Summary
Objective
Many fundamental patterns of coinfection (multi-species infections) are undescribed, including the relative frequency of coinfection by various pathogens, differences between single-species infections and coinfection, and the burden of coinfection on human health. We aimed to address the paucity of general knowledge on coinfection by systematically collating and analysing data from recent publications to understand the types of coinfection and their effects.
Methods
From an electronic search to find all publications from 2009 on coinfection and its synonyms in humans we recorded data on i) coinfecting pathogens and their effect on ii) host health and iii) intensity of infection.
Results
The most commonly reported coinfections differ from infections causing highest global mortality, with a notable lack of serious childhood infections in reported coinfections. We found that coinfection is generally reported to worsen human health (76% publications) and exacerbate infections (57% publications). Reported coinfections included all kinds of pathogens, but were most likely to contain bacteria.
Conclusions
These results suggest differences between coinfected patients and those with single infections, with coinfection having serious health effects. There is a pressing need to quantify the tendency towards negative effects and to evaluate any sampling biases in the coverage of coinfection research.
Keywords: Coinfection, Concomitant disease, Host health, Integrated control, Parasite infracommunity, Pathogen abundance, Pathogen–host interactions, Pathogen diversity, Polymicrobial infection, Within-host parasite ecology
Introduction
The many pathogens that infect humans (e.g., viruses, bacteria, protozoa, fungal parasites, helminths) often co-occur within individuals.1–5 Helminth coinfections alone are thought to occur in over 800 million people,6 and are especially prevalent among the global poor.7–9 Other coinfections involve globally important diseases such as HIV,10 tuberculosis,11 malaria,12 hepatitis,13 leishmaniasis,14 and dengue fever.15 It seems likely, therefore, that the true prevalence of coinfection exceeds one sixth of the global population and often involves infectious diseases of pressing human concern.
Improved understanding of coinfection prevalence is greatly needed,16 partly because coinfecting pathogens can interact either directly with one another or indirectly via the host’s resources or immune system.3 Compared to infections of single pathogen species, these interactions within coinfected hosts can alter the transmission, clinical progression and control of multiple infectious diseases.17–19 Establishing the nature and consequences of coinfection requires integrated monitoring and research of different infectious diseases,1 but such data are rare.9,20,21
Reviews of coinfection have emphasised that coinfection requires further research, especially in humans,2,3,20,22 where coinfection outnumbers single infection in many communities2,23 and where helminth coinfections appear to worsen human health.20 Coinfection involves a range of pathogens and can have various effects on coinfected hosts.3 There are many individual studies concerning coinfection, but these use various approaches and are often narrowly focused. We aimed to gain a coherent picture of the nature and consequences of coinfection in humans. We surveyed the published literature for the occurrence of coinfecting pathogens and their effects on other infecting organisms and human health. We found that coinfections involve a huge variety of pathogens, and most studies report negative effects on human health. However, current coinfection research rarely focuses on pathogens with highest global mortality.
Materials and methods
Literature search
We searched the published literature for studies of coinfection (i.e. multi-species infections) in humans using the Advanced Search facility on the largest online citation database, Scopus (Elsevier Ltd.). Many disciplines study infectious diseases and various terms are used to describe coinfection. We therefore searched for coinfection, concomitant infection, multiple infection, concurrent infection, simultaneous infection, double infection, polymicrobial, polyparasitism, or multiple parasitism in the Title, Abstract, or Keywords of publications in the Life and Health Sciences before 2010. In June 2011 this search returned 12,963 results; an equivalent search on an alternative online citation database, Web of Science [Thomson Reuters], yielded similar trends in publications through time, but fewer results. Due to the large number of publications matching the search terms, we chose to focus on publications from 2009. Furthermore, publications concerning non-human hosts, non-infectious diseases or multiple genotypes of only one pathogen species were excluded.
For each publication we collected data on the identity of coinfecting pathogens, journal, study type and maximum number of pathogen species found per person. Study types included experiments treating each infection, observational studies, and reviews/meta-analyses. Observational studies were either case notes on particular patients, studies of patient groups, or epidemiological surveys among human communities.
Many publications reported the stated effect of one pathogen on the abundance of coinfecting pathogens (i.e. proxies for the intensity of infection, e.g. from measures of viral load, faecal egg counts, antibody response, bacterial cultures etc.) and/or host health (e.g. survival, recovery time, anaemia, liver fibrosis, immune cell counts). These effects of coinfection are relative to conditions observed under infections of single pathogen species. Where these effects were reported we recorded the pair of coinfecting pathogens involved, the quality of measurement (rated as low [e.g. anecdotal], adequate [e.g. correlation] and high [i.e. full reporting of appropriate statistical test supported by theoretical mechanisms]) and other data (see below). Data from review-type publications, case notes and from publications not mentioning the effects of coinfection (120 publications for pathogen abundance and 110 for host health) were excluded to avoid double counting, undue influence of individual cases and the inclusion of irrelevant publications. Reported effects based on low quality evidence (10 publications for pathogen abundance and 24 for host health) were also omitted.
