Abstract
Wnt morphogens control embryonic development and homeostasis in adult tissues. In vertebrates the N-terminal WIF domain (WIF-1WD) of Wnt inhibitory factor 1 (WIF-1) binds Wnt ligands. Our crystal structure of WIF-1WD reveals a previously unidentified binding site for phospholipid; two acyl chains extend deep into the domain, and the head group is exposed to the surface. Biophysical and cellular assays indicate that there is a WIF-1WD Wnt-binding surface proximal to the lipid head group but also implicate the five epidermal growth factor (EGF)-like domains (EGFs I–V) in Wnt binding. The six-domain WIF-1 crystal structure shows that EGFs I–V are wrapped back, interfacing with WIF-1WD at EGF III. EGFs II–V contain a heparan sulfate proteoglycan (HSPG)-binding site, consistent with conserved positively charged residues on EGF IV. This combination of HSPG- and Wnt-binding properties suggests a modular model for the localization of WIF-1 and for signal inhibition within morphogen gradients.
INTRODUCTION
The fine control of short-range and long-range communication between cells requires mechanisms to create and modulate gradients of secreted signaling molecules (morphogens). The Wnt morphogens are secreted glycoproteins that bind to cell surface receptors so as to activate intracellular signaling pathways that are associated with embryonic development, oncogenesis and the differentiation of stem cells1, 2. The most studied Wnt pathway, Wnt–β-catenin, is initiated when Wnt forms a ternary complex with the receptor Frizzled (Fzd)3 and low-density lipoprotein receptor-related proteins 5 or 6 (LRP5/6)4. Wnt signaling is modulated by two broad classes of secreted extracellular proteins that interfere with the formation of the Fzd–Wnt–LRP5/6 complex5. Members of the Dickkopf (Dkk) class bind to LRP5/6 and prevent binding of the receptor to Wnt, whereas Cerberus, WIF-1 and members of the secreted Fzd-related protein (sFRP) family bind directly to Wnts and prevent them from triggering signaling.
WIF-1 is present across vertebrate families and consists of an N-terminal secretion signal sequence, the WIF domain (WD, 143 amino acid residues), five EGF-like domains (31–33 residues each) and a hydrophilic C terminus (Fig. 1a). Human WIF-1 binds through its WIF domain to eight Wnts (3a, 4, 5a, 7a, 9a, 11 (ref. 6), Wingless and Xenopus Wnt8 (ref. 7)) and a protein involved in neuronal differentiation, Olfactomedin 1 (ref. 8). The three-dimensional structure of refolded WIF-1WD expressed in Escherichia coli was determined by NMR spectroscopy and revealed an interaction between a surface binding site on the WIF domain and alkyl chains of the detergent used during protein refolding9. This raised the question of whether the WIF domain can sequester Wnt3a-linked acyl chains10, 11. The role of the WIF-1 EGF-like domains is unknown. During embryogenesis in Xenopus and zebrafish, expression of WIF-1 is first detectable at the start of somitogenesis in the paraxial mesoderm7. WIF-1 expression continues in adults in the heart, lungs and cartilage-mesenchyme interfaces of various species6, 7. Deletion of the mouse WIF-1 gene accelerates the development of radiation-induced osteosarcomas in vivo12. Downregulation of WIF-1 expression by promoter methylation has been reported in human cancers13, and overexpression of WIF-1 inhibits the growth of cells from lung and bladder cancers14, 15.
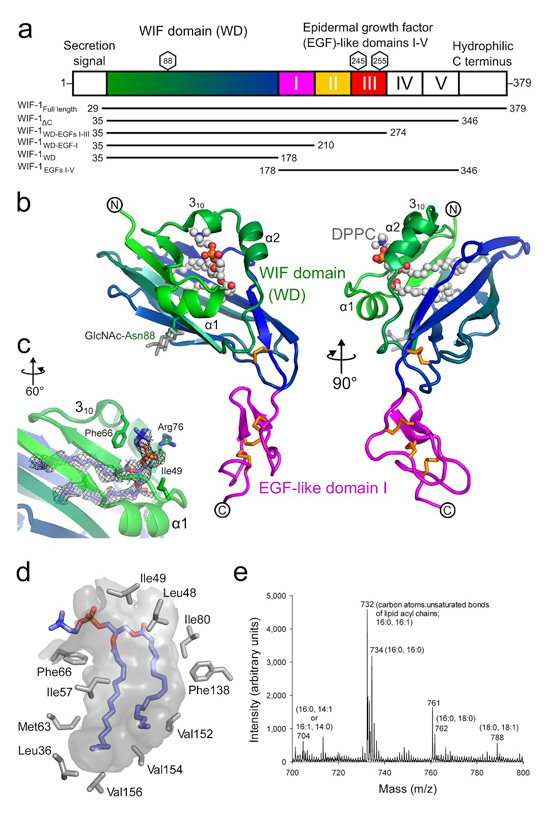
Figure 1.
The WIF-1WD-EGF-I structure and lipid-binding cavity. (a) Schematic domain organization of human WIF-1. Glycosylation sites linked to Asn88, Asn245 and Thr255 are marked by hexagons. The WIF domain is colored in green-to-blue transition, and EGFs I–III are colored magenta, yellow and red, respectively. The amino acid sequence boundaries of the WIF-1 constructs are shown below. (b) Ribbon diagram of WIF-1WD-EGF-I in two views that differ by a 90° rotation about a vertical axis. WIF-1WD–EGF-I is colored as in a. Asn88-linked N-acetylglucosamine (GlcNAc, gray) and disulfide bridges (orange) are shown as sticks. DPPC is shown as spheres (C, gray; N, blue; O, red; P, orange). (c) The phosphatidylcholine head group of DPPC (shown as sticks) exposed to the solvent with color coding as in b (except C in light blue). The composite omit electron density map (contoured at 1σ) is shown as gray mesh. (d) The WIF domain lipid-binding pocket (gray surface; DPPC shown as sticks with color coding as in c). Residues within 4.5 Å of DPPC are shown as gray sticks. (e) MALDI-TOF mass spectrum of WIF-1–bound ligands. The length and saturation of the lipid acyl chains are shown in parentheses.
How gradients of secreted morphogens such as Wnts and Hedgehogs (Hhs) are established and maintained is a key question16. Models of this process include covalent lipid modification co-localizing morphogens to cell membranes or lipoprotein particles, and morphogens interacting with the heparan sulfate glycosaminoglycans of HSPGs to modulate diffusion17, 18. The molecular mechanisms by which Wnt signaling is inhibited in lipoprotein particle- and HSPG-shaped morphogen gradients have remained poorly understood because lipid-modified Wnts19 and their antagonists are difficult to purify in sufficient quantities for biochemical studies.
We set out to dissect the mechanisms by which WIF-1 interacts with Wnt3a and morphogen gradients using a series of WIF-1 constructs expressed in human embryonic kidney (HEK293) cells. When we determined the structure of four natively folded, glycosylated WIF domain–containing constructs, we found a tightly bound diacyl phosphatidylcholine-type lipid in the core of the WIF domain. Our structural analyses also suggested that the EGF-like domains can adopt a specific (wrapped back) position relative to the WIF domain. Structure-guided, site-directed mutagenesis in combination with biophysical and cellular assays showed that Wnt binds both to the WIF domain, in the vicinity of the phospholipid head group, and to the EGF-like domains, which also provided a HSPG-binding site. These findings lead us to conclude that the structure of WIF-1 allows the interactive characteristics of the WIF domain and EGF-like domains to synergize, and suggest a modular model for the function of WIF-1 in HSPG-shaped morphogen gradients.
