Abstract
Functional impairment of DNA damage response pathways leads to increased genomic instability. Here we describe the centrosomal protein CEP152 as a new regulator of genomic integrity and cellular response to DNA damage. Using homozygosity mapping and exome sequencing, we identified CEP152 mutations in Seckel syndrome and showed that impaired CEP152 function leads to accumulation of genomic defects resulting from replicative stress through enhanced activation of ATM signaling and increased H2AX phosphorylation.
Maintenance of genomic integrity is required for cell survival and function. As the accumulation of DNA damage has an important effect on cell viability, organisms have evolved mechanisms to protect the integrity of DNA by inducing DNA damage responses and cell cycle arrest or by triggering apoptosis1. Disruption of these mechanisms can lead to increased genomic instability, which is a key factor for the development of cancer and is involved in aging. Two important regulators of DNA-damage responses are the phosphoinositide 3-kinase–related serine/threonine kinases ATM (Ataxia-Telangiectasia Mutated) and ATR (Ataxia-Telangiectasia and Rad3 related)1,2. Whereas ATM is activated in response to DNA double-strand breaks, ATR is involved mainly in responses to single-stranded DNA induced by ultraviolet light damage or replication arrest. ATM and ATR both trigger an overlapping set of cellular responses that promote cell cycle arrest and DNA repair3.
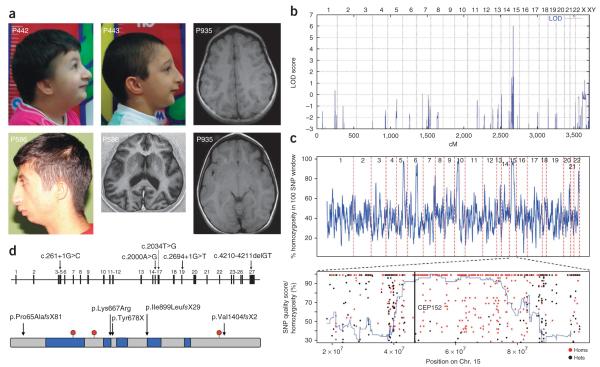
Recently, it was reported that a hypomorphic ATR mutation is associated with embryonic replicative stress, accelerated aging in mice and Seckel syndrome in humans4,5. Seckel syndrome (MIM210600) is a heterogeneous autosomal recessive disorder characterized by a proportionate short stature, severe microcephaly and mental retardation, and a typical ‘bird-head’ facial appearance6. Initially, we clinically evaluated five consanguineous families with Seckel syndrome originating from an isolated rural area in Turkey (Fig. 1, Supplementary Figs. 1 and 2 and Supplementary Table 1). We genotyped DNA from four affected members of three families using the 250K SNP Array (Affymetrix) and obtained a single maximum log10 odds (LOD) score of 6.03 for a region between rs1598206 and rs2330591 on chromosome 15q21.1–q21.2 (Fig. 1b). Subsequent fine mapping using microsatellite markers confirmed shared homozygosity and a founder haplotype in a 3.4-Mb region between markers D15S123 and D15S1017. This genomic region contains 28 known and predicted genes. Microcephalic osteodysplastic primordial dwarfism type II (MOPDII, MIM210720), which shows overlapping features with Seckel syndrome, was recently associated with mutations in PCNT, the centrosomal pericentrin gene7,8. Hence, we considered CEP152, the centrosomal protein 152 gene located in the critical region, to be a highly relevant candidate gene.
Figure 1.
Clinical and molecular characterization of CEP152 Seckel subjects. (a) Clinical characteristics of subjects 442, 443 and 586 presenting with microcephaly, sloping forehead, high nasal bridge, beaked nose and retrognathia. Informed consents to publish the photographs were obtained from the subjects’ parents. Cranial magnetic resonance imaging of subjects 586 and 935 showing simplified gyri. (b) Genome-wide graphical view of LOD scores using SNP array homozygosity mapping in four affected subjects, 442, 443, 586 and 633, indicated significant linkage to chromosome 15q21.1–q21.2. (c) Above, homozygosity (blue line) was measured as the percent of homozygous sites within a sliding window of 100 variant sites, relative to the reference genome, obtained from the exome sequencing data. CEP152 is located on chromosome 15, which harbors one of the longest stretches of homozygosity in this genome. Below, the chromosomal locations of all single nucleotide variants called on chromosome 15 are plotted against the genotype quality for that variant. Homozygous variants are plotted in red, and heterozygous variants are plotted in black. Homozygosity (blue line) is measured as the fraction of homozygous sites within a sliding window of 50 variant sites (relative to the reference genome) called from the exome sequencing data. (d) Above, the genomic structure of human CEP152. The position of each mutation is shown on the coding DNA level. Below, the protein structure of CEP152 with predicted coiled-coil domains (blue boxes) and Thr/Ser-phosphorylation sites (red). The position and the predicted effects of the mutations on CEP152 are marked by arrows.