Analyses of the effects of coinfection
There was considerable heterogeneity in the reporting of the effects of coinfection, both in terms of the response variable and in terms of the quantitative measure given (e.g. odds ratios, adjusted odds ratios, P-values, hazards ratios, raw comparisons). Furthermore, many publications gave qualitative statements of effect direction. Among publications quantifying effect size, diverse measures were given across publications. We focused on the direction of reported effects (positive, negative and no-effect) to maximise the data available. Reported directions of the effects on both pathogen abundance and host health for each pair of coinfecting pathogens was coded +1 for positive effect, 0 for neutral, −1 for negative effects, and NA if no information about effect direction was given. The resulting dataset includes some repeated measures because some publications reported multiple pairs of coinfecting pathogens and some coinfections were reported in multiple publications. We created two independent datasets containing the mean effect direction (i) per publication and (ii) per coinfection to eliminate these sources of pseudoreplication. A negative mean implied a predominance of negative effects; a positive mean implied a dominance of positive effects. A mean close to 0 could result from either many neutral effects (whereby a pathogen consistently had no discernible effect) and/or equal numbers of positive and negative effects (whereby a pathogen had different, possibly context-dependent effects). In either case, there is no clear indication of these pathogens having a consistent effect on each other (or on host health), so we adopt the most conservative interpretation and assume there is no-effect. These means were converted into three categories: negative (−1 to −⅓), neutral (−1 to +⅓) and positive (+⅓ to +1). Chi-squared tests24 based on double log-likelihood values25,26 were done to establish whether totals in each category differed from those expected from two different null hypotheses (random and no-effect). The random null model was of equal proportions of positive, neutral and negative effects, while the no-effect null model was that coinfecting pathogens do not interact, allowing for a 5% error rate (hence 2.5% negative, 2.5% positive, and 95% neutral reported effects). This constitutes a recommended vote-counting method deriving continuous parameters analysed against confidence intervals (α = 0.05).27
Finally, we explored the potential influence of the missing data (NAs) on the effects of coinfection in the analysis (56 for pathogen abundance, 79 for host health). These values represent reported coinfections where the effect on either pathogen abundance or host health was not reported, despite the possibility that these coinfecting pathogens did interact with each other and/or influence host health. We therefore assessed how potential interactions from these unreported effects may alter the overall patterns of coinfection effects. To determine their potential impact on the estimated overall effects, NAs were assigned one of three values at random (+1, 0, −1). The mean effect was then calculated per publication or coinfection pair as before, and a grand mean taken across all publications or coinfection-pairs. The grand mean represents an estimate of overall effect of coinfection on either host health or pathogen abundance across either publications or coinfections, given a particular random assignment of −1, 0, +1 to NAs. Repeating this random assignment 1000 times produced a distribution of grand means.
Comparison with WHO data
We examined whether recent coinfection research focuses on the pathogens causing the highest global mortality. We obtained global totals for the number of deaths (both sexes, all ages) in 2009 under every category of infection collated by the World Health Organisation (obtained from the Global Burden of Disease section of the Global Health Observatory website).28 We compared the ten categories causing most global deaths in 2009 with total reports of coinfection involving these infections. Comparing the top ten infection categories by mortality with their morbidity measures (DALYs) yielded similar trends, so we present only data from the mortality comparison.
