Highlights
► Development of small cantilever. ► Speed increase by a factor of ten using small cantilevers on a commercial AFM. ► Force sensitivity increase by a factor of five using small cantilever prototypes for force spectroscopy measurements.
Keywords: Small cantilever, High resolution imaging, Fast AFM imaging, Ultra-sensitive molecular recognition force spectroscopy
Abstract
In this study, we demonstrate the increased performance in speed and sensitivity achieved by the use of small AFM cantilevers on a standard AFM system. For this, small rectangular silicon oxynitride cantilevers were utilized to arrive at faster atomic force microscopy (AFM) imaging times and more sensitive molecular recognition force spectroscopy (MRFS) experiments. The cantilevers we used had lengths between 13 and 46 μm, a width of about 11 μm, and a thickness between 150 and 600 nm. They were coated with chromium and gold on the backside for a better laser reflection. We characterized these small cantilevers through their frequency spectrum and with electron microscopy. Due to their small size and high resonance frequency we were able to increase the imaging speed by a factor of 10 without any loss in resolution for images from several μm scansize down to the nanometer scale. This was shown on bacterial surface layers (s-layer) with tapping mode under aqueous, near physiological conditions and on nuclear membranes in contact mode in ambient environment. In addition, we showed that single molecular forces can be measured with an up to 5 times higher force sensitivity in comparison to conventional cantilevers with similar spring constants.
1. Introduction
Since the atomic force microscope was the first time presented by Binnig et al. (1986), it has evolved into a powerful, widely used tool in nanoscience and nanotechnology. The applications span the range from material science and biology to quality control in semiconductor industry. Particularly important for biological sciences is that the AFM enables recording images under aqueous, near physiological conditions with nanometer (Kamruzzahan et al., 2004; Kienberger et al., 2005, 2004) or even sub-nanometer (Muller et al., 1995) resolution. Chemical functionalization (Ebner et al., 2007a,b; Riener et al., 2003) of the AFM tip allows the measurement of pico-Newton forces (Hinterdorfer et al., 1996; Tang et al., 2007; Wielert-Badt et al., 2002), as well as simultaneously topography and recognition imaging (Ebner et al., 2005; Leitner et al., 2011; Stroh et al., 2004; Tang et al., 2008b). Combination of AFM and optical (Madl et al., 2006) or fluorescence microscopy (Duman et al., 2010) has also been realized. A main – if not the main – limitation for a much wider use of this technique, e.g. industrial biotechnology, pharmacology, and medicine, is the long time (several minutes) that is required to take a high resolution AFM image. The foremost factors for this speed limitation in AFM imaging are the scanner, the feedback loop, and the cantilever. That it is possible to speed up the AFM has been demonstrated by several research groups since the early 1990s (Ando et al., 2001, 2008; Barrett and Quate, 1991; Fantner et al., 2010; Hillner et al., 1992; Hobbs et al., 2001; Humphris et al., 2005; Sulchek et al., 2000a; Viani et al., 2000). During this time a lot of progress has been made in the field, especially in the development of small and sensitive low noise cantilevers for imaging (Chand et al., 2000; Hosaka et al., 2000, 1999; Viani et al., 1999a; Walters et al., 1996; Yang et al., 2005), force spectroscopy (Hosaka et al., 2000; van der Mei et al., 2003; Viani et al., 1999a,b), and force mapping (Schaffer and Jiao, 2001), as well as in their detailed characterization (Schaffer and Hansma, 1998). Some groups also fabricated special cantilevers with integrated actuators and/or sensors (Kim et al., 2003; Manalis et al., 1996; Pedrak et al., 2003; Rogers et al., 2003) for faster AFM imaging. With respect to scanners (Fukuma et al., 2008; Kodera et al., 2005; Schitter et al., 2007), optimized feedback (Ying et al., 2008), controlling (Ando, 2008; Kodera et al., 2006; Schitter et al., 2004; Schitter and Stemmer, 2003; Sulchek et al., 2000b; Tien et al., 2004; Uchihashi et al., 2006), data acquisition (Fantner et al., 2005) or even complete high speed AFM system (Fantner et al., 2006; Humphris et al., 2005), a lot of effort has been undertaken, too. Nonetheless, most of these high speed AFM components are limited to a certain number of research laboratories, because the developed instruments are highly specialized.
Cantilevers which combine high resonance frequency, high bandwidth, and low spring constant have been developed here, in which the cantilever design was optimized for several applications to achieve optimal performance. For example, molecular recognition force spectroscopy requires a particular soft cantilever, whereas a cantilever for high speed imaging needs a high resonance frequency. With cantilever lengths between 13 and 46 μm, widths of about 10 μm, and thicknesses of 150–600 nm, we were able to speed up a commercially available AFM by a factor of 10 in both ambient and aqueous conditions. We also detected diminished thermal force fluctuations, when performing single molecular recognition force spectroscopy.
2. Materials and methods
2.1. Tip preparation
All used chemicals were purchased from Sigma Aldrich and were of highest quality.
AFM tip functionalization for molecular recognition force microscopy: two functionalization methods, the ethanolamine. HCL (Ebner et al., 2007a) and the gas phase silanization using aminopropyltriethoxysilane (APTES) (Ebner et al., 2007a) coating were tested. For ethanolamine-hydrochloride coating the small silicon oxynitride AFM cantilevers were cleaned with ethanol (3 × 5 min) and chloroform (3 × 5 min) and dried in a gentle stream of nitrogen. Afterwards the tips were incubated with 0.5 g/mL ethanolamine-hydrochloride in DMSO in the presence of molecular sieve (3 Å) at room temperature, to generate reactive amino groups on the tip surface.
For APTES coating the tips were cleaned as described before. APTES was freshly distilled under vacuum. For removal of air and moisture a desiccator was flooded with argon. Two small plastic trays were placed in the desiccator. 30 μl of APTES and 10 μl triethylamine were pippeted into the two trays and the AFM tips were placed nearby. After 120 min of incubation in the closed desiccator, APTES and triethylamine were removed and the desiccator again flooded with argon.
The Biotin-PEG-NHS crosslinker was covalently bound to the generated amino groups on the tips. For this, 1 mg of Biotin-PEG-NHS crosslinker was dissolved in 0.5 mL chloroform and the cantilevers were placed in this solution. As a catalyst 10 μl triethylamine was added to start the reaction. After 2 h, the tips were washed with chloroform (3 × 5 min) und dried with nitrogen. The cantilevers were stored in PBS Buffer (150 mM NaCl, and 5 mM NaH2PO4) at 4 °C and used within three days.