Sequencing of all 27 exons of CEP152 (Supplementary Table 4) revealed a homozygous splice donor-site mutation in intron 4, c.261+1G>C, which co-segregated with the founder haplotype and the disease in all affected family members (Supplementary Fig. 3) and was not found in 250 healthy Turkish control individuals. The c.261+1G>C mutation completely disrupted the splice donor site, as shown through RT-PCR analysis of RNA from affected individuals. We found four different aberrant transcripts likely to cause loss of protein function though partial functional activity of one mutant protein, Val86_Asn87del, could not be excluded (Supplementary Fig. 4).
Independently, we identified CEP152 as the causative gene in a French individual with Seckel syndrome of Turkish origin born to consanguineous parents through the use of an exome sequencing strategy (Supplementary Methods). We identified seven new and homozygous nonsense and essential splice-site variants that were expected to have a severe impact on gene function (Supplementary Table 2). Of these seven variants, a G>C transversion at the +1 position of a donor site was embedded within a ~35-Mb tract of homozygosity on chromosome 15, which was the longest tract of homozygosity observed on any autosome in this individual (Fig. 1c). None of the other variants were associated with a large region of homozygosity. This mutation is the same c.261+1G>C Turkish founder mutation in CEP152 described above. These data demonstrated that mutation identification through exome sequencing, in conjunction with simultaneous analysis of homozygous stretches, can be used as an efficient approach to gene identification in autosomal recessive disorders.
In addition, sequencing CEP152 in further subjects with Seckel syndrome identified two individuals that were compound heterozygous for likely loss-of-function mutations. One subject of Italian origin from Germany was compound heterozygous for a nonsense mutation, c.2034T>G (p.Tyr678X), and an intron 19-splice donor-site mutation, c.2694+1G>T, leading to retention of the entire intron 19 in the CEP152 mRNA (r.2694G_ins3581, Ile899LeufsX29) (Supplementary Fig. 5). Because the phenotype of this subject was as severe as the phenotypes seen in the Turkish families, it is likely that the c.261+1G>C Turkish founder mutation has similar functional consequences as the mutation seen here. A subject from South Africa was compound heterozygous for a paternally inherited 2-bp deletion, c.4210–4211delGT (p.Val1404fsX2, exon 27), and a maternally inherited missense mutation, c.2000A>G (exon 15), affecting the highly conserved lysine at position 667 (p.Lys667Arg) (Fig. 1d and Supplementary Fig. 3). Very recently, mutations in CEP152 have been associated with microcephaly in an Eastern Canadian subpopulation9.
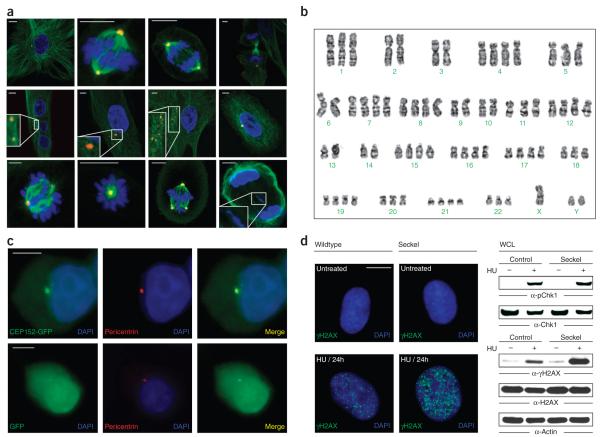
CEP152 encodes a 1,654 amino acid protein that was originally identified in a proteomic screen of human centrosomes10. Analysis of the subcellular localization of asl (asterless), the Drosophila ortholog of CEP152, has shown it to be associated with the periphery of centrioles, where it is involved in the initiation of centriole duplication11. Expression of CEP152 in HEK293T cells revealed fluorescence staining of the centrosomes, where it co-localized with pericentrin (Fig. 2)12. To determine the functional effect of CEP152 deficiency on genomic stability, we analyzed the morphology of CEP152-deficient Seckel fibroblasts during interphase and during different stages of mitosis. A substantial number of Seckel fibroblasts contained multiple, differently sized nuclei and centrosomes, micronuclei and fragmented centrosomes during interphase (Fig. 2a and Supplementary Fig. 6). Metaphase karyotyping of CEP152-deficient Seckel lymphocytes showed aneuploidy in 15 out of a total 109 metaphase spreads (Fig. 2b). During metaphase, we observed incorrectly aligned chromosomes, monopolar spindles with a single large centrosome, triple spindles with differently sized and structurally compromised centrosomes, and prematurely separated sister chromatids in Seckel cells. Statistical analysis revealed that CEP152 deficiency leads to an increased number of cells containing multiple nuclei and centrosomes, fragmented centrosomes and an increased frequency of aberrant cell divisions.
