Abstract
Little is known about the nature of floral inductive cues in day-neutral plants that are insensitive to photoperiod variations and, therefore, rely on endogenous signals to initiate reproductive growth. The INDETERMINATE1 (ID1) transcription factor is a key regulator of the transition to flowering in day-neutral maize. The ID1 gene is expressed exclusively in developing leaves, where it controls the production or transmission of leaf-derived florigenic signals. Florigen-producing source leaves were compared with mature leaves of late-flowering id1 plants, and metabolite and gene expression differences associated with the floral transition in maize were observed. While id1 mutants have a similar capacity for photosynthesis to wild-type siblings, id1 source leaves show quantitative differences in carbohydrate allocation prior to the floral transition stage, with a marked increase in sucrose and other soluble sugars, accompanied by a decrease in tricarboxylic acid (TCA) cycle organic acids. Importantly, source leaves of autonomous-flowering maize are typified by a higher transitory starch to sucrose ratio and a transcript profile enriched for sucrose synthesis and starch metabolism-related gene function. Finally, similar changes in transitory starch and sucrose are not observed in teosinte, the tropical progenitor of maize that requires short-day photoperiods to induce flowering. Together, these data define a transcript and metabolite signature associated with the autonomous floral transition in temperate maize leaves.
Key words: Autonomous floral transition, indeterminate1, maize, source leaf, teosinte, transcript and metabolite
Introduction
Flowering plants integrate multiple external and endogenous signals to optimize the timing of the transition from vegetative to reproductive growth. The coordination between vegetative growth for biomass accumulation and timely transition to flowering is critical for reproductive success. Floral inductive cues are perceived or originate in mature leaves, and mobile signals travel long distances to the shoot apical meristem (SAM) to activate floral identity genes and initiate inflorescence development. Floral evocation in response to environmental signals, such as photoperiod and vernalization, has been described in detail (Jaeger et al., 2006; Amasino, 2010; Tsuji et al., 2011). However, the nature of endogenous floral induction mechanisms, which involve physiological readiness to allocate resources to floral organ formation, are less well understood for two main reasons. First, autonomous signals are controlled developmentally, rather than environmentally, making it difficult to tease apart physiological responses specific to the floral transition. Secondly, while most plants coordinate both environmental and endogenous cues to induce flowering, relatively few model species are strictly day neutral.
Domestication of modern maize (Zea mays ssp mays) from its tropical progenitor teosinte (Zea mays ssp parviglumis) necessitated adaptations to optimize flowering time at higher latitudes. Thus, whereas teosinte has a strict dependence on short-day photoperiods to induce flowering (Emerson, 1924), temperate maize is largely day neutral; that is it flowers after initiating a particular number of leaves, regardless of day length. Despite evidence of residual photoperiod sensitivity in modern maize accessions (Chardon et al., 2004; Sheehan et al., 2007), temperate maize is one of a few model species that relies mainly on endogenous indicators of the plant’s developmental stage to cue the floral transition. The SAM of temperate maize commits to inflorescence development once a particular number of leaves are initiated, which reflect plant age and physiological status (Irish and Nelson, 1991; Irish and Jegla, 1997). For example, maize B73 inbred plants consistently initiate 16–18 leaf primordia before ultimately producing a terminal male inflorescence, the tassel. Quantitative trait loci (QTLs) with significant effects on the timing of the floral transition in maize have been described, but few underlying genes have been identified (Chardon et al., 2004; Salvi et al., 2007; Buckler et al., 2009). Maize orthologues of the Arabidopsis florigen-encoding gene FLOWERING LOCUS T (FT) and the transcription regulator FLOWERING LOCUS D (FD) have been identified (Muszynski et al., 2006; Lazakis et al., 2011; Meng et al., 2011). The maize FT/FD module probably integrates both environmental and endogenous leaf-derived signals to regulate flowering time. Indeed, the discovery that an FT orthologue in tomato, SFT (SINGLE FLOWER TRUSS), promotes flowering in day-neutral tomato and tobacco plants points to the universality of FT proteins as leaf-derived, phloem-mobile florigens whose functions are not dependent exclusively on photoperiod input (Lifschitz et al., 2006). However, the molecular and biochemical nature of autonomous florigenic signals acting upstream of conserved elements of the FT/FD module is largely unknown.
Mutation of the maize indeterminate1 (id1) gene, which encodes a putative zinc finger transcription factor, causes an extreme late-flowering phenotype, with mutant plants producing ≥27 leaves in the B73 background before initiating inflorescence development (Colasanti et al., 1998; Kozaki et al., 2004; Colasanti and Muszynski, 2008). id1 is expressed exclusively in non-photosynthetically active leaves at early stages of development, with the highest expression at the floral transition in B73 seedlings with seven visible leaves (denoted as V7). Importantly, id1 transcript is not detected in mature leaves or at the SAM and subtending leaf primordia (Colasanti et al., 1998), and ID1 protein remains localized to nuclei of the same tissues in which the gene is expressed (Wong and Colasanti, 2007). Orthologues of id1 in other grasses, such as sorghum and rice, also show protein accumulation in developing leaves (Colasanti et al., 2006). Interestingly, similar to maize id1, the rice orthologue of id1, called OsId1 (also RID1 or Ehd2), appears to function at least partly through the autonomous pathway, since mutant and knock-down OsId1 plants are late flowering in both inductive short days and non-inductive long days (Matsubara et al., 2008; Park et al., 2008; Wu et al., 2008). In extreme cases, loss of OsId1 results in plants that are unable to initiate the floral transition regardless of photoperiod (Wu et al., 2008), indicating that ID1 and its orthologues probably define an important grass- or monocot-specific endogenous flowering time regulatory module.
In an earlier study, expression profiles of normal and id1 mut ant plants were compared to define possible regulatory networks in immature, developing leaves; that is the tissue where ID1 is expressed. The pattern of expression differences that emerged suggested a link between ID1 activity and energy metabolism in this sink tissue (Coneva et al., 2007). Although no clear orthologue of id1 exists in Arabidopsis, recent characterization of mutations in several INDETERMINATE DOMAIN genes (AtIDD) suggest a link to the regulation of sucrose and/or starch metabolism (Morita et al., 2006; Tanimoto et al., 2008; Feurtado et al., 2011; Seo et al., 2011a, b). This connection between sucrose and starch metabolism and the floral transition is intriguing, especially in light of long-standing physiological studies that link floral induction to carbohydrate changes in mature leaves (Sachs and Hackett, 1983; Bodson and Outlaw, 1985; Lejeune et al., 1993). Collectively, these data lend support to the notion that the autonomous floral induction pathway may involve a coordinated change in carbohydrate metabolism in order to support the process of floral organ formation and subsequent reproductive development.
Here, late-flowering id1 mutants are used to analyse the physiological basis of autonomous flowering-time regulation in day-neutral maize by comparing transcript and metabolite levels in florigen-producing wild-type source leaves and mature leaves of non-flowering mutants just prior to the normal floral transition. It was found that id1 mutant leaves show significant transcriptional and metabolite changes, whereas overall carbon assimilation capacity is unaffected. Specifically, leaves of non-flowering id1 plants have a lower ratio of transitory starch to sucrose as well as lower levels of major tricarboxylic acid cycle (TCA) organic acids. Together these changes define a transcript and metabolite signature associated with the autonomous floral transition in maize.
Materials and methods
Plant growth conditions and sample collection
The id1-m1 allele backcrossed 10 times to maize inbred B73 was used for all experiments. Plants were grown in Conviron controlled-condition growth chambers under a light intensity of 1000 µmol m–2 s–1 in 15/9h light cycles at 27 °C during the light period, and 23 °C during the dark period. Teosinte plants were grown in short days (SD), 9/15h light cycles, or night break (NB) where 15h of darkness were interrupted by a single hour of light. Carbon dioxide (CO2) was maintained at ~400 ppm and plants were fertilized weekly with 20–20–20 (nitrogen–phosphorus–potassium). At the two leaf-tip stage, plants were genotyped for id1 using PCR primers IdF, IdR4, and DsR (Supplementary Table S6 available at JXB online) as described previously (Wong and Colasanti, 2007). At the seven leaf-tip stage (V7), entire leaf blades of the youngest fully expanded leaf, leaf 5 (L5), were sampled for starch and sucrose quantification at Zeitgeber time 0, 8, and 14h after dawn (ZT0, ZT8, and ZT14). Similarly, leaf 7 (L7) was sampled from plants at the ninth visible leaf-tip stage (V9). Five biological samples were collected per time point and the experiment was repeated three times. For gas chromatography–mass spectrometry (GC-MS) analysis and gene expression experiments, L5 leaf blades from V7 plants were collected at ZT8. Immature leaves for GC-MS analysis were collected at the V7 stage by removing the leaf sheaths of all but the last two visible leaves and excising a 7cm section starting above the SAM.
Expression analysis
For microarray analysis, entire leaf blades (L5) from three wild-type plants and three id1 mutants were collected as specified above and ground in liquid nitrogen. RNA was extracted from 150mg of leaf tissue per plant using TRIzol® reagent (Invitrogen) and cleaned up on RNeasy columns (Qiagen) as per the manufacturers’ instructions. For global expression analysis, the GeneChip Maize Genome array (Affymetrix) with 17 555 probe sets was used to interrogate ~14 850 Z. mays transcripts. Labelling, hybridization, and scanning of the chips were carried out by the London Regional Genomics Center facility (London, Ontario, Canada) according to company protocols. The .cel files were loaded into GeneSpring GX 11 (Agilent Technologies) and the GC-RMA normalization algorithm was applied. A minimum 2-fold change between normalized average wild-type and id1 samples was applied, and statistical significance was assessed with a t-test (Benjamini–Hochberg-corrected P < 0.05). MapMan pathway analysis was used to assign statistically significant probe sets to functional bins (Thimm et al., 2004). AgriGO analysis for Gene Ontology (GO) term enrichment was carried out using Singular Enrichment Analysis with maize Affymetrix Array as a background reference (Du et al., 2010). A hypergeometric distribution was used for the analysis, and the Yekutieli multiple testing correction was applied.
