Abstract
Polyphenol oxidases (PPOs) are copper-binding enzymes of the plant secondary metabolism that oxidize polyphenols to quinones. Although PPOs are nearly ubiquitous in seed plants, knowledge on their evolution and function in other plant groups is missing. This study reports on the PPO gene family in the moss Physcomitrella patens (Hedw.) B.S.G. asan example for an early divergent plant. The P. patens PPO multigene family comprises 13 paralogues. Phylogenetic analyses suggest that plant PPOs evolved with the colonization of land and that PPO duplications within the monophyletic P. patens paralogue clade occurred after the separation of the moss and seed plant lineages. PPO functionality was demonstrated for recombinant PPO6. P. patens was analysed for phenolic compounds and six substances were detected intracellularly by LC-MS analysis: 4-hydroxybenzoic acid, p-cumaric acid, protocatechuic acid, salicylic acid, caffeic acid, and an ester of caffeic acid. Targeted PPO1 knockout (d|ppo1) plants were generated and plants lacking PPO1 exhibited only ~30% of the wild-type PPO activity in the culture medium, thus suggesting extracellular localization of PPO1, which is in contrast to the mostly plastidic PPO localization in seed plants. Further, d|ppo1 lines formed significantly more gametophores with a reduced areal plant size, which could be related to an increase of endogenously produced cytokinins and indicates an impact of PPO1 on plant development. d|ppo1 plants were less tolerant towards applied 4-methylcatechol compared to the wild type, which suggests a role of extracellular PPO1 in establishing appropriate conditions by the removal of inhibitory extracellular phenolic compounds.
Key words: Cytokinins, gene family, gene replacement, knockout, phylogeny, Physcomitrella patens, polyphenol oxidase, PPO
Introduction
Polyphenol oxidases are copper-binding enzymes that oxidize various polyphenolic plant compounds to the corresponding quinones in the presence of molecular oxygen (Mayer, 2006). The extended group of polyphenol oxidases is widespread among all groups of organisms and can be divided into three subgroups based on their substrate preferences: o-diphenol oxidases (PPO, EC 1.10.3.1), which oxidize o-diphenols to o-diquinones; laccases (LAC, EC. 1.10.3.2), which oxidize p-diphenols, and tyrosinases (TYR, EC 1.14.18.1), which are catechol oxidases with an additional function for hydroxylation of monophenols to o-diphenols prior to the diphenol oxidation.
TYRs occur typically in bacteria, fungi, and animals, but in plants they are rarely found (Mayer, 2006). PPOs and LACs are nearly ubiquitous in the plant kingdom, although for Arabidopsis thaliana no o-diphenol oxidase-encoding genes have been found in the genome (Sullivan et al., 2004). For Arabidopsis, only the presence of a large LAC gene family has been reported (McCaig et al., 2005).
PPOs are nuclear-encoded proteins (Lax et al., 1984) and PPO cDNAs have been characterized for numerous seed plant species, including Solanum tuberosum (Hunt et al., 1993; Thygesen et al., 1995), Solanum esculentum (Newman et al., 1993), Prunus armeniaca (Chevalier et al., 1999), Vitis vinifera (Dry and Robinson, 1994), Musa cavendishii (Gooding et al., 2001), Triticum aestivum (Demeke and Morris, 2002), and Populus trichocarpa (Tran and Constabel, 2011). In most plant species, PPOs are encoded by multigene families with no or a low number of introns.
Although many studies have been published for seed plants, the specific function of PPOs, which are in most cases located in plastids, is generally not well understood. PPO transcript levels are usually highest in young and meristematic tissues and decline during further development (Dry and Robinson, 1994; Thygesen et al., 1995). In late developmental stages, PPO expression is often not detectable, as found for Prunus armeniaca (Chevalier et al., 1999). Moreover, PPO gene family members from seed plants exhibit different temporal and spatial gene expression patterns in vegetative and reproductive organs, for example as described for the tomato PPO gene family (Thipyapong et al., 1997). Also, in Trifolium pratense, PPO genes are differently expressed, each being predominant in a certain stage: for example TpPPO1 in young leaves and TpPPO2 in flowers and petioles (Sullivan et al., 2004). Although being differentially expressed during plant development, it is unclear whether PPOs themselves are involved in the modulation of developmental processes. Galuszka et al. (2005) found evidence in maize that apoplastic laccases could play a role in enhancing cytokinin breakdown by reoxidation of the cytokinin catabolic enzyme cytokinin oxidase/dehydrogenase.
PPO transcript levels were found to be induced by biotic stress factors. In hybrid Populus trichocarpa × Populus deltoides, PPO expression was induced especially in young leaves by wounding, spraying with methyl jasmonate, and forest tent caterpillars (Malacosoma disstria) feeding on the plants (Constabel et al., 2000). Promoter::GUS fusions revealed that the tomato PPO gene F was induced in young leaves in response to wounding and after infection by Alternaria solani and Pseudomonas syringae, presumably in order to protect juvenile tissues from subsequent attacks by pathogens and pests (Thipyapong and Steffens, 1997).
So far, little information exists on transcriptional changes in PPO expression following the onset of abiotic stress conditions. Tomato PPO genes B and D are transcriptionally upregulated in abscission zones of leaf petioles in response to water stress, and it was proposed that this upregulation is correlated with the onset of apoptosis during water stress (Thipyapong et al., 2004). Moreover, tomato PPO genes B and F are ethylene inducible, and expression of the PPO gene F was found to be absent during water stress (Thipyapong and Steffens, 1997; Thipyapong et al., 2004). Recently, analysis of tomato PPO B promotor by GUS fusions in transgenic tomato confirmed the ethylene responsiveness for various tissues and cell types (Newman et al., 2011).
One widespread property of plastidic PPO activity is latency, meaning that PPOs are bound to thylakoid membranes in an inactive form and become active after tissue and membrane disintegration (Steffens et al., 1994; Yoruk and Marshall, 2003), once contact with the vacuolar substrates is possible. PPOs can possess a pronounced persistence throughout growth and development: in apricot, PPO protein is still present and active at a late stage of fruit development, whereas its mRNA is no longer detectable (Chevalier et al., 1999). In contrast, Thipyapong et al. (1997) suggested that PPO protein accumulation is primarily transcriptionally controlled by mRNA levels, because, in tomato, PPO accumulation patterns reflect PPO transcript patterns.
Known PPO proteins contain two highly conserved copper-binding domains, CuA and CuB, that are responsible for interaction with molecular oxygen and phenolic substrates. Each copper-binding domain harbours three histidines that bind one copper atom (Steffens et al., 1994). The subdivision of the extended group of polyphenol oxidases into three subgroups is also supported by the structure of the copper-binding domains: o-diphenol oxidases (PPO) and tyrosinases (TYR) are type-3 copper proteins, whereas laccases (LAC) contain a combination of type-2 and type-3 copper centres (reviewed by Gerdemann et al., 2002). Crystal structure of the active form of a catechol oxidase from Ipomoea batatas revealed that the secondary structure is dominated by α-helical regions and that the catalytic copper centre is situated within four α-helices in a hydrophobic pocket close to the enzyme surface (Klabunde et al., 1998).
As PPO substrates, phenolic compounds play several and diverse roles in seed plants. They can be involved in defence mechanisms against herbivores and pathogens, UV radiation protection, and blossom pigmentation, and they also possess antibiotic activity against bacteria and fungi (reviewed by Hahlbrock and Scheel, 1989; Waterman and Mole, 1994). Interest in phenolic compounds has grown enormously in relation to human nutrition as the compounds have antioxidative properties as radical scavengers (Rice-Evans et al., 1995) and are considered to exert protective effects against cancer and cardiovascular diseases. PPO and polyphenols contribute to flavour-generating processes, for example in the fermentation of wine, tea, coffee, and cocoa. In contrast, during the production of numerous vegetable products such as fruit juices and potato chips, PPO-mediated oxidation of phenolic compounds is undesirable and attempts are made to inhibit it.
With the scarcity of functional knowledge on plant PPOs outside of the group of seed plants, this study addressed the question of possible PPO functions in early divergent land plants. The PPO gene family of the moss Physcomitrella patens was phylogenetically characterized in the context of PPOs of other sequenced genomes. Functional conclusions on PPO1 as a major P. patens PPO isoform are drawn from a study of a targeted gene knockout and the effects on overall PPO activity, growth, and differentiation, content of endogenous cytokinins, and resistance to an externally applied polyphenol are described. Results are discussed with respect to original PPO functions at the emergence of land plants.
Materials and methods
Plant material and culture/growth conditions
P. patens (Hedw.) BSG wild type (Bryophyta, Funariales, Funariaceae), derived from the strain ‘Gransden 2004’ (Rensing et al., 2008), was used in all experiments. Moss tissue was cultivated axenically in growth chambers (RUMED Typ 1602) at 25±1 °C and white light (fluorescent tubes, TLM 18W/840, Philips) under 16/8 light/dark conditions with a flux of 50 µmol m–2 s–1 (Schulz et al., 2001). P. patens tissue grown in liquid medium (Wang et al., 1980) was used for PPO activity determination, vitality tests, determination of cytokinins, and determination of isopentenyladenine depletion. For maintenance of liquid cultures, the tissue was disintegrated weekly using an Ultraturrax blender (IKA), separated from old medium, extensively washed with fresh medium, and subsequently transferred to fresh medium (day 0). Under standard conditions, protonema status was maintained by supplementation of medium with 5mM di-ammonium tartrate. Tissue from liquid cultures transferred to agar plates (Knight et al., 1988) was used for growth tests and phenotypical observations.