Results
Overall trends in coinfection publications
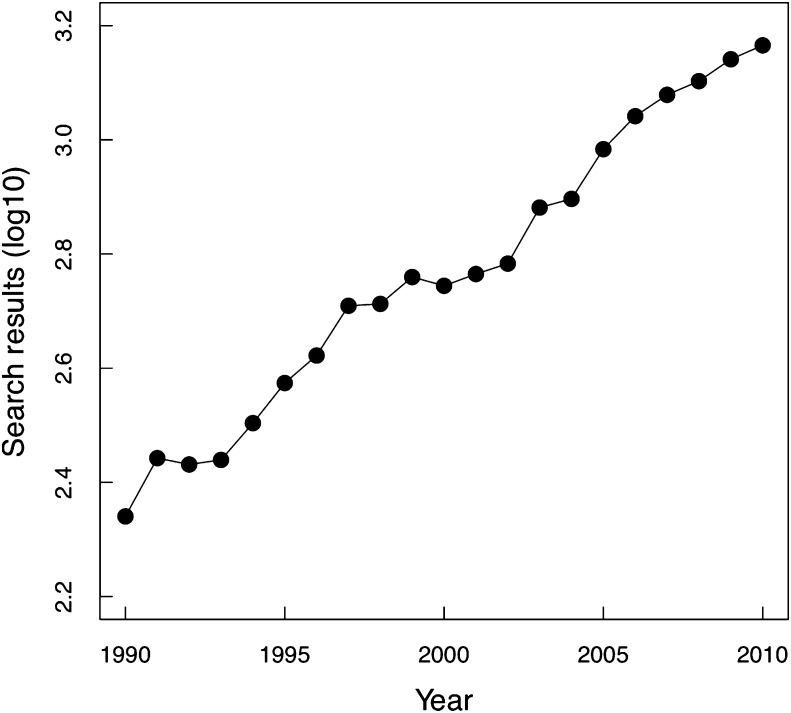
Hundreds of publications on coinfection are published annually and have increased from 219 publications in the first year of search results to 1464 publications in 2009 (Fig. 1). This increase includes studies of both human and non-human hosts. Of the 1464 publications retrieved for 2009, 309 reported multiple pathogen species coinfecting humans. Publications came from 192 journals, with most (136 of 192 journals, 70.8%) publishing a single coinfection article in 2009.
Figure 1.
Annual coinfection publications (log10) from initial Scopus search. See the Methods section for search criteria.
The majority of relevant publications from 2009 were observational studies (234 of 309, 75.0%), of which 159 (67.9%) involved patient groups, 60 (25.6%) were case notes and 18 (7.7%) surveyed a population. Three observational studies (1.3%) analysed death records. Seventy publications (22.4%) were reviews or metaanalyses. Five publications (1.6%) were experimental, whereby treatment and controls were applied to both singly infected and coinfected groups. A majority of the relevant publications concerned coinfections by two pathogen species (249 of 309, 80.5%), but more pathogen species per individual were occasionally reported; the mean number of pathogens was 2.4 and a maximum of 13 pathogens was reported twice in a venous leg ulcer29 and a periodontal infection.30
Reported coinfecting pathogens
A total of 270 pathogen taxa were reported in coinfection publications from 2009, across 1265 reports of coinfections comprising 933 different pairs of coinfecting pathogen taxa. All pathogen types (viruses, bacteria, protozoa, fungal parasites, helminths) were reported in coinfections; the most common pathogen group was bacteria (Table 1). In terms of specific pairs of reported coinfecting pathogens there was high diversity, but HIV and hepatitis viruses featured relatively highly (Table 1).
Table 1.
Number of reports of each type of pathogen and the five most reported pairs of coinfecting pathogens among 2009 coinfection publications.
| Pathogen Type | Frequency (%) | Coinfecting pathogens | Frequency (%) |
|---|---|---|---|
| Bacteria | 1351 (53.4) | HCV-HIV | 82 (6.5) |
| Viruses | 877 (34.7) | HBV-HIV | 31 (2.4) |
| Protozoa | 117 (4.6) | HBV-HCV | 30 (2.4) |
| Helminths | 78 (3.1) | HIV-Mtb | 28 (2.2) |
| Fungi | 81 (3.2) | HIV-HPV | 27 (2.1) |
HBV = Hepatitis B Virus, HCV = Hepatitis C Virus, HIV = Human Immunodeficiency Virus, Mtb = Mycobacterium tuberculosis, HPV = Human Papillomavirus.
Effects of coinfection on pathogen abundance and human health
Effects of coinfection on pathogen abundance and host health were sampled across 173 suitable publications according to pathogen abundance and host health. These publications covered 827 coinfecting pairs of pathogens, involving 183 pathogen species. Among these coinfections, 203 (24.5%) measured the size or direction of effects on pathogen abundance and 191 (23.1%) measured the size or direction of effects on host health. The remainder of coinfections had no reports of the effects of coinfection in suitable publications.