Further gene expression analysis was carried out using real-time reverse transcription-PCR (qRT-PCR). RNA was extracted as detailed above from five biological replicates per genotype. cDNA was prepared using qScript cDNA SuperMix (Quanta Biosciences) according to the manufacturer’s instructions. A full list of qRT-PCR primers is available in Supplementary Table S6 at JXB online. A PerfeCTa SYBR Green SuperMix (Quanta Biosciences) and an Applied Biosystems 7300 Real Time PCR instrument were used, and resulting data were analysed by the 2–∆∆Ct method to obtain fold difference in expression between id1 and the wild type for each gene (Livak and Schmittgen, 2001). Glyceraldehyde-3-phosphate dehydrogenase (ZmGAPDH) was used for normalization. Statistical significance is reported for Student’s t-tests with P < 0.05.
Transmission electron microscopy
L5 was harvested between the 10th and 15th cm from the leaf tip of V7 plants at ZT0, ZT8, and ZT14. Leaf tissue was fixed in 3% glutaraldehyde, 2% acrolein solution in 25mM phosphate buffer and then post-fixed in 2% osmium tetroxide. The fixed tissue was dehydrated through an ethanol series (30–100%) and then incubated in propylene oxide before embedding in Spurr’s resin (SPI-Chem) according to the manufacturer’s instructions. Sectioned material was examined with a Philips CM10 electron microscope.
Carboxyfluorescein diacetate (CFDA) studies
The blade of L5 of plants at the V7 stage was cut under water and submerged in a 50 µg ml–1 (5-,6-) carboxyfluorescein diacetate (CFDA; Invitrogen) solution to evaluate phloem transport of the dye as previously described (Ma et al., 2009). Three plants of each genotype were tested and the experiment was repeated twice. Cross-sections of L5 sheaths were examined 3h after CFDA application and observed with a Leica DM RE confocal microscope using an argon 488nm laser for excitation, and emission settings for carboxyfluorescein in the 495–535nm range and in the 620–670nm range for chlorophyll autofluorescence.
Photosynthesis measurements
A Li-6400 instrument (LiCor, Lincoln, NE, USA) was used for photosynthesis measurements. L5 for V7 stage plants, L7 for V9, or L9 for V11 was clamped into a 6cm2 leaf chamber (6400-02B) with a red/blue LED light source. Measurements for five plants of each genotype were obtained starting 2h after the onset of the light period (ZT2) and the experiment was repeated three times. All measurements were performed using an airflow rate of 400 µl s–1 and auto-programs with standard stability definitions. Matching of the sample and reference IRGAs was performed for ∆CO2 <20. Statistical significance was evaluated using a Student’s t- test with P < 0.05.
Chlorophyll assay
For chlorophyll quantification, entire leaf blades of L5 at the V7 stage were collected and ground in liquid nitrogen using a Retsch® MM301 ball mill. Chlorophylls were extracted using 80% aqueous acetone solution and quantified based on absorbance measurements at 645nm and 663nm. Chlorophylls a and b, as well as total chlorophyll were determined as previously described (Arnon, 1949).
Sucrose and starch quantification
Entire leaf blades were collected, lyophilized, and then ground to a fine powder with a Retsch® MM301 ball mill. Methanol-soluble compounds were extracted at 70 °C. Phase separation was carried out using methanol:chloroform:water (5:3:7, v/v/v) and the polar fraction was anlysed further for sucrose and total protein content. Sucrose was quantified using treatment with invertase and hexokinase, and finally measuring ∆NADPH concentration at 340nm upon addition of glucose-6-phosphate dehydrogenase. Glucose was quantified in parallel and then subtracted from the invertase-treated sample. The pellet remaining after methanol extraction of the dry leaf samples was lyophilized and suspended in water. Total starch was determined using a starch quantification kit (Megazyme, Ireland) as specified by the manufacturer.
Total soluble protein was quantified in polar fractions after dissolution of lyophilized pellets in 20mM TRIS-HCl buffer (pH 7.2) containing 300mM sodium chloride. Bradford reagent was added and the samples were analysed spectrophotometrically as previously described (Bradford, 1976). Protein amounts were interpolated from a bovine serum albumin (BSA) standard curve.
Metabolite profiling using GC-MS
Blades of the mature fifth leaf and immature leaves of V7 stage plants were prepared as described above. In addition, ribitol was added to methanol and used in the extraction of dry ground plant material. An experimentally optimized aliquot of the polar fraction was lyophilized and derivatized using methoxyamine and N-methyl-N-trimethylsilyl-trifluoroacetamide as previously described (Roessner et al., 2000). A 1 µl aliquot of derivatized sample was injected into the splitless injection port of a Varian 1200 GC-MS system (Varian). Chromatography was performed using an Rtx-5MS column (Chromatographic Specialties, Brockville, Ontario, Canada). Mass spectra were recorded with an m/z scanning range of 50–650. Data analysis was performed using the automated mass spectral deconvolution and identification system (AMDIS, http://chemdata.nist.gov/mass-spc/amdis). The resulting components were filtered in GASP (Nuin, 2004) using a signal to noise ratio of 5 as a cut-off requirement. Component data were normalized to ribitol, and average normalized component area was compared between genotypes for the same tissue type using a t-test at α < 0.1 or α < 0.05. The Golm Metabolite Database was used for component identification (Kopka et al., 2005). Sucrose was present in mature leaf extract samples at significantly higher levels relative to all other metabolites and was, therefore, outside of the limit of quantification.
Results
Use of indeterminate1 maize mutants as a late-flowering model in a day-neutral species
Late-flowering id1 maize affords a unique system to study the endogenous floral induction pathway since the developmental signals necessary to initiate a normal transition to flowering are absent in the mutants. While ID1 is presumed to function in developing, heterotrophic leaves, florigen signals originate in fully expanded photosynthetically active leaf blades (Colasanti et al., 1998; Wong and Colasanti, 2007). Thus, it was reasoned that ID1 might act during early leaf development to produce quantifiable changes in mature, florigen-producing leaves. Maize plants homozygous for the id1-m1 mutant allele (Colasanti et al., 1998) and their normal-flowering siblings were compared at the same stage of growth that corresponds to the floral transition point in wild-type plants (Fig. 1A). Normal B73 plants (henceforth referred to as ‘wild type’) at the V7 stage are approaching floral transition, but shoot apical meristems of id1 and wild-type plants are indistinguishable at this stage; that is they have the dome-shape typical of vegetative meristems (Fig. 1B). Although no morphological changes are evident in wild-type apices at this stage, qRT-PCR analysis showed that expression of ZCN8, a maize orthologue of the Arabidopsis FT florigen-encoding gene, is significantly up-regulated in L5 of wild-type plants compared with the same leaf in the late-flowering id1 mutants (Supplementary Fig. S1 at JXB online).
Fig. 1.
Normal-flowering (wt) and indeterminate1 (id1) mutant plants in the B73 maize inbred. (A) Wild-type (wt) and id1 plants at the seven visible leaf-tip developmental stage (V7). L5, leaf five is the youngest leaf with a visible collar and the leaf sampled for subsequent analysis. (B) Shoot apical meristems (SAMs) of wt and id1 plants at the V7, V9, and V11 developmental stages. Scale bars represent 0.1mm. Asterisks show the position of tassel branch meristems. (This figure is available in colour at JXB online.)
At the V9 stage, which occurs ~5 d after V7 under the growth conditions used here, wild-type apices have undergone the transition to flowering as is evident from elongation of the wild-type SAM. At V11, ~10 d after the floral transition, the differentiating apical meristems of wild-type plants have greatly increased in size and exhibit clear tassel primordium morphology, including branch meristems (Fig. 1B).
Thus, the V7 stage is an ideal developmental point to assess physiological changes associated with the floral transition since it coincides with the period of floral induction signalling but precedes morphological changes of the SAM. At this stage, physiological, metabolite, and transcriptional differences were profiled in mature leaves of wild-type and id1 plants.
