Abstract
Leaves develop as planar organs, with a morphology that is specialized for photosynthesis. Development of a planar leaf requires genetic networks that set up opposing adaxial and abaxial sides of the leaf, which leads to establishment of dorsoventral polarity. While many genes have been identified that regulate adaxial and abaxial fate there is little information on how this is integrated with cellular function. EMBRYO DEFECTIVE DEVELOPMENT1 (EDD1) is a nuclear gene that encodes a plastid and mitochondrial localized glycyl-tRNA synthetase. Plants with partial loss of EDD1 function have changes in patterning of margin and distal regions of the leaf. In combination with mutations in the MYB domain transcription factor gene ASYMMETRIC LEAVES1 (AS1), partial loss of EDD1 function results in leaves with reduced adaxial fate. EDD1 may influence leaf dorsoventral polarity through regulating the abaxial fate genes KANADI1 (KAN1) and ETTIN (ETT)/AUXIN RESPONSE FACTOR3 (ARF3) since these genes are upregulated in the edd1 as1 double mutant. SCABRA3 (SCA3), a nuclear gene that encodes the plastid RNA polymerase is also required for leaf adaxial fate in the absence of AS1. These results add a novel component to networks of genetic regulation of leaf development and suggest that organelles, particularly plastids, are required in leaf patterning. Potentially, signalling from organelles is essential for coordination of different cell fates within the developing leaf.
Key words: Arabidopsis, AS1, chloroplast, EDD1, leaf patterning, mitochondria, plastid, tRNA synthetase, Arabidopsis, Arabidopsis
Introduction
Plant shoots are initiated during embryogenesis, when the single cell zygote undergoes cell divisions to form a basal root meristem and a shoot apical meristem. During vegetative development, cells on the flanks of the shoot apical meristem are recruited into the production of leaves. In Arabidopsis, initiating leaves develop into determinate, flattened structures with morphologically distinct dorsoventral polarity. The adaxial (dorsal) side of the leaf is specialized for capture of light energy and the abaxial (ventral) side of the leaf is specialized for gas exchange, so that leaf dorsovental polarity is optimized for photosynthesis.
Development of leaf dorsoventral polarity requires juxtaposition of adaxial and abaxial fates and, in the absence of either fate, leaves develop as radial organs. Dorsoventral polarity is determined by distinct adaxial and abaxial factors. Class III HD-ZIP transcription factor genes REVOLUTA (REV), PHABULOSA (PHB), and PHAVULOTA (PHV) are expressed on the adaxial side of lateral organs and act redundantly in adaxial fate (McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003; Prigge et al., 2005). While loss of PHB and PHV has no phenotypic effect, loss of REV results in a reduced number of lateral branches and floral organs, as well as vascular patterning defects (Talbert et al., 1995; Zhong and Ye, 1999; Otsuga et al., 2001; Emery et al., 2003; Prigge et al., 2005). Loss of these three class III HD-ZIP genes in triple mutants results in radial organs (Emery et al., 2003; Prigge et al., 2005). Dominant mutations, which disrupt posttranscriptional regulation of REV, PHB, and PHV by mir165/166, result in radial, adaxialized leaves, and in the dominant phb-1d mutant PHB expression is expanded to the abaxial side of the leaf (McConnell et al., 2001; Emery et al., 2003; Kidner and Martienssen, 2004; Zhong and Ye, 2004). Class III HD-ZIP gene expression is also restricted to the adaxial side of developing leaves through a genetic pathway involving KANADI (KAN) family genes. KANADI genes, which encode GARP-domain transcription factors, are expressed on the abaxial side of lateral organs and act redundantly to promote abaxial fate. Loss of KANADI1 (KAN1) causes mild dorsoventral patterning defects such as upward curled leaves and precocious development of abaxial trichomes (Eshed et al., 2001, 2004; Kerstetter et al., 2001). The extent of leaf adaxialization is increased as more of the KANADI gene family members (KAN2, KAN3, and KAN4) are mutated (Eshed et al., 2001, 2004). In kan1 kan2, for example, ectopic patches of adaxial fate on the abaxial side of the leaf leads to ectopic abaxial lamina protrusions. In addition, ectopic expression of KAN1 throughout the leaf results in development of radial, abaxial organs consistent with a requirement for KAN1 for abaxial fate (Eshed et al., 2001; Kerstetter et al., 2001). Abaxial fate also requires the AUXIN RESPONSE FACTOR (ARF) family genes ETTIN (ETT)/AFR3 and ARF4. Loss of both ETT and ARF4 in the ett arf4 double mutant results in leaves resembling kan1 kan2 and kan mutants are enhanced by loss of either ETT or ARF4. KANADI and ETT/ARF4 appear to cooperate in specification of abaxial fate, and this could be in part mediated by protein interaction between KAN1 and ETT (Pekker et al., 2005; Kelley et al., 2012).