Sample preparation for molecular recognition force spectroscopy: avidin with a concentration of 0.1 mg/mL in PBS-Buffer was incubated on freshly cleaved mica for 20 min. After the incubation step the mica was extensively cleaned by rinsing 50 times with 1/10 PBS-Buffer (15 mM NaCl, 0.5 mM NaH2PO4) and used immediately.
2.2. Nucleopore complex preparation
The nuclear pore complex samples used were nuclear membranes prepared from Xenopus laevis oocytes. The oocytes were extracted from X. laevis during operation. A single oocyte was put into a BSA coated Petri dish with KHP (nuclear membrane puffer, 90 mM KCl, 10 mM NaCl, 2.2 mM MgCl2, 1.1 mM EGTA, and 10 mM HEPES, pH 7.4 adjusted with KOH). The oocyte was opened using two needles so that the nucleus was spilled out. The nucleus was cleaned in KHP for 5 min to remove the cytosol. The cleaned nucleus was transferred to a cell-TAK modified class slide in ultra-pure water. Afterwards the nucleus was opened with the two preparation needles to dispose the chromatin. The membrane was spanned onto the cell-TAK modified position of the glass slide with the help of the needles. The glass slide with the nucleus membrane was dried and used within 5 days.
2.2.1. Class slide modification using cell-TAK
The class slides were washed with 70% ethanol, rinsed with ultra-pure water and dried in a gently steam of nitrogen. Cell-TAK was diluted 1:6 with KHP Buffer. A 3 μl droplet of the diluted cell-TAK was put on a glass slide and dried for 2 h. The modified glass slide was washed with 70% ethanol, rinsed two times with ultra-pure water and air-dried. The cell-TAK modified class slides were used immediately.
2.3. Bacterial surface layer preparation
All bacterial surface layer measurements were performed on the wildtype protein purified from Bacillus sphaericus CCM2177. For this, 1 mg of protein was dissolved in 1 mL GHCl Buffer (5 M GHCl, 50 mM Tris, pH adjusted to 7.2). The obtained solution was dialyzed in 3 L of ultra-pure water for 3 h by changing the water every hour. After dialysis the monomeric solution was centrifuged for 10 min at 4 °C to remove unsolved particles. The concentration of the protein was adjusted to 0.1 mg/mL using ultra-pure water. To start the recrystallization process 10 mM of CaCl2 were added. A cleaned silicon chip (5 × 5 mm) was incubated to 1 mL recrystallization solution at room temperature. After 3 h of incubation the silicon chips were gently rinsed with ultra-pure water and stored in physiological PBS-Buffer (1.8 mM KH2PO4, 10.1 mM Na2HPO4, 2.7 mM KCl, and 140 mM NaCl, pH adjusted to 7.2). 3 μL of NaN3 was added the next day to protect the samples from bacteria. The samples were used within 5 days.
2.3.1. Cleaning silicon chips
5 mm × 5 mm sized silicon chips were cleaned with piranha (3:7 = H2O2:H2SO4) for 30 min. Afterwards they were cleaned with ultra-pure water (3 × 5 min) and air-dried. On the next day they were immersed in ethanol (30 min), air-dried and immediately used for incubation.
2.4. AFM imaging
All AFM imaging measurements were performed on a Pico Plus setup (Pico SPM II 5500, Agilent, Chandler, USA), all the MRFS measurements were performed on a Pico SPM I setup (Agilent, Chandler, USA). The molecular recognition force microscopy measurements were performed in liquid with sweep duration of 1 s at a z-range of 300 nm. Bacterial surface layer measurements were performed in liquid environment using both tapping and contact mode. The nucleopore complex measurements were performed using contact mode in dry state. Tapping mode measurements were performed with different acceleration frequencies using cantilevers with different sizes and characteristics.
3. Results
3.1. Characterization of small cantilevers
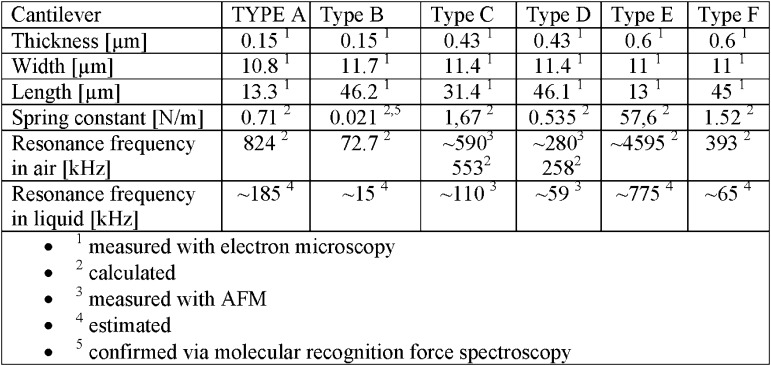
The used cantilevers were significantly smaller than conventional cantilevers, but still large enough to be used with the optical detection system of a conventional AFM system (width of 11 μm for all prototypes). A long 150 nm thick cantilever (Type B) with a low spring constant (about 21 pN/nm) was specially tailored for high sensitivity molecular recognition force spectroscopy (MRFS). In contrast, long 430 nm thick cantilevers (Types C and D) and short 150 nm thick cantilevers (Type A) were designed for imaging of soft biological samples under aqueous and dry condition, both for tapping and contact mode. The latter represented a good compromise between high resonance frequency and low spring constant. Prototypes E and F of 600 nm thickness were mainly used for characterization by electron microscopy. In particular type E turned out too stiff for biological samples and its resonance frequency exceeded the range for tapping mode imaging with our commercial AFM.
First, the silicon oxynitride (Ohashi et al., 1997) prototypes were characterized using electron microscopy (EM) so as to determine length, width, thickness, tip shape and radius (see Fig. 1A and B, and table in Fig. 2). Using these dimensions, the spring constant and resonance frequencies were calculated (Fig. 2, for equations see supporting information). Because of the reduced dimensions of these cantilevers the mass of the tip and the different material constants of the reflex coating cannot be neglected. The shape (cone), the height, and the diameter at the base were extracted from the EM measurements. For coating a 10 nm film of chromium as adhesive agent and a 20 nm thick gold layer for laser reflection were used. The main results are summarized in the table of Fig. 2 and the calculations are detailed in the supporting information. Calculated resonance frequencies were compared with values measured using the AFM frequency plot (Fig. 2, Type C and D) and they slightly disagree due to several assumptions made for the calculations. More precisely, the tip was treated as point mass at the outer end of the cantilever and the cone shaped tip is partially hollow, so that the exact mass is difficult to determine. In addition the thicknesses of the chromium and gold layer can only be estimated from fabrication parameters. Unfortunately, measuring the spring constant using the more accurate thermal noise method was not possible with the used equipment, because the cantilevers’ resonance frequency exceeded the bandwidth of the A/D converter.