Application of phenolic compounds
A sterile filtered stock solution of 4-methylcatechol (4-MC) or caffeic acid (CA) was added to 5-day-old liquid cultures to a final concentration of 50 (4-MC) or 100 µM (4-MC/CA) and cultures were further cultivated under standard growth conditions. For growth inhibitory tests, CA was also added to solid culture medium to final concentrations of 50, 100, 500, and 1000 µM.
Application of tritiated isopentenyladenine
To determine in vivo cytokinin oxidase/dehydrogenase activity of liquid-cultured protonemata, feeding experiments were performed with tritiated isopentenyladenine ([2-3H]-iP) at 25 °C and constant white light (c.30 µmol m–2s–1, fluorescent tubes, 15W, Osram) according to Schwartzenberg et al. (2003, 2007). [2-3H]-iP (specific activity 1.29 Tbq mmol–1) was added at a final concentration of 5 pM to concentrated 6-day-old protonema suspensions (day 0). After 2, 4, and 8 hours, 50 µl samples of the culture medium were taken and overall radioactivity (disintegrations per minutes) was determined by liquid scintillation counting (liquid scintillation cocktail Optisafe ‘HiSafe’ 2, Tri-Carb 2800 TR, PerkinElmer).
Phylogenetic analysis
The P. patens PPO1 amino acid sequence was used in a BLAST query to retrieve PPO sequences from other completely sequenced plant genomes. Based on the length of the PFAM domain PF00264 (228 aa), which is considered necessary for a functional PPO, only sequences with at least 190 aa alignment length and 30% identity were taken into account. Upon initial alignment and phylogenetic tree inference, some species were removed for brevity: namely, from the Poales, the species with the highest (Sorghum bicolor, eight) and the lowest number of PPOs (Oryza sativa, three) were kept, while Zea mays (six), Setaria italica (five), and Brachypodium distachyon (five) were removed. The same scheme was applied to the Rosales, where Malus domestica (16) and Fragaria vesca (six) were kept and Prunus persica (seven) removed. No PPO could be detected in the genomes of the Brassicaceae A. thaliana and Arabidopsis lyrata. In the case of P. patens, the two PPOs that are considered pseudogenes were not included for phylogenetic inference. As an outgroup, three laccases each from P. patens and A. thaliana were added. The alignment was generated using MAFFT linsi (Katoh et al., 2005) and contained 2093 columns. By manual curation using Jalview (Clamp et al., 2004), all columns were removed that had a low quality score or contained only a single sequence, resulting in a clipped alignment of 489 positions. From this alignment, all sequences lacking continuous stretches >160 columns (1/3 of alignment length) were removed to avoid problems during phylogenetic inference. Such sequences are probably due to fragmentary gene models; in particular, this affected three sequences from Malus domestica and one each from Eucalyptus grandis, Glycine max, Carica papaya, and O. sativa. As an exception from this rule, the P. patens Pp1s559_8V6.1 (PPO5) sequence was kept, although it lacked ~220 positions from the C-terminus. It should be noted that some other remaining sequences lacked continuous stretches up to 100 columns from this core alignment. The minimum overlap between sequences remaining in the alignment was ~140 columns, i.e. ~30% of the alignment length. Low support of some nodes was probably due to these remaining fragmentary sequences. These sequences were kept in the alignment as otherwise some interesting information, e.g. about C. papaya and V. vinifera, would have been lost. The best-suited evolutionary model was selected using ProtTest (Abascal et al., 2005) and was found to be WAG (Whelan and Goldman, 2001) with gamma distributed rates. Bayesian inference of phylogenetic relationships was carried out with MrBayes (Ronquist and Huelsenbeck, 2003) and visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
Isolation of genomic DNA
A simplified CTAB method was used for isolation of genomic DNA from moss tissue. Part of a young green gametophore was transferred to a 1.5ml tube containing 400 µl of 2× CTAB buffer (2%, w/v, CTAB, 100mM TRIS-HCl pH 8.0, 1.4M NaCl, 20mM EDTA pH 8.0) and ground with a small plastic pestle. The homogenate was incubated for 1 hour at 60 °C in a water bath, subsequently extracted with an equal volume of chloroform/isoamylalcohol (25:1) and centrifuged at 16,000 g for 10min. The upper aqueous phase was transferred to a new tube and an equal volume of 2-propanol (approx. 300–350 µl) was added, followed by a second centrifugation to precipitate the genomic DNA. The supernatant was discarded and the DNA pellet was washed with 70% ethanol and dissolved in 50 µl TE buffer containing 1 µl of 1mg RNaseA ml–1. For PCR, 0.5–2.0 µl DNA were used as a template in a total reaction volume of 25 µl.
Vector construction
The vector for heterologous expression of His-tagged PPO6 was created by amplification of the PPO6 sequences encoding for the predicted mature protein PPO6 from a cDNA library (Reski et al., 1995) with the primers cPPO6_forw and cPPO6_rev (Supplementary Material, available at JXB online) and ligation into the plasmid pTrcHis2_TOPO (Invitrogen) in frame to a sequence encoding for a C-terminal His-tag.
For cloning of a PPO1 knockout construct, the entire coding region of PpPPO1 (Richter et al., 2005) was amplified with the primers cPPO1_forw and cPPO1_rev (Supplementary Material) from a protonema cDNA library with a proofreading polymerase, cut with SalI and NcoI, and cloned into the SalI/NcoI-cut plasmid pET28a (Novagen by Merck). In a second cloning step, an nptII resistance cassette (neomycin phosphotransferase II gene under control of the 35S promoter, terminated by the nos terminator) was released from the vector pHP23 (Paszkowski et al., 1988) by digestion with EcoRI and blunt-end inserted into the coding sequence of PpPPO1 digested with Ecl136II and Bsp1407.
Transformation of Physcomitrella
The PPO1 knockout cassette (nptII cassette flanked by 730 and 853bp of PPO1 coding sequence) was amplified from the knockout construct using the primers cPPO1_forw and cPPO1_rev and used for PEG-mediated transfection of P. patens protoplasts. Generation of protoplasts from protonema tissue cultivated for 5 days in liquid medium and transformation was performed as described by Hohe et al. (2004) with slight modifications.
Heterologous expression of recombinant His-tagged PPO6 in Escherichia coli
To obtain recombinant PPO from PPO6 expressing E. coli clones (strain TOP10, Invitrogen), 200ml LB culture supplemented with the appropriate antibiotic was inoculated with a starter culture. Cultures were grown in 1 l flasks with 200rpm shaking at room temperature to an OD600 of 0.4, and transcription was induced by the addition of 1mM IPTG. Cultures were further supplemented with 20 µM CuCl2 and grown for additional 6 hours at room temperature. Additionally, control expressions were performed with TOP10 clones expressing a His-tagged lacZ protein as well as with BL21(DE3) clones expressing His-tagged PPO1 of Trifolium pratense (TpPPO1) (Sullivan et al., 2004). Protein extracts obtained from these expression systems served as positive controls for proper expression (lacZ:his and TpPPO1:his) and as negative (lacZ:his) as well as positive controls (TpPPO1:his) for PPO activity determinations from E. coli protein extracts.
Protein isolation/extraction and PPO enzyme activity assay
Crude protein extracts from P. patens tissue were prepared as described by Richter et al. (2005) with slight modifications. Sample tissue was frozen in liquid nitrogen and subsequently homogenized in a small volume of ice-cold phosphate buffer composed of KH2PO4 and Na2HPO4 (67mM each) at pH 6.4 using the FastPrep FP120 homogenizer (MP Biomedicals). To remove cell debris, the homogenate was centrifuged for 10min at 16,000 g and 4 °C.
Protein extracts from culture medium were prepared by lyophilization of the medium at –20 °C. The lyophilized powder was resuspended in ice-cold phosphate buffer and insoluble compounds were sedimented by centrifugation at 16,000 g for 5min. The supernatant was subsequently desalted by application on an NAP 25 column (GE Healthcare), and the proteins were eluted in ice-cold phosphate buffer.
Protein extracts from E. coli TOP10 expressing P. patens PPO6 were prepared prepared by using the pTrcHis2-TOPO-TA expression kit (Invitrogen). Harvested bacteria were incubated for 15min at room temperature with 100mg lysozyme ml–1 and extract was further treated as described in Richter (2009). Recombinant His-tagged PPO was purified by column chromatography on His-bind Ni-NTA-columns (Novagen by Merck) following the manufacturer’s instructions. The purified protein was desalted and finally eluted in phosphate buffer.
Protein contents were determined according to Bradford (1976) using bovine serum albumin as a standard. For quantification of low-concentration protein extracts (prepared from culture medium), the NanoOrange protein quantitation kit (Invitrogen) was used according to the manufacturer’s instructions and fluorescence was determined with a luminescence spectrometer (LS 55, Perkin Elmer).
Total in vitro PPO activity was determined polarographically according to the method of Lieberei and Biehl (1976) as described in Richter et al. (2005).
Determination of cell viability
Cell vitality of P. patens was determined by fluorescein diacetate staining. Protonema culture (500 µl) was incubated for 5min at 20 °C with 2 µl of fluorescein diacetate (FDA) stock solution (10mg ml–1 in DMSO). Green fluorescence of living cells, resulting from de-esterification of FDA to fluorescein by esterase activity (Power and Chapman, 1985), was examined under blue light excitation.
Extraction of cytokinins from tissue and culture medium and analysis by ultra-performance LC-MS/MS
Concentration of endogenous cytokinins were determined by ultra-performance LC (UPLC)-MS/MS analysis as described in Schwartzenberg et al. (2007) including modifications described by Novák et al. (2008). Samples of both tissues and culture media were freeze-dried and stored at –20°C until analysis.