RESULTS
WIF domain structure contains a lipid-binding pocket
We produced a series of human WIF-1 constructs (WIF-1WD, WIF-1WD-EGF-I, WIF-1WD–EGFs I–III, WIF-1ΔC, WIF-1Full length and WIF-1EGFs I-V; Fig. 1a and Supplementary Figs. 1 and 2). The crystal structures of WIF-1WD and WIF-1WD–EGF-I were determined to 1.85-Å resolution (see Online Methods and Table 1).
Table 1.
Data collection and refinement statistics.
| WIF-1ΔC | WIF-1WD-EGF-I | WIF-1WD-EGF-I Met77Trp | WIF-1WD | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | I213 | C222 | C222 | C222 |
| Cell dimensions | ||||
| a, b, c (Å) | 178.0, 178.0, 178.0 | 51.0, 134.3, 60.4 | 50.8, 134.2, 60.2 | 49.9, 134.1, 60.4 |
| a, b, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 40.00-3.95 (4.09-3.95) |
40.00-1.85 (1.99-1.85) |
29.32-2.22 (2.28-2.22) |
30.00-1.85 (1.93- 1.85) |
| R merge | 10.0 (87.4) | 14.4 (94.5) | 9.4 (57.3) | 10.3 (70.0) |
| I/σI | 27.2 (2.8) | 15.5 (1.9) | 12.7 (2.3) | 16.3 (1.4) |
| Completeness (%) | 99.9 (99.8) | 100.0 (100.0) | 99.4 (92.0) | 98.8 (91.3) |
| Redundancy | 11.1 (10.9) | 7.3 (7.2) | 6.4 (6.0) | 5.3 (3.5) |
| Refinement | ||||
| Resolution (Å) | 39.80-3.95 | 37.42-1.85 | 29.32-2.22 | 27.53-1.85 |
| No. reflections | 8,048 | 17,107 | 10,431 | 16,407 |
| Rwork/Rfree | 29.7/33.1 | 18.4/21.7 | 18.6/22.9 | 20.4/22.4 |
| No. atoms | ||||
| Protein | 1864 | 1424 | 1426 | 1169 |
| Ligand/ion | 105 | 51 | 61 | 54 |
| Water | 0 | 151 | 111 | 83 |
| B-factors | ||||
| Protein | 222 | 35 | 50 | 43 |
| Ligand/ion | 382 | 53 | 76 | 75 |
| Water | - | 40 | 46 | 49 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.014 | 0.015 | 0.015 | 0.015 |
| Bond angles (°) | 2.1 | 2.1 | 2.2 | 2.2 |
One crystal was used for each data set. Values in parentheses are for highest-resolution shell.
The WIF domain is essentially identical in the WIF-1WD and WIF-1WD–EGF-I structures (r.m.s. deviation of 0.17 Å between equivalent Cα atoms for 144 residues). The WIF domain fold consists of a β-sandwich splayed apart along one edge by insertion of three short helices (one 310 and two α-helices; Fig. 1b and Supplementary Fig. 3). A ~10-Å gap between the α1 and 310 helices provides a gateway into a large cavity (1,515 Å3, calculated with CASTp20), lined by primarily hydrophobic residues, which penetrates deep into the core of the WIF domain. Thus, the WIF domain lacks the close-packed interior of a standard β-sandwich. The β-sheets are locked together at the base of the sandwich by a disulfide bridge (Cys140–Cys177). Sequence alignments suggest this disulfide is conserved between species and is a canonical feature of the WIF domain fold when present in other proteins (Supplementary Fig. 3b). In the WIF-1WD–EGF-I structure, the EGF-like domain (EGF I) is connected to the WIF domain by a 4-residue linker and extends away from the WIF domain into solvent. The cysteine knot topology of EGF I is stabilized by the canonical three disulfide bridges. However, the structure of this domain appears relatively disordered compared to that of the WIF domain (as judged from crystallographic B-factors and quality of electron density), probably as a result of orientational flexibility relative to the WIF domain.
The crystal structures of the WIF domain described here differ markedly (r.m.s. deviation of 2.64 Å for 144 equivalent Cα atoms) from the previously reported WIF-1 WIF domain structure determined by solution NMR9 (Supplementary Fig. 4). The WIF domain topology is largely identical between crystal and solution structures, but there is no cavity within the β-sandwich in the NMR structure: the two β-sheets form a close packed protein core. By contrast, the electron density maps for the WIF-1WD and WIF-1WD–EGF-I crystal structures show clear evidence for a lipid-like molecule bound as an integral part of the WIF domain core (Fig. 1c, d). This additional molecule fills the large hydrophobic cavity, stabilizing the splayed-out conformation of the WIF domain β-sandwich. We used no supplemental lipids during protein production and crystallization, so the molecule must have been incorporated into the WIF-1 WIF domain during mammalian expression (the NMR structure was determined from protein expressed as inclusion bodies in E. coli and refolded9). We extracted hydrophobic ligands from WIF-1ΔC expressed in HEK293 cells and identified them by MS as a series of phosphatidylcholines (Fig. 1e) that differed in acyl chain length (C14–C18) and degree of saturation (C16:0–1, C18:0–1). The main species consists of two acyl chain lipids with 16 carbon atoms each—a chemistry that is typified by the common mammalian cell membrane lipid 1,2-dipalmitoylphosphatidylcholine (DPPC). DPPC refined successfully as part of the WIF-1WD and WIF-1WD–EGF-I crystal structures. The acyl chains fitted in an extended conformation snugly within the WIF domain core, filling the available volume of the pocket, whereas the phosphatidylcholine head group was exposed to the surface and interacted with three residues (Fig. 1b–d). The ethylene group of the head group formed a hydrophobic interaction with Phe66, the glycerol backbone packed against Ile49 and the phosphatidyl oxygen made a hydrogen bond with Arg76 (Fig. 1c).
The solution structure of the refolded human WIF domain indicated an association between the WIF domain and the alkyl chain–containing detergent Brij-35 used in the refolding protocol9. This observation prompted the suggestion that the WIF-1 WIF domain might interact with the palmitoyl or palmitoleoyl chains linked to Cys77 and Ser209 of Wnt3a, respectively10, 11. To assess the lipid-binding properties of mammalian, cell-expressed WIF-1, we incubated purified WIF-1ΔC with a fluorescent lipid, 16:0 Liss Rhod PE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) ammonium salt). Gel filtration revealed that WIF-1ΔC and 16:0 Liss Rhod PE coeluted for a sample incubated in the presence of 1% CHAPS, suggesting that phosphatidylcholines bound within the core of the WIF domain, such as DPPC, could be exchanged in the presence of detergent (Supplementary Fig. 5). Under similar conditions (including 1% CHAPS), we found no binding between WIF-1ΔC (50 μM) and a fluorescent head group of 16:0 Liss Rhod PE alone (sulforhodamine B, 160 μM), consistent with the idea that the diacyl lipid chains of 16:0 Liss Rhod PE mediated its binding to WIF-1ΔC. Finally, in the light of the detergent interaction mapped to the surface of the WIF-1 WIF domain in the solution NMR analysis9, we probed WIF-1ΔC for surface-exposed lipid-binding sites using a lipid strip. We observed no binding of WIF-1ΔC to any of 15 biologically abundant lipids in the absence of 1% CHAPS (Supplementary Fig. 5f, g). These analyses, in combination with the WIF domain structures of mammalian cell–expressed constructs, show that DPPC and closely related phosphatidylcholines act as physiological components of WIF-1 and saturate the lipid-binding capacity of the WIF domain. Sequence alignments indicate that the hydrophobic nature, and, in general, the size of the residues that line the lipid-binding pocket in the WIF-1 WIF domain are conserved in other WIF domain–containing proteins (Supplementary Fig. 3b), which suggests that phosphatidylcholines such as DPPC could be integral to WIF domains in general.