Expression profiling of mature source leaves
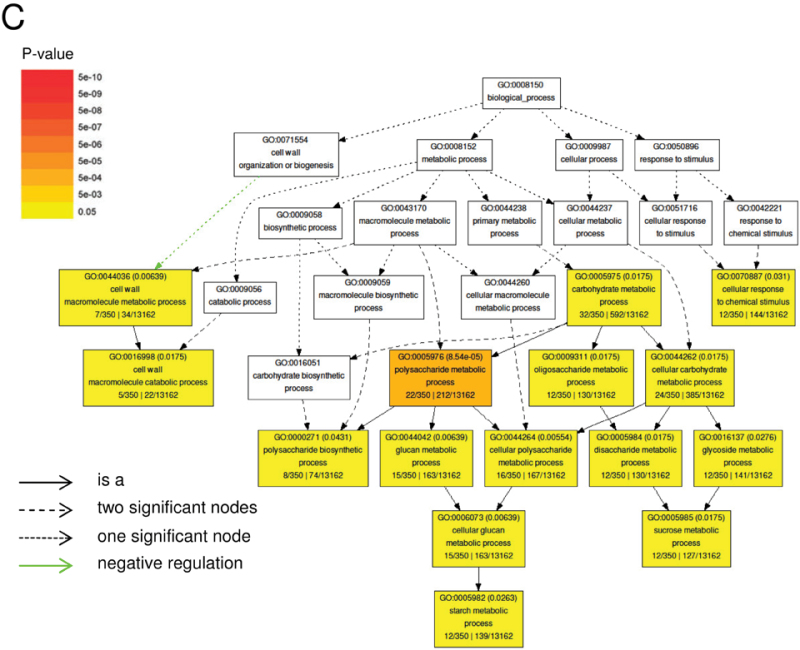
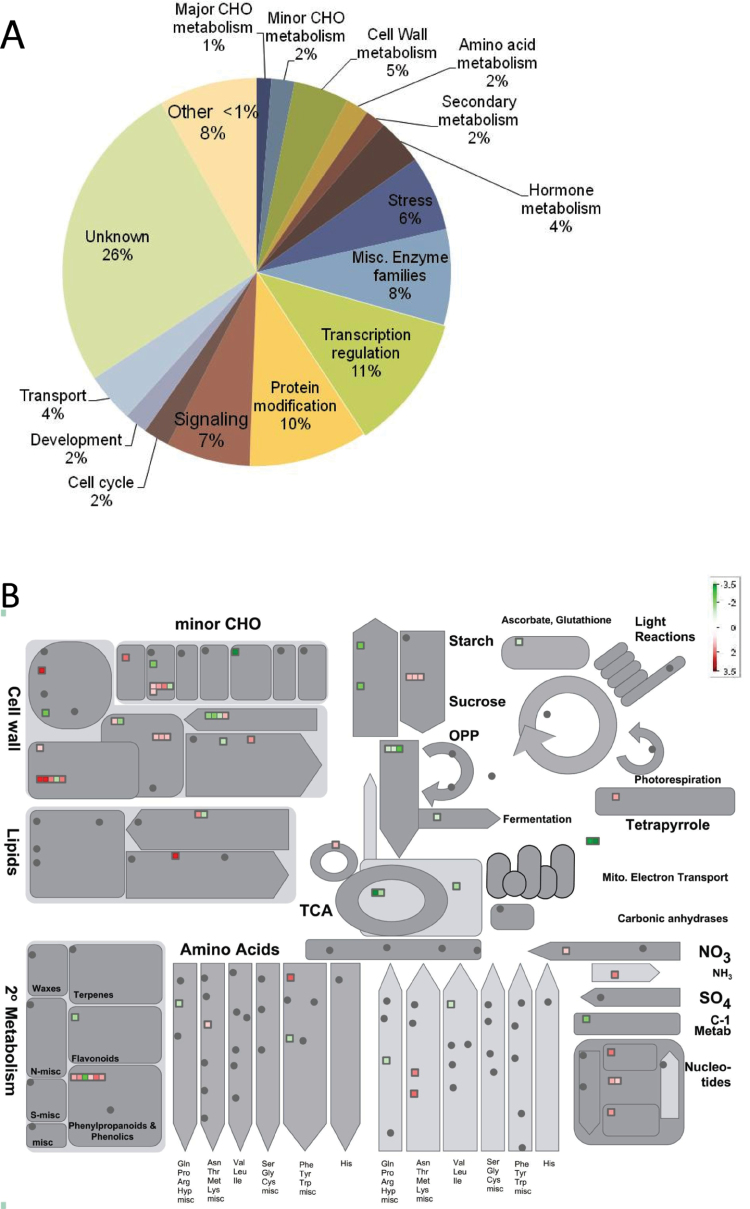
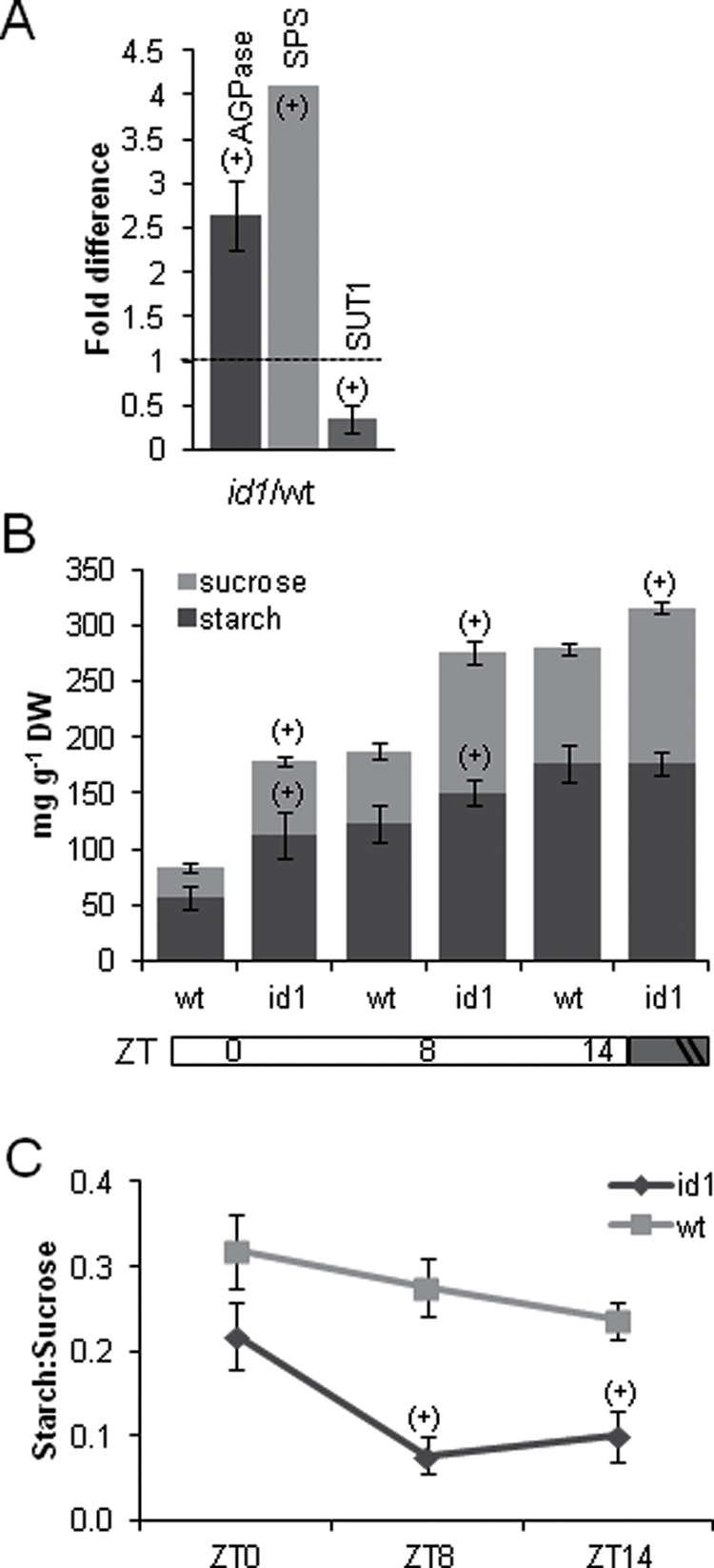
Microarray data analysis interrogating about a third of known maize genes yielded a list of 460 entities with a minimum 2-fold change in expression between L5 of V7 stage wild-type and id1 plants significant at the 0.05 false discovery rate level. A full list of statistically significant expression differences is shown in Supplementary Table S1 at JXB online. MapMan analysis, which groups probe sets into hierarchical functional categories, termed bins, based on putative biological function (Thimm et al., 2004), was used. Thus, 414 of 460 probe sets representing differentially expressed genes were assigned to 35 bins in the ‘Overview’ visualization pathway using Affymetrix mapping for maize (Supplementary Table S2 at JXB online). Several major functional categories were identified by calculating the relative proportions of probe sets in each bin (Fig. 2A). Transcriptional changes in genes with putative functions in metabolism were visualized using the ‘Metabolism overview’ pathway (Fig. 2B). Primary metabolic processes, including minor and major carbohydrate metabolism, cell wall biosynthetic processes, energy-generating processes, amino acid, and lipid, and nucleotide metabolism, constitute 14.5% of all transcriptional changes (Fig. 2B). To ascertain if any GO terms are statistically enriched in the data set of differentially expressed entities, agriGO analysis was performed (Du et al., 2010). Fifteen GO terms that belong to the ‘Biological Process’ ontology are significantly over-represented in this data set (P < 0.05) (Fig. 2C). The majority of statistically enriched terms belong to the ‘Carbohydrate metabolic process’ node including ‘Polysaccharide metabolic process’, which is enriched most significantly. Specifically, several genes encoding key proteins in starch and sucrose metabolism have altered expression levels in id1 mutant leaves. For example, transcripts encoding the small subunit of an ADP glucose pyrophosphorylase (AGPase; NM_001111708) along with a putative sucrose phosphate synthase (SPS; BF728690), key enzymes in starch and sucrose synthesis, respectively, are more abundant in id1 mutant leaves (Fig. 3A). SUT1 (Sucrose Transporter1), encoding the maize sucrose transporter responsible for loading of sucrose in source leaves for export to sinks (Aoki et al., 1999; Slewinski et al., 2009), is expressed at lower levels in id1 source leaves relative to the wild type (Fig. 3A). Further, the microarray data show that id1 mature leaves have higher transcript levels for predicted glycolysis and TCA cycle enzyme-encoding genes such as UDP-glucose pyrophosphorylase (UGPase; NM_001137270), 6-phosphofructokinase (PFK; NM_001137275), phosphoenol pyruvate carboxylase (PEPCase; NM_001158445), citrate synthase4 (NM_001153134), and aconitase (NM_001138804) (Fig. 2B; Supplementary Table S2).
Fig. 2.
Microarray analysis of mature leaves of floral transition stage wild-type and id1 mutant plants resulted in 460 probe sets with at least a 2-fold change in expression (P < 0.05) using three biological replicates per genotype. (A) Differentially expressed genes were assigned to 35 bins in the ‘Overview’ visualization pathway of MapMan using Affymetrix mapping for maize. The frequency of each bin is reported as a percentage (%) of the total number of mapped changes (414). (B) The ‘Metabolism overview’ MapMan pathway was used to visualize transcriptional changes in genes with putative functions in metabolism. Green represents higher expression in id1 mutants, while red denotes lower expression in id1 relative to the wild type, with darker shading indicating increasing magnitude of log2 expression fold change as specified by the scale. (C) AgriGO analysis of expression differences between the wild type and id1 for statistically enriched GO terms in the ‘Biological process’ ontology. P-values <0.05 are reported in parentheses. Colouring of GO term nodes is proportional to their significance as indicated by the scale.