Extracts prepared with Bieleski’s reagent were purified using a cation (SCX-cartridge), an anion exchanger (DEAE-Sephadex/C18 cartidge, and an immunoaffinity chromatography (IAC) based on wide-range specific monoclonal antibodies against cytokinins (Novák et al., 2003). By comparison of the specific multireaction monitoring transition and chromatographic retention times with those of authentic standards, the identity of all measured cytokinin metabolites was verified.
Extraction of phenolic compounds from tissue andquantification by UPLC-MS/MS
Freeze-dried tissue samples were homogenized with 10% methanol using an oscillation ball mill (MM 301, Retsch, Haan, Germany). The extract was centrifuged for 10min at 20,000 g, the supernatant was filtered through a 0.2 µm Micro-Spin nylon membrane filter (GRACE, Columbia, MD, USA) and directly analysed by UPLC-MS/MS as described by Gruz et al. (2008).
Video-based online growth monitoring
For online growth monitoring, protonemata were cultivated at 25°C in continuous light on agar plates containing standard medium supplemented with 500 µM caffeic acid (CA). Growth measurements took place in a growth chamber equipped with a camera for recording of two-dimensional growth based on increase of pixel number, as described by Roleda et al. (2004).
Statistical analysis
For statistical analysis, the software package Statistica version 9.1 (StatSoft, Tulsa, OK, USA) was used. Either t-test or ANOVA with post-hoc test according to Tukey was used with P-values <0.5, <0.05, or <0.01, as indicated.
Results
Physcomitrella PPOs are encoded by a multigene family
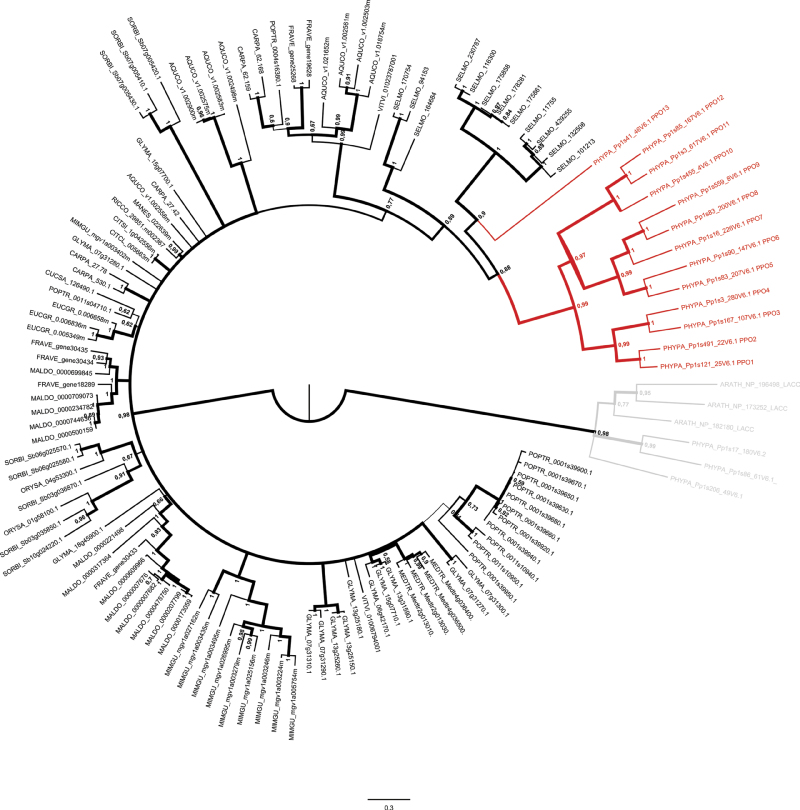
After the P. patens genome sequence became available (Rensing et al., 2008), evidence was found that P. patens PPOs are encoded by a multigene family. BLAST analysis using the derived amino acid sequence of PpPPO1 (Richter et al., 2005) as a query for BLASTp in the P. patens V1.6 protein database and for tBLASTn in the P. patens V1.6 genome database (www.cosmoss.org), identified 15 loci possessing similarities to PPO1 (cut off 35% identity over a length of at least 80 amino acids). To complete the analysis, BLAST searches with PPOs from sequenced plant genomes were also performed and no additional loci were found. The 15 loci and their predicted gene models (version 1.6) with their intron/exon structure were evaluated according to the presence of the essential central domain binding two copper ions (PFAM Tyr PF00264), transcript evidence, and homology to published PPO sequences. PPO-related sequences that did not contain the complete CuA and CuB domains were excluded from experimental analysis. From 13 selected loci (Supplementary Table S1), PPO1 (Richter et al., 2005) was found to be the most basal isoform of the P. patens-specific PPO (Fig. 1).
Fig. 1.
Bayesian inference of the PPO gene family phylogeny. Branch width and numbers at nodes correspond to posterior probabilities. The tree is outgroup rooted with laccases (shown in grey). The P. patens PPOs are shown in red. Coding sequences were obtained from cosmoss.org version V1.6 (see Supplementary Table S1). AQUCO, Aquilegia coerulea; ARATH, Arabidopsis thaliana; CARPA, Carica papaya; CITCL, Citrus clementina; CITSI, Citrus sinensis; CUCSA, Cucumis sativus; EUCGR, Eucalyptus grandis; FRAVE, Fragaria vesca; GLYMA, Glycine max; MALDO, Malus domesticus; MANES, Manihot esculenta; MEDTR, Medicago truncatula; MIMGU, Mimulus guttatus; ORYSA, Oryza sativa; PHYPA, Physcomitrella patens; POPTR, Populus trichocarpa; RICCO, Ricinus cummunis; SELMO, Selaginella moellendorffii; SORBI, Sorghum bicolor; VITVI, Vitis vinifera.
Sequence properties of the derived amino acid sequences of PPO1–PPO13
PPO5 and PPO8 were found to be located tail to tail on the same scaffold as a tandem array, separated by about 15 kbp. Despite their close localization, the protein sequences derived from the V1.6 gene models showed only 51% identity (not shown). PPO4 and PPO11 were found located head to head about 1.89 Mbp apart from each other and exhibited only 29% identity. The differences in the distance of these arrayed PPO loci was reflected by their relative distance in the phylogenetic tree. Percentage identities of pairwise alignment (not shown) of the overall amino acid sequences ranged from 27% (PPO11 and PPO13) to 72% (PPO1 and PPO2). Also PPO8 and PPO9 shared a high degree of identical amino acids (71%).
Physcomitrella PPOs are monophyletic
The PPO gene family tree (Fig. 1) suggests that the extant PPO sequences in the non-seed plants P. patens (13 genes) and Selaginella moellendorffii (12 genes) to be monophyletic, but subject to significant lineage specific expansion. The relationships between seed plant PPO clades are unclear and do not allow to reconstruct the detailed history of gene loss and gain. However, lineage specific expansion of PPO genes is evident in several seed plant taxa as well, for example within the Rosales, Fabales, and Poales. Within the seed plants, the number of PPO genes per species ranges from zero (e.g. A. thaliana) to 16 (Malus domestica). While most genomes harbour more than one PPO gene, several seem to encode only a single PPO, e.g. the two Citrus species, Manihot esculenta, and Cucumis sativus. Interestingly, the C. papaya (Brassicales) genome encodes PPOs, evidencing loss of PPO genes probably within the Brassicaceae or in the genus Arabidopsis.
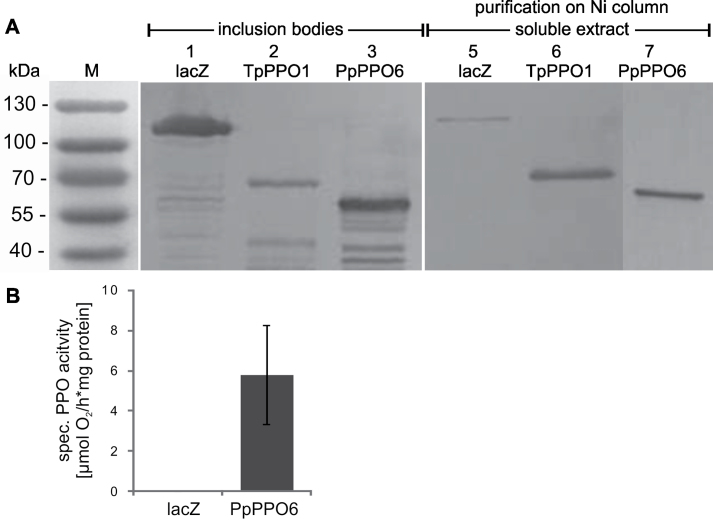
Physcomitrella PPO6 is a functional o-diphenol oxidase
To test the functionality of one member of the P. patens PPO gene family, this study randomly chose PPO6 for heterologous expression in E. coli under the control of the IPTG-inducible trc-promoter. The expression vector pTrc_PpPPO6-his was generated inserting a PCR product encoding for the putative coding sequence of PPO6. Expression of PPO6:his was induced and protein extracts were purified using Ni-NTA-columns. Western blot analysis using an anti-His-tag antibody revealed that a major portion of the expressed PPO6:his protein was apparently insoluble and accumulated in inclusion bodies, but a small part of soluble PPO6:his protein could be enriched by purification via Ni-NTA-column. Subsequently, PPO functionality of the recombinant His-tagged PPO6 protein was confirmed in a polarographic enzyme assay using 4-methylcatechol (4-MC) as a substrate. In contrast, control protein extracts for lacZ:his in the same expression system had no measurable in vitro PPO activity (Fig. 2). PPO enzyme activities of PPO6:his protein extracts could be inhibited by the addition of KCN to the reaction mixture, which is a necessary prerequisite for a PPO activity assay. Although it was extremely difficult to obtain active recombinant PPO protein from P. patens, strong evidence for the functionality of one exemplary PPO gene product was found using PPO6.