Wnt3a binding and inhibition by WIF and EGF-like domains
The WIF domain of WIF-1 is sufficient to inhibit Wnt signalling7, but the function of the five EGF-like domains (EGFs I–V) has remained obscure. We therefore used our series of WIF-1 constructs (Fig. 1a) in a Wnt response assay21 to investigate the roles of the domains of WIF-1 in inhibition of Wnt3a. Wnt3a signaling was inhibited in a dose-dependent manner (33–500 nM; Fig. 2a) by WIF-1ΔC. WIF-1WD and WIF-1WD–EGF-I were also inhibitory, consistent with the previous report that the WIF domain in isolation can inhibit Wnt signaling7, but both these constructs were notably less effective than the molecules that contained the WIF domain and EGFs I–V. Thus, although EGFs I–V in isolation (construct WIF-1EGFs I–V) did not show pronounced inhibitory activity (Fig. 2a), the results of the cellular assays suggest that the EGF-like domains are involved in Wnt3a signal inhibition.
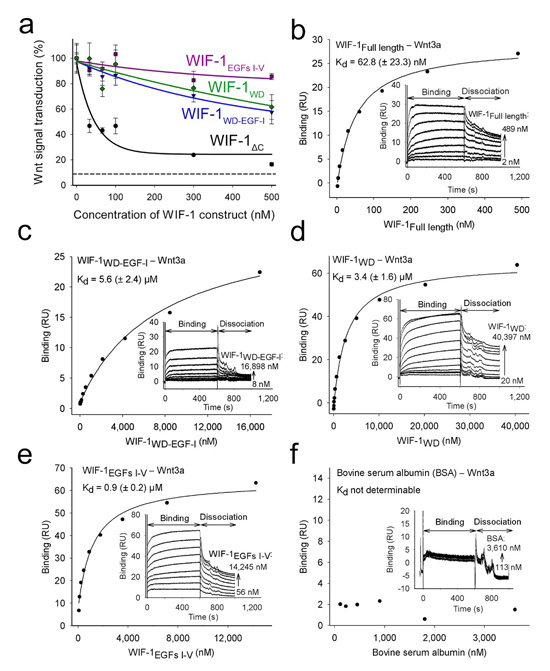
Figure 2.
The contributions of the WIF and EGF-like domains to Wnt inhibition. (a) The concentration-dependent inhibition of Wnt3a signaling in a cellular assay by four constructs of WIF-1: WIF-1ΔC (black), WIF-1WD–EGF-I (blue), WIF-1WD (green) and WIF-1EGFs I–V (magenta). The concentration of human Wnt3a was kept constant at 4.7 nM. The dashed line indicates endogenous Wnt signaling in HEK293T cells without added Wnt3a. Experiments were performed in triplicate, and error bars show s.d. (b–f) Binding of WIF-1 constructs and BSA (control) to mouse Wnt3a. Shown are WIF-1Full length (b); WIF-1WD–EGF-I (c); WIF-1WD (d); WIF-1EGFs I–V (e); and BSA (f). Different concentrations of WIF-1 constructs were injected over surface coupled with mouse Wnt3a. Insets, representative plots of the binding response (response units (RU)) derived from the sensorgrams as a function of WIF-1 construct (or BSA) concentration. Kd values (±s.d.) calculated from three independent experiments are shown.
To analyze the biophysical basis of the difference in efficiency of Wnt3a inhibition by the WIF domain and molecules containing the WIF domain plus EGFs I–V, we probed the interactions between the different WIF-1 domains and Wnt3a using a surface plasmon resonance (SPR) binding assay (Fig. 2b–f and Table 2). In agreement with the cellular assay, binding was tightest between Wnt3a and WIF-1ΔC, with a dissociation constant (Kd) of 21.6 ± 8.0 nM. The full-length WIF-1 construct has a comparable binding affinity (Kd 62.8 ± 23.3 nM), indicating that the hydrophilic C-terminal region makes no contribution to Wnt3a binding, consistent with a sequence-based prediction of flexibility (data not shown) and evolutionary divergence (Supplementary Fig. 1). These SPR-based dissociation constants agree with a previously reported Kd of 16 nM derived from plate-based affinity assays for the interaction of Xenopus Wnt8 with human WIF-1 (ref. 7). WIF-1WD and WIF-1WD–EGF-I bound much more weakly to Wnt3a (Kd 3.4 ± 1.6 and 5.6 ± 2.4 μM, respectively). There was substantial binding affinity between WIF-1EGFs I–V and Wnt3a (Kd 0.9 ± 0.2 μM), suggesting that the differences in binding in the cellular assays resulted from a direct interaction between EGFs I–V and Wnt3a in addition to the interaction with the WIF domain. To provide further evidence of this additive effect, we tested an additional construct, WIF-1WD–EGFs I–III, in the cellular and SPR assays (Fig. 3a and Table 2). Again, binding and inhibition of Wnt3a were lower than for WIF-1ΔC. This suggests that EGFs IV and V are necessary for the contribution of the EGF I–V unit to Wnt3a binding, but we cannot exclude the possibility that the reduced Wnt3a binding is an indirect effect arising from perturbation of EGF III because of the truncation at the EGF III–EGF IV interface (Supplementary Fig. 2c). Together, the cellular and SPR binding assays are consistent with a modular model in which both the WIF domain and the EGF-like domains of WIF-1 interact with Wnt3a and contribute to signal inhibition.
Table 2.
Dissociation constants of human WIF-1 constructs to mouse Wnt3a.
| WIF-1 construct | Dissociation constant, Kd (nM) |
|---|---|
| WIF-1WD | 3400 ± 1600 |
| WIF-1WD-EGF-I | 5600 ± 2400 |
| WIF-1WD-EGFs I-III | 3210 ± 670 |
| WIF-1ΔC | 21.6 ± 8.0 |
| WIF-1EGFs I-V | 900 ± 200 |
| WIF-1Full length | 62.8 ± 23.3 |
| WIF-1Full length with 2 mM CaCl2 pH 8.0 (buffer A) | 190.0 ± 34.9 |
| WIF-1Full length with 2 mM EDTA pH 8.0 (buffer A) | 167.6 ± 12.4 |
| WIF-1WD-EGF-I (buffer B) | 9860 ± 1150 |
| WIF-1WD-EGF-I with 2 mM phosphocholine chloride (buffer A) | 15900 ± 930 |
| WIF-1WD-EGF-I with 2 mM phosphocholine chloride (buffer B) | 16900 ± 4400 |
| WIF-1WD-EGF-I with 2 mM CaCl2 pH 7.8 (buffer B) | 16800 ± 1970 |
| WIF-1ΔC Met77Trp | 217.0 ± 26.0 |
| WIF-1WD-EGF-I Met77Trp | 2860.0 ± 530.0 |
| Bovine serum albumin | Not determinable |
Measurements were done by SPR in PBS with 1% CHAPS buffer unless stated otherwise (see Supplementary Methods).