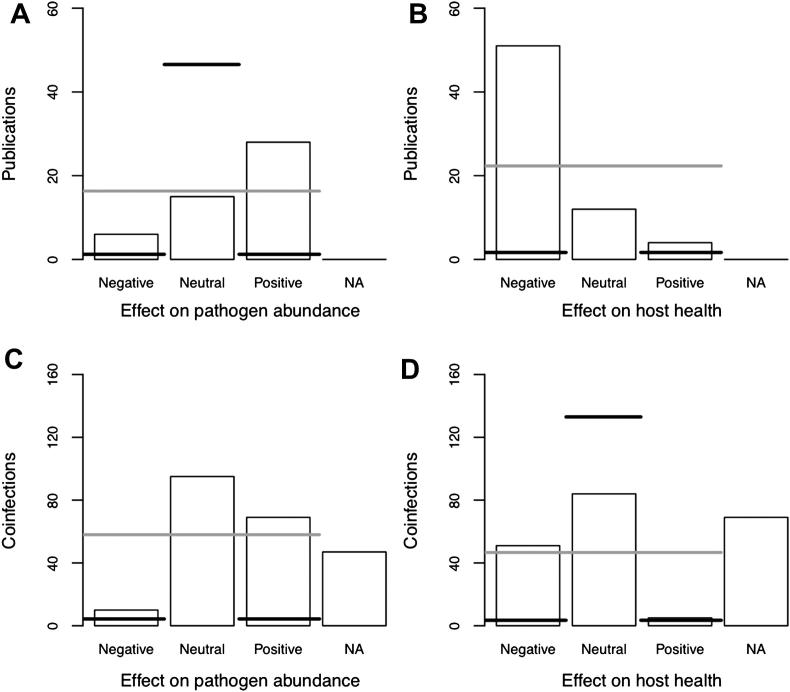
Overall, positive effects of coinfection on pathogen abundance were the most common reported across publications (6 negative, 15 neutral, 28 positive reports across 49 publications; Fig. 2A). Among specific pairs of coinfecting pathogens neutral effects exceeded positive effects (10 negative, 95 neutral, 69 positive across 174 unique pathogen pairs; Fig. 2C). In both cases these patterns were strongly significantly different from both the random null model (grey line on Fig. 2, by publication [X2 = 15.6, d.f. = 2, P < 0.001] and by coinfection [X2 = 82.6, d.f. = 2, P < 0.001]) and from the no-effect null model (black line on Fig. 2, by publication [X2 = 160.3, d.f. = 2, P < 0.001] and by coinfection [X2 = 292.8, d.f. = 2, P < 0.001]).
Figure 2.
Direction of reported effects of coinfection on the abundance of infecting pathogens and host health averaged across publications and coinfections published in 2009. Horizontal lines indicate expected values of null hypotheses (black = no-effect, grey = random).
Regarding the impact of coinfection on host health, there was a much greater number of negative effects reported in publications than either positive, neutral or NA categories (51 negative, 12 neutral, 4 positive across 67 publications; Fig. 2B). When data were aggregated by specific pathogen pairs the neutral effects exceed the negative effects (51 negative, 84 neutral, 5 positive across 140 unique pathogen pairs; Fig. 2D). In both cases these patterns were significantly different from both the random null model (grey line, by publication [X2 = 55.6, d.f. = 2, P < 0.001, Fig. 2B] and by coinfection [X2 = 85.5, d.f. = 2, P < 0.001, Fig. 2D]) and from the no-effect null model (black line, by publication [X2 = 315.4, d.f. = 2, P < 0.001, Fig. 2A] and by coinfection [X2 = 199.6, d.f. = 2, P < 0.001, Fig. 2C]).
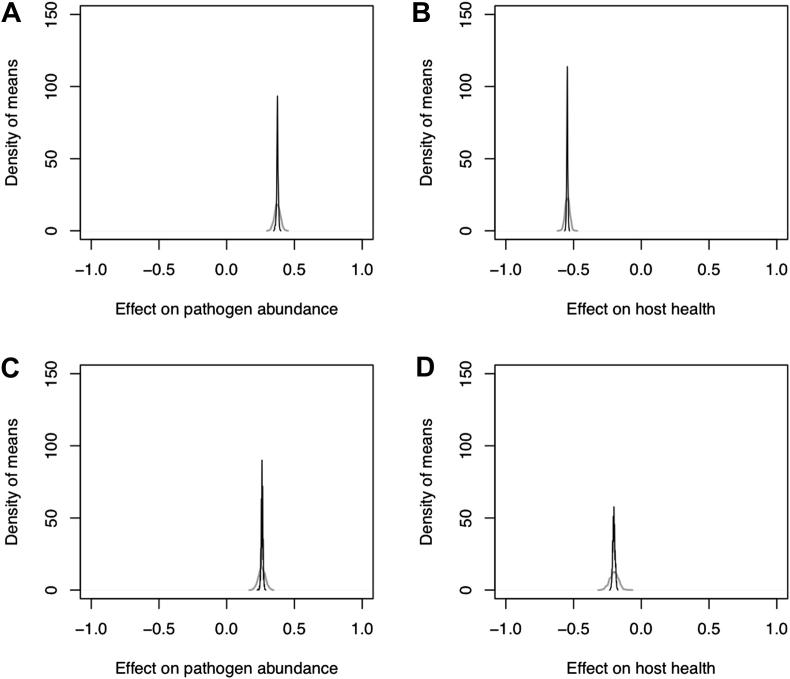
It is unlikely that these patterns of the effects of coinfection would be changed by knowledge of the unreported effects (the NAs in Fig. 2). Even after NA values were assigned predominantly to the neutral category (i.e. under the no-effect null model), the distribution of the grand mean effect was positive for the effects on pathogen abundance (Fig. 3A and C), and negative for effects on host health (Fig. 3B and D). None of the distributions of grand means overlapped zero (Fig. 3).