Fig. 3.
Transcriptional and metabolite level differences in sucrose and starch metabolism of id1 and wild-type leaf 5 at V7. (A) Quantitative RT-PCR of maize genes encoding a small unit of ADP-glucose pyrophosphorylase (AGPase) and the Sucrose Transporter1 (SUT1) genes. A putative Sucrose Phosphate Synthase (SPS) transcript is up-regulated in id1 leaves based on microarray analysis (no error bars). All fold change values are statistically significant [(+), t-test, P < 0.05]. (B) Starch and sucrose levels in mg metabolite per g leaf dry weight (DW) at Zeitgeber time points (ZT) 0, 8, or 14h after dawn. Horizontal bars below the x-axis denote light/dark cycles, with vertical slashes indicating that the length of the dark cycle is not drawn to scale. (C) Relative ratio of starch to sucrose at the same ZT as B. The ratios of log10 of each metabolite in mg g–1 leaf DW for wild type (light grey) and id1 (dark grey) are shown. Statistical significance between id1 and the wild type at each ZT based on a t-test (P < 0.05) is indicated with (+). Error bars represent the SD (n = 5).
Expression differences for selected genes were confirmed using real-time RT-PCR analysis. Relative expression of genes mapping to bins that reflect several functional categories of expression change were confirmed in all cases (Supplementary Table S3 at JXB online).
Major alterations in transitory starch and sucrose in id1 leaves
To follow up on the finding of expression level differences for major sucrose and starch metabolism genes in source leaves of late-flowering id1 maize (Figs 2, 3A), sucrose and transitory starch were quantified in pre-transition V7 as well as post-transition V9 mature leaf blades. Leaf samples collected at three ZT points: dawn (ZT0), midday (ZT8), and the end of a long day (ZT14), were analysed to account for diurnal fluctuations in starch and sucrose accumulation. Overall, sucrose and starch levels are higher in id1 mutant leaves relative to wild-type plants, especially at midday (Fig. 3B). The total carbohydrate amount (starch plus sucrose) is significantly higher in late-flowering plants when expressed both as a ratio to soluble leaf protein and relative to dry leaf mass (Supplementary Fig. S2A at JXB online). While maximum starch accumulation in leaves at the end of a long day is not significantly different between mutants and the wild type, the amount of starch detected at the end of the dark period (ZT0) is higher in id1 mutant leaves (Fig. 3B). However, expression level differences were not detected in genes encoding enzymes required for starch degradation, such as β-amylase and isoamylase, or in genes that facilitate the export of transitory starch degradation products, such as mex1 (maltose excess1; EU963397) and GlcT (Glucose Translocator; AF215854) (Supplementary Table S1, Fig. S2B) (Weise et al., 2004; Kikuchi et al., 2009; Pin et al., 2010).
The relative ratio of transitory starch to sucrose was also examined in id1 and wild-type leaves. Mutant plants at the V7 stage have a markedly decreased starch to sucrose ratio during the day (Fig. 3B). It is also intriguing that, while wild-type leaves have higher daytime starch to sucrose ratios at V9 compared with V7, the ratio remains unchanged within this developmental time frame in the id1 mutant (Supplementary Fig. S2C at JXB online).
Photosynthetic rate and leaf morphology are unchanged in id1 mutant leaves
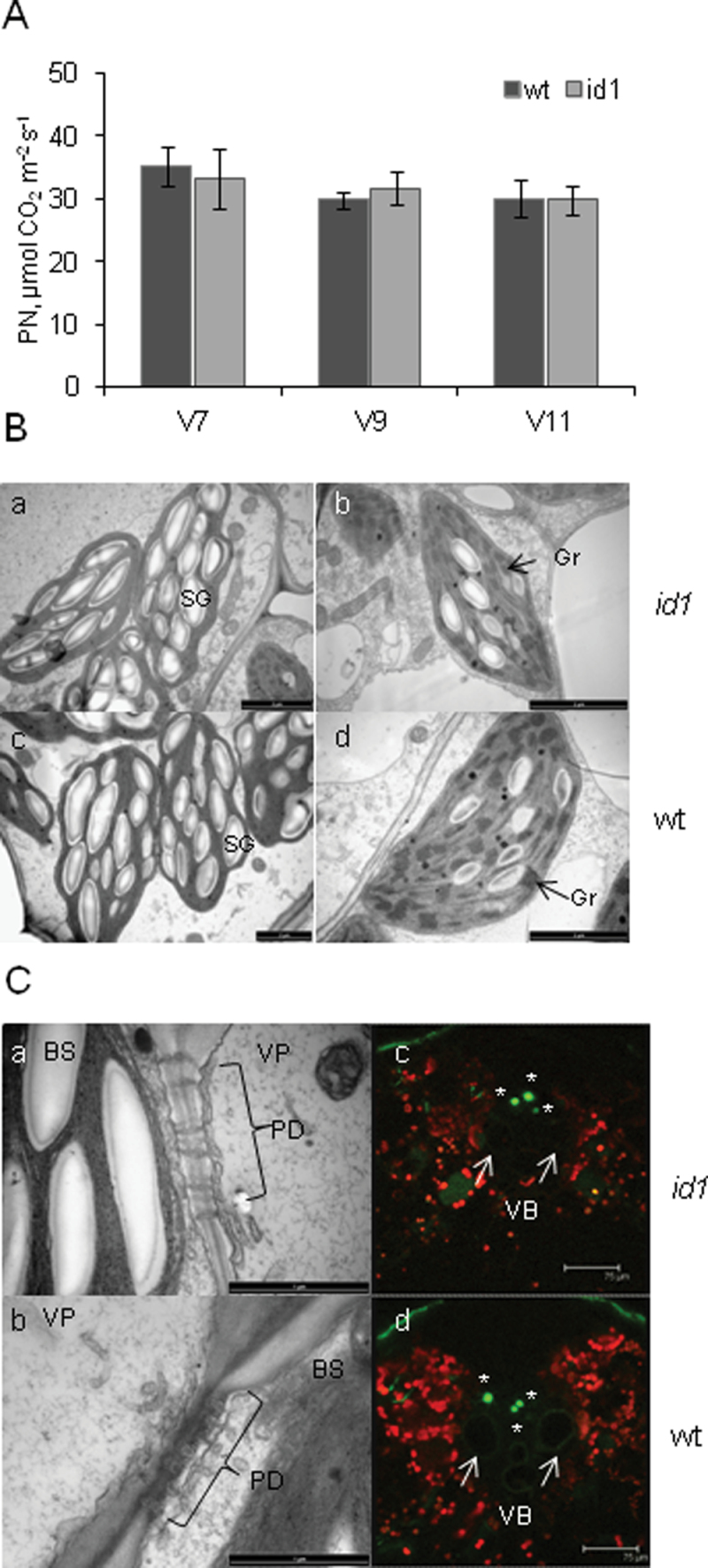
To investigate whether the observed higher levels of sucrose and starch in mature leaves of id1 mutant plants might be due to increased carbohydrate assimilation, photosynthetic rate and capacity were compared in mutant and wild-type plants. Plants at V7, V9, and V11 developmental stages were included in the analysis. No alteration in the photosynthetic rate of id1 mutant leaves at the V7 stage was detected (Fig. 4A; Supplementary Fig. S3A at JXB online) and leaf chlorophyll content was similar in id1 and normal-flowering plants at this stage (Supplementary Fig. S3B). Cross-sections of fully expanded leaves of the mutant and wild-type transition stage plants were also examined at key time points in the diurnal cycle (ZT0, ZT8, and ZT14) using transmission electron microscopy (TEM). No qualitative differences were apparent in mesophyll or bundle sheath chloroplast distribution or morphology, nor in the appearance or size of bundle sheath chloroplast starch grains (Fig. 4B). In light of the observed differences in sucrose levels, the gross morphology of cell–cell connectivity along the symplastic route of sugar loading in leaves was also evaluated. No major differences were observed in the distribution or overall appearance of plasmodesmata connections between bundle sheath and vascular parenchyma cells (Fig. 4C). Further, the ability of source leaves to transport the phloem mobile fluorescent tracer CFDA from blades to sheaths was assessed. CFDA has been used to study phloem solute transport of maize leaves impaired in carbohydrate loading (Ma et al., 2009). Carboxyfluorescein was always detected in sieve elements of the phloem of both id1 mutant and wild-type leaf sheaths (Fig. 4C), indicating that id1 leaves are similar to the wild type in their ability to transport the dye through the leaf vasculature. Moreover, analysis of carbohydrate levels in immature leaves, which are sinks for sucrose unloading, showed that these heterotrophic tissues in id1 mutants accumulate similar levels of glucose, sucrose, and starch to wild-type plants at this stage (Supplementary Fig. S4 at JXB online). Together, these data show that the elevated sucrose levels detected in id1 mutant leaves cannot be attributed to apparent defects in cell–cell connectivity along the symplastic sucrose transport route or along the phloem stream, although minor alterations that are undetectable by these methods cannot be ruled out.
Fig. 4.