Fig. 2.
Recombinant PPO proteins and enzyme activity. (A) Western blot analysis of insoluble (left panel), and soluble protein extracts (right panel) enriched and purified on a Ni-column. Protein extracts of 6h IPTG-induced E. coli cultures expressing His-tagged proteins were separated on a 12.5% SDS gel and detected by an anti-His-tag antibody: lacZ:his (120kDa), TpPPO1:his (59kDa, migrates at 65kDa according to Sullivan et al., 2004), PPO6:his (62kDa). (B) Specific PPO activity of enriched and purified soluble protein extracts determined polarographically using 4-methylcatechol as a substrate. Bar, standard deviation (n = 3).
Searching for phenolic compounds from Physcomitrella as putative PPO substrates
To investigate which phenolic compounds could be potential substrates, P. patens tissue was analysed for the presence and composition of polyphenolic compounds using UPLC-MS/MS. The compounds protocatechuic acid, 4-hydroxybenzoic acid, salicylic acid, CA, and 4-coumaric acid were detected at concentrations ranging from 1 to 6.5 nmol (g dry weight)–1 (Supplementary Fig. S1). In addition, an unknown ester of CA was detected in tissue samples. Based on the spectral data, the ester was tentatively identified as 2-O-caffeoylthreonic acid (Supplementary Fig. S2).
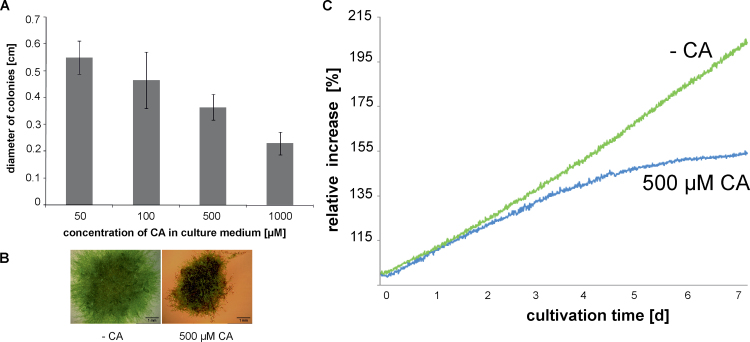
Caffeic acid causes growth reduction
As caffeic acid was shown to be an endogenous polyphenolic compound, it was chosen as an example to examine the effects of phenolics on P. patens growth and PPO activity. Protonema was cultured on agar medium supplemented with concentrations of CA ranging from 50 to 1000 µM. As indicated in Fig. 3A, plant diameters were lower with increasing CA concentrations. In Fig. 3B, examples of P. patens cultivated with 500 µM CA are displayed along with the control showing growth reduction caused by CA. Optical online growth monitoring displaying the relative increase of plant area (Fig. 3C) revealed that substantial growth reduction was apparent 2 days after the exposure to CA.
Fig. 3.
Growth inhibition of P. patens wild type by caffeic acid (CA). (A) Diameter of plants grown on solid culture medium supplemented with different concentrations of CA measured after 4 weeks of cultivation (n = 10, bars represent standard deviation). (B) Growth with 500 µM CA (right) and without CA (left) measured after 4 weeks. (C) Optical online monitoring of arial plant growth displaying the relative area increase of a moss plant cultivated with 500 µM CA and without CA (this figure is available in colour at JXB online).
Additionally, PPO activity, determined polarographically, had a tendency for decreased values in tissue cultivated with CA [227.95±28.91 nmol O2 h–1 (mg protein)–1] compared to tissue cultivated in standard culture medium [287.60±13.93 nmol O2 h–1 (mg protein)–1] (n = 6).
Generation and molecular characterizationof PPO1 knockout plants
To obtain information on the biological functions of PPO in P. patens, targeted knockout lines (Schaefer and Zryd, 1997) were generated. For this experiment, PPO1 was chosen as it had been previously characterized (Richter et al., 2005) and because it is well expressed in protonemata. A PPO1-knockout vector was constructed by placing a 730-bp 5′- and a 853-bp 3′-genomic fragment flanking a 35S-NPTII resistance cassette. From this, the knockout cassette was amplified by PCR and then used for PEG-based transfection. The expected gene disruption caused by integration of the resistance cassette into the genomic PPO1 locus was demonstrated by PCR (Supplementary Fig. S3A, B) and subsequent sequencing (not shown). All five selected PPO1 knockout strains (d|ppo1) had undergone a recombination event at the 5′ end of PPO1. Plants of d|ppo1 lines 1, 3, and 5 showed additionally recombination at the 3′ end, indicating that a gene replacement had occurred. d|ppo1 lines 5 and 6 had undergone targeted knockout but no gene replacement occurred as the recombination did not take place in the 3′ region of PPO1. The expected absence of the PPO1 transcript in the stable and haploid d|ppo1 lines 1, 3, 5, 6, and 8 was demonstrated by reverse-transcription PCR (Supplementary Fig. S3C). d|ppo1 lines 1 and 5 were arbitrarily chosen for physiological experiments.
PPO1 knockout plants possess a reduced extracellular PPO activity and exhibit a decreased tolerance towards 4-methylcatechol
The only slight decrease in PPO enzymatic activity between tissue extracts of wild type and d|ppo1 lines 1 and 5 revealed that PPO1, if at all, accounts only for a small portion of the overall intracellular PPO activity (Table 1). However, the strong difference in PPO activity between wild type and knockout strains in the extracellular compartment demonstrated that PPO1 is a major extracellular isoform, providing 68% of the overall PPO activity released into the medium. Additionally, this result provides indirect evidence that PPO1 is, besides PPO6, also a functional PPO accepting 4-MC as a substrate.
Table 1.
In vitro PPO activity measured in culture medium and tissue of wild type and d|ppo1 lines.
Protein extracts were prepared from tissue and culture medium from cultures grown under standard conditions for 7 days and PPO activity was determined polarographically using 4-methylcatechol as a substrate (n = 3). Values are µmol O2 h–1 (mg protein)–1. Different letters within rows indicate significant differences according to ANOVA with post-hoc test after Tukey using P < 0.01.
| Wild type | d|ppo1 line 1 | d|ppo1 line 5 | |
| Culture medium | 12.17±3.26a | 3.93±0.57b | 3.78±1.42b |
| Tissue | 0.19±0.03a | 0.15±0.01a | 0.15±0.04a |
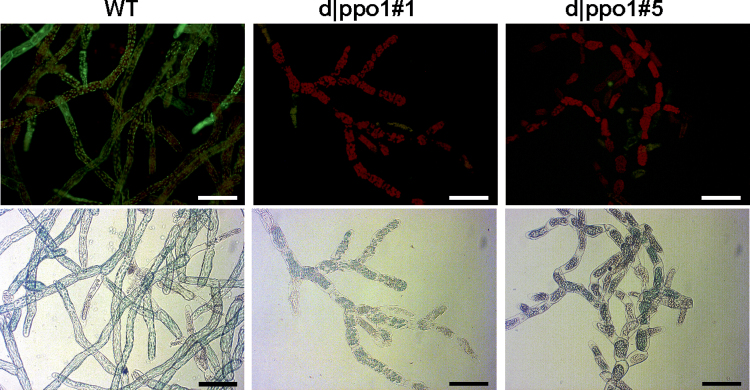
In order to obtain indications about possible functions of PPO1, the wild type and d|ppo1 lines 1 and 5 were examined for tolerance towards externally applied 4-MC, being a substrate for PPO1. Concentrations as low as 50 µM 4-MC induced a severe die back of mutant tissue and mutant cells lacked fluorescence when FDA staining for vitality was performed. Wild-type protonema, however, was able to resist the 4-MC treatment, and FDA staining indicated that cells were alive and metabolically active (Fig. 4).
Fig. 4.
Effect of 4-methylcatechol (4-MC) application on protonemata of PPO1 knockout mutants and wild type. Tissue was cultivated under standard conditions in liquid medium supplemented with 50 µM 4-MC and cell vitality was displayed by FDA staining and microscopical observation after 48h of incubation (upper panel) and bright light microscopy (lower panel). Bars, 100 µm.
Growth assays in which d|ppo1 strains were exposed to various concentrations of CA did not result in significant differences in plant size when compared to wild type (not shown).