Figure 3.
Distribution of grand mean effects of coinfection including simulations of missing values according to the random (grey line) and no-effect (black line) null models. Lines generated by a Gaussian kernel estimator (smoothing bandwidths: random = 5.1 × 10−3, no-effect = 1.2 × 10−3).
Do coinfection studies focus on the most important infectious diseases?
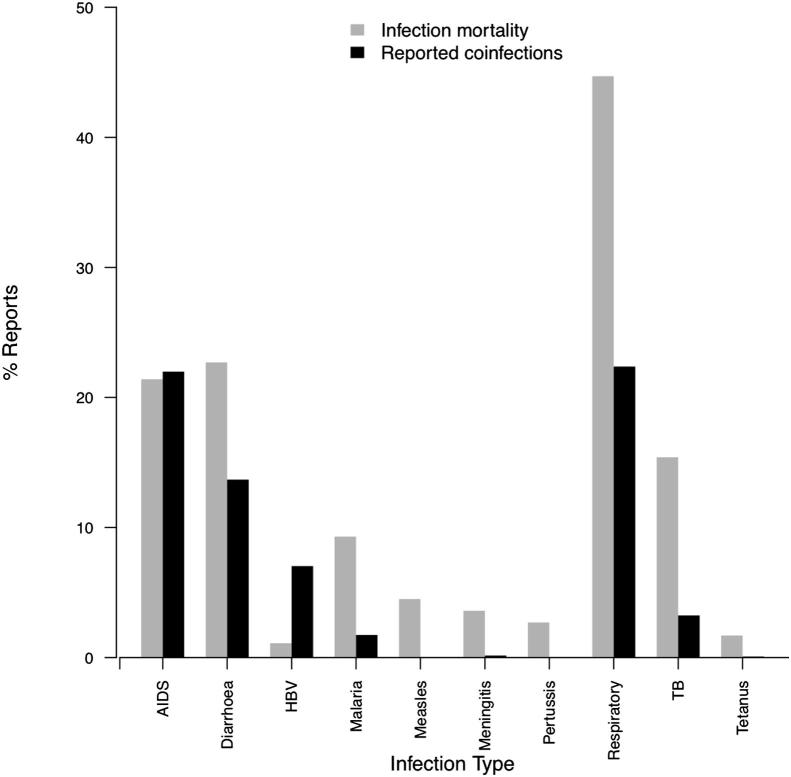
We found notable differences between the most commonly reported coinfecting pathogens and the infections causing the greatest global health burden (Fig. 4). The largest infectious causes of mortality are respiratory infections, causing 44.7% of these deaths with the next greatest causes, diarrhoea and HIV/AIDS, causing half as many deaths. Other important infections by global mortality are tuberculosis, malaria and childhood infections (measles, meningitis, whooping cough and tetanus). The tenth biggest infectious cause of mortality worldwide, HBV, is the only hepatitis virus featuring in the top ten infectious causes of mortality, causing 1.1% of infectious disease deaths. In comparison, hepatitis viruses featured in one fifth of reported coinfections (286 of 1265, 22.6%). The top ten pathogen species reported in coinfections were HIV (in 266 [21.9%] of 1265 coinfections), HCV (11.4%), HBV (7.04%), Staphylococcus aureus (4.58%), Escherichia coli (4.43%), Pseudomonas aeruginosa (3.72%), Mycobacterium tuberculosis (5.9%), HPV (3.16%), unidentified Streptococcus spp. (3.00%), and unidentified Staphylococcus spp. (3.00%). Some of the most common reported coinfecting pathogens (HCV, Staphylococcus, HPV, and Streptococcus) contribute relatively little to global infection mortality. Perhaps surprisingly, four of the most important infectious causes of mortality (all of them childhood infections) received very few or no reports of coinfection in 2009 publications.
Figure 4.
Top ten infections from global mortality data (28) (grey bars), compared with percentage of times the infections were reported in coinfections in 2009 publications (black bars).