Figure 2.
Characterization of CEP152 Seckel cells. (a) Mitotic morphology of CEP152 Seckel fibroblasts. Immunofluorescence staining of CEP152 Seckel fibroblasts carrying the c.261+1G>C mutation with antibodies against α-tubulin (green), pericentrin (red) and DAPI staining of DNA (blue). Above, from left to right, control fibroblasts showing normal mitotic morphology of interphase, metaphase, anaphase and telophase. Middle, Seckel interphase cells containing three equally sized nuclei and two centrosomes without astral microtubules (inset, tenfold magnification), two unseparated centrosomes per nucleus without asters (inset, threefold magnification), fragmented centrosomes without asters and micronuclei (inset, twofold magnification), and partially depolymerized microtubules together with micronuclei in addition to a main nucleus. Below, abnormal Seckel metaphases showing incorrectly aligned chromosomes on the metaphase plate, a monopolar spindle with a large centrosome and reduced spindle, a tripolar spindle with differently sized and structurally compromised centrosomes, and an abnormal Seckel telophase showing defects in cytokinesis (inset, twofold magnification). Scale bars, 5 μm. (b) Aneuploid metaphase karyotype of a CEP152 Seckel lymphocyte. (c) Centrosomal localization of wildtype CEP152 in HEK293T cells expressing either GFP-tagged wildtype CEP152 (above) or GFP as a control (below). Additional staining was with pericentrin (red) and DAPI (blue). (d) DNA-damage response in wildtype and Seckel fibroblasts. H2AX phosphorylation of wildtype and CEP152 Seckel primary fibroblasts after treatment with hydroxyurea (HU) (left). Protein blot analysis of HU-induced phosphorylation of CHK1 (Ser345) and H2AX (Ser139) (right). Equal protein loading was confirmed by re-probing of the membranes with antibodies against CHK1 or H2AX and actin antibodies.
Moreover, the number of CEP152-deficient cells in telophase was decreased. Most strikingly, CEP152-deficient cells appeared to be arrested at early anaphase, reflected by an increased number of early anaphase figures in fixed Seckel populations when compared to the wildtype population (Supplementary Fig. 6f). This block may have resulted from problems with chromatid alignment, uneven pulling forces in the spindle, or activation of a checkpoint that responds to weakly attached or misaligned chromosomes. There did not appear to be additional apoptosis at early anaphase. Taken together, these data clearly showed centrosomal and mitotic aberrations caused by CEP152 mutations.
In addition, Seckel cells showed an overall increased sensitivity to oxidative stress and responded to this stress with increased apoptosis (Supplementary Fig. 7). Cell cycle analysis in CEP152 knockdown cells using short hairpin RNA suggested that CEP152 deficiency delays S-phase entry. Furthermore, fewer Seckel cells progressed to the G2/M phase and an increased proportion of Seckel cells stayed in G0/G1. These findings suggest altered ATR-mediated checkpoint activity and increased replicative stress in CEP152-deficient cells.