PPO1 knockout lines exhibit altered differentiation
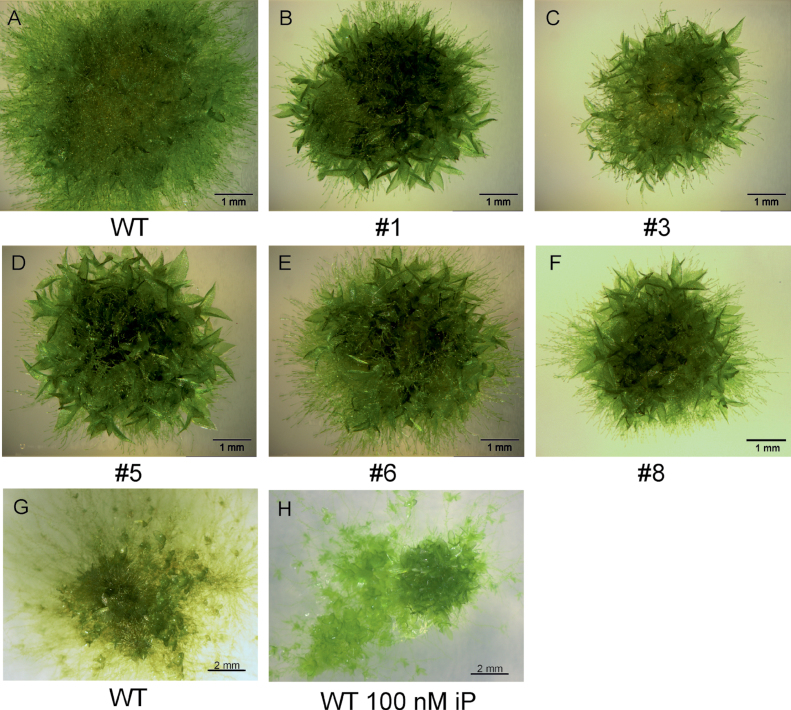
Phenotypical observations revealed that the d|ppo1 lines exhibited abnormal protonema growth with shorter and more rounded chloronema cells (Supplementary Fig. S4). Moreover, these transgenic plants were significantly smaller with more gametophores cm–2 than wild type (Fig. 5, Table 2). Reduced areal size with enhanced gametophore formation could be partially mimicked with wild-type tissue grown in the presences of the cytokinin isopentenyladenine (100nM) (Fig. 5 G, H), thus suggesting that the altered phenotype might be related to changes in the homeostasis of cytokinins.
Fig. 5.
Phenotypes of PPO1 knockout plants and wild type. (A–F) Wild type and d|ppo1 lines 1, 3, 5, 6, and 8 cultivated under standard growth conditions for 17 days on the same agar plate. Bars, 1mm. (G, H) Wild type cultivated for 14 days without and with isopentenyladenine (100nM). Bars, 2mm.
Table 2.
Gametophore formation in wild-type and d|ppo1 plants
Freshly disintegrated protonema tissue was transferred to agar plates (day 0) and cultivated for 7 days under standard conditions. d|ppo1 lines showed an enhanced production of gametophores compared to wild type. Values are means with standard deviations (n = 8). Different letters indicate significant differences according to ANOVA with post-hoc test after Tukey using P < 0.01.
| Line | Gametophores after 7 days (cm–2) |
| Wild type | 0.57±0.18a |
| d|ppo1 line 1 | 4.17±0.83b |
| d|ppo1 line 5 | 4.52±1.27b |
| d|ppo1 line 6 | 3.25±0.75b |
| d|ppo1 line 8 | 3.80±0.91b |
PPO1- knockout causes alterations in cytokinin profile
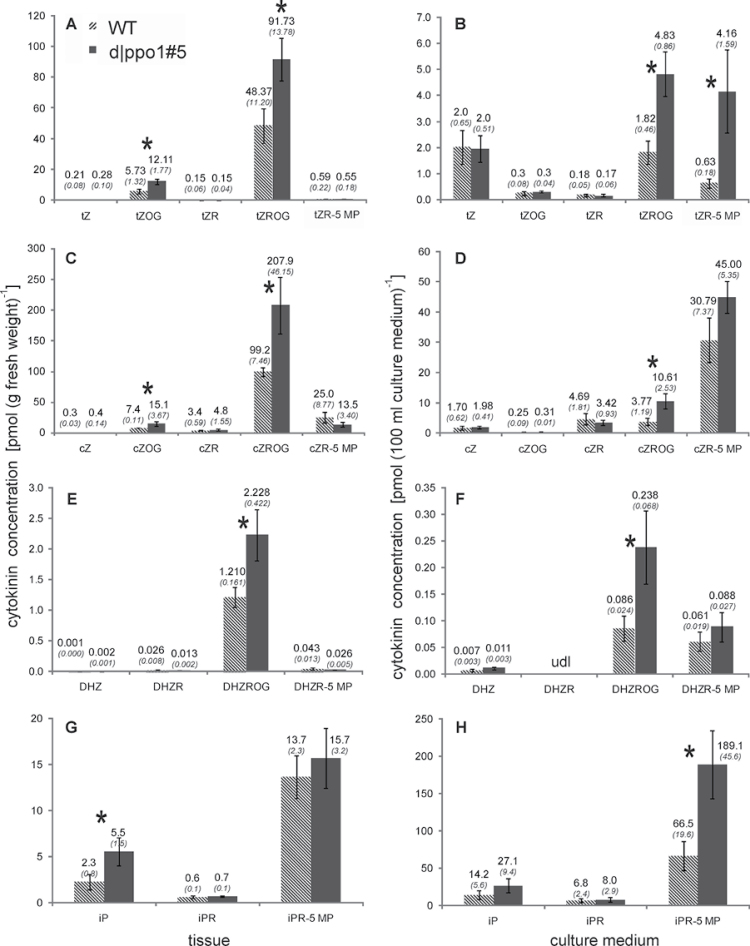
It is well known that gametophore formation in mosses is induced by cytokinins (reviewed in von Schwartzenberg, 2009). To examine whether changes of the cytokinin profile could be a cause for the enhanced differentiation in PPO1-deficient plants, this study carried out a LC-MS based cytokinin analysis in tissue and culture medium and compared d|ppo1 line 5 with the wild type. d|ppo1 line 5 contained increased intracellular amounts of the cytokinin forms trans-zeatin-O-glucoside, trans-zeatin riboside-O-glucoside, cis-zeatin-O-glucoside, cis-zeatin riboside-O-glucoside, dehydrozeatin riboside-O-glucoside and isopentenyladenine (Fig. 6A, 6C, 6E, 6G). In each cytokinin family, at least one compound was significantly increased (t-test, P < 0.05).
Fig. 6.
Cytokinins in wild type and PPO1 knockout strain d|ppo1 line 5 in tissue (A, C, E, G) and culture medium (B, D, F, H): (A, B) trans-zeatins, (C, D) cis-zeatins, (E, F) dihydroxyzeatins, (G, H) isopentenyladenine-type cytokinins. Cytokinins were determined from tissue and the corresponding culture medium of wild type and d|ppo1 line 5 after 15 days of culture under standard conditions. Cytokinins were determined from three biological replicates for each genotype (n = 3) by UPLC-MS displayed here as pmol (g fresh weight)–1 or pmol (100ml culture medium)–1. Values are means ± standard deviations; numbers above each column are the numeric values. * indicate significant differences in t-test (P < 0.05). tZ, trans-zeatin; tZOG, trans-zeatin-O-glucoside; tZR, trans-zeatin riboside; tZROG, trans-zeatin riboside-O-glucoside; tZR-5 MP, trans-zeatin-riboside-5’-monophosphate; cZ, cis-zeatin; cZOG, cis-zeatin-O-glucoside; cZR, cis-zeatin riboside; cZROG, cis-zeatin-riboside-O-glucoside; cZR-5 MP, cis-zeatin-riboside-5’-monophosphate; DHZ, dihydrozeatin; DHZROG, dihydrozeatin riboside-O-glucoside; DHZR-5 MP, dihydrozeatin riboside-5’-monophosphate; iP, isopentenyladenine; iPR, isopentenyladenosine; iPR-5 MP, isopentenyladenosine-5’-monophosphate.
Moreover, analysis of the culture media also revealed increased extracellular concentrations of isopentenyladenine, isopentenyladenosine-5′-monophosphate, cis-zeatin-riboside- 5′-monophosphate, and dihydrozeatin riboside-O-glucoside in knockout cultures compared to the wild type (Fig. 6B, 6D, 6F, 6H). Also for culture media, at least one compound per family of cytokinin was significantly increased.
PPO1 knockout plants show slower degradationof extracellular radiolabelled isopentenyladenine
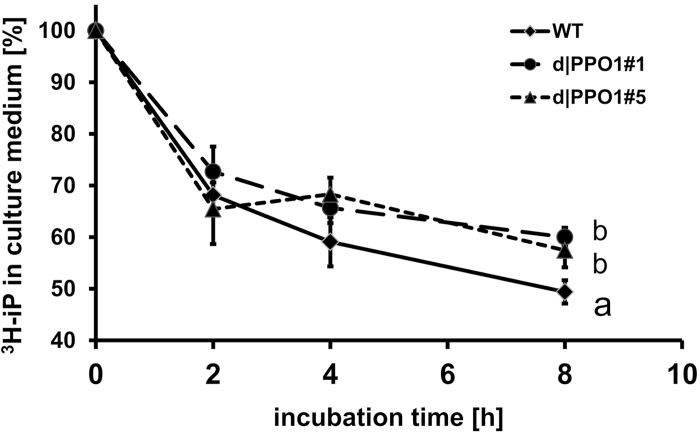
To determine if the increase of cytokinin levels found in d|ppo1 line 5 tissue and medium could be a consequence of a reduction in cytokinin breakdown, in vivo metabolism studies were carried out by adding [2-3H]-iP to liquid cultures of d|ppo1 lines 1 and 5 and the wild type. From previous studies, it was known that short-term breakdown of [2-3H]-iP in P. patens cultures can be easily monitored by counting the radioactivity in the culture medium, which contains 90% of the applied isopentenyladenine (Schwartzenberg et al., 1993, 2007).
The amount of the applied [2-3H]-iP (5 pmol ml–1) was comparable to the usual amount of extracellular isopentenyladenine occuring in a P. patens wild-type culture (~10 pmol ml–1 in 10-day-old liquid cultures, according to Schwartzenberg et al., 2007). Liquid scintillation counting in cultures of d|ppo1 lines 1and 5 revealed a reduced loss of the applied [2-3H]-iP (Fig. 7), which was found to be significant at 8h incubation time. HPLC analysis coupled to online liquid scintillation counting (not shown) confirmed the reduction of [2-3H]-iP depletion in theknockout lines, which is likely to be causally connected to the increase of cytokinins and the enhanced differentiation inthe PPO1 knockout lines.