Discussion
Interest in coinfection has increased in recent years, with publications on human coinfection involving hundreds of pathogen taxa across all major pathogen groups. Recent publications tend to show that negative effects of coinfection on human health are more frequent than no-effect or positive effects. However, the most commonly reported coinfecting pathogens differ from those infections causing highest global mortality. These results raise questions concerning the occurrence and study of coinfection in humans and their implications for effective infectious disease management.
The overall consequence of reported coinfections was poorer host health and enhanced pathogen abundance, compared with single infections. This is strongly supported by significant statistical differences in the reported direction of effects (P < 0.001) from expectations of either no-effect or of random distributions, and by the robustness of these trends in the face of missing values and by diversity in the types of publications in which these coinfections were reported. Moreover the tendency for positive effects on pathogen abundance corroborates the negative effects on host health because larger infections are a mechanism by which disease can be exacerbated. The consistency of these detrimental coinfection effects across a wide range of pathogens suggests a general incidence of interactions between coinfections. The long-term effects among survivors of coinfections can be varied and in some cases severe, including blindness, chronic diarrhoea, chronic inflammation, carcinoma, immunosuppression, liver fibrosis, meningitis, renal failure, rheumatic fever, etc.31
The direction of reported coinfection effects could have at least two explanations. The first is that coinfection may be more likely in individuals of poor health, which in turn leads to poorer prognosis among coinfected cases. The relative paucity of experimental studies of coinfection in humans means sampling biases towards people of poorer health is possible, but impossible to account for in our analyses. The second explanation is that coinfecting pathogens interact synergistically with each other, for example via the host’s immune system, so that the presence of one enhances the abundance and/or virulence of the other. A clear example of this is HIV, which causes immunosuppression, increasing the likelihood of additional infections and occurred in two fifths of reported coinfections (Fig. 4).
Differences between reported coinfections and global mortality figures may also suggest important interactions between coinfecting pathogens. Coinfections that were more commonly reported than their relative contribution to global mortality may involve particular synergistic pathogen–pathogen interactions, such as among herpes viruses like CMV or HSV infection enhancing the risk of HPV coinfection.32 Conversely, infections that cause high mortality but had relatively few reports of coinfection could result from antagonistic interactions, reducing the likelihood of such coinfections occurring and being reported, like P. aeruginosa exoproduct limiting S. aureus colony formation.33 An alternative and possibly more likely explanation of the discrepancies between reported coinfections and global mortalities from infections could be greater funding availability (e.g. HIV/AIDS research), higher interests of virologists in coinfection and/or easier observations or more routine screening compared with other pathogens, for instance the greater difficulty of detecting intestinal helminths in coinfection research. The lack of coinfection publications reporting on major infectious causes of childhood mortality remains unexplained. While some publications do study childhood coinfection and find coinfection to be more common in children,34 current coinfection research does not include the infections that kill the most infants globally. Fewer than 1 in 20 publications reported coinfections involving helminths, despite hundreds of millions of helminth coinfections globally,6 which could arise from limited published research on helminthiases. To what extent disparities between global mortality data reflect actual epidemiology or biases in research attention remains to be established, in part hindered by current inadequacies in coinfection surveillance.
The disparity between infections that feature highly in global mortality statistics and those receiving most attention in published coinfection studies poses a challenge to infectious disease research. A general understanding of the effects of coinfection is important for appropriate control of infectious diseases.4,7,8,35 Poor or uncertain observational data regarding coinfection hinders efforts to improve health strategies for infectious disease in at-risk populations.9 For example, global infectious disease mortality data28 report only single causes of death, even if comorbidities were identified. If health statistics better represent coinfection, published coinfection research could be better evaluated. Moreover there is a lack of coherence in coinfection literature, with a variety of synonyms being used for the same phenomenon, which is multi-species infection (see the Methods for examples). The term polymicrobial, while commonplace, is restricted to coinfections involving microbes. Coinfection is a broader term encompassing all pathogen types including interactions between the same kinds of pathogens as well as cross-kingdom coinfections between, say, bacteria and helminths. Ultimately decisions over which term to prefer (if any) need to be made by a consensus of the diverse research communities concerned with this phenomenon. True patterns of coinfection remain unknown21 and our results suggest that it may be starkly different from existing data on important infectious diseases.