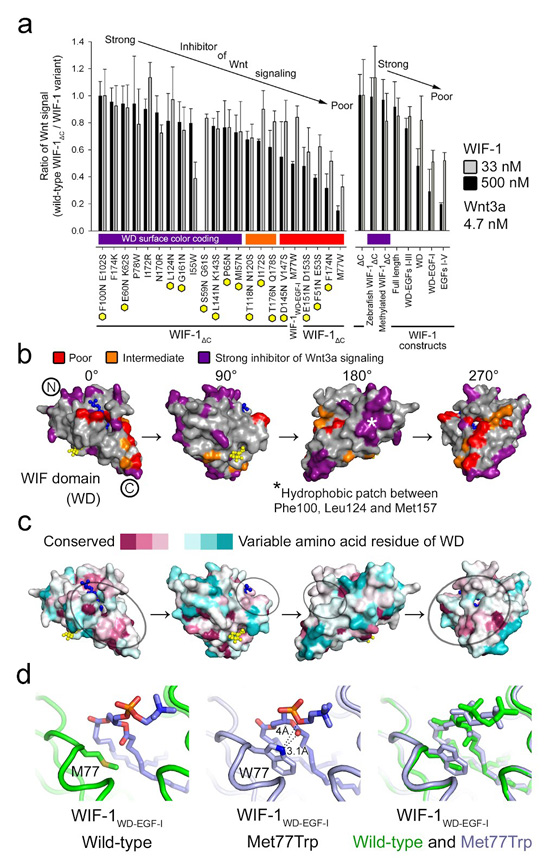
Figure 3.
A discontinuous Wnt-binding site on the WIF domain. (a) Inhibition of Wnt3a signaling by mutation-bearing (left) and wild-type WIF-1 constructs (right). Inhibition of each WIF-1 construct was compared to the wild-type WIF-1ΔC in the cellular assay. Higher ratio (0.8–1) of Wnt signal (wild-type WIF-1ΔC/WIF-1 variant) corresponds to strong inhibition and lower (0.2–0.5) to poor inhibition. Inhibition of Wnt3a (4.7 nM) signaling was tested in the presence of 33 nM (gray bar) and 500 nM (black bar) of the WIF-1 variant. Yellow hexagons indicate mutations predicted to introduce N-linked glycosylation sites. (b) Residues tested in the cellular assay a for Wnt3a signaling inhibition mapped on the surface of the WIF domain. DPPC (blue) and GlcNAc (yellow) are shown as balls and sticks. Residues for which mutations resulted in poor, intermediate and strong inhibition of Wnt3a signaling are colored red, orange and purple, respectively; Phe174 and Ile172 are colored red and orange, respectively. A hydrophobic patch on the surface of the WIF domain (180° view) is marked with an asterisk. (c) The surface of the WIF domain colored by residue conservation (conserved, magenta; variable, cyan). Fourteen Wnt-binding WIF domains (Supplementary Fig. 3b) are included in the sequence conservation analysis. WIF domains from Hh-binding Shifted are excluded. A potential Wnt3a-binding surface is circled. (d) WIF-1WD–EGF-I wild-type (left) and WIF-1WD–EGF-I Met77Trp (middle) crystal structures are superimposed on each other (right). DPPC is shown as sticks.
The Wnt3a-binding site on the WIF domain
We used our high-resolution crystal structures to identify suitable residues for mutagenesis to map out areas of the WIF domain surface that were involved in Wnt3a recognition. We individually mutated 15 surface-exposed sequences to introduce asparagine-linked glycosylation sites (Supplementary Fig. 6) and evaluated any resulting hindrance to WIF-1–Wnt3a recognition using the cellular assay (Fig. 3a and Supplementary Fig. 7a). We also used six additional single-residue mutations for finer-scale investigation of potential binding surfaces (Fig. 3a and Supplementary Fig. 7b). Out of the 21 WIF-1ΔC variants assayed, Phe174Asn and Phe51Asn Glu53Ser, for which the mutations were predicted to introduce N-linked glycosylation sites, and Met77Trp, a single point mutation, were the weakest inhibitors of Wnt3a signaling. In the context of WIF-1ΔC, the Met77Trp mutation also showed a ten-fold decrease in binding affinity to Wnt3a in the SPR assay (Table 2).
The chemical conversion of lysines to dimethyllysines22 did not impair WIF-1 function (Fig. 3a and Supplementary Fig. 7c), suggesting that this modification does not interfere with the binding of Wnt3a. We also assessed the effect of sequence variation between species and found that zebrafish WIF-1 inhibited Wnt3a signaling as efficiently as the human ortholog (Fig. 3a and Supplementary Fig. 7d), consistent with the Wnt3a-binding surface on WIF-1 being highly conserved. The combined results of the mutagenesis, lysine methylation and species variation analyses highlight a discontinuous but discrete area of the WIF domain surface as a putative Wnt3a-binding site (Fig. 3b, c).
Within the putative Wnt3a-binding site, the effects of the Phe174Asn and Met77Trp variations on function highlight the region in the vicinity of the lipid head group (Fig. 3a, b). Met77 is partially buried at the entrance to the lipid-binding cavity. To assess changes introduced by this mutation, we determined the crystal structure of WIF-1WD–EGF-I Met77Trp at 2.22-Å resolution (Table 1). Superposition of the wild-type and mutant WIF-1WD–EGF-I structures revealed no major long-range perturbations (r.m.s. deviation of 0.22 Å for 170 equivalent Cα atoms), and the acyl chains of the DPPC lipid remained buried within the core of the WIF domain. However, unlike the side chain of Met77, which makes no direct contact with the lipid in the wild-type structure, the tryptophan side chain in WIF-1WD–EGF-I Met77Trp formed a hydrogen bond with a carbonyl oxygen of the palmitoyl tail group and the oxygen atom of the phosphatidylcholine head group, and the indole ring provided stacking interactions that further restricted the orientation of the lipid head group (Fig. 3d). We assessed whether lipid exchange was still possible in the presence of these stacking interactions and found that 16:0 Liss Rhod PE could still exchange the natively bound phosphatidylcholines (data not shown). Thus, the notable reduction in the ability of the WIF-1 Met77Trp mutant to inhibit Wnt3a signaling suggests that Met77 and the protein surface proximal to the lipid head group are involved in the Wnt3a-binding site. The involvement of the lipid may be indirect and the contribution of the head group is certainly limited; we could not inhibit the binding of wild-type WIF-1WD–EGF-I to Wnt3a in SPR experiments using a 40-fold excess of the lipid head group analog phosphocholine (Table 2).