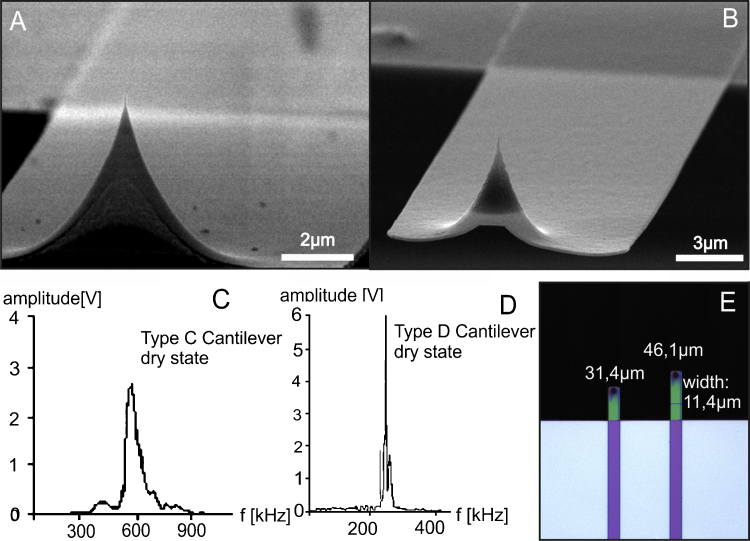
Fig. 1.
Electron microscope images of prototype cantilevers. (A and B) Typical images used for further characterization. The horn shaped and partly hollow tip from the front side. The upper side of the silicon oxynitride cantilever is coated with chromium as adhesive agent and gold for laser reflection. The tip was visualized as cone with a height of about 3 μm and a base radius of about 2 μm. The radius of the curvature is about 5–10 nm for the sharpest cantilevers. (C) The measured resonance frequency for the short 430 nm cantilever (Type C) is about 600 kHz. (D) The measured resonance frequency is about 280 kHz for the long 430 nm (Type D) cantilever. (E) Image of the 430 nm thick cantilevers (Type C and D).
Fig. 2.
The chart in Fig. 2 shows a summary of the results of the different characterized cantilever prototypes. The cantilever prototypes combine low spring constant with a high resonance frequency when compared to common cantilevers. The long 430 nm thick (Type D) and the short 150 nm thick (Type A) cantilevers were used for high speed imaging at nanometer resolution in contact and tapping mode. The long 150 nm thick (Type B) cantilever was used for MRFS on avidin/biotin test system, showing lower thermal fluctuations then conventional cantilevers with similar spring constant. The long and short 600 nm (Type E and F) cantilever was only used for EM imaging and characterization. The difference in the measured and calculated frequencies for Type C and D cantilever is mainly due to meanderings of the cantilever dimensions, the reflex coating, the tip mass and the shape of the tip.
3.2. Molecular recognition force spectroscopy
AFM allows for the measurement of ultra-low inter- and intra- molecular forces on the single molecule level. In molecular recognition force spectroscopy (MRFS) (Florin et al., 1994; Hinterdorfer et al., 1996; Lee et al., 1994) a force is exerted on a receptor/ligand complex and the dissociation process is followed over time. So as to perform this type of measurement between a ligand on the tip and a surface-bound receptor, a careful tip sensor design is required. Therefore the AFM tip had to be chemically modified (see Section 2) and, as a first step, amino groups were generated using either APTES (Ebner et al., 2007a) or ethanolamine coating (Ebner et al., 2007a). Subsequently, a flexible PEG-Biotin linker was covalently bound to the amino groups on the tip. The suchlike biotin-functionalized AFM sensor was then moved towards the sample containing the cognate receptors (avidin) and subsequently retracted (Fig. 3). During contact time the biotin on the tip eventually bound to the avidin on the sample surface. Upon retraction the cantilever was bended downwards and a force signal with a distinct shape occurred. This force increased until the ligand receptor bond was broken and the cantilever flipped back into its resting position. From the unbinding event, the binding force between tip tethered ligand and surface bond receptor was determined. Importantly, comparing our prototype cantilever with a typical commercially available force spectroscopy cantilever, the force noise was much lower using the small prototype cantilever (Fig. 3).
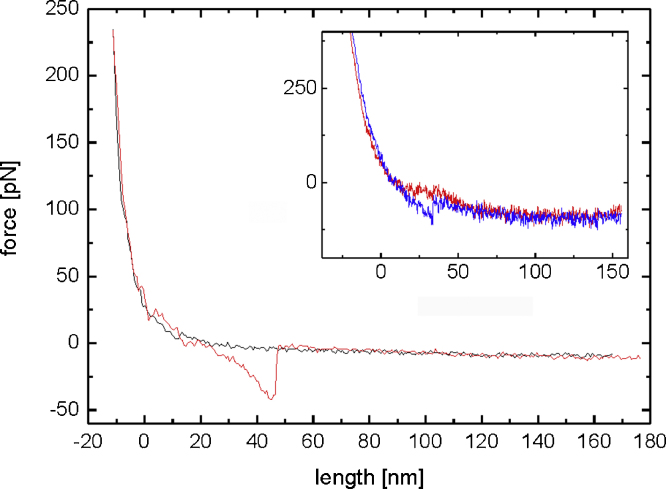
Fig. 3.
Fig. 3 shows a typical force distance cycle with a specific unbinding event using a chemically modified cantilever prototype (Type B). Force distance cycles were recorded on avidin adsorbed to freshly cleaved mica. The inset shows a typical force distance cycle recorded on the same system using a Bruker MSCT C (10 pN/nm) cantilever. Obviously it can be seen that the force noise is much lower using the small prototype cantilever.
For a clear discrimination between noise and unbinding, the thermal force fluctuation of the cantilever needs to be significantly lower than the measured forces. Thus the force–distance cycles recorded on the avidin/biotin system were analyzed regarding their thermal fluctuation by extracting 50 nm intercepts from the non-contact regime of the retraction curve. So as to evaluate the quantities of the prototype cantilevers, they were compared to commercially available standard force spectroscopy cantilevers (AUHW MSCT Type B-E, Bruker, USA; OMCL-TR400PSA and BioLever, Olympus, Japan). The spring constant of typical MRFS cantilevers is specified between 6 and 40 pN/nm. More precisely, Type B cantilever showed a spring constant of about 21 pN/nm. The BRUKER AUSHM MSCT B-D ranged from 10 to 30 pN/nm (B = 20, C = 10, D = 30), whereas the Olympus BioLever was the softest available cantilever with a spring of 6 pN/nm. Finally, BRUKER AUHW MSCT Type E, which is usually not used for MFRS, showed a typical spring constant of 100 pN/nm and was compared to Type D cantilever. All measurements were performed using the same setup, same scanner, and same photodiode to minimize instrumental influences.