Overall recently published reports of coinfection in humans show coinfection to be detrimental to human health. Understanding the nature and consequences of coinfection is vital for accurate estimates of infectious disease burden. In particular, more holistic data on infectious diseases would help to quantify the size of the effects on coinfection on human health. Improved knowledge of the factors controlling an individual’s risk of coinfection, circumstances when coinfecting pathogens interact, and the mechanisms behind these pathogen–pathogen interactions, especially from experimental studies, will also aid the design and evaluation of infectious disease management programmes. To date, most disease control programs typically adopt a vertical approach to intervention, dealing with each pathogen infection in isolation. If coinfecting pathogens generally interact to worsen human health, as suggested here, control measures may need to be more integrated and specialist treatments developed for clinical cases of coinfection. Further research is needed to identify the role of predisposed risks to coinfection.
Acknowledgements
We thank one anonymous reviewer for their helpful suggestions. We are also grateful to various organisations for funding. This report is independent research arising from ECG’s Ph.D. studentship attached to a Natural Environment Research Council grant [grant number NE/G007349/1]. OLP was part funded by the Royal Society and University of Zürich. ABP has a Wellcome Trust Centre for Immunity, Infection and Evolution Advanced Fellowship. None of the funding organisations played any role in the planning, implementation or documentation of this research.
References
- 1.Esch G.W., Shostak A., Marcogliese D.J., Goater T.M. London: Chapman& Hall; 1990. Patterns and processes in helminth parasite communities: an overview. [Google Scholar]
- 2.Petney T.N., Andrews R.H. Multiparasite communities in animals and humans: frequency, structure and pathogenic significance. International Journal for Parasitology. 1998;28(3):377–393. doi: 10.1016/s0020-7519(97)00189-6. [DOI] [PubMed] [Google Scholar]
- 3.Cox F.E.G. Concomitant infections, parasites and immune responses. Parasitology. 2001;122:S23–S38. doi: 10.1017/s003118200001698x. [DOI] [PubMed] [Google Scholar]
- 4.Brogden K.A., Guthmiller J.M., Taylor C.E. Human polymicrobial infections. The Lancet. 2005;365(9455):253–255. doi: 10.1016/S0140-6736(05)17745-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rigaud T., Perrot-Minnot M.J., Brown M.J.F. Parasite and host assemblages: embracing the reality will improve our knowledge of parasite transmission and virulence. Proceedings of the Royal Society B: Biological Sciences. 2010;277(1701):3693–3702. doi: 10.1098/rspb.2010.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotez P.J., Molyneux D.H., Fenwick A., Kumaresan J., Sachs S.E., Sachs J.D. Control of neglected tropical diseases. The New England Journal of Medicine. 2007;357(10):1018–1027. doi: 10.1056/NEJMra064142. [DOI] [PubMed] [Google Scholar]
- 7.Boraschi D., Alemayehu M., Aseffa A., Chiodi F., Chisi J., Prete G.D. Immunity against HIV/AIDS, malaria, and tuberculosis during co-infections with neglected infectious diseases: recommendations for the european union research priorities. PLoS Neglected Tropical Diseases. 2008;2 doi: 10.1371/journal.pntd.0000255. e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hotez P.J. Mass drug administration and integrated control for the world’s high-prevalence neglected tropical diseases. Clinical Pharmacology & Therapeutics. 2009;85:659–664. doi: 10.1038/clpt.2009.16. [DOI] [PubMed] [Google Scholar]
- 9.Steinmann P., Utzinger J., Du Z.W., Zhou X.N. Multiparasitism: a neglected reality on global, regional and local scale. Important Helminthic Infections in Southeast Asia. 2010;73:21–50. doi: 10.1016/S0065-308X(10)73002-5. [DOI] [PubMed] [Google Scholar]
- 10.Lawn S.D. AIDS in africa: the impact of coinfections on the pathogenesis of HIV-1 infection. Journal of Infection. 2004;48(1):1–12. doi: 10.1016/j.jinf.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Resende T., Hirsch C.S., Toossi Z., Dietze R., Ribeiro-Rodrigues R. Intestinal helminth co-infection has a negative impact on both anti-Mycobacterium tuberculosis immunity and clinical response to tuberculosis therapy. Clinical and Experimental Immunology. 2007;147:45–52. doi: 10.1111/j.1365-2249.2006.03247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muturi E.J., Mbogo C.M., Mwangangi J.M., Ng’ang’a Z.W., Kabiru E.W., Mwandawiro C. Concomitant infections of Plasmodium falciparum and Wuchereria bancrofti on the kenyan coast. Filaria Journal. 2006;5:8. doi: 10.1186/1475-2883-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sagnelli E., Pasquale G., Coppola N., Scarano F., Marrocco C., Scolastico C. Influence of chronic coinfection with hepatitis B and C virus on liver histology. Infection. 2004;32(3):144–148. doi: 10.1007/s15010-004-3080-6. [DOI] [PubMed] [Google Scholar]