In order to determine possible defects in DNA repair mechanisms, we measured the mitomycin-C–induced sister chromatid exchange frequency. We observed a substantial increase in chromosome instability in CEP152-deficient lymphocytes (Supplementary Table 3). In a yeast two-hybrid (Y2H) screen using an N-terminal deletion construct of CEP152 as bait, the CDK2-interacting protein (CINP) was identified as an interaction partner of CEP152 in three independent hits. We confirmed the CEP152-CINP interaction by using complementary immunoprecipitation approaches, and we showed that CEP152 constitutively binds to CINP (Supplementary Fig. 8). We observed a weak nuclear staining for both CEP152 and CINP (data not shown). CINP has recently been identified as a genome maintenance protein13 that interacts with the ATR-interacting protein (ATRIP) and has an important function in ATR-mediated checkpoint signaling. Silencing of CINP causes increased γH2AX foci formation, suggesting that CINP contributes to the formation of a functional unit in DNA damage response. The identification of CEP152 as a binding partner of CINP provides additional evidence for an important role of CEP152 in DNA-damage response and genome maintenance.
ATR-mediated phosphorylation of CHK1 and H2AX are the initial steps in response to DNA damage and increased replicative stress14,15. We did not observe a detectable alteration in hydroxyurea-induced phosphorylation of CHK1 in CEP152-deficient cells (Fig. 2d). However, we found that replicative stress leads to enhanced CHK2 phosphorylation in Seckel cells (Supplementary Fig. 8), indicating an increased activation of ATM signaling pathways. Furthermore, we found that formation of hydroxyurea-induced γH2AX foci in CEP152-deficient cells was increased. Protein blot analysis confirmed that the levels of γH2AX were considerably higher in hydroxyurea-treated CEP152-deficient cells as compared to wildtype cells. There are likely to be additional mechanisms by which CEP152 might affect ATR checkpoint control and subsequently lead to increased replicative stress, which causes a compensatory activation of CHK2 and γH2AX. Recent analysis of H2AX phosphorylation in the ATRs/s Seckel mouse model indicated that γH2AX has been substantially increased throughout embryonic development, particularly in tissues with a high mitotic index5. In agreement with our results, a previous study suggested that accumulated replicative stress can activate ATM-dependent DNA damage response and thereby lead to increased H2AX phosphorylation5.
In conclusion, we identified the centrosomal protein CEP152 as a new protein involved in the maintenance of genomic integrity and in the ability to respond to DNA damage. Impaired CEP152 function leads to genomic instability and increased H2AX phosphorylation, a measure of accumulated replicative stress. Our data further support the essential link between genomic instability, as seen in individuals with Seckel syndrome, and the activation of histone H2AX as response to increased replicative stress.
Supplementary Material
ACKNOWLEDGMENTS
We thank all family members who participated in this study, E. Milz, A. Alver and A. Coffey for excellent technical assistance, F. Kokocinski and A. Palotie for experimental design, and C. Kubisch, B. Wirth and K. Boss for critical reading the manuscript. The CINP antibody was a kind gift from D. Cortez (Nashville, Tennessee, USA). This work was supported by the German Federal Ministry of Education and Research (BMBF) by grant numbers 01GM0880 (SKELNET) and 01GM0801 (E-RARE network CRANIRARE) to B.W., the Karadeniz Technical University Research Fund by grant numbers 2008.114.001.02 and 2008.114.001.12 to E.K., the Medical Research Council and Lister Institute to A.P.J. and by the Wellcome Trust, grant 077014/Z/05/Z.
Footnotes
Accession codes. The CEP152 reference sequence is deposited in the NCBI nucleotide database under the accession code NM_014985.2.
Note: Supplementary information is available on the Nature Genetics website.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Annu. Rev. Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 2.Cimprich KA, Cortez D. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonner WM, et al. Nat. Rev. Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. Nat. Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 5.Murga M, et al. Nat. Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majewski F, Goecke T. Am. J. Med. Genet. 1982;12:7–21. doi: 10.1002/ajmg.1320120103. [DOI] [PubMed] [Google Scholar]
- 7.Rauch A, et al. Science. 2008;319:816–819. doi: 10.1126/science.1151174. [DOI] [PubMed] [Google Scholar]
- 8.Griffith E, et al. Nat. Genet. 2008;40:232–236. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guernsey DL, et al. Am. J. Hum. Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andersen JS, et al. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 11.Blachon S, et al. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- 13.Lovejoy CA, et al. Proc. Natl. Acad. Sci. USA. 2009;106:19304–19309. doi: 10.1073/pnas.0909345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Q, et al. Genes Dev. 2000;70:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 15.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. Proc. Natl. Acad. Sci. USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.