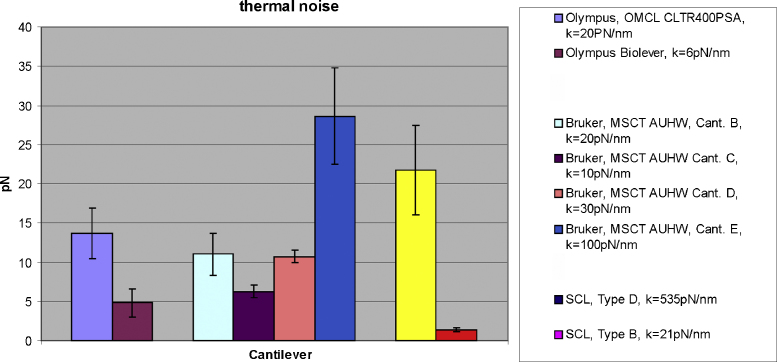
Evidently, the thermal force fluctuations decreased with lower spring constants (Fig. 4). Comparing commercial reference cantilevers to our prototypes (SCL) clearly indicated that small cantilevers with similar spring constants have much lower force fluctuations (Figs. 4 and 5) under the same experimental conditions. In detail, the thermal fluctuation for the Type B cantilever turned out to be 5–6 times lower than BRUKER cantilever B and D. When we compared Type B cantilever to BRUKER C and Olympus BioLever, the fluctuation was still 2–3 times lower, even though the reference cantilevers showed a lower spring constant. Finally, the thermal force fluctuation of Type D, with a value of about 21 pN, was comparable BRUKER E, despite the considerably higher the spring constant (535 pN/nm) (Figs. 4 and 5).
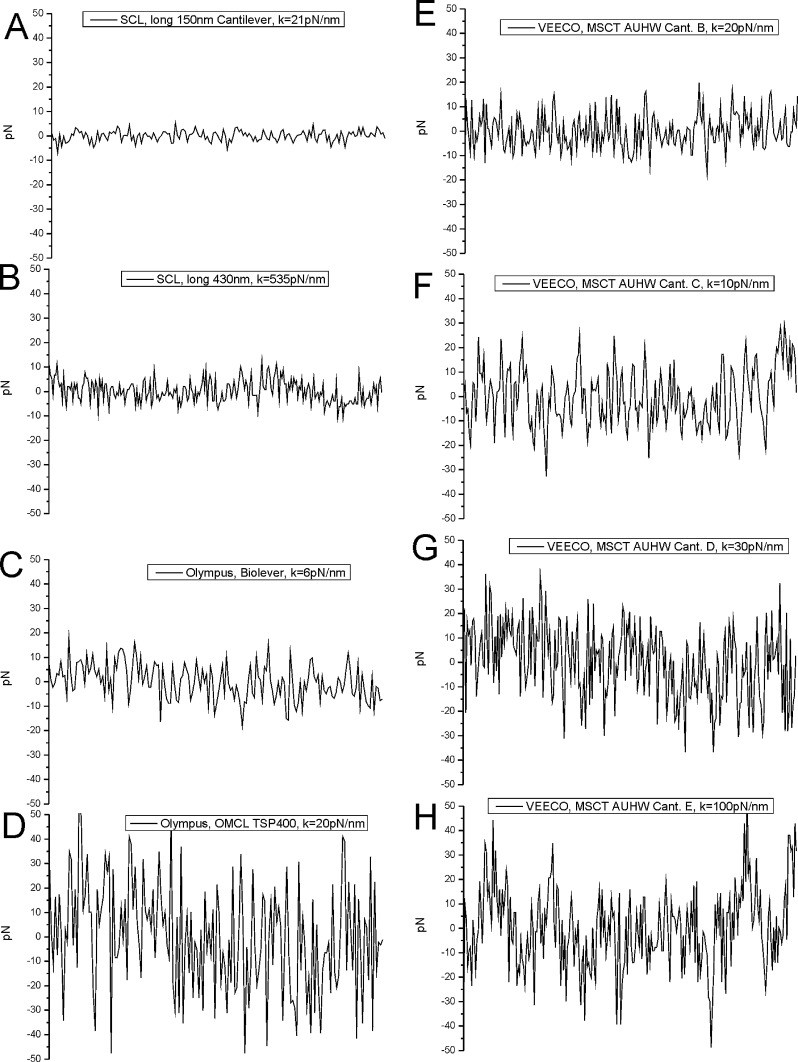
Fig. 4.
Fig. 4 shows force noise traces of different cantilevers which are common in molecular recognition force spectroscopy (MRFS). All curves have been extracted from force distance cycles, showing 50 nm ranges of the non-contact region of the retrace at the same loading rate (1 s sweep duration, 600 nm force distance cycles, 2000 data points). (A and B) Force noise traces of the Type B and Type D prototype cantilevers. The 150 nm cantilever with a spring constant of 21 pN/nm showed the lowest thermal force noise. BRUKER Cant. (B) (Fig. 4E, 20 pN/nm), (C) (Fig. 4F, 10 pN/nm) and (D) (Fig. 4G, 30 pN/nm) and the Olympus BioLever (Fig. 4C, 6 pN/nm) and TSP400 (Fig. 4D, 20 pN/nm) cantilever showed higher thermal force fluctuations.
Fig. 5.
The diagram in Fig. 5 summarizes the thermal force fluctuation analysis for common MRFS cantilever and the prototype cantilevers. Type D cantilever showed a thermal fluctuation of about 22 pN which is comparable to Bruker cantilever E (∼28 pN). Type B cantilever showed a thermal fluctuation of about 1.4 pN which is about 5 times lower when compared with Bruker cantilever B, and about 3 times lower when compared with Bruker cantilever C and Olympus BioLever.
3.3. Imaging of biological samples under different conditions with higher speed
The developed cantilever prototypes were utilized for imaging of several biological samples under aqueous and ambient conditions in both tapping and contact mode. As test system for high resolution imaging in liquid bacterial surface layer proteins were separated from B. sphaericus and recrystallized on cleaned silicon chips. For imaging in dry state we used cell nuclear membranes from X. laevi oocytes so as to visualize the embedded nuclear pore complexes.