Leaf photosynthetic capacity and morphology of the wild type and id1. (A) Photosynthetic rate (PN) of the youngest fully expanded leaf of V7, V9, and V11 id1 (dark grey bars) and wild-type (light grey bars) plants. Error bars represent the SD (n = 5). (B) TEM images of the fifth leaf of V7 stage id1 (a, b) and wild-type (c, d) plants. (a and c) Chloroplasts in bundle sheath cells; SG, starch grains. (b and d) Chloroplasts in mesophyll cells; Gr, grana. (C) (a and b) The interface between bundle sheath (BS) and vascular parenchyma (VP) cells in a vascular bundle of leaf 5 shows symplastic connectivity between these cell types via plasmodesmata (PD); (c and d) cross-sections of leaf sheaths show carboxyflourescein fluorescence (green, asterisk) in cells of the vascular bundle (VB). Red indicates chlorophyll autofluorescence. Arrows indicate mature xylem vessels.
Finally, since no major alterations in photosynthetic capacity and gross defects with sucrose export were observed, the expression levels of several maize sucrose transporter-encoding genes were examined to assess possible changes at the molecular level. In addition to ZmSUT1 (Fig. 3A), ZmSUT2, ZmSUT4, and ZmSUT5 expression was quantitatively compared in sink and source leaves of id1 mutants and normal-flowering maize. Interestingly, ZmSUT2 expression is down-regulated in source leaves of id1 mutants relative to the wild type, while ZmSUT4 and ZmSUT5 transcripts are detected at decreased levels in id1 sink leaves (Table 1).
Table 1.
Expression of ZmSUT2, ZmSUT4 and ZmSUT5 transcripts in fully expanded source leaf blades and immature sink leaves represented as fold up-regulation in the wild type relative to id1 mutants at the V7 stage
| Gene | Tissue | RQ wild type/id1 | P-value |
| ZmSUT2 | Sink leaves | 1.08 | NS |
| Source leaf blade | 1.81 | 0.007 | |
| ZmSUT4 | Sink leaves | 1.34 | 0.038 |
| Source leaf blade | 1.34 | 0.069 | |
| ZmSUT5 | Sink leaves | 1.56 | 0.026 |
| Source leaf blade | Not detected | NA |
Data are based on qRT-PCR with five biological replicates; fold change values with statistical significance at the P < 0.05 level are reported in bold.
Primary metabolite alterations in mature leaves associated with the floral transition
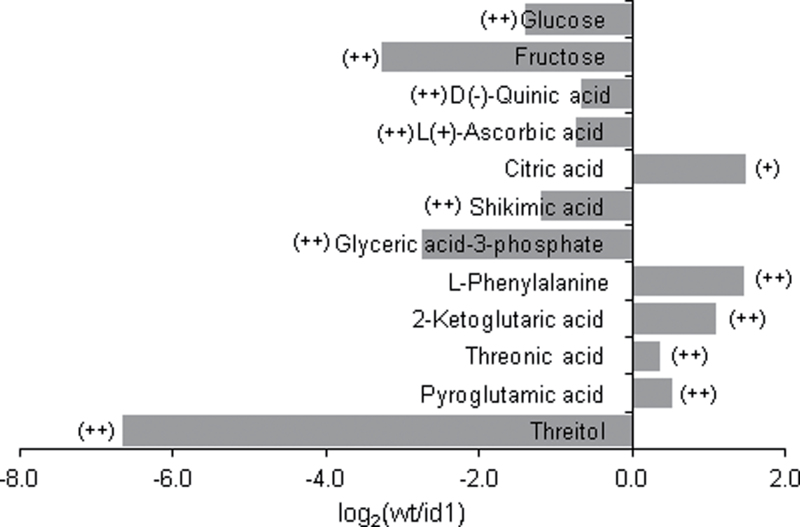
To follow up on the observed transcript and carbohydrate alterations in id1 V7 leaves, wild-type and id1 leaf extracts were analysed using GC-MS. Approximately 120 components (peaks) were detected per sample, and the average normalized peak area was significantly different between wild-type and id1 leaves for 26 peaks (t-test, P < 0.05). Putative identities were assigned for 13 of these peaks based on the best mass spectrum match to metabolites in the Golm Metabolite Database (Fig. 5; Supplementary Table S4 at JXB online). In addition to sucrose (Fig. 3B), glucose, fructose and glyceric acid-3-phosphate are among the primary metabolites present at significantly higher levels in id1 mutant leaves, while pyroglutamic acid and the TCA cycle organic acids citric acid and α-ketoglutaric acid are lower in id1 leaves (Fig. 5). Metabolite levels were also compared in extracts of immature leaves, a sink tissue where id1 is expressed, at the onset of floral induction (V7). With the exception of elevated levels of fructose, as well as a strong candidate for trehalose, relatively few differences were detected between the immature leaves of id1 and wild-type plants (Supplementary Table S5).
Fig. 5.
Comparison of metabolite levels in the fifth leaf of wild-type and id1 mutant plants at the V7 stage based on GC-MS analysis. The log2 ratio of the wild type to id1 normalized component area (n = 5) is shown. (++) represents ratios significant at the P < 0.05 level, while (+) indicates metabolites with a significant difference between genotypes at the P < 0.1 level based on t-tests.
Starch and sucrose levels in flowering versus non-flowering teosinte
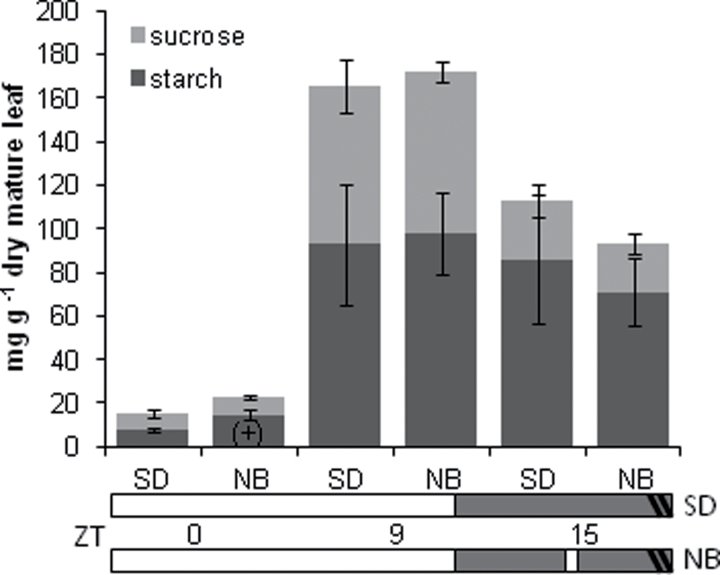
The progenitor of autonomously flowering temperate maize is teosinte, a tropical maize that requires short-day photoperiods to induce the floral transition (Emerson, 1924). Levels of transitory starch and sucrose were compared between florally induced and uninduced V7 teosinte leaves to delineate whether similar alterations in the levels of major carbohydrates observed in id1 mutants are associated with the floral transition in general or may be specific to the ID1-regulated autonomous pathway in maize. For this experiment, teosinte was grown under inductive SD photoperiods and under night break (NB) conditions where short-day conditions were interrupted by exposure to light. It was determined that as little as 1h of light in the middle of a 15h night causes 100% of teosinte plants to remain in a vegetative growth state, which was confirmed by excision and observation of shoot meristems at V7 and V9 (not shown). Comparison of plants grown in SD and NB conditions minimizes the effect of large photoperiod differences. It was found that, with the exception of higher levels of transitory starch at ZT0 in the NB treatment, no other significant differences in carbohydrate accumulation were detected in mature leaf blades associated with florally induced teosinte plants (Fig. 6). Therefore, changes in carbohydrate levels are associated with flowering in autonomous day-neutral maize but not in photoperiod-dependent teosinte.
Fig. 6.
Sucrose and starch levels in leaf 5 of flowering (SD) versus non-flowering (NB) teosinte plants at ZT 0, 9, and 15h after dawn. Error bars represent the SE for three experiments. (+) denotes statistical significance between SD- and NB-grown teosinte plants at a particular ZT.
Discussion
Expression differences in leaves of id1 maize highlight a central role for carbohydrate metabolism at the autonomous floral transition
Primary metabolic processes constitute 14.5% of all transcriptional differences between wild-type and id1 source leaves (Fig. 2B), and GO enrichment analysis showed that ‘carbohydrate metabolic process’-related nodes are over-represented among transcriptional changes (Fig. 2C). Specifically, genes involved in starch and sucrose metabolism are up-regulated in id1 source leaves (Figs 2B, 3A), while genes encoding enzymes involved in glycolysis and the TCA cycle are down-regulated (Supplementary Table S2 at JXB online). These results provide evidence of a close association between carbohydrate metabolism and the floral transition in florigen-producing maize leaves. Interestingly, microarray profiling experiments that aimed to capture broad functional categories of transcriptional changes associated with the transition to flowering in other species, such as rice, soybean, and Arabidopsis, have also reported a prominent role for metabolism-related transcripts indicating that metabolic status is closely linked with the floral transition (Wilson et al., 2005; Chen and Wang, 2008; Wong et al., 2009).