Fig. 7.
Time-course of changes in the amount of applied [2-3H]-isopentenyladenine in cultures of d|ppo1 plants compared to wild type (n = 3). Total radioactivity in the culture medium was determined by liquid scintillation counting and is displayed here as percentage radiolabelled isopentenyladenine (t0 = 100%). Different letters indicate significant difference at measurement after 8h according to ANOVA with post-hoc test after Tukey using P < 0.05.
Discussion
Phylogenetic analysis suggests emergence of PPOs during the conquest of land by plants
Phylogenetic analysis revealed that P. patens possesses 13 paralogous PPO genes. Twelve PPO genes (1–12) form a monophyletic clade and this PPO gene family seems to have established independently from the vascular plant lineage.
PPO13 represents an additional gene located on a separate branch together with PPOs from the lycophyte Selaginella moellendorffii. It appears likely that the last common ancestor of P. patens and the vascular plant lineage possessed at least two PPO genes, one giving rise to the multigene family with PPO1–12 and one which resulted in PPO13. Regarding the PPO gene families of seed plants, it can be assumed that their last common ancestor harboured two PPO genes as well, the descendents of which evolved independently in different plant lineages.
PPO gene duplication within the P. patens genome occurred most likely several times. The pairs of PPO genes evident in the phylogenetic tree (PPO1/PPO2, 3/4, 5/6, 8/9, and 11/12; Fig. 1) probably were retained after the whole-genome duplication, approximately 45 million years ago (Rensing et al., 2007). As indicated by the branch length within the phylogenetic tree, the formation of PPO gene families by gene and genome duplications occurred independently in Selaginella moellendorffii and seed plants.
When screening the sequenced genomes of the green algae Chlamydomonas reinhardtii and Ostreococcus tauri, neither o-diphenol oxidases and laccases nor tyrosinases were found. Hence, it is most likely that PPO-encoding genes newly occurred during the evolution of the earliest land plants. However, there is a lack of sequenced genomes on the level of basal streptophytes like charopycean and zygnematophycean algae. Organisms of these clades need to be screened for PPO activity and corresponding genes. Analysis of algae adapted to non-aquatic habitats (Lewis, 2007) for PPO would also enable a better estimation of the exact evolutionary stage when PPOs first occurred. Sherman et al. (1991) analysed the distribution of PPO activity in a broad variety of aquatic and terrestrial plant species and found indication for PPO activity in the streptophytes Coleochaete and Nitella. The occurrence of PPOs in streptophytes other than seed plants might point towards an early function of PPOs that has developed further in higher organization forms and differentially evolved to perform diverse functions in different plant species.
Lang et al. (2008) pointed out that the adaptation of the first land plants to harsh conditions with high solar (UV) radiation and cycles of flooding and desiccation must have caused substantial changes in morphology and regulatory processes leading to the development of newly generated pathways in P. patens. Thus, characterization of the PPO gene family of P. patens, which, as a model system, occupies an important position in the land plant phylogeny, opens up new possibilities to obtain information on potential and probably more original functions of PPOs.
Although o-diphenol oxidases are likely to have evolved within the streptophyte clade, many tyrosinase and laccase genes have been characterized from several fungi, such as Trichoderma reesei (Selinheimo et al., 2006) and Pycnoporus sanguineus (Halaouli et al., 2006). Genome analysis has revealed that P. patens also possesses three putative laccase-encoding genes but in the present study’s database searches, no tyrosinase-encoding genes have been found in the P. patens genome. Consequently, P. patens possesses two types of enzymes from the extended group of polyphenol oxidases, three p-diphenol oxidases (laccases), and 13 o-diphenol oxidases.
The functional gene knockout of PPO1 with the concomitant strong decrease of extracellular PPO activity (Table 1) provides indirect evidence that PPO1 is secreted to the culture medium and contributes as much as 68% of the total extracellular PPO activity (in 8-day-old protonema cultures). For unknown reasons, this study was unable to express sufficiently high amounts of a PPO1:GFP fusion protein in P. patens in order to directly confirm the extracellular targeting of PPO1 (not shown).
However, the indirect evidence of PPO1 being an extracellular enzyme is in clear contrast to PPO localization in seed plants, which is mostly plastidic (Steffens et al., 1994; Yoruk and Marshall, 2003). Ono et al. (2006) reported on a vacuolar flavonoid biosynthetic PPO and recently Tran and Constabel (2011) describe PtPPO13 from Populus trichocarpa as targeted to the vacuole thus underlining that other than plastidic localizations have to be taken into account. Extracellular localization of PpPPO1 is also supported by prediction of a putative cleavage site (between position 23 and 24, SignalP 4.0, http://www.cbs.dtu.dk/services/SignalP/) and of localization in the secretory pathway (YLoc+, http://abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi).
Preliminary quantitative real-time PCR-based analysis of PPO gene expression have revealed that different isoforms of the PPO family are differentially expressed and that expression is in addition differentially influenced by factors like culture age, stress by strong-light irradiation or treatment by CA (Richter, 2009). The PpPPO5 gene has been shown to be induced upon irradiation with UV-B (Wolf et al., 2010). These results suggest that different PPO family members have different functions in P. patens.
Heterologous expression of PPO6
From sequence comparison and phylogenetic analysis, it seemed likely that the identified putative PPO genes from P. patens encode for o-diphenol oxidases. Repeated experiments to express recombinant PPO1 in E. coli failed for unknown reasons. However, for PPO6, small amounts of the soluble protein could be purified after heterologous expression in E. coli and the o-diphenol oxidase activity was confirmed in polarographic assays using 4-MC as a substrate (Fig. 2). The extracts exhibited only low PPO activities not sufficient to allow substrate specificity tests.
In earlier work, Sullivan et al. (2004) also could obtain only relatively small amounts of soluble Trifolium pratense PPO (TpPPO1) expressed in E. coli. Bacterial clones expressing TpPPO1 were also used as a positive control monitoring proper expression conditions in this study, and protein extracts exhibited in vitro PPO activity under identical experimental conditions to those used for PpPPO6 expression. Nevertheless, as already observed by Sullivan et al. (2004), TpPPO1, like PpPPO6, exhibited only low PPO activities relative to the amount of protein used for the assay. Therefore, it seems that PPOs from P. patens, like those from seed plants, require appropriate post-translational modification to generate a highly active PPO enzyme, although PPO with limited levels of enzyme activity can be produced in a prokaryotic expression system. In spite of this, functional evidence that one exemplary member of the P. patens PPO gene family encodes for an o-diphenol oxidase was clearly given by the heterologous expression of PPO6.
Phenols from Physcomitrella
P. patens possesses chalcone synthases, catalysing the first step in flavonoid biosynthesis and also phenylalanine ammonia-lyase (PAL) homologues are present in the P. patens genome (Wolf et al., 2010). Consequently, as PAL is a key enzyme in the polyphenol synthesis (Boudet, 2007), P. patens possesses the general ability and a set of enzymes to synthesize simple phenolic compounds and flavonoids, underlined by the detection of quercetin by Wolf et al. (2010).
Using UPLC-MS, this study detected and quantified the phenolic compounds protocatechuic acid, 4-hydroxybenzoic acid, salicylic acid, CA, p-coumaric acid, and an ester of CA (Supplementary Figs. S1 and S2). So far, nothing is known about possible functions of these compounds in the bryophyte P. patens and a comparison of the amounts of phenolic compounds found in the wild type with those in d|ppo1 plants (not shown) did not allow conclusions on the substrate preference of PPO1. Attempts to determine substrate specificity of recombinant protein of PPO1 and PPO6 were not successful because of limitations in protein quantity.
Effects of caffeic acid applicationon Physcomitrella growth
The application of CA (50 µM) to the culture medium of wild type inhibited protonemal growth and caused browning of the culture medium. Although a much higher than the endogenous CA concentration was used in these experiments, a considerable extracellular CA consumption in protonemal cultures was monitored spectrophotometrically within 3 days of cultivation (data not shown). These observations suggest that CA applied to the culture medium can be metabolized by certain PPOs produced (and presumably secreted) by P. patens.
Bollag et al. (1988) reported that the exogenously applied growth-inhibiting phenols 2,6-xylenol and p-cresol can be detoxified by an extracellular laccase of the fungus Rhizoctonia praticola. Moreover, transgenic Arabidopsis seedlings expressing a secreted laccase from Gossypium arboreum exhibited an enhanced resistance to certain growth-inhibiting phenolic compounds. This pointed to a role for laccase in transforming phenolics ex planta without uptake of the substance by the plant (Wang et al., 2004). The possibility arises that PPOs may play a similar role in P. patens.
Effects of PPO1 knockout and tolerance of 4-methylcatechol
From measurements of extracellular PPO activity in concentrated medium extracts of PPO1 knockout plants and wild type (Table 1), this study concluded that PPO1 is a major extracellular PPO with a potential role in detoxification of growth-inhibiting phenolic compounds. The fact that P. patens plants lacking PPO1 exhibited a reduced extracellular (in vitro) PPO activity using the substrate 4-MC provided indirect functional evidence that PPO1 encodes a functional o-diphenol oxidase. It seems therefore likely that 4-MC, already identified as a substrate for PPO6, is also a substrate for PPO1. Consequently, applied phenolic compounds, potentially toxic for P. patens tissue, would be metabolized and removed by an extracellular PPO1-mediated oxidation.