3.3.1. Tapping mode imaging
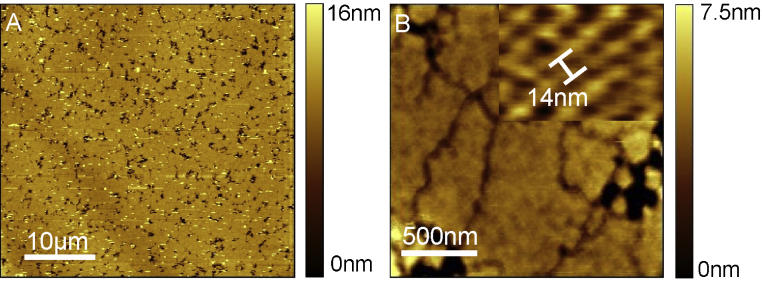
Bacterial surface layers comprise the outer surface layer of some bacteria (Sleytr and Messner, 1983). They consist of proteins which have the ability to self-assemble into monolayers on various surfaces (e.g. silicon), resulting in well defined, regular two-dimensional lattice structures. Depending on the bacterial strain these layers from 5 to 20 nm thick oblique, square or hexagonal structures with a center-to-center spacing of 2.5–35 nm (Sleytr, 1997; Sleytr et al., 2007, 1994). Here B. sphaericus CCM2177 (Ilk et al., 2002; Meyer and Neide, 1904) was used that forms a 2-D square lattice (p-4 symmetry) with a spacing of about 14 nm and a thickness of 7 nm. As this layer is extremely flat and displays nm-sized features, it is perfectly suited to determine the lateral resolution at different imaging speeds. The overview image in 6A shows a silicon chip densely packed with bacterial surface layer patches (for preparation see Section 2) recorded in contact mode at aqueous conditions using Type D cantilever. Image 6B was recorded in tapping mode in the same environment. The typical square lattice structure with 14 nm (Tang et al., 2008a) can be clearly resolved at larger magnifications (Fig. 6B inset). The achieved resolution of the recorded images was comparable to images recorded with conventional cantilevers at low speeds (5 μm/s) (Tang et al., 2008a). The maximum tip velocity, where nm resolution could be observed with Type D cantilever was about 55 μm/s. Therefore the usage of the small cantilever prototypes resulted in a speed increase by a factor of 10, reducing the total recording time of a 512 × 512 pixel high resolution image to less than a minute.
Fig. 6.
Bacterial surface layer from Bacillus Sphaericus were imaged with up to ten times higher speed compared to common cantilevers. (A) Overview image recorded in contact mode using Type D cantilever. Imaging speed was about 55 μm/s (∼10 lines/s). (B) Larger magnification image. The typical lattice structure with a constant of 14 nm constant is clearly visible (see inset). Image recorded in tapping mode with nm resolution using the Type C Cantilever.
3.3.2. Contact mode imaging
Contact mode imaging in ambient conditions was performed on nuclear membranes in which the nuclear pore complexes (NPC) (Kaftan et al., 2002; Bednenko and Gerace, 2003; Stewart et al., 2001) are embedded. The NPCs are large protein assemblies consisting of about 30 different proteins, the nucleoporines (Nups), each of which exists in several copies (Rout et al., 2000). In higher eukaryotic cells an NPC assembles 120 of these proteins in total. It reveals an eight-fold rational symmetry normal to the membrane and an asymmetric assembly with respect to the horizontal plane of the membrane (Hinshaw et al., 1992).
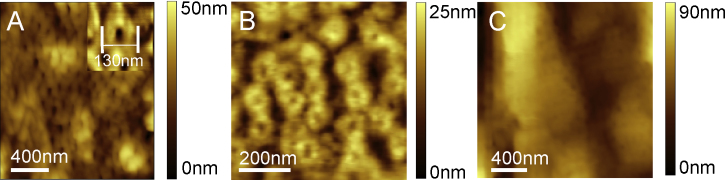
The NPCs were chosen because they represent an example of a rougher protein surface of several tens of nm in z scale. Due to their stability they can also be prepared and imaged in ambient conditions. Fig. 7A shows a nuclear membrane in which the embedded nuclear pore complexes (NPC) (Kaftan et al., 2002) are clearly visible. The image was recorded using Type D cantilever in contact mode at ambient conditions. The maximum tip velocity for high resolution NPC imaging where the structures could be clearly observed was 33 μm/s (Fig. 7A). Fig. 7B shows NPC's recorded with a conventional cantilever at 2.5 μm/s. Evidently, the lateral resolution is comparable. In contrast, using a conventional cantilever at high speed (25 μm/s) yields images with much less resolution (Fig. 7C).
Fig. 7.
(A) Nuclear pore complexes imaged with Type D cantilever using contact mode at ambient conditions. (B) The imaging speed of 12 lines per second was about ten times higher than in (A) using the same cantilever. (C) Conventional cantilever at a speed of 10 lines/s yielded images with much less resolution.
4. Discussion
Speeding up AFM imaging often requires tedious electronic developments and usage of special scanners. In this study we aimed at increasing the force sensitivity and the imaging speed of a conventional AFM without any changes to the setup. Enhancements were accomplished by using small and sensitive cantilevers. The force resolution for MRFS experiments was 3–5 times higher than with usual cantilevers. Furthermore we achieved an increase of the imaging speed by a factor of 10, while at the same time retaining the tip sample interaction forces sufficiently low for obtaining high resolution images soft biological samples in fluid. This reduced the time required for recording a 512 × 512 pixel image to less than 1 min. It should be noted that this is still quite far away from video AFM imaging reported by several other groups (Ando et al., 2008; Anwar and Rousso, 2005; Fantner et al., 2006; Kobayashi et al., 2007; Miyagi et al., 2008; Picco et al., 2008, 2007), but nevertheless the achieved increase in scan speed will allow the investigation of the dynamics of many biological processes.
The main challenge of using our prototype cantilevers in a conventional AFM is the small laser reflection area of the cantilever (∼11 μm) compared to the relatively large laser spots of the AFM laser unit (∼30 μm). This makes the detected sum signal up to 10 times lower compared to a conventional cantilever; improvements were achieved by the usage of an amplifying photodiode. Another limitation was the line-rate speed limit of the software used (13 scan lines/s). At much larger line rates however, the resonances of the scanner become a limiting factor. Efforts for reducing these vibrations through input shaping have yielded good results (Schitter et al., 2004; Schitter and Stemmer, 2003, 2004). Optimization of imaging speed to video rate requires significant changes and improvements of the whole AFM instrument (Ando et al., 2001). We believe, however, that the 10-fold increase in performance due to the use of the small cantilevers is sufficient for many interesting biological processes and constitutes a significant improvement for general AFM users.
Acknowledgments
This work was supported by Fabican (project 819637) which is part of the project cluster Austrian Science Foundation (project N00104-NAN), by FFG MNT.era.net (project 823980), the Swiss National Science Foundation (205321_134786) and the Schrödinger foundation. We thank Christian Rankl for help in data evaluation.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.micron.2012.05.007.