In a previous transcriptome analysis of immature leaves of id1 mutants that aimed to identify downstream targets of ID1 activity, prominent transcriptional differences were detected in carbohydrate metabolism-related genes (Coneva et al., 2007). In the current study id1 mutants were use as a tool and advantage was taken of the developmental gradient characteristic of grass leaves to compare mature leaves that produce florigen with mature leaves at the same developmental stage that do not produce florigen. Collectively the findings suggest that changes in primary metabolism-related gene function are extended to later stages of leaf development and are associated with mature, florigen-producing maize leaves.
id1 source leaves have a distinct metabolite signature indicative of changes in carbohydrate utilization
Examination of florigen-producing mature leaves at the developmental stage preceding the floral transition in normal maize showed that id1 mutants accumulate significantly higher levels of sucrose, other soluble sugars, and, to a lesser degree, starch, during the day than the wild type (Figs 3B, 35). The metabolite profile of id1 leaves is also characterized by lower levels of TCA cycle organic acids, such as citric acid and α-ketoglutaric acid, compared with wild-type plants (Fig. 5). In the absence of an increase in photosynthetic capacity in id1 mutants relative to wild-type plants (Fig. 4A; Supplementary Fig. S3A at JXB online), it was hypothesized that the increased sucrose levels in id1 leaves may be due to a change in the relative proportion of major carbohydrate metabolites in the leaf. Indeed, id1 mutants have a significantly decreased transitory starch to sucrose ratio (Fig. 3B) and this ratio may be a more accurate indicator of the physiological status of the plant. Indeed, sucrose level differences alone may not be sufficient to explain flowering-time effects, and some studies find that elevated sucrose is associated with an inhibitory effect on flowering time (Ohto et al., 2001; Seo et al., 2011b). Several lines of evidence support the hypothesized importance of the balance between transitory starch and sucrose in the leaf. First, an increase in the starch to soluble carbohydrate ratio is associated with floral promotion in day-neutral tobacco plants (Rideout et al., 1992). Further, tomato plants overexpressing SPS direct an increased proportion of newly assimilated carbon into sucrose synthesis at the expense of transitory starch accumulation. While a moderate increase in sucrose levels in SPS overexpression plants led to a stimulation of flowering and fruit yield, plants with a dramatically decreased starch to sucrose ratio did not have increased yield (Sharkey et al., 2004; Raines and Paul, 2006). Moreover, QTL analyses in Arabidopsis support a negative correlation between soluble sugar levels in early stages of development and flowering, while transitory starch accumulation is associated with early flowering (El-Lithy et al., 2010). Finally, pgm1 (phosphoglucomutase1) mutants, which accumulate 10-fold higher levels of soluble carbohydrates than non-mutant plants but are unable to synthesize starch, do not complete the floral transition, even under inductive photoperiods (Corbesier et al., 1998). These various studies show a connection between altered carbon assimilation and the floral transition, although whether this relationship is causal or correlative remains to be determined.
In addition to changes in the relative abundance of sucrose and starch, id1 source leaves are typified by lower levels of some organic acids (Fig. 5). QTL analysis and metabolite profiling have uncovered strong correlations between the levels of primary metabolites and growth rate, biomass accumulation, and flowering (Meyer et al., 2007; Gibon et al., 2009; El-Lithy et al., 2010). Based on the observed metabolite and transcriptional profiles, it is suggested that id1 mutant leaves favour carbohydrate utilization over storage in source leaves (Fig. 7). This hypothesis is supported by the metabolic profile of pea embryos in which the Sucrose non-fermenting1 related kinase1 gene, SnRK1, is repressed. Similar to id1 leaves, SnRK1 antisense embryos have increased sucrose and a decreased starch to sucrose ratio, as well as lower TCA cycle organic acids and amino acids (Radchuk et al., 2010). Storage activity and seed maturation are strongly impaired in SnRK1 antisense lines (Radchuk et al., 2006), illustrating the importance of concerted carbohydrate allocation for normal development.
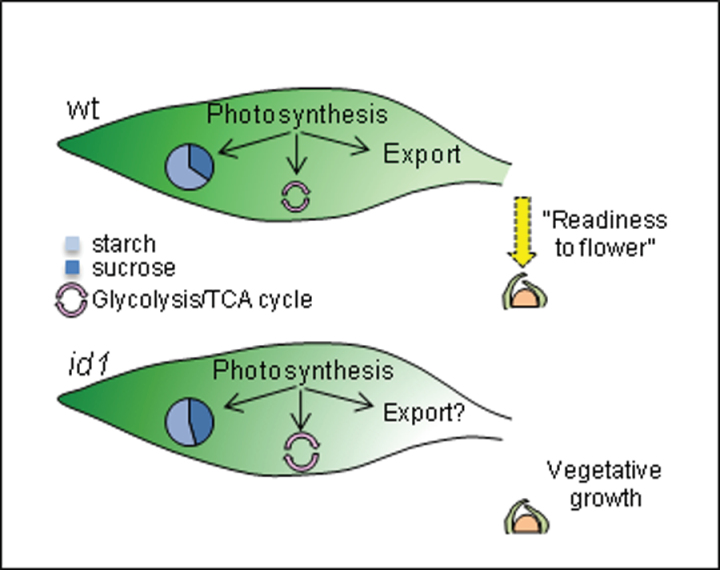
Fig. 7.
The metabolic signatures of source leaves associated with ‘readiness to flower’ (yellow arrow) in wild-type versus vegetative growth in id1 at V7. Transcript and metabolite profiles of id1 leaves reflect changes in the ratio of transitory starch to sucrose (pie charts) as well as sucrose utilization within the leaf (circular arrows). The balance between carbohydrate storage and utilization is shifted in id1 source leaves, resulting in the absence of a ‘readiness to flower’ signal and, thus, vegetative growth. The dome structure represents the SAM.
A possible role for transitory starch accumulation as a flowering-time cue in maize
The finding that late-flowering id1 mutants have a significantly decreased starch to sucrose ratio (Fig. 3C) and enrichment of ‘polysaccharide metabolic process’ and related GO terms among expression differences in id1 source leaves (Fig. 2C) highlights a possible role for transitory starch at the floral transition. This association is also evident in a number of Arabidopsis mutants where late flowering results from defects in starch accumulation. For example, in addition to pgm1, pgi1 (phosphoglucose isomerase1) and adg1 (AGPase) mutants are unable to produce transitory starch in leaves and are late flowering, especially in short days (Corbesier et al., 1998). Moreover, analysis of starch content in rice leaves shows that a peak in starch accumulation coincides with heading (Chen and Wang, 2008). However, as with sucrose levels, it is probably not starch levels per se that influence flowering time, but rather the control of carbohydrate allocation in the leaf. This idea is supported by the observation that both starch-overaccumulating Arabidopsis mutants, such as sex1 (starch excess1) and cam1 (carbohydrate accumulation1), and the starch-deficient mutants discussed above are late-flowering (Eimert et al., 1995). Collectively, these data show a correlation between the ability to flower at the proper time and both sufficient transitory starch accumulation and adequate and timely mobilization of leaf starch. Moreover, expression differences were not observed in genes encoding amylases, or a putative chloroplast maltose translocator mex1 in id1 leaves, suggesting that starch mobilization is not affected (Supplementary Table S1, Fig. S2B at JXB online). However, because current knowledge of the mechanisms that control transitory starch degradation in C4 plants such as maize is limited (Weise et al., 2011), dissection of the observed association between transitory starch and the floral transition awaits a more complete understanding of this process.
Is sucrose export linked to the floral transition in maize?
Carbohydrate export to sink tissues is an essential prerequisite for floral induction (i.e. sucrose is required to fuel the metabolic requirements of the rapidly growing inflorescence). In maize, sucrose travels along a symplastic route from chloroplast-containing cells to vascular parenchyma cells and is then exported to the apoplasm before being loaded onto sieve elements via the ZmSUT1 symporter (Slewinski and Braun, 2010). Thus, sucrose export from maize leaves consists of both symplastic and apoplastic steps. Defects in the symplastic portion of sucrose export, such as structurally modified plasmodesmata in the sxd1 (sucrose export defective1) mutant (Russin et al., 1996; Botha et al., 2000), as well as the sut1 mutant which is impaired in loading of sucrose from the apoplast (Aoki et al., 1999; Slewinski et al., 2009), exhibit carbohydrate hyperaccumulation in leaves accompanied by reduced vegetative growth and delayed flowering. While no obvious obstruction of the symplastic route or the phloem stream was observed in id1 mutants at the floral transition (Fig. 4B), SUT1 expression is down-regulated in mutant leaves (Fig. 2A), pointing to the possibility that the apoplastic step of sucrose export from leaves may be affected in id1 mutants. Alternatively, decreased SUT1 levels in id1 mutants may be due to feedback regulation as a consequence of elevated sucrose levels (Fig. 2B).
Interestingly, in addition to ZmSUT1, ZmSUT2, 4, and 5 are also down-regulated in leaves of id1 mutants (Table 1). Although the function of these transporters requires further investigation, phylogenetic analysis suggests that ZmSUT2 belongs to the same group of SUT proteins as potato StSUT4, which has been demonstrated to regulate flowering time and tuberization by coordinating light and carbohydrate signals (Chincinska et al., 2008). The notion that ZmSUT2 may have an analogous function to StSUT4 is consistent with the observation that ZmSUT2 expression is lower in source leaves of id1 mutants, which display carbohydrate allocation alterations and lower ZmSUT1 levels. An earlier report speculated that Arabidopsis members related to ZmSUT4 may have a role in sucrose sensing (Barker et al., 2000). The ZmSUT5 clade of proteins are uncharacterized, but many members have high expression in sink organs (Braun and Slewinski, 2009). These data suggest that, although no alterations in major carbohydrate levels were detected in sink tissues of id1 mutants (Supplementary Fig. S4 at JXB online), the possibility of subtle and/or transient adjustments in whole-plant carbohydrate allocation cannot be ruled out. An association between transcript levels of SUT1 transporters in source leaves and flowering time has been reported for potato and day-neutral tobacco (Burkle et al., 1998; Leggewie et al., 2003; Chincinska et al., 2008). Further, the expression of several sucrose transporters, including the Arabidopsis orthologue of SUT1, AtSUC2, is affected in the late-flowering idd8 mutant (Seo et al., 2011b). However, the possibility that sucrose export is dramatically altered in id1 mutants is inconsistent with the observation that lowered SUT1 expression is associated with fewer leaves at flowering and decreased growth (Burkle et al., 1998; Slewinski et al., 2009). These phenotypes contrast with the vigorous vegetative growth of id1 mutants and with the lack of appreciable changes in photosynthetic activity (Fig. 4A). Additionally, although SUT1 levels are decreased by ~50% in id1 source leaves, sucrose levels in id1 immature leaves, a sink tissue, are comparable with the wild type (Supplementary Fig. S4).