The WIF-1 structure and orientation of the EGF-like domains
To investigate further the modular mechanism of Wnt signal inhibition, we determined the crystal structure of WIF-1ΔC to 3.95-Å resolution (Table 1).The structure showed the lipid-containing WIF domain loosely wrapped by EGFs I–III, with EGFs IV and V extending away into the solvent (Fig. 4a, b). The WIF domain retained the structure of WIF-1WD (r.m.s. deviation of 0.53 Å for 143 equivalent Cα atoms) and DPPC again provided a suitable model for bound lipid density. The extended conformation of the acyl tails in the WIF domain lipid-binding cavity was conserved between the WIF-1WD, WIF-1WD–EGF-I and WIF-1ΔC structures, but the position of the DPPC head group in WIF-1ΔC deviated markedly from that observed for WIF-1WD, WIF-1WD–EGF-I and WIF-1WD–EGF-I Met77Trp. The head group was oriented toward the C terminus of the WIF domain and the linker to EGF I in the WIF-1ΔC structure, whereas it was oriented in essentially the opposite direction in the crystal structures of the shorter constructs (Fig. 4c). This reorientation seems to occur because of differences in the packing of neighboring molecules in the various crystal lattices, suggesting that the orientation of the lipid head group can alter in response to external interactions.
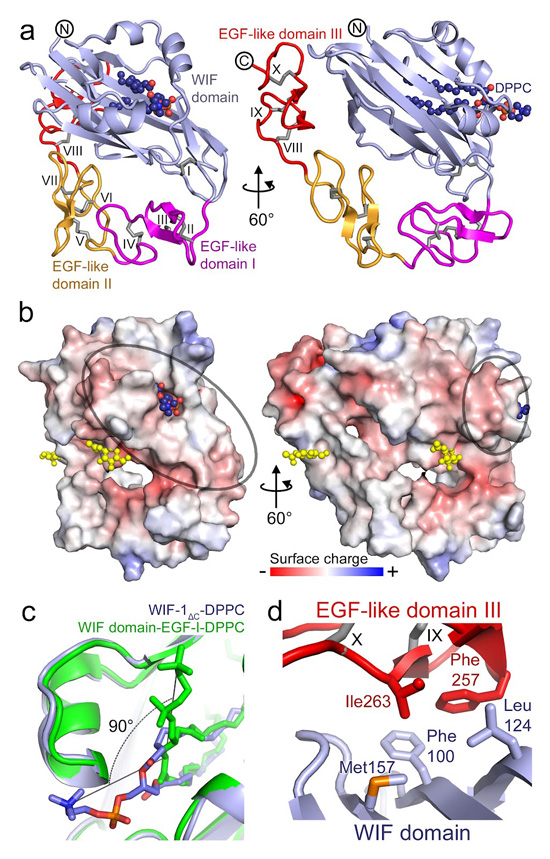
Figure 4.
The structure of WIF-1ΔC. (a) Ribbon diagram of WIF-1ΔC. The orientation of the WIF domain is the same as in Fig. 1b. The WIF domain and EGFs I–III are colored light blue, magenta, yellow and red, respectively. Ten disulfide bridges are shown as gray sticks and marked with Roman numerals. DPPC is shown as spheres. (b) Electrostatic properties of WIF-1ΔC. The protein is shown as solvent-accessible surface colored by electrostatic potential contoured at ± 8 kT/e (red, acidic; blue, basic). GlcNAc moieties on Asn88 (WIF domain) and Asn245 (EGF III) are shown as yellow spheres. A potential Wnt3a-binding surface is circled. (c) Orthogonal relative orientation of the DPPC head group between superimposed structures of WIF-1ΔC (light blue; DPPC colored as in Fig. 1c, d) and WIF-1WD–EGF-I (green). (d) Hydrophobic contact formed between the WIF domain hydrophobic patch (marked with an asterisk in Fig. 3b, 180° view) and EGF III. Interacting residues are shown as sticks.
The second notable difference between the WIF-1ΔC and WIF-1WD–EGF-I structures is the relative orientation of EGF I and the WIF domain: EGF I was rotated by some 30° toward the WIF domain in WIF-1ΔC. With this orientation, EGFs I–V wrap back along the WIF domain (Fig. 4a, b) so that residues Phe257 and Ile263 of EGF III make hydrophobic interactions with WIF domain residues Phe100, Leu124 and Met157 (Fig. 3b, asterisk on the 180° view; Fig. 4d). Three variants of WIF-1ΔC with mutations designed to disrupt this interface by introducing N-linked glycosylation sites (Phe100Asn Glu102Ser, Leu124Asn and Met157Asn) did not show impaired inhibitory function (Figs. 3a, 4d), arguing against the idea that the hydrophobic patch is directly involved in Wnt3a recognition. The interface between the WIF domain and EGF III is not extensive (total buried surface area 260 Å2, calculated with PISA23), but it does serve to define a preferred relative orientation between the WIF domain and EGFs I–V. Several observations suggest that EGFs I–V would have substantial inter-domain flexibility. The different orientation of EGF I in the WIF-1WD–EGF-I structure indicates that the linker between the WIF domain and EGF I is flexible, and the electron density for EGFs IV–V in the WIF-1ΔC crystal structure was fragmentary, consistent with considerable flexibility, so we omitted these domains from the final refined coordinates for WIF-1ΔC. The WD–EGF III interface stabilizes the ‘wrapped-back’ orientation of the EGF-like domains in the WIF-1ΔC crystal, and the juxtaposition of the WIF domain and EGFs I–V is consistent with both regions contributing to an extended Wnt3a-binding surface. Thus the structure of WIF-1ΔC, in combination with the results of the cellular Wnt inhibition assay and solution binding studies, suggest a modular mechanism for Wnt binding that combines sites on the WIF domain and EGFs I–V.
WIF-1 binds glycosaminoglycans via the EGF-like domains
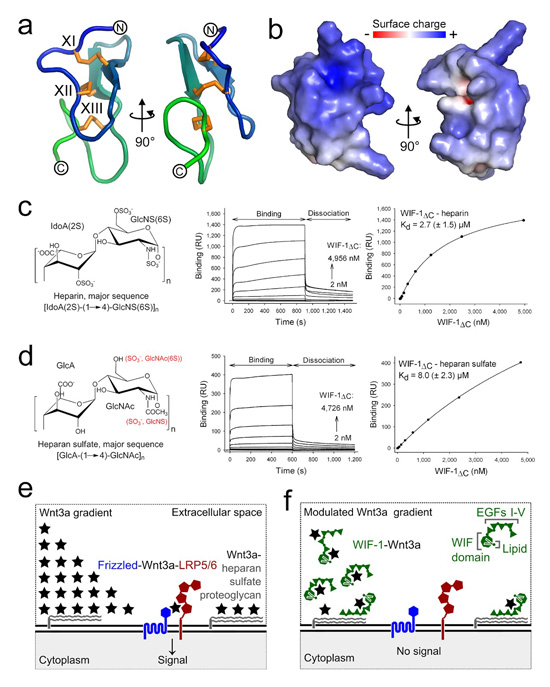
We hypothesized that, in addition to contributing to Wnt binding, EGFs I–V might have a further, distinctive role in Wnt inhibition. Eight evolutionarily conserved arginines and lysines in the sequence of EGF IV (Supplementary Fig. 8a), when mapped into the modeled structure of the domain, generate a highly positively charged surface (Fig. 5a, b), suggesting that WIF-1 might bind to highly sulfated, negatively charged glycosaminoglycans (GAGs). We studied interactions between WIF-1ΔC, WIF-1WD–EGF-I and four GAGs using the SPR binding assay. Binding was tightest between WIF-1ΔC and the most sulfated GAG, heparin (Kd 2.7 ± 1.5 μM; Fig. 5c). Binding was weaker between WIF-1ΔC and the second most sulfated GAG, heparan sulfate (Kd 8.0 ± 2.3 μM; Fig. 5d). We detected no binding between WIF-1WD–EGF-I and any of the GAGs, suggesting that interactions between GAGs and WIF-1 are mediated by EGFs II–V.
