y, Zhang W, Du H, Li S, Zheng Y, Li Q, Chen X, Liao B, Xiao G, Wang W, Sun X. Derivation and characterization of human embryonic stem cell lines from poor quality embryos. J Genet Genomics. 2009;36:229–39. doi: 10.1016/S1673-8527(08)60110-1. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Ma W, Hou Y, Sun XF, Sun QY, Wang WH. Improved isolation and culture of embryonic stem cells from Chinese miniature pig. J Reprod Dev. 2004;50:237–44. doi: 10.1262/jrd.50.237. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, Long X, Yin Y, Jiang Y, Chen X, Liu W, Zhang W, Du H, Li S, Zheng Y, Kong S, Pang Q, Shi Y, Huang Y, Huang S, Liao B, Xiao G, Wang W. Similar biological characteristics of human embryonic stem cell lines with normal and abnormal karyotypes. Hum Reprod. 2008;23:2185–93. doi: 10.1093/humrep/den137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor CJ, Bolton EM, Pocock S, Sharples LD, Pedersen RA, Bradley JA. Banking on human embryonic stem cells: estimating the number of donor cell lines needed for HLA matching. Lancet. 2005;366:2019–25. doi: 10.1016/S0140-6736(05)67813-0. [DOI] [PubMed] [Google Scholar]
- 20.Baharvand H, Ashtiani SK, Taee A, Massumi M, Valojerdi MR, Yazdi PE, Moradi SZ, Farrokhi A. Generation of new human embryonic stem cell lines with diploid and triploid karyotypes. Dev Growth Differ. 2006;48:117–28. doi: 10.1111/j.1440-169X.2006.00851.x. [DOI] [PubMed] [Google Scholar]
- 21.Oh SK, Kim HS, Ahn HJ, Seol HW, Kim YY, Park YB, Yoon CJ, Kim DW, Kim SH, Moon SY. Derivation and characterization of new human embryonic stem cell lines: SNUhES1, SNUhES2, and SNUhES3. Stem Cells. 2005;23:211–9. doi: 10.1634/stemcells.2004-0122. [DOI] [PubMed] [Google Scholar]
- 22.Simon C, Escobedo C, Valbuena D, Genbacev O, Galan A, Krtolica A, Asensi A, Sanchez E, Esplugues J, Fisher S, Pellicer A. First derivation in Spain of human embryonic stem cell lines: use of long-term cryopreserved embryos and animal-free conditions. Fertil Steril. 2005;83:246–9. doi: 10.1016/j.fertnstert.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Zacharias DG, Nelson TJ, Mueller PS, Hook CC. The science and ethics of induced pluripotency: what will become of embryonic stem cells? Mayo Clin Proc. 2011;86:634–40. doi: 10.4065/mcp.2011.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sathananthan AH, Tarin JJ, Gianaroli L, Ng SC, Dharmawardena V, Magli MC, Fernando R, Trounson AO. Development of the human dispermic embryo. Hum Reprod Update. 1999;5:553–60. doi: 10.1093/humupd/5.5.553. [DOI] [PubMed] [Google Scholar]
- 25.Tarin JJ, Trounson AO, Sathananthan H. Origin and ploidy of multipronuclear zygotes. Reprod Fertil Dev. 1999;11:273–9. doi: 10.1071/RD99057. [DOI] [PubMed] [Google Scholar]
- 26.Rosenbusch BE. Mechanisms giving rise to triploid zygotes during assisted reproduction. Fertil Steril. 2008;90:49–55. doi: 10.1016/j.fertnstert.2007.06.031. [DOI] [PubMed] [Google Scholar]
- 27.Kola I, Trounson A, Dawson G, Rogers P. Tripronuclear human oocytes: altered cleavage patterns and subsequent karyotypic analysis of embryos. Biol Reprod. 1987;37:395–401. doi: 10.1095/biolreprod37.2.395. [DOI] [PubMed] [Google Scholar]
- 28.Rosenbusch B, Schneider M, Sterzik K. The chromosomal constitution of multipronuclear zygotes resulting from in-vitro fertilization. Hum Reprod. 1997;12:2257–62. doi: 10.1093/humrep/12.10.2257. [DOI] [PubMed] [Google Scholar]
- 29.Peura T, Bosman A, Chami O, Jansen RP, Texlova K, Stojanov T. Karyotypically normal and abnormal human embryonic stem cell lines derived from PGD-analyzed embryos. Cloning Stem Cells. 2008;10:203–16. doi: 10.1089/clo.2007.0062. [DOI] [PubMed] [Google Scholar]