Alternatively, elevated leaf concentrations of sucrose in id1 mutants may down-regulate the sucrose transporter transcript level similar to what has been reported for SUT1 transcription in sugar beet (Chiou and Bush, 1998; Vaughn et al., 2002; Ainsworth and Bush, 2011). Ultimately, the causal relationship between ID1, SUT1 expression, and sucrose levels in source leaves of maize is an interesting area for further investigation, especially in light of the report by Kühn and Frommer (1995) showing that the potato IDD gene PCP1 is able to rescue a sucrose transport deficiency in yeast.
Autonomous flowering signals and the mechanism of ID1 action
The transcriptional and metabolite data suggest that the absence of ID1 activity is marked by significant carbohydrate allocation changes in source leaves that alter the balance between utilization and storage of resources that is required to signal ‘readiness to flower’. Thus, id1 mutants continue to grow vegetatively (Fig. 7). The major alterations in starch and sucrose levels in source leaves of id1 mutants and wild-type siblings were not observed when comparing florally induced with vegetatively growing teosinte (Fig. 6). These findings support the notion that carbohydrate allocation changes in mature maize leaves are associated specifically with the ID1-regulated autonomous floral transition in temperate maize.
A question that remains is how the autonomous pathway integrates known flowering-time components with leaf-derived florigen signals. The Arabidopsis FT gene, and its corresponding orthologues in diverse species, encodes a protein that acts as a florigen (Corbesier et al., 2007; Lin et al., 2007; Tamaki et al., 2007; Lazakis et al., 2011; Meng et al., 2011). Recent evidence suggests that maize has at least one FT orthologue, ZCN8, and the expression of this florigen-encoding gene is dependent on id1 (Lazakis et al., 2011; Meng et al., 2011). It is intriguing to speculate that ZCN8 (and potentially other ZCN genes) integrates signals from the autonomous and photoperiod pathways. In this scenario, ID1 acts to establish an epigenetic blueprint in developing leaves that is manifested as a physiological programme in mature leaves that results in activation of genes required for florigen production. Ultimately, these changes result in a metabolic signature in mature leaves that converges with other leaf-derived signals to control the expression or mobility of ZCN8 or other undiscovered florigens. It is speculated that during the rapid evolution of day-neutral temperate maize from teosinte, the ID1-regulated autonomous floral induction pathway overshadowed the ancestral obligate SD photoperiod pathway, thus allowing maize growth to expand into a broader range of latitudes.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. qRT-PCR analysis of ZCN8 expression level in mature leaf five of V7 wild-type plants relative to id1 mutants at the same developmental stage.
Figure S2. (A) Total carbohydrate (sucrose plus starch) to soluble protein ratio in id1 and wild-type mature leaf 5 at V7. (B) Relative expression levels of a putative maize maltose transporter gene (maltose excess1, mex1) and a glucose translocator gene (GlcT) in wild-type and id1 mature leaves. (C) Starch to sucrose ratio in source leaves of id1 and wild-type plants after the floral transition (V9).
Figure S3. (A) CO2 response curve for id1 and wild-type leaf five at the V7 stage. (B) Chlorophyll quantification of id1 and wild-type leaf five at the V7 stage.
Figure S4. Comparison of starch, sucrose, and glucose levels in immature sink leaves of id1 mutants relative to normal-flowering V7 maize plants.
Table S1. Full list of 460 probe sets differentially expressed at the 2-fold, P < 0.05 level between wild-type and id1 mutant source leaves (L5) at the V7 developmental stage.
Table S2. Full list of 414 MapMan ‘Overview’ bin assignments of differentially expressed probe sets.
Table S3. List of differentially expressed genes from microarray analysis of mature leaves of id1 mutants and wild-type plants confirmed by real-time (q)RT-PCR.
Table S4. List of statistically significant (P < 0.05) leaf five metabolite differences between mature leaves of id1 and wild-type plants prior to the floral transition (V7 stage).
Table S5. List of statistically significant (P < 0.05) metabolite differences between immature leaves of id1 and wild-type plants at V7.
Table S6. List of primers used for id1 genotyping and qRT-PCR experiments.
Supplementary Material
Acknowledgements
We thank Mike Mucci and Tannis Slimmon at the Guelph Phytotron facility for expert plant care and advice. Dr R. Tremblay and Dr S. Humbert provided helpful suggestions on the manuscript. We thank David Carter at the London Affymetrix facility for microarray hybridization and data retrieval. Dr J. Greenwood provided expertise in sample preparation for TEM analysis. We thank Dr B. Micallef and Dr H. Earl, who generously provided LiCor-6400 instruments. This research is supported by a Natural Sciences and Engineering Research Council (NSERC) Discovery grant and the Ontario Research Fund (to JC and SR). VC is a recipient of an NSERC doctoral student fellowship.
References
- Ainsworth EA, Bush DR. Carbohydrate export from the leaf: a highly regulated process and target to enhance photosynthesis and productivity. Plant Physiology. 2011;155:64–69. doi: 10.1104/pp.110.167684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amasino R. Seasonal and developmental timing of flowering. The Plant Journal. 2010;61:1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- Aoki N, Hirose T, Takahashi S, Ono K, Ishimaru K, Ohsugi R. Molecular cloning and expression analysis of a gene for a sucrose transporter maize (Zea mays L.) Plant and Cell Physiology. 1999;40:1072–1078. doi: 10.1093/oxfordjournals.pcp.a029489. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts—polyphenoloxidase in Beta vulgaris. Plant Physiology. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker L, Kühn C, Weise A, Schulz A, Gebhardt C, Hirner B, Hellmann H, Schulze W, Ward JM, Frommer WB. SUT2, a putative sucrose sensor in sieve elements. The Plant Cell. 2000;12:1153–1164. doi: 10.1105/tpc.12.7.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodson M Outlaw WH Jr.1985Elevation in the sucrose content of the shoot apical meristem of Sinapis alba at floral evocation Plant Physiology 79420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botha CEJ, Cross RHM, van Bel AJE, Peter CI. Phloem loading in the sucrose-export-defective (SXD-1) mutant maize is limited by callose deposition at plasmodesmata in bundle sheath–vascular parenchyma interface. Protoplasma. 2000;214:65–72. [Google Scholar]
- Bradford M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein–dye-binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Braun DM, Slewinski TL. Genetic control of carbon partitioning in grasses: roles of sucrose transporters and Tie-dyed loci in phloem loading. Plant Physiology. 2009;149:71–81. doi: 10.1104/pp.108.129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler ES Holland JB Bradbury PJ , et al.2009The genetic architecture of maize flowering time Science 325714–718. [DOI] [PubMed] [Google Scholar]
- Burkle L, Hibberd JM, Quick WP, Kühn C, Hirner B, Frommer WB. The H+-sucrose cotransporter NtSUT1 is essential for sugar export from tobacco leaves. Plant Physiology. 1998;118:59–68. doi: 10.1104/pp.118.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon F, Virlon B, Moreau L, Falque M, Joets J, Decousset L, Murigneux A, Charcosset A. Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome. Genetics. 2004;168:2169–2185. doi: 10.1534/genetics.104.032375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HJ, Wang SJ. Molecular regulation of sink–source transition in rice leaf sheaths during the heading period. Acta Physiologiae Plantarum. 2008;30:639–649. [Google Scholar]
- Chincinska IA, Liesche J, Kruegel U, Michalska J, Geigenberger P, Grimm B, Kühn C. Sucrose transporter StSUT4 from potato affects flowering, tuberization, and shade avoidance response. Plant Physiology. 2008;146:515–528. doi: 10.1104/pp.107.112334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR. Sucrose is a signal molecule in assimilate partitioning. Proceedings of the National Academy of Sciences, USA. 1998;95:4784–4788. doi: 10.1073/pnas.95.8.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Muszynski MG. 2008. The maize floral transition. In: Hake SC, Bennetzen JL, eds. Handbook of maize: its biology, Vol. 1. Berlin: Springer Science; 41 55 [Google Scholar]
- Colasanti J, Tremblay R, Wong AYM, Coneva V, Kozaki A, Mable BK. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics. 2006;7:1–15. doi: 10.1186/1471-2164-7-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasanti J, Yuan Z, Sundaresan V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell. 1998;93:593–603. doi: 10.1016/s0092-8674(00)81188-5. [DOI] [PubMed] [Google Scholar]
- Coneva V, Zhu T, Colasanti J. Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. Journal of Experimental Botany. 2007;58:3679–3693. doi: 10.1093/jxb/erm217. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Lejeune P, Bernier G. The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta. 1998;206:131–137. doi: 10.1007/s004250050383. [DOI] [PubMed] [Google Scholar]
- Corbesier L Vincent C Jang SH , et al.2007FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis Science 3161030–1033. [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimert K, Wang SM, Lue WL, Chen JC. Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in Arabidopsis. The Plant Cell. 1995;7:1703–1712. doi: 10.1105/tpc.7.10.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Lithy ME, Reymond M, Stich B, Koornneef M, Vreugdenhil D. Relation among plant growth, carbohydrates and flowering time in the Arabidopsis Landsberg erecta×Kondara recombinant inbred line population. Plant, Cell and Environment. 2010;33:1369–1382. doi: 10.1111/j.1365-3040.2010.02155.x. [DOI] [PubMed] [Google Scholar]
- Emerson RA. Control of flowering in teosinte. Journal of Heredity. 1924;15:41–48. [Google Scholar]