Figure 5.
HSPG binding contributes to a modular mechanism for WIF-1 function. (a) Homology model of the human WIF-1 EGF IV as a ribbon diagram. Disulfide bridges XI–XIII are shown as orange sticks. (b) Electrostatic properties of WIF-1 EGF IV model. The protein is shown as solvent-accessible surface colored by electrostatic potential contoured at ±8 kT/e (red, acidic; blue, basic). (c, d) Chemical structures of heparin (c) and heparan sulfate (d; left), binding sensorgrams of WIF-1ΔC to heparin and heparan sulfate (middle), and representative plots of the binding response (response units (RU)) derived from the sensorgram as a function of WIF-1ΔC concentration (right). Kd values (± s.d.) calculated from three independent experiments are shown. (e) The Wnt–β-catenin pathway is initiated at the cell surface when secreted Wnt3a (black star) forms a ternary complex with the cell-surface receptors Frizzled (blue) and LRP5/6 (red). This results in downstream signal transduction and activation of Wnt target genes. HSPGs (gray) on the cell surface bind to Wnts and modulate their extracellular gradient. (f) The combination of HSPG- and Wnt-binding properties provides a modular mechanism for controlling the localization of WIF-1 (green) and hence Wnt-signal inhibition within the Wnt concentration gradient.
We used WIF-1WD–EGFs I–III to further define the individual contributions of the EGFs, measuring binding to heparin and heparan sulfate in the SPR assay. The resulting sensorgrams (Supplementary Fig. 8b, c) showed qualitatively different binding characteristics from these of WIF-1ΔC and could not be fitted to a simple 1:1 binding model to give an accurate measure of Kd. WIF-1ΔC did not show detectable binding to the least sulfated GAGs, chondroitin sulfate and chondroitin 4-sulfate. The correlation between higher GAG sulfation and tighter binding to WIF-1ΔC suggests that electrostatic interactions between negatively charged GAG sulfate groups and positively charged EGF residues are important. Consistent with the SPR binding assay, EGFs I–V and WIF-1ΔC (but not WIF-1WD–EGF-I) bound to a heparin column (Supplementary Fig. 8d). This binding was disrupted under conditions of low ionic strength (250 mM and 300 mM NaCl for WIF-1ΔC and EGFs I–V, respectively; Supplementary Fig. 8d), consistent with the idea that electrostatic interactions are the main drivers of GAG binding. Within EGFs II–V the conserved cluster of lysines and arginines on EGF IV provides the probable focus for the HSPG-binding properties of WIF-1.
DISCUSSION
The WIF domain lipid-binding cavity and interaction with Wnt
A solution structure of the refolded human WIF-1 WIF domain provided evidence for a surface interaction between the WIF domain and the single alkyl chain detergent Brij-35 (C12H25(OCH2CH2)nOH, n ≈ 23) that was used in the refolding protocol9. This observation raised the possibility that the WIF-1 WIF domain interacted with Wnt3a-linked palmitoyl or palmitoleoyl chains10, 11. Our results show a deep hydrophobic pocket in the WIF domain but argue against a binding mode involving insertion of Wnt3a-linked lipid; exchange of the integral WIF-1–bound diacyl lipid with a single-chain Wnt3a-linked lipid would be energetically very unfavorable in the aqueous extracellular environment. Conversely, the WIF domain with diacyl lipid bound lacks an external hydrophobic surface suitable for lipid binding, and we observed no binding of WIF-1ΔC to 15 different, biologically abundant lipids immobilized on hydrophobic membranes (including examples that were structurally similar to Wnt3a-linked moieties); these observations do not support the existence of interactions between the WIF-1 surface and Wnt3a-linked lipids.
Thus, the WIF-1 WIF domain does not seem to share the characteristics of lipoprotein particles17 that can sequester Wnt3a-linked lipids. So what is the role of the diacyl lipid that is an integral part of the mammalian cell-expressed WIF-1 WIF domain? The lipid clearly contributes to the structural integrity of the WIF domain in the crystal structure, but the NMR structure shows that the WIF domain fold is stable in the absence of diacyl phospholipid, albeit in a somewhat collapsed form. Thus, the role of the lipid may not be limited to stabilization of the WIF domain fold. It has been shown that a deletion of two residues (Glu-Leu) in a conserved region of the WIF domain of the Caenorhabditis elegans Wnt receptor RYK LIN-18 abolishes its activity24. At the equivalent positions human WIF-1 has Asp54 and Ile55, which mark the entry into the lipid-binding cavity (Supplementary Fig. 3b). This two-residue deletion might severely disturb the WIF domain structure or lipid binding (or both). Our mapping of the Wnt3a-binding surface is consistent with previous studies24 in implicating an area of the WIF domain surface close to the lipid head group. The involvement of the lipid in Wnt3a binding might be indirect, through the stabilization of the appropriate protein surface; our results do not provide evidence that the lipid head group makes the major contribution to direct interactions with Wnt3a. One possibility is that the head group contributes to the binding properties of the WIF-1 WIF domain indirectly by providing conformational flexibility, allowing exposure of the appropriate surface on ligand binding.
Our results argue for the role of the WD-lipid unit in the protein interactions mediated by WIF-1. Is this likely to be a general characteristic of WD-containing molecules? Sequence analyses have identified WDs in a rather limited number of proteins: WIF-1 in vertebrates, the RYKs (Wnt receptors related to tyrosine kinases), and Shifted (a Hh-binding, rather than Wnt-binding, ortholog of WIF-1 in Drosophila25, 26) (Supplementary Fig. 3b). For each of these proteins except Shifted, the WD has been identified as a site for protein-protein interactions. Sequence analysis points to conservation of the WD lipid-binding cavity in these proteins, however, further studies will be required to establish whether this lipid-mediated contribution to protein-protein recognition is maintained across the WD family.
The contributions of the EGF-like domains to WIF-1 function
EGFs I–V are required for some aspects of WIF-1 function in vivo. Human WIF-1 blocks Wnt8-dependent axis induction in early Xenopus embryos, and the WIF domain in isolation is as effective as full-length WIF-1 (ref. 7). However, injections of RNA for Chordin (an inhibitor of bone morphogenetic protein function) and full-length WIF-1 produce different morphological effects in the developing Xenopus embryo from injections of Chordin and the WIF domain, suggesting that EGFs I–V are essential for the full activity of WIF-1 (ref. 7). The molecular mechanism(s) by which the EGF-like domains contribute to WIF-1 function have remained obscure. Our results reveal two mechanisms by which EGFs I–V might act. First, EGFs I–V bind directly to Wnt3a in SPR experiments and WIF-1 constructs lacking EGFs I–V, II–V or IV–V inhibit Wnt3a signaling less effectively in a luciferase reporter assay than constructs containing both the WIF domain and EGFs I–V. Thus the EGF-like domains may either contribute a second component to an extended Wnt-binding surface or provide a second, independent site for Wnt binding (Fig. 5e, f). Second, WIF-1 binds directly to heparin and heparan sulfate through EGFs II–V, consistent with a highly conserved cluster of positively charged residues on EGF IV. These observations suggest a potential mechanism by which to link WIF-1 localization and inhibitory activity to morphogen gradients (Fig. 5e, f).