Supporting Information Available: Detailed calculations for the cantilever characterization
Appendix A. Supplementary data
The following is the supplementary data to this article.
References
- Ando T. Control techniques in high-speed atomic force microscopy. American Control Conference. 2008:3194–3200. [Google Scholar]
- Ando T., Kodera N., Takai E., Maruyama D., Saito K., Toda A. A high-speed atomic force microscope for studying biological macromolecules. Proceedings of the National Academy of Sciences of the United States of America. 2001:12468–12472. doi: 10.1073/pnas.211400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T., Uchihashi T., Fukuma T. High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Progress in Surface Science. 2008;83:337–437. [Google Scholar]
- Anwar M., Rousso I. Atomic force microscopy with time resolution of microseconds. Applied Physics Letters. 2005;86 pp. 014101–014103. [Google Scholar]
- Barrett R.C., Quate C.F. High-speed, large-scale imaging with the atomic force microscope. Journal of Vacuum Science & Technology B: Microelectronics and Nanometer Structures. 1991;9:302–306. [Google Scholar]
- Bednenko J., Gerace G.C.L. Nucleocytoplasmic Transport: Navigating the Channel. Traffic. 2003:127–135. doi: 10.1034/j.1600-0854.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- Binnig G., Quate C.F., Gerber C. Atomic force microscope. Physical Review Letters. 1986;56:930. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Chand A., Viani M.B., Schaffer T.E., Hansma P.K. Microfabricated small metal cantilevers with silicon tip for atomic force microscopy. Journal of Microelectromechanical Systems. 2000;9:112–116. [Google Scholar]
- Duman M., Pfleger M., Zhu R., Rankl C., Chtcheglova L.A., Neundlinger I., Bozna B.L., Mayer B., Salio M., Shepherd D., Polzella P., Moertelmaier M., Kada G., Ebner A., Dieudonne M., Schütz G.J., Cerundolo V., Kienberger F., Hinterdorfer P. Improved localization of cellular membrane receptors using combined fluorescence microscopy and simultaneous topography and recognition imaging. Nanotechnology. 2010:21. doi: 10.1088/0957-4484/21/11/115504. [DOI] [PubMed] [Google Scholar]
- Ebner A., Hinterdorfer P., Gruber H.J. Comparison of different aminofunctionalization strategies for attachment of single antibodies to AFM cantilevers. Ultramicroscopy. 2007;107:922–927. doi: 10.1016/j.ultramic.2007.02.035. [DOI] [PubMed] [Google Scholar]
- Ebner A., Kienberger F., Kada G., Stroh C.M., Geretschlager M., Kamruzzahan A.S.M., Wildling L., Johnson W.T., Ashcroft B., Nelson J., Lindsay S.M., Gruber H.J., Hinterdorfer P. Localization of single avidin–biotin interactions using simultaneous topography and molecular recognition imaging. ChemPhysChem. 2005;6:897–900. doi: 10.1002/cphc.200400545. [DOI] [PubMed] [Google Scholar]
- Ebner A., Wildling L., Kamruzzahan A.S.M., Rankl C., Wruss J., Hahn C.D., Holzl M., Zhu R., Kienberger F., Blaas D., Hinterdorfer P., Gruber H.J. A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconjugate Chemistry. 2007;18:1176–1184. doi: 10.1021/bc070030s. [DOI] [PubMed] [Google Scholar]
- Fantner G.E., Barbero R.J., Gray D.S., Belcher A.M. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nature Nanotechnology. 2010;5:280–285. doi: 10.1038/nnano.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantner G.E., Hegarty P., Kindt J.H., Schitter G., Cidade G.A.G., Hansma P.K. Data acquisition system for high speed atomic force microscopy. Review of Scientific Instruments. 2005:76. [Google Scholar]
- Fantner G.E., Schitter G., Kindt J.H., Ivanov T., Ivanova K., Patel R., Holten-Andersen N., Adams J., Thurner P.J., Rangelow I.W., Hansma P.K. Components for high speed atomic force microscopy. Ultramicroscopy. 2006;106:881–887. doi: 10.1016/j.ultramic.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Florin E.L., Moy V.T., Gaub H.E. Adhesion forces between individual ligand-receptor pairs. Science. 1994:415–417. doi: 10.1126/science.8153628. [DOI] [PubMed] [Google Scholar]
- Takeshi Fukuma, Yasutaka Okazaki, Noriyuki Kodera, Takayuki Uchihashi, Toshio Ando. American Institute of Physics; Melville, NY, ETATS-UNIS: 2008. High Resonance Frequency Force Microscope Scanner Using Inertia Balance Support. [Google Scholar]
- Hillner P.E., Gratz A.J., Manne S., Hansma P.K. Atomic-scale imaging of calcite growth and dissolution in real time. Geology. 1992:359–362. [Google Scholar]
- Hinshaw J.E., Carragher B.O., Milligan R.A. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Hinterdorfer P., Baumgartner W., Gruber H.J., Schilcher K., Schindler H. Detection and localization of individual antibody–antigen recognition events by atomic force microscopy. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs J.K., Humphris A.D.L., Miles M.J. In situ atomic force microscopy of polyethylene crystallization. 1. Crystallization from an oriented backbone. Macromolecules. 2001;34:5508–5519. [Google Scholar]
- Hosaka S., Etoh K., Kikukawa A., koyanagi H. Megahertz silicon atomic force microscopy (AFM) cantilever and high-speed readout in AFM-based recording. Journal of Vacuum Science and Technology B. 2000:18. [Google Scholar]
- Hosaka S., Etoh K., Kikukawa A., Koyanagi H., Itoh K. 6.6 MHz silicon AFM cantilever for high-speed readout in AFM-based recording. Microelectronic Engineering. 1999;46:109–112. [Google Scholar]
- Humphris A.D.L., Miles M.J., Hobbs J.K. A mechanical microscope: high-speed atomic force microscopy. Applied Physics Letters. 2005;86 pp. 034103–034106. [Google Scholar]