- Feurtado JA, Huang DQ, Wicki-Stordeur L, Hemstock LE, Potentier MS, Tsang EWT, Cutler AJ. The Arabidopsis C2H2 zinc finger INDETERMINATE DOMAIN1/ENHYDROUS promotes the transition to germination by regulating light and hormonal signaling during seed maturation. The Plant Cell. 2011;23:1772–1794. doi: 10.1105/tpc.111.085134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Pyl ET, Sulpice R, Lunn JE, Hohne M, Gunther M, Stitt M. Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant, Cell and Environment. 2009;32:859–874. doi: 10.1111/j.1365-3040.2009.01965.x. [DOI] [PubMed] [Google Scholar]
- Irish E, Jegla D. Regulation of extent of vegetative development of the maize shoot meristem. The Plant Journal. 1997;11:63–71. [Google Scholar]
- Irish EE, Nelson TM. Identification of multiple stages in the conversion of maize meristems from vegetative to floral development. Development. 1991;112:891–898. [Google Scholar]
- Jaeger KE, Graf A, Wigge PA. The control of flowering in time and space. Journal of Experimental Botany. 2006;57:3415–3418. doi: 10.1093/jxb/erl159. [DOI] [PubMed] [Google Scholar]
- Kikuchi R, Kawahigashi H, Ando T, Tonooka T, Handa H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiology. 2009;149:1341–1353. doi: 10.1104/pp.108.132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J Schauer N Krueger S , et al. 2005. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics 21 1635 1638 [DOI] [PubMed] [Google Scholar]
- Kozaki A, Hake S, Colasanti J. The maize ID1 flowering time regulator is a zinc finger protein with novel DNA binding properties. Nucleic Acids Research. 2004;32:1710–1720. doi: 10.1093/nar/gkh337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn C, Frommer W. A novel zinc finger protein encoded by a couch potatohomologue from Solanum tuberosumenables a sucrose transport-deficient yeast strain to grow on sucrose. Molecular and General Genetics. 1995;247:759–763. doi: 10.1007/BF00290408. [DOI] [PubMed] [Google Scholar]
- Lazakis CM, Coneva V, Colasanti J. ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. Journal of Experimental Botany. 2011;62:4833–4842. doi: 10.1093/jxb/err129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggewie G Kolbe A Lemoine R , et al.2003Overexpression of the sucrose transporter SoSUT1 in potato results in alterations in leaf carbon partitioning and in tuber metabolism but has little impact on tuber morphology Planta 217158–167. [DOI] [PubMed] [Google Scholar]
- Lejeune P, Bernier G, Requier MC, Kinet JM. Sucrose increases during floral induction in the phloem sap collected at the apical part of the shoot of the long-day plant Sinapis-alba L. Planta. 1993;190:71–74. [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proceedings of the National Academy of Sciences, USA. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MK Belanger H Lee YJ , et al.2007FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits The Plant Cell 191488–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T) (-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ma Y, Slewinski TL, Baker RF, Braun DM. Tie-dyed1 encodes a novel, phloem-expressed transmembrane protein that functions in carbohydrate partitioning. Plant Physiology. 2009;149:181–194. doi: 10.1104/pp.108.130971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, Yano M. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiology. 2008;148:1425–1435. doi: 10.1104/pp.108.125542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Muszynski MG, Danilevskaya ON. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. The Plant Cell. 2011;23:942–960. doi: 10.1105/tpc.110.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC Steinfath M Lisec J , et al.2007The metabolic signature related to high plant growth rate in Arabidopsis thaliana Proceedings of the National Academy of Sciences, USA 1044759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita MT, Sakaguchi K, Kiyose SI, Taira K, Kato T, Nakamura M, Tasaka M. A C2H2-type zinc finger protein, SGR5, is involved in early events of gravitropism in Arabidopsis inflorescence stems. The Plant Journal. 2006;47:619–628. doi: 10.1111/j.1365-313X.2006.02807.x. [DOI] [PubMed] [Google Scholar]
- Muszynski MG, Dam T, Li B, Shirbroun DM, Hou ZL, Bruggemann E, Archibald R, Ananiev EV, Danilevskaya ON. Delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiology. 2006;142:1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuin P, Weretilnyk EA, Summers PS, Guevara DR, Golding GB. 2004. GASP: GC/MS Analysis Software Package
- Ohto M, Onai K, Furukawa Y, Aoki E, Araki T, Nakamura K. Effects of sugar on vegetative development and floral transition in arabidopsis. Plant Physiology. 2001;127:252–261. doi: 10.1104/pp.127.1.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ Kim SL Lee S , et al.2008Rice Indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (Early heading date 1) regardless of photoperiod The Plant Journal 561018–1029. [DOI] [PubMed] [Google Scholar]
- Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJL, Nilsson O. An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science. 2010;330:1397–1400. doi: 10.1126/science.1197004. [DOI] [PubMed] [Google Scholar]
- Radchuk R, Radchuk V, Weschke W, Borisjuk L, Weber H. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiology. 2006;140:263–278. doi: 10.1104/pp.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk R, Emery RJN, Weier D, Vigeolas H, Geigenberger P, Lunn JE, Feil R, Weschke W, Weber H. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. The Plant Journal. 2010;61:324–338. doi: 10.1111/j.1365-313X.2009.04057.x. [DOI] [PubMed] [Google Scholar]
- Raines CA, Paul MJ. Products of leaf primary carbon metabolism modulate the developmental programme determining plant morphology. Journal of Experimental Botany. 2006;57:1857–1862. doi: 10.1093/jxb/erl011. [DOI] [PubMed] [Google Scholar]
- Rideout JW, Raper CDJ, Miner GS. Changes in ratio of soluble sugars and free amino nitrogen in the apical meristem during floral transition in tobacco. International Journal of Plant Sciences. 1992;153:78–88. doi: 10.1086/297008. [DOI] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L. Simultaneous analysis of metabolites in potato tuber by gas chromatography–mass spectrometry. The Plant Journal. 2000;23:131–142. doi: 10.1046/j.1365-313x.2000.00774.x. [DOI] [PubMed] [Google Scholar]
- Russin WA, Evert RF, Vanderveer PJ, Sharkey TJ, Briggs SP. Modification of a specific class of plasmodesmata and loss of sucrose export ability in the sucrose export defective1 maize mutant. The Plant Cell. 1996;8:645–658. doi: 10.1105/tpc.8.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs RM, Hackett WP. 1983. Source–sink relationships and flowering. In: Meudt WJ, ed. Strategies of plant reproduciton Totowa, NJ: Allenheld-Osmun; 263 272 [Google Scholar]
- Salvi S Sponza G Morgante M , et al.2007Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize Proceedings of the National Academy of Sciences, USA 10411376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Kim MJ, Ryu J-Y, Jeong E-Y, Park C-M. Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nature Communications. 2011a;2 doi: 10.1038/ncomms1303. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Ryu J, Kang SK, Park CM. Modulation of sugar metabolism by an INDETERMINATE DOMAIN transcription factor contributes to photoperiodic flowering in Arabidopsis. The Plant Journal. 2011b;65:418–429. doi: 10.1111/j.1365-313X.2010.04432.x. [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Laporte M, Lu Y, Weise S, Weber APM. Engineering plants for elevated CO2: a relationship between starch degradation and sugar sensing. Plant Biology. 2004;6:280–288. doi: 10.1055/s-2004-817911. [DOI] [PubMed] [Google Scholar]
- Sheehan MJ, Kennedy LM, Costich DE, Brutnell TP. Subfunctionalization of PhyB1 and PhyB2 in the control of seedling and mature plant traits in maize. The Plant Journal. 2007;49:338–353. doi: 10.1111/j.1365-313X.2006.02962.x. [DOI] [PubMed] [Google Scholar]
- Slewinski TL, Braun DM. Current perspectives on the regulation of whole-plant carbohydrate partitioning. Plant Science. 2010;178:341–349. [Google Scholar]
- Slewinski TL, Meeley R, Braun DM. Sucrose transporter1 functions in phloem loading in maize leaves. Journal of Experimental Botany. 2009;60:881–892. doi: 10.1093/jxb/ern335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tanimoto M, Tremblay R, Colasanti J. Altered gravitropic response, amyloplast sedimentation and circumnutation in the Arabidopsis shoot gravitropism 5 mutant are associated with reduced starch levels. Plant Molecular Biology. 2008;67:57–69. doi: 10.1007/s11103-008-9301-0. [DOI] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Current Opinion in Plant Biology. 2011;14:45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Harrington GN, Bush DR. Sucrose-mediated transcriptional regulation of sucrose symporter activity in the phloem. Proceedings of the National Academy of Sciences, USA. 2002;99:10876–10880. doi: 10.1073/pnas.172198599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise SE, van Wijk KJ, Sharkey TD. The role of transitory starch in C(3), CAM, and C(4) metabolism and opportunities for engineering leaf starch accumulation. Journal of Experimental Botany. 2011;62:3109–3118. doi: 10.1093/jxb/err035. [DOI] [PubMed] [Google Scholar]
- Weise SE, Weber APM, Sharkey TD. Maltose is the major form of carbon exported from the chloroplast at night. Planta. 2004;218:474–482. doi: 10.1007/s00425-003-1128-y. [DOI] [PubMed] [Google Scholar]
- Wilson IW, Kennedy GC, Peacock JW, Dennis ES. Microarray analysis reveals vegetative molecular phenotypes of arabidopsis flowering-time mutants. Plant and Cell Physiology. 2005;46:1190–1201. doi: 10.1093/pcp/pci128. [DOI] [PubMed] [Google Scholar]
- Wong AYM, Colasanti J. Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. Journal of Experimental Botany. 2007;58:403–414. doi: 10.1093/jxb/erl206. [DOI] [PubMed] [Google Scholar]
- Wong CE, Singh MB, Bhalla PL. Molecular processes underlying the floral transition in the soybean shoot apical meristem. The Plant Journal. 2009;57:832–845. doi: 10.1111/j.1365-313X.2008.03730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CY, You CJ, Li CS, Long T, Chen GX, Byrne ME, Zhang QF. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proceedings of the National Academy of Sciences, USA. 2008;105:12915–12920. doi: 10.1073/pnas.0806019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.