- 14.Alvar J., Aparicio P., Aseffa A., Den Boer M., Canavate C., Dedet J.P. The relationship between leishmaniasis and AIDS: the second 10 years. Clinical Microbiology Reviews. 2008;21(2):334–359. doi: 10.1128/CMR.00061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pancharoen C., Thisyakorn U. Coinfections in dengue patients. Pediatric Infectious Disease Journal. 1998;17(1):81–82. doi: 10.1097/00006454-199801000-00019. [DOI] [PubMed] [Google Scholar]
- 16.Brooker S., Hotez P.J., Bundy D.A.P. The global atlas of helminth infection: mapping the way forward in neglected tropical disease control. PLoS Neglected Tropical Diseases. 2010;4(7):e779. doi: 10.1371/journal.pntd.0000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedersen A.B., Fenton A. Emphasizing the ecology in parasite community ecology. TRENDS in Ecology & Evolution. 2007;22:133–139. doi: 10.1016/j.tree.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Chiodini P.L. Chemotherapy for patients with multiple parasitic infections. Parasitology. 2001;122:S83–S89. doi: 10.1017/s0031182000017674. [DOI] [PubMed] [Google Scholar]
- 19.Sternberg E.D., Lefèvre T., Rawstern A.H., de Roode J.C. A virulent parasite can provide protection against a lethal parasitoid. Infection, Genetics and Evolution. 2011;11:399–406. doi: 10.1016/j.meegid.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 20.Pullan R., Brooker S. The health impact of polyparasitism in humans: are we under-estimating the burden of parasitic diseases? Parasitology. 2008;135(7):783–794. doi: 10.1017/S0031182008000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooker S., Utzinger J. Integrated disease mapping in a polyparasitic world. Geospatial Health. 2007;2:141–146. doi: 10.4081/gh.2007.262. [DOI] [PubMed] [Google Scholar]
- 22.Holmes J.C., Price P.W. Communities of parasites. In: Anderson D.J., Kikkawa J., editors. Community ecology: pattern and process. Blackwell Scientific Publications; Oxford: 1986. [Google Scholar]
- 23.Raso G., Luginbuhl A., Adjoua C.A., Tian-Bi N. Multiple parasite infections and their relationship to self-reported morbidity in a community of rural cote d’ivoire. International Journal of Epidemiology. 2004;33:1092–1102. doi: 10.1093/ije/dyh241. [DOI] [PubMed] [Google Scholar]
- 24.Bushman B. Vote-counting procedures in meta-analysis. In: Cooper H., Hedges L., editors. The handbook of research synthesis; chap. 14. Russell Sage Foundation; New York: 1994. [Google Scholar]
- 25.Crawley M.J. John Wiley & Sons; Chichester: 2007. The R Book. [Google Scholar]
- 26.Sokal R., Rohlf F. 2nd ed. WH Freeman; San Francisco: 1981. Biometry. [Google Scholar]
- 27.Hedges L., Olkin I. Academic Press; New York: 1985. Statistical methods for meta-analysis. [Google Scholar]
- 28.World Health Organisation . 2009. Global health observatory.http://apps.who.int/ghodata URL. [Google Scholar]
- 29.Wolcott R.D., Gontcharova V., Sun Y., Dowd S.E. Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded flx and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiology. 2009;9 doi: 10.1186/1471-2180-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He X.S., Shi W.Y. Oral microbiology: past, present and future. International Journal of Oral Science. 2009;1(2):47–58. doi: 10.4248/ijos.09029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fratamico P., Smith J., Brogden K. ASM Press; 2009. For Microbiology A.S. Sequelae and long-term consequences of infectious diseases. [Google Scholar]
- 32.Baldauf J.J., Dreyfus M., Ritter J., Meyer P., Philippe E., Obert G. A PCR study on the coexistence of herpes simplex virus, cytomegalovirus and human papillomavirus DNAs in cervical neoplasia. International Journal of Gynecological Cancer. 1996;6(5):389–395. [Google Scholar]
- 33.Hoffman L.R., Déziel E., D’Argenio D.A., Lépine F., Emerson J., McNamara S. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences. 2006;103(52):19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plata-Nazar K., Kamiaska B., Jurczyk J. Prevention of hospital infections in pediatric department. Przeglad Pediatryczny. 2009;39(1):35–38. [Google Scholar]
- 35.Laserson K.F., Wells C.D. Reaching the targets for tuberculosis control: the impact of HIV. Bulletin of the World Health Organization. 2007;85(5):377–386. doi: 10.2471/06-035329. [DOI] [PMC free article] [PubMed] [Google Scholar]