- Ilk N., Völlenkle C., Egelseer E.M., Breitwieser A., Sleytr U.B., Sára M. Molecular characterization of the S-layer gene, sbpA, of Bacillus sphaericus CCM 2177 and production of a functional S-layer fusion protein with the ability to recrystallize in a defined orientation while present in the fused allergen. Applied and Environmental Microbiology. 2002;68:3251–3260. doi: 10.1128/AEM.68.7.3251-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaftan D., Brumfeld V., Nevo R., Scherz A., Reich Z. From chloroplasts to photosystems: in situ scanning force microscopy on intact thylakid membranes. EMBO Journal. 2002;21:6146–6153. doi: 10.1093/emboj/cdf624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamruzzahan A.S.M., Kienberger F., Stroh C.M., Berg J., Huss R., Ebner A., Zhu R., Rankl C., Gruber H.J., Hinterdorfer Imaging morphological details and pathological differences of red blood cells using tapping-mode AFM. Biological Chemistry. 2004;385:955–960. doi: 10.1515/BC.2004.124. [DOI] [PubMed] [Google Scholar]
- Kienberger F., Rankl C., Pastushenko V., Zhu R., Blaas D., Hinterdorfer P. Visualization of single receptor molecules bound to human rhinovirus under physiological conditions. Structure. 2005;13:1247–1253. doi: 10.1016/j.str.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Kienberger F., Zhu R., Moser R., Blaas D., Hinterdorfer P. Monitoring RNA release from human rhinovirus by dynamic force microscopy. Journal of Virology. 2004;78:3203–3209. doi: 10.1128/JVI.78.7.3203-3209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-S., Nam H.-J., Cho S.-M., Hong J.-W., Kim D.-C., Bu J.U. PZT cantilever array integrated with piezoresistor sensor for high speed parallel operation of AFM. Sensors and Actuators A: Physical. 2003;103:122–129. [Google Scholar]
- Kobayashi M., Sumitomo K., Torimitso K. Real-time imaging of DNA–streptavidin complex formation in solution using a high-speed atomic force microscope. Ultramicroscopy. 2007;107:184–190. doi: 10.1016/j.ultramic.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Kodera N., Sakashita M., Ando T. Dynamic proportional-integral-differential controller for high-speed atomic force microscopy. Review of Scientific Instruments. 2006;77 083704–083707. [Google Scholar]
- Kodera N., Yamashita H., Ando T. Active damping of the scanner for high-speed atomic force microscopy. Review of Scientific Instruments. 2005;76 pp. 053705–053708. [Google Scholar]
- Lee G.U., Kidwell D.A., Colton R.J. Sensing discrete streptavidin–biotin interactions with atomic force microscopy. Langmuir. 1994:354–357. [Google Scholar]
- Leitner M., Mitchell N., Kastner M., Gruber H., Hinterdorfer P., Howorka S., Ebner A. Single-molecule AFM characterization of individual chemically tagged DNA tetrahedra. ACS Nano. 2011 doi: 10.1021/nn201705p. [DOI] [PubMed] [Google Scholar]
- Madl J., Rhode S., Stangl H., Stockinger H., Hinterdorfer P., Schutz G.J., Kada G. A combined optical and atomic force microscope for live cell investigations, ultramicroscopy proceedings of the seventh international conference on scanning probe microscopy. Sensors and Nanostructures. 2006;106:645–651. doi: 10.1016/j.ultramic.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Manalis S.R., Minne S.C., Atalar A., Quate C.F. High-speed atomic force microscopy using an integrated actuator and optical lever detection. Review of Scientific Instruments. 1996;67:3294–3297. [Google Scholar]
- Meyer A., Neide E. Botanische Beschreibung einiger sporenbildender Bakterien. Zentralblatt für bakteriologie, Parasitenkunde. Infektionskrankheiten und Hygiene. 1904:12. [Google Scholar]
- Miyagi A., Tsunaka Y., Uchihashi T., Mayanagi K., Hirose S., Morikawa K., Ando T. Visualization of intrinsically disordered regions of proteins by high-speed atomic force microscopy. ChemPhysChem. 2008;9:1859–1866. doi: 10.1002/cphc.200800210. [DOI] [PubMed] [Google Scholar]
- Muller D.J., Schabert F.A., Buldt G., Engel A. Imaging purple membranes in aqueous solutions at sub-nanometer resolution by atomic force microscopy. Biophysical Journal. 1995;68:1681–1686. doi: 10.1016/S0006-3495(95)80345-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi M., Nakamura K., Hirao K., Toriyama M., Kanzaki S. Factors affecting mechanical properties of silicon oxynitride ceramics. Ceramics International. 1997;23:27–37. [Google Scholar]
- Pedrak R., Ivanov T., Ivanova K., Gotszalk T., Abedinov N., Rangelow I.W., Edinger K., Tomerov E., Schenkel T., Hudek P. Micromachined atomic force microscopy sensor with intergrated piezoresistive sensor and thermal bimorph actuator for high-speed tapping-mode atomic force microscopy phase-imaging in higher eigenmodes. Journal of Vacuum Science Technology B. 2003;21(November/December (6)):3102–3107. [Google Scholar]
- Picco L.M., Dunton P.G., Ulcinas A., Engledew D.J., Hoshi O., Ushiki T., Miles M.J. Institute of Physics; Bristol, ROYAUME-UNI: 2008. High-speed AFM of Human Chromosomes in Liquid. [DOI] [PubMed] [Google Scholar]
- Picco L.M., Bozec L., Ulcinas A., Engledew D.J., Antognozzi M., Horton M.A., Miles M.J. IOP Publishing; 2007. Breaking the Speed Limit with Atomic Force Microscopy. [Google Scholar]
- Riener C.K., Kienberger F., Hahn C.D., Buchinger G.M., Egwim I.O.C., Haselgrübler T., Ebner A., Romanin C., Klampfl C., Lackner B., Prinz H., Blaas D., Hinterdorfer P., Gruber H.J. Heterobifunctional crosslinkers for tethering single ligand molecules to scanning probes. Analytica Chimica Acta. 2003;497:101–114. [Google Scholar]
- Rogers B., Sulchek T., Murrary K., York D., Jones M., Manning L., Malekos S., Beneschott B., Adams J.D., Cavazos H., Minne S.C. American Institute of Physics; Melville, NY, ETATS-UNIS: 2003. High Speed Tapping Mode Atomic Force Microscopy in Liquid Using an Insulated Piezoelectric Cantilever. [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. The yeast nuclear pore complex: composition. Architecture, and Transport Mechanism. 2000:635–652. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer T.E., Hansma P.K. American Institute of Physics; Melville, NY, ETATS-UNIS: 1998. Characterization and Optimization of the Detection Sensitivity of an Atomic Force Microscope for Small Cantilevers. [Google Scholar]
- Schaffer T.E., Jiao Y.K. High speed force mapping using small cantilevers. Biophysical Journal. 2001;80:303A. [Google Scholar]
- Schitter G., Astrom K.J., DeMartini B.E., Thurner P.J., Turner K.L., Hansma P.K. Design and modeling of a high-speed AFM-scanner. IEEE Transactions on Control Systems Technology. 2007;15:906–915. [Google Scholar]