Genetic studies on Shifted, the Drosophila ortholog of WIF-1, suggest that EGFs I–V mediate interactions with Hh and HSPGs and are required for their colocalization25, 26. HSPGs provide a mechanism for localizing proteins and assemblies (for example, lipoprotein particles associated with Wnts and Hhs17) near the target cell surface. Extracellular concentration and Wnt signaling are reduced in Drosophila cells defective for EXT glycosyltransferases, which are required for the synthesis of heparan sulfate27, and mutations in human orthologs cause hereditary diseases28. Injection of the heparin-degrading enzyme heparinase into wild-type Drosophila embryos results in the degradation of heparin-like GAGs and a wingless-like phenotype29.
The ability of WIF-1 to bind GAGs through evolutionarily conserved EGFs provides a mechanism for the localization of WIF-1 and Shifted within Wnt and Hh morphogen gradients, respectively. Several Wnt signaling inhibitors bind to heparin, including Sclerostin30, Dkk-1 (ref. 31) and sFRP-1 (ref. 32). Similarly to WIF-1, the two-domain sFRP-1 has been reported to use the N-terminal (cysteine-rich) domain for Wnt binding33 and the C-terminal (netrin-like) domain for heparin binding34. Our structural and functional studies suggest a modular model for human WIF-1 that exemplifies how the interplay between Wnt- and GAG-binding sites in Wnt antagonists might link inhibition to the morphogen gradient.
METHODS
Protein expression and purification
We cloned human (Swiss-Prot Q9Y5W5; isoform AAH18037, Gln166Lys; WIF-1WD, residues 35–178; WIF-1WD–EGF-I, residues 35–210; WIF-1WD–EGFs I–III, residues 35–274; WIF-1ΔC, residues 35–346; WIF-1Full length, residues 29–379; WIF-1EGFs I–V, residues 178–346) and zebrafish (residues 33–336) WIF-1 constructs into the pHLsec vector35, resulting in secreted proteins with N-terminal Glu-Thr-Gly and C-terminal Gly-Thr-Lys-His6 tags. Proteins were expressed by transient transfection in GnTI(−) HEK293S cells36 and purified from dialyzed (against PBS buffer) conditioned medium by immobilized Co2+-affinity chromatography. For crystallization, proteins were partially deglycosylated with endo-β-N-acetylglucosaminidase F1 (ref. 37); glycosylation was preserved for cellular and SPR assays. Lysine methylation22 was carried out to optimize crystal growth for WIF-1WD, WIF-1WD–EGF-I, and WIF-1WD–EGF-I Met77Trp. Proteins were concentrated and further purified by size-exclusion chromatography in 20 mM Tris-HCl, pH 8.0, 300 mM NaCl. Mouse Wnt3a was purified as described19 with minor modifications: conditioned medium was dialyzed before blue Sepharose fractionation, Cu2+-affinity chromatography was replaced with Ni2+-affinity chromatography, and a heparin-affinity purification step was omitted.
Crystallization and structure determination
WIF-1 constructs were crystallized at 21 °C (WIF-1WD in 30% (w/v) PEG 8000, 200 mM sodium acetate, 100 mM sodium cacodylate, pH 6.5; WIF-1WD–EGF-I, 1.3 M diammonium tartrate, 100 mM bis-tris propane, pH 7.0; WIF-1WD–EGF-I Met77Trp, 1.1 M diammonium tartrate pH 7.0, 20 mM spermidine, 0.5 mM CaCl2; WIF-1ΔC, 0.75 M sodium succinate, pH 5.7, 35 mg ml−1 sucrose octasulfate) using nanoliter-scale automated pipetting procedures38. Crystals were flash frozen by immersion of the crystal into a reservoir solution containing 25% (WIF-1ΔC) or 30% v/v (WIF-1WD, WIF-1WD–EGF-I, WIF-1WD–EGF-I Met77Trp) glycerol followed by transfer to liquid nitrogen. The crystals were kept at 100 K during X-ray diffraction data collection. Data were collected at beamlines BM14 (WIF-1WD, λ = 0.978 Å) and ID23.2 (WIF-1 WD–EGF-I, λ = 0.873 Å) at the European Synchrotron Radiation Facility (France) and at beamlines I03 (WIF-1ΔC, λ = 0.976 Å) and I24 (WIF-1WD-EGF-I Met77Trp, λ = 0.978 Å) at the Diamond Light Source (UK). Structures were phased by molecular replacement using standard protocols (Supplementary Fig. 9). Ramachandran statistics for the refined coordinates (residues in favored region (%), number of outliers): WIF-1ΔC (93.7, 0), WIF-1 WD–EGF-I (98.8, 0), WIF-1WD–EGF-I Met77Trp (95.4, 0), WIF-1WD (97.8, 0). Further information is provided in Supplementary Methods. Crystallographic statistics are shown in Table 1.
Site-directed mutagenesis
Mutations were generated by a two-step overlapping PCR. PCR products were cloned into the pHLsec vector. All constructs were verified by DNA sequencing. Proteins were expressed and purified as described for the wild-type constructs.
Binding studies and cellular assays
SPR binding studies used a Biacore T100 with WIF-1 constructs as analytes and either Wnt3a or GAGs immobilized on the sensor chip. Kd values were calculated for a 1:1 Langmuir model. For mass spectroscopy (MS) analysis, WIF-1–bound lipids were extracted into CHCl3/CH3OH and analyzed by MALDI-TOF MS. Interactions between WIF-1ΔC and Liss Rhod PE, sulforhodamine B and 15 biologically abundant lipids were evaluated by fluorescence intensity or western blotting and chemiluminescence. The Wnt3a-responsive cellular assay was performed using the Super-TOPFlash plasmid21. Further details for all these analyses are in the Supplementary Methods.
Accession codes
Coordinates and structure factors for WIF-1ΔC (2YGQ), WIF-1WD–EGF-I (2YGO), WIF-1WD–EGF-I Met77Trp (2YGP) and WIF-1WD (2YGN) have been deposited in Protein Data Bank under the accession codes indicated in parentheses.