Interesting and significant effects on d|ppo1 protonema were observed after incubation with the PPO substrate 4-MC, which caused an earlier die back in comparison to the wild type, suggesting that this compound is (more) toxic for d|ppo1 plants(Fig 4). Although catechol and its methyl derivatives arewidespread phenolic componds in seed plants (Quideau et al., 2011), it was not found in the current LC-MS based analyses of P. patens tissue and medium.
As already mentioned above, Wang et al. (2004) observed that transgenic Arabidopsis seedlings expressing a secreted laccase from G. arboreum exhibited an enhanced resistance to certain phenolic compounds. HPLC analysis indicated that the growth-inhibiting phenols were detoxified ex planta. The importance of phenolics and their oxidation products in plant–plant interactions and the role of plant PPOs to detoxify exuded phenolics, and thereby decrease their allelopathic action, has been reviewed by Li et al. (2010). The increased sensitivity of d|ppo1 plants towards 4-MC supports the view that also PPO1 has a function in detoxifying extracellular phenolic compounds in P. patens.
Knockout experiments suggest influence of PPO1on cytokinin mediated differentiation
All analysed d|ppo1 plants exhibited a reduced areal growth of protonemata when grown on agar connected to a significantly higher production of gametophores cm–2 (Fig. 5, Table 2). Growth experiments using hormonal application showed that this increased differentiation to gametophores can be caused by external application of isopentenyladenine to the wild type(Fig. 5G, 5H).
In the work reported in Newman et al. (2011), the responsiveness of tomato PPO B to ethylene is well documented and also partial homologies of the PPO B promotor with responsive elements for gibberellic acid, abscisic acid, and jasmonic acid suggest that PPO B might be under control of various plant hormones. The current study demonstrated that a PPO1 knockout causes a significant increase of endogenously produced cytokinins, including intra- and extracellular isopentenyladenine (Fig. 6), which is known to be a key regulator of budding and gametophore production in P. patens (Schwartzenberg et al., 2007). The current study also demonstrated that PPO1 knockout plants show a reduced depletion of externally applied [2-3H]-iP (Fig. 7). At this point, the mechanisms that lead from PPO1 deficiency to alterations in cytokinin homeostasis and the resulting developmental changes can only be speculated upon. Lowered cytokinin breakdown, which is likely to be the cause of the reduced isopentenyladenine depletion in the culture medium of the PPO1-deficient plants, is consistent with the proposal that PPOs could be involved in reoxidation of cytokinin oxidase/dehydrogenase, responsible for the cleavage of the isopentenyl side chain (Galuszka et al., 2005). These authors collected evidence that the products of the PPO action on phenolics, the quinones, act as electron acceptors for the reoxidation of cytokinin oxidase/dehydrogenase, thus promoting cytokinin breakdown. This implies that, in the case of P. patens plants lacking PPO1, lower PPO activities would lead to less quinones and therefore to lower rates of cytokinin oxidase/dehydrogenase reoxidation. As a result, less cytokinin degradation would occur in plants that have a reduced PPO activity, leading to an excess of active cytokinins and consequently causing an increased bud and gametophore production.
Alternatively, the increase of cytokinins in PPO1-deficient plants could also be more distantly related to polyphenol metabolism, so the observed phenotype could be interpreted as a general and rather unspecific response to stress. Wolf et al. (2010) observed similarly reduced sizes of colonies and higher gametophore production as photomorphogenetic changes after UV-B irradiation of P. patens wild type and discussed this phenomenon as an avoidance reaction, in which increased gametophore occurrence reduces the exposure to UV-B by shadowing basal parts of the plant.
Lower amounts of protonemata with increased gametophore production also minimize the contact surface to the substrate and could be an answer to chemically induced stress. It cannot be excluded that PPO1 deficiency in P. patens leads to a type of internal metabolic stress, e.g. change in the general redox status, being at the origin of observed morphogenetic changes. In terms of reproduction and formation of persisting spores, it makes sense that a plant sensing stress accelerates the transition to generative reproduction, the first step of which would be cytokinin-induced gametophore initiation.
Although the phylogenetic analysis suggests that the occurrence of PPOs is connected to the evolution of the land plant lineage, a single and specific function cannot be attributed to these enzymes. The importance of PPOs is underlined by the high number of PPO gene family members, which most probably take over different functions. A characteristic phenotype of d|ppo1 plants, besides the developmental changes, is the strongly increased susceptibility towards 4-MC, suggesting that extracellular PPO1 is involved in the detoxification of external phenolics. Ongoing work on other PPO gene family members will elucidate in how far their function and properties differ from the ones of PPO1 and which role PPOs take over in stress adaptation of P. patens and other bryophytes.
Supplementary Material
Supplementary material
Supplementary data are available at JXB online.
Supplementary information. Primers for PpPPO1 and PpPPO6 and deduced and edited amino acid sequence for PPO9
Properties of PPO gene family members
Supplementary Fig. S1. Quantification of phenolic compounds in P. patens wild type tissue
Supplementary Fig. S2. ESI(-)-MS spectrum of the unknown ester of caffeic acid
Supplementary Fig. S3. Characterization of PPO1 knockout (d|ppo1) plants
Supplementary Fig. S4. Protonema growth of PPO1 knockout plants and wild type
Acknowledgements
KvS, RL, and HR acknowledge funding by the DFG (SCHW687/5). HR acknowledges the German Academic Exchange Service (DAAD; D/04/34906) and the ‘Graduiertenförderung’ of the University of Hamburg for financial support. MS and ON acknowledge funding by the Centre of the Region Haná for Biotechnological Agricultural Research (grant no. ED0007/01/01).
The authors thank M. Sullivan (USDA-ARS, Wisconsin) for the plasmid harbouring TpPPO1, D. Hanelt (University of Hamburg) for help with the online monitoring of P. patens growth and C. Reisdorff (University of Hamburg) for advice in statistical analyses. The skilful technical help of S. Bringe, V. Schwekendiek (University of Hamburg), and H. Martinkova (Palacký University) is acknowledged.
References
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution Bioinformatics 21 2104–2105 [DOI] [PubMed] [Google Scholar]
- Bollag JM, Shuttleworth KL, Anderson DH. 1988. Laccase-mediated detoxification of phenolic compounds Applied and Environmental Microbiology 54 3086–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet AM. 2007. Evolution and current status of research in phenolic compounds Phytochemistry 68 2722–2735 [DOI] [PubMed] [Google Scholar]
- Bradford M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding Analytical Biochemistry 72 248–254 [DOI] [PubMed] [Google Scholar]
- Chevalier T, Rigal D, Mbéguié-A-Mbéguié D, Gauillard F, Richard-Forget F, Fils-Lycaon B. 1999. Molecular cloning and characterization of apricot fruit polyphenol oxidase Plant Physiology 11 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. 2004. The Jalview Java alignment editor Bioinformatics 20 426–427 [DOI] [PubMed] [Google Scholar]
- Constabel CP, Yip L, Patton JJ, Christopher ME. 2000. Polyphenol oxidase from hybrid poplar. Cloning and expression in response to wounding and herbivory Plant Physiology 124 285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeke T, Morris CF. 2002. Molecular characterization of wheat polyphenol oxidase (PPO) Theoretical and Applied Genetics 104 813–818 [DOI] [PubMed] [Google Scholar]
- Dry IB, Robinson SP. 1994. Molecular cloning and characterisation of grape berry polyphenol oxidase Plant Molecular Biology 26 495–502 [DOI] [PubMed] [Google Scholar]
- Galuszka P, Frébortová J, Luhová L, Bilyeu KD, English JT, Frébort I. 2005. Tissue localization of cytokinin dehydrogenase in maize: possible involvement of quinone species generated from plant phenolics by other enzymatic systems in the catalytic reaction Plant Cell Physiology 46 716–728 [DOI] [PubMed] [Google Scholar]
- Gerdemann C, Eicken C, Krebs B. 2002. The crystal structure of catechol oxidase: new insight into the function of type-3 copper proteins Accounts of Chemical Research 35 183–191 [DOI] [PubMed] [Google Scholar]
- Gooding PS, Bird C, Robinson SP. 2001. Molecular cloning and characterisation of banana fruit polyphenol oxidase Planta 213 748–757 [DOI] [PubMed] [Google Scholar]
- Gruz J, Novák O, Strnad M. 2008. Rapid analysis of phenolic acids in beverages by UPLC-MS/MS Food Chemistry 111 789–794 [Google Scholar]
- Hahlbrock K, Scheel D. 1989. Physiology and molecular biology of phenylpropanoid metabolism Annual Review of Plant Physiology and Plant Molecular Biology 40 347–369 [Google Scholar]
- Halaouli S, Record E, Casalot L, Hamdi M, Sigoillot J.-C, Asther M, Lomascolo A. 2006. Cloning and characterization of a tyrosinase gene from the white-rot fungus Pycnoporus sanguineus, and overproduction of the recombinant protein in Aspergillus niger Applied Microbiology and Biotechnology 70 580–589 [DOI] [PubMed] [Google Scholar]