- Schitter G., Stark R.W., Stemmer A. Fast contact-mode atomic force microscopy on biological specimen by model-based control. Ultramicroscopy. 2004;100:253–257. doi: 10.1016/j.ultramic.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Schitter G., Stemmer A. Model-based signal conditioning for high-speed atomic force and friction force microscopy. Microelectronic Engineering. 2003;67–68:938–944. [Google Scholar]
- Schitter G., Stemmer A. Identification and open-loop tracking control of a piezoelectric tube scanner for high-speed scanning-probe microscopy. IEEE Transactions on Control Systems Technology. 2004;12:449–454. [Google Scholar]
- Sleytr U.B. I. Basic and applied S-layer research: an overview. FEMS Microbiology Reviews. 1997;20:5–12. [Google Scholar]
- Sleytr U.B., Huber C., Ilk N., Pum D., Schuster B., Egelseer E.M. S-layers as a tool kit for nanobiotechnological applications. FEMS Microbiology Letters. 2007;267:131–144. doi: 10.1111/j.1574-6968.2006.00573.x. [DOI] [PubMed] [Google Scholar]
- Sleytr U.B., Messner P. Crystalline surface-layers on bacteria. Annual Review of Microbiology. 1983;37:311–339. doi: 10.1146/annurev.mi.37.100183.001523. [DOI] [PubMed] [Google Scholar]
- Sleytr U.B., Sara M., Messner P., Pum D. Two-dimensional protein crystals (S-layers): fundamentals and applications. Journal of Cellular Biochemistry. 1994;56:171–176. doi: 10.1002/jcb.240560209. [DOI] [PubMed] [Google Scholar]
- Stewart M., Baker R.P., Bayliss R., Clayton L., Grant R.P., Littlewood T., Matsuura Y. Molecular mechanism of translocation through nuclear pore complexes during nuclear protein import. FEBS Letters. 2001;498:145–149. doi: 10.1016/s0014-5793(01)02489-9. [DOI] [PubMed] [Google Scholar]
- Stroh C.M., Ebner A., Geretschlager M., Freudenthaler G., Kienberger F., Kamruzzahan A.S.M., Smith-Gill S.J., Gruber H.J., Hinterdorfer P. Simultaneous topography and recognition imaging using force microscopy. Biophysical Journal. 2004;87:1981–1990. doi: 10.1529/biophysj.104.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulchek T., Hsieh R., Adams J.D., Minne S.C., Quate C.F., Adderton D.M. High-speed atomic force microscopy in liquid. Review of Scientific Instruments. 2000:71. [Google Scholar]
- Sulchek T., Hsieh R., Adams J.D., Yaralioglu G.G., Minne S.C. High-speed tapping mode imaging with active Q control for atomic force microscopy. Applied Physics Letters. 2000;76 [Google Scholar]
- Tang J., Ebner A., Ilk N., Lichtblau H., Huber C., Zhu R., Pum D., Leitner M., Pastushenko V., Gruber H.J., Sleytr U.B., Hinterdorfer P. High-affinity tags fused to s-layer proteins probed by atomic force microscopy. Langmuir. 2008:1324–1329. doi: 10.1021/la702276k. [DOI] [PubMed] [Google Scholar]
- Tang J., Krajcikova D., Zhu R., Ebner A., Cutting S., Gruber H.J., Barak I., Hinterdorfer P. Atomic force microscopy imaging and single molecule recognition force spectroscopy of coat proteins on the surface of Bacillus subtilis spore. Journal of Molecular Recognition. 2007;20:483–489. doi: 10.1002/jmr.828. [DOI] [PubMed] [Google Scholar]
- Tang J.L., Ebner A., Badelt-Lichtblau H., Vollenkle C., Rankl C., Kraxberger B., Leitner M., Wildling L., Gruber H.J., Sleytr U.B., Ilk N., Hinterdorfer P. Recognition imaging and highly ordered molecular templating of bacterial S-layer nanoarrays containing affinity-tags. Nano Letters. 2008;8:4312–4319. doi: 10.1021/nl802092c. [DOI] [PubMed] [Google Scholar]
- Tien S., Qingze Z., Devasia S. Control of dynamics-coupling effects in piezo-actuator for high-speed AFM operation. Proceedings of the 2004 American Control Conference. 2004;3114:3116–3121. [Google Scholar]
- Uchihashi T., Kodera N., Itoh H., Yamashita H., Ando T. Feed-forward compensation for high-speed atomic force microscopy imaging of biomolecules. Japanese Journal of Applied Physics. 2006 [Google Scholar]
- van der Mei H.C., van de Belt-Gritter B., Pouwels P.H., Martinez B., Busscher H.J. Cell surface hydrophobicity is conveyed by S-layer proteins—a study in recombinant lactobacilli. Colloids and Surfaces B: Biointerfaces. 2003;28:127–134. [Google Scholar]
- Viani M.B., Schaffer T.E., Paioczi G.T., Pietrasanta L.I., Smith B.L., Thompson J.B., Richter M., Rief M., Gaub H.E., Plaxco K.W., Cleland A.N., Hansma H.G., Hansma P.K. American Institute of Physics; Melville, NY, ETATS-UNIS: 1999. Fast Imaging and Fast Force Spectroscopy of Single Biopolymers with a New Atomic Force Microscope Designed for Small Cantilevers. [Google Scholar]
- Viani M.B., Pietrasanta L.I., Thompson J.B., Chand A., Gebeshuber I.C., Kindt J.H., Richter M., Hansma H.G., Hansma P.K. Probing protein–protein interactions in real time. Nature Structural & Molecular Biology. 2000;7:644–647. doi: 10.1038/77936. [DOI] [PubMed] [Google Scholar]
- Viani M.B., Schäffer T.E., Chand A., Rief M., Gaub H.E. Small cantilevers for force spectroscopy of single molecules. Journal of Applied Physics. 1999:86. [Google Scholar]
- Walters D.A., Cleveland J.P., Thomson N.H., Hansma P.K., Wendman M.A., Gurley G., Elings V. American Institute of Physics; Melville, NY, ETATS-UNIS: 1996. Short cantilevers for atomic force microscopy. [Google Scholar]
- Wielert-Badt S., Hinterdorfer P., Gruber H.J., Lin J.-T., Badt D., Wimmer B., Schindler H., Kinne R.K.H. Single molecule recognition of protein binding epitopes in brush border membranes by force microscopy. Biophysical Journal. 2002;82:2767–2774. doi: 10.1016/S0006-3495(02)75617-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.L., Despont M., Drechlser U., Hoogenboom B.W., Frederix P.L. Miniaturized single-crystal silicon cantilevers for scanning force microscopy. Applied Physics Letters. 2005:86. [Google Scholar]
- Ying W., Qingze Z., Chanmin S. A current cycle feedback iterative learning control approach to AFM imaging. American Control Conference. 2008:2040–2045. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.