Supplementary Material
Acknowledgments
We thank the staff of the European Synchrotron Radiation Facility and Diamond Light Source for assistance with data collection; T.S. Walter and K. Harlos for help with crystallization; B.M. Kessler and K. di Gleria for MS analysis; B.J.C. Janssen for help with solvent flattening; and E. Seiradake, Y. Zhao and M.A. Jones for help with tissue culture. T.M. and E.Y.J. are funded by Cancer Research UK, C.S. by the Wellcome Trust and A.R.A. by the UK Medical Research Council.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
REFERENCES
- 1.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 4.Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 5.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 6.Surmann-Schmitt C, Widmann N, Dietz U, Saeger B, Eitzinger N, Nakamura Y, Rattel M, Latham R, Hartmann C, von der Mark H, Schett G, von der Mark K, Stock M. Wif-1 is expressed at cartilage-mesenchyme interfaces and impedes Wnt3a-mediated inhibition of chondrogenesis. J. Cell Sci. 2009;122:3627–3637. doi: 10.1242/jcs.048926. [DOI] [PubMed] [Google Scholar]
- 7.Hsieh JC, Kodjabachian L, Rebbert ML, Rattner A, Smallwood PM, Samos CH, Nusse R, Dawid IB, Nathans J. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- 8.Nakaya N, Lee HS, Takada Y, Tzchori I, Tomarev SI. Zebrafish olfactomedin 1 regulates retinal axon elongation in vivo and is a modulator of Wnt signaling pathway. J. Neurosci. 2008;28:7900–7910. doi: 10.1523/JNEUROSCI.0617-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liepinsh E, Bányai L, Patthy L, Otting G. NMR structure of the WIF domain of the human Wnt-inhibitory factor-1. J. Mol. Biol. 2006;357:942–950. doi: 10.1016/j.jmb.2006.01.047. [DOI] [PubMed] [Google Scholar]
- 10.Malinauskas T. Docking of fatty acids into the WIF domain of the human Wnt inhibitory factor-1. Lipids. 2008;43:227–230. doi: 10.1007/s11745-007-3144-3. [DOI] [PubMed] [Google Scholar]
- 11.Bazan JF, de Sauvage FJ. Structural ties between cholesterol transport and morphogen signaling. Cell. 2009;138:1055–1056. doi: 10.1016/j.cell.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Kansara M, Tsang M, Kodjabachian L, Sims NA, Trivett MK, Ehrich M, Dobrovic A, Slavin J, Choong PF, Simmons PJ, Dawid IB, Thomas DM. Wnt inhibitory factor 1 is epigenetically silenced in human osteosarcoma, and targeted disruption accelerates osteosarcomagenesis in mice. J. Clin. Invest. 2009;119:837–851. doi: 10.1172/JCI37175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazieres J, He B, You L, Xu Z, Lee AY, Mikami I, Reguart N, Rosell R, McCormick F, Jablons DM. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–4720. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, You L, Xu Z, Kuchenbecker K, Raz D, He B, Jablons D. Wnt inhibitory factor inhibits lung cancer cell growth. J. Thorac. Cardiovasc. Surg. 2007;133:733–737. doi: 10.1016/j.jtcvs.2006.09.053. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y, Simoneau AR, Liao WX, Yi G, Hope C, Liu F, Li S, Xie J, Holcombe RF, Jurnak FA, Mercola D, Hoang BH, Zi X. WIF1, a Wnt pathway inhibitor, regulates SKP2 and c-myc expression leading to G1 arrest and growth inhibition of human invasive urinary bladder cancer cells. Mol. Cancer Ther. 2009;8:458–468. doi: 10.1158/1535-7163.MCT-08-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabata T, Takei Y. Morphogens, their identification and regulation. Development. 2004;131:703–712. doi: 10.1242/dev.01043. [DOI] [PubMed] [Google Scholar]
- 17.Panáková D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435:58–65. doi: 10.1038/nature03504. [DOI] [PubMed] [Google Scholar]
- 18.Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb. Perspect. Biol. 2009;1:a002493. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, 3rd, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 20.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DasGupta R, Kaykas A, Moon RT, Perrimon N. Functional genomic analysis of the Wnt-wingless signaling pathway. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 22.Walter TS, Meier C, Assenberg R, Au KF, Ren J, Verma A, Nettleship JE, Owens RJ, Stuart DI, Grimes JM. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Inoue T, Oz HS, Wiland D, Gharib S, Deshpande R, Hill RJ, Katz WS, Sternberg PW. C. elegans LIN-18 is a Ryk ortholog and functions in parallel to LIN-17/Frizzled in Wnt signaling. Cell. 2004;118:795–806. doi: 10.1016/j.cell.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 25.Glise B, Miller CA, Crozatier M, Halbisen MA, Wise S, Olson DJ, Vincent A, Blair SS. Shifted, the Drosophila ortholog of Wnt inhibitory factor-1, controls the distribution and movement of Hedgehog. Dev. Cell. 2005;8:255–266. doi: 10.1016/j.devcel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Gorfinkiel N, Sierra J, Callejo A, Ibañez C, Guerrero I. The Drosophila ortholog of the human Wnt inhibitor factor Shifted controls the diffusion of lipid-modified Hedgehog. Dev. Cell. 2005;8:241–253. doi: 10.1016/j.devcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 27.Takei Y, Ozawa Y, Sato M, Watanabe A, Tabata T. Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development. 2004;131:73–82. doi: 10.1242/dev.00913. [DOI] [PubMed] [Google Scholar]
- 28.Ahn J, LÜdecke HJ, Lindow S, Horton WA, Lee B, Wagner MJ, Horsthemke B, Wells DE. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1) Nat Genet. 1995;11:137–143. doi: 10.1038/ng1095-137. [DOI] [PubMed] [Google Scholar]
- 29.Binari RC, Staveley BE, Johnson WA, Godavarti R, Sasisekharan R, Manoukian AS. Genetic evidence that heparin-like glycosaminoglycans are involved in wingless signaling. Development. 1997;124:2623–2632. doi: 10.1242/dev.124.13.2623. [DOI] [PubMed] [Google Scholar]
- 30.Veverka V, Henry AJ, Slocombe PM, Ventom A, Mulloy B, Muskett FW, Muzylak M, Greenslade K, Moore A, Zhang L, Gong J, Qian X, Paszty C, Taylor RJ, Robinson MK, Carr MD. Characterization of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation. J. Biol. Chem. 2009;284:10890–10900. doi: 10.1074/jbc.M807994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fedi P, Bafico A, Nieto Soria A, Burgess WH, Miki T, Bottaro DP, Kraus MH, Aaronson SA. Isolation and biochemical characterization of the human Dkk-1 homologue, a novel inhibitor of mammalian Wnt signaling. J. Biol. Chem. 1999;274:19465–19472. doi: 10.1074/jbc.274.27.19465. [DOI] [PubMed] [Google Scholar]
- 32.Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, Rudikoff S, Aaronson SA, Varmus HE, Rubin JS. Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc. Natl. Acad. Sci. USA. 1997;94:6770–6775. doi: 10.1073/pnas.94.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bafico A, Gazit A, Pramila T, Finch PW, Yaniv A, Aaronson SA. Interaction of frizzled related protein (FRP) with Wnt ligands and the frizzled receptor suggests alternative mechanisms for FRP inhibition of Wnt signaling. J. Biol. Chem. 1999;274:16180–16187. doi: 10.1074/jbc.274.23.16180. [DOI] [PubMed] [Google Scholar]
- 34.Üren A, Reichsman F, Anest V, Taylor WG, Muraiso K, Bottaro DP, Cumberledge S, Rubin JS. Secreted frizzled-related protein-1 binds directly to Wingless and is a biphasic modulator of Wnt signaling. J. Biol. Chem. 2000;275:4374–4382. doi: 10.1074/jbc.275.6.4374. [DOI] [PubMed] [Google Scholar]
- 35.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 36.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang VT, Crispin M, Aricescu AR, Harvey DJ, Nettleship JE, Fennelly JA, Yu C, Boles KS, Evans EJ, Stuart DI, Dwek RA, Jones EY, Owens RJ, Davis SJ. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15:267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter TS, Diprose JM, Mayo CJ, Siebold C, Pickford MG, Carter L, Sutton GC, Berrow NS, Brown J, Berry IM, Stewart-Jones GB, Grimes JM, Stammers DK, Esnouf RM, Jones EY, Owens RJ, Stuart DI, Harlos K. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr. D Biol. Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.