- Hohe A, Egener T, Lucht J, Holtorf H, Reinhard C, Schween G, Reski R. 2004. An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene knockouts in a moss Physcomitrella patens. Current Genetics 44 339–347 [DOI] [PubMed] [Google Scholar]
- Hunt MD, Eannetta NT, Yu H, Newman SM, Steffens JC. 1993. cDNA cloning and expression of potato polyphenol oxidase Plant Molecular Biology 21 59–68 [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment Nucleic Acids Research 33 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde T, Eicken C, Sacchettini JC, Krebs B. 1998. Crystal structure of a plant catechol oxidase containing a dicopper center Nature Structural Biology 5 1084–1090 [DOI] [PubMed] [Google Scholar]
- Knight CD, Cove DJ, Boyd PJ, Ashton NW. 1988. The isolation of biochemical and developmental mutants in Physcomitrella patens . In: Glime JM, ed, Methods in Bryology. Nichinan, Miyazaki, Japan: The Hattori Botanical Laboratory; pp 47–58 [Google Scholar]
- Lang D, Zimmer AD, Rensing SA, Reski R. 2008. Exploring plant biodiversity: the Physcomitrella genome and beyond Trends in Plant Science 13 542–549 [DOI] [PubMed] [Google Scholar]
- Lax AR, Vaughn KC, Templeton GE. 1984. Nuclear inheritance of polyphenol oxidase in Nicotiana The Journal of Heredity 75 285–287 [Google Scholar]
- Lewis LA. 2007. Chlorophyta on land. Independent lineages of green eukaryotes from arid lands. In: Seckbach J, editor, Algae and cyanobacteria in extreme environments: cellular origin, life in extreme habitats and astrobiology, volume 11, part 6, 569–582 Berlin: Springer; [Google Scholar]
- Li Z-H, Wang Q, Ruan X, Pan C-D, Jiang D-A. 2010. Phenolics and plant allelopathy Molecules 15 8933–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberei R, Biehl B. 1976. Freisetzung und Activierung von Polyphenoloxydasen aus Thylakoid Membranen der Spinat-Chloroplasten Berichte der Deutschen Botanischen Gesellschaft 89 663–676 [Google Scholar]
- Mayer AM. 2006. Polyphenol oxidases in plants and fungi: going places? A review Phytochemistry 67 2318–2331 [DOI] [PubMed] [Google Scholar]
- McCaig BC, Meagher RB, Dean JFD. 2005. Gene structure and molecular analysis of the laccase-like multicopperoxidase (LMCO) gene family in Arabidopsis thaliana Planta 221 619–636 [DOI] [PubMed] [Google Scholar]
- Newman SM, Eannetta NT, Yu H, Prince JP, de Vicente MC, Tanksley SD, Steffens JC. 1993. Organisation of the tomato polyphenol oxidase gene family Plant Molecular Biology 21 1035–1051 [DOI] [PubMed] [Google Scholar]
- Newman SM, Tantasawat P, Steffens JC. 2011. Tomato polyphenol oxidase B is spatially and temporally regulated during development and in response to ethylene Molecules 16 493–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O, Hauserová E, Amakorová P, Doležal K, Strnad M. 2008. Cytokinin profiling in plant tissues using ultra-performance liquid chromatography – electrospray tandem mass spectrometry Phytochemistry 69 2214–2224 [DOI] [PubMed] [Google Scholar]
- Novák O, Tarkowski P, Tarkowská D, Doležal K, Lenobel R, Strnad M. 2003. Stanovení cytokininú v rostlinách metodou imunoafinitní chromatografie a kapalinové chromatografie-hmotnostní spektrometrie Chemické listy 97–302 [Google Scholar]
- Ono E, Hatayama M, Isono Y, et al. 2006. Localization of a flavonoid biosynthetic polyphenol oxidase in vacuoles The Plant Journal 45 133–143 [DOI] [PubMed] [Google Scholar]
- Paszkowski J, Baur M, Bogucki A, Potrykus I. 1988. Gene targeting in plants The EMBO Journal 7 4021–4026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JB, Chapman JV. 1985. Isolation, culture and genetic manipulation of plant protoplasts. In: Dixon RA, ed, Plant cell culture – a practical approach Oxford, UK: IRL Press; pp 37–66 [Google Scholar]
- Quideau S, Deffieux D, Douat-Casassus C, Pouységu L. 2011. Plant polyphenols: chemical properties, biological activities, and synthesis Angewandte Chemie International Edition 50 586–621 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Ick J, Fawcett JA, Lang D, Zimmer A, Van de Peer Y, Reski R. 2007. An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens BMC Evolutionary Biology 7–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants Science 319 64–69 [DOI] [PubMed] [Google Scholar]
- Reski R, Reutter K, Karsten B, Faust M, Kruse S, Gorr G, Strepp R, Abel WO. 1995. Molecular analysis of chloroplast division. In: Terzi M, Cella R, Falavigna A, editors, Current issues in plant molecular and cellular biology Dordrecht, NL: Kluwer Academic Publishers; 291–296 [Google Scholar]
- Rice-Evans CA, Miller NJ, Bolwell PG, Bramley PM, Pridham JB. 1995. The relative antioxidant activities of plant-derived polyphenolic flavonoids Free Radical Research 22 375–383 [DOI] [PubMed] [Google Scholar]
- Richter H. 2009. Organisation and transcriptional regulation of the polyphenol oxidase (PPO) multigene family of the moss Physcomitrella patens (Hedw.) B.S.G. and functional gene knockout of PpPPO1. PhD thesis,University of Hamburg; Germany: http://ediss.sub.uni-hamburg.de/volltexte/2009/4027/ [Google Scholar]
- Richter H, Lieberei R, Schwartzenberg v. K. 2005. Identification and characterisation of a bryophyte polyphenol oxidase encoding gene from Physcomitrella patens Plant Biology 7 283–291 [DOI] [PubMed] [Google Scholar]
- Roleda MY, Hanelt D, Kräbs G, Wiencke C. 2004. Morphology, growth, photosynthesis and pigments in Laminaria ochroleuca (Laminariales, Phaeophyta) under ultraviolet radiation Phycologia 43 603–613 [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models Bioinformatics 19 1572–1574 [DOI] [PubMed] [Google Scholar]
- Schaefer D, Zryd JP. 1997. Efficient gene targeting in the moss Physcomitrella patens The Plant Journal 11 1195–1206 [DOI] [PubMed] [Google Scholar]
- Schulz P, Hofmann A, Russo V, Hartmann E, Laloue M, v Schwartzenberg K. 2001. Cytokinin overproducing ove mutants of Physcomitrella show increased riboside to base conversion Plant Physiology 126 1224–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzenberg v K, Pethe C, Laloue M. 2003. Cytokinin metabolism in Physcomitrella patens – differences and similarities to higher plants Plant Growth Regulation 39 99–106 [Google Scholar]
- Schwartzenberg v K, Fernández Núñez M, Blaschke H, Dobrev PI, Novák O, Motyka V, Strnad M. 2007. Cytokinins in the bryophyte Physcomitrella patens: Analyses of activity, distribution and cytokinin oxidase/dehydrogenase overexpression reveal the role of extracellular cytokinins Plant Physiology 145 786–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinheimo E, Saloheimo M, Ahola E, Westerholm-Parvinen A, Kalkkinen N, Buchert J, Kruus K. 2006. Production and characterization of a secreted, C-terminally processed tyrosinase from the filamentous fungus Trichoderma reesei FEBS Journal 273 4322–4335 [DOI] [PubMed] [Google Scholar]
- Sherman TD, Vaughn KC, Duke SO. 1991. A limited survey of the phylogenetic distribution of polyphenol oxidase Phytochemistry 30 2499–2506 [Google Scholar]
- Steffens JC, Harel E, Hunt MD. 1994. Polyphenol oxidase. In: Ellis BE, Stafford HA, editors, Recent advances in phytochemistry, genetic engineering of plant secondary metabolism, vol. 28. New York: Plenum Press; pp 275–312 [Google Scholar]
- Sullivan ML, Hatfield RD, Thoma SL, Samac DA. 2004. Cloning and characterisation of red clover polyphenol oxidase cDNAs and expression of active protein in Escherichia coli and transgenic alfalfa Plant Physiology 136 3234–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thipyapong P, Joel DM, Steffens JC. 1997. Differential expression and turnover of the tomato polyphenol oxidase gene family during vegetative and reproductive development Plant Physiology 113 707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thipyapong P, Melkonian J, Wolfe DW, Steffens JC. 2004. Suppression of polyphenol oxidases increases stress tolerance in tomato Plant Science 167 693–703 [Google Scholar]
- Thipyapong P, Steffens JC. 1997. Tomato polyphenol oxidase: differential response of the polyphenol oxidase F promoter to injuries and wound signals Plant Physiology 115 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thygesen PW, Dry IB, Robinson SP. 1995. Polyphenol oxidase in potato – a multigene family that exhibits differential expression patterns Plant Physiology 109 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LT, Constabel CP. 2011. The polyphenol oxidase gene family in poplar: phylogeny, differential expression and identification of a novel, vacuolar isoform Planta 234 799–813 [DOI] [PubMed] [Google Scholar]
- von Schwartzenberg K. 2009. Hormonal regulation by auxin and cytokinin in moss Annual Plant Reviews 36 246–281 [Google Scholar]
- Wang GD, Li QJ, Luo B, Chen XY. 2004. Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase Nature Biotechnology 22 893–897 [DOI] [PubMed] [Google Scholar]
- Wang TL, Cove DJ, Beutelmann P, Hartmann E. 1980. Isopentenyladenine from mutants of the moss Physcomitrella patens Phytochemistry 19 1103–1105 [Google Scholar]
- Waterman PG, Mole S. 1994. Analysis of phenolic plant metabolites Oxford, UK: Blackwell Scientific Publications; [Google Scholar]
- Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach Molecular Biology and Evolution 18 691–699 [DOI] [PubMed] [Google Scholar]
- Wolf L, Rizzini L, Stracke R, Ulm R, Rensing SA. 2010. The molecular and physiological responses of Physcomitrella patens to unltraviolet-B radiation Plant Physiology 153 1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoruk R, Marshall MR. 2003. Physicochemical properties and function of plant polyphenol oxidase: a review Journal of Food Biochemistry 27 361–422 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.