Initiation of organ primordia involves the transition of cells from an indeterminate to a determinate cell fate. Two genes involved in this transition are the MYB domain transcription factor gene ASYMMETRIC LEAVES1 (AS1) and the LOB/ASL-domain transcription factor gene ASYMMETRIC LEAVES2 (AS2) (Byrne et al., 2000, 2002; Iwakawa et al., 2002; Lin et al., 2003). AS1 is expressed throughout developing leaves whereas AS2 expression is restricted to the adaxial side of the leaf (Byrne et al., 2000; Iwakawa et al., 2002, 2007). AS1 and AS2 act as a heterodimer, which may serve to limit the activity of these two proteins to the adaxial side of the leaf (Xu et al., 2003). AS1 and AS2 repress the expression of meristem homeodomain transcription factor class I KNOX genes in determinate organs (Byrne et al., 2000; Iwakawa et al., 2002; Lin et al., 2003). Loss of either AS1 or AS2 results in similar phenotypes with changes of leaf shape to short, round and weakly epinastic leaves. Although this phenotype does not suggest a prominent role in leaf dorsoventral polarity, the function of AS2 in adaxial fate is indicated by the phenotype of plants overexpressing AS2, which have leaves similar to kan1 kan2, and AS2 is a direct target of KAN1 regulation (Lin et al., 2003; Wu et al., 2008). In addition, orthologues of AS1 in other dicotyledonous species, including PHANTASTICA (PHAN) in Antirrhinum, CRISPA in pea, and NSPHAN in tobacco, demonstrate a role for these AS1-related genes in adaxial fate since loss or reduced function of these genes results in abaxialized and radial leaves (Waites and Hudson, 1995; Waites et al., 1998; McHale and Koning, 2004; Tattersall et al., 2005). Therefore in some species AS1 has a prominent role in leaf adaxial fate. This function has either been reduced in Arabidopsis or alternate pathways mask the contribution of AS1 to leaf dorsoventral polarity.
Several genes act in parallel with AS1 and AS2 in leaf adaxial fate and enhance the polarity defect of as1 and as2 mutants resulting in trumpet-shaped or radial leaves. These genes encode for proteins involved in a diverse range of biological processes. These include trans-acting siRNA components that are involved in small RNA-mediated cleavage of ETT and ARF4 transcripts; ARGONAUTE1 (AGO1); the histone deacetylases HDT1/HD2A and HDT2/HD2B; proteasome complex proteins; and Elongator complex proteins (Li et al., 2005; Garcia et al., 2006; Huang et al., 2006; Yang et al., 2006; Ueno et al., 2007; Kojima et al., 2011). Mutations in ribosomal protein genes also enhance as1 leaf dorsoventral polarity defects (Pinon et al., 2008; Yao et al., 2008; Horiguchi et al., 2011; Szakonyi and Byrne, 2011). In the case of the piggyback (pgy) ribosomal protein gene mutants, the as1 pgy double mutants have ectopic lamina outgrowths on the adaxial side of the leaf. These ribosomal protein mutants enhance the adaxial defect of class III HD-ZIP mutants and suppress the abaxial defect of mutants in KANADI genes, suggesting that ribosomal proteins or the ribosome have a specific function in leaf adaxial fate (Pinon et al., 2008; Yao et al., 2008). How ribosome function influences leaf dorsoventral polarity is not known. Possibly reduced ribosome function selectively affects expression of regulatory genes involved in leaf dorsoventral polarity.
In this study, EMBRYO DEFECTIVE DEVELOPMENT1 (EDD1) has been identified as a gene that acts with AS1 in leaf adaxial fate. EDD1 encodes a glycyl-tRNA synthetase localized to plastids and mitochondria, and loss-of-function mutations in EDD1 are embryo lethal (Uwer et al., 1998; Duchene et al., 2001; Berg et al., 2005). However, the partial loss-of-function mutant edd1-3 is viable, revealing a role for organelles in leaf development. Single edd1 mutants alter patterning of marginal and distal regions of leaves whereas edd1 as1 double mutants have trumpet-shaped and radial abaxial leaves. Genetic interactions and gene expression analysis indicate that adaxial fate is sensitive to organelle function and suggest that EDD1 may influence leaf dorsoventrality in part through KAN1 and ETT.
Materials and methods
Plant material and growth conditions
edd1-3 was an ethylmethane sulphonate (EMS)-induced mutation generated in an as1-1 background, as described previously (Byrne et al., 2002). edd1-4 was a Ds transposon insertion allele (GT_5_108612) obtained from the European Arabidopsis Stock Centre (NASC) (Scholl et al., 2000). pgy2-1, rev-6, kan1-2, and kan2-1 were obtained as described previously (Pinon et al., 2008). sca3-1 was obtained from José Micol. All mutants were in the Landsberg erecta (Ler) background. For complementation analysis, a 10.5-kb genomic region encompassing the EDD1 gene was cloned into the pMDC123 binary vector and transformed into edd1-3 as1/+ plants by the floral dip method (Clough and Bent, 1998; Curtis and Grossniklaus, 2003).
Molecular biology
DNA and RNA techniques were carried out using standard methods. Genotyping for edd1-3 was performed by PCR using the primers 5'-GCAGGTAGTGGATTGTTCAAGT-3' and 5'-CGCTCTGCTAGGACAGACC-3', followed by restriction digestion of the product with DdeI, which cleaves the mutant allele but not the wild-type allele. Quantitative reverse-transcription PCR (qRT-PCR) analysis was carried out as previously described (Pinon et al., 2008). Total RNA was extracted from 20 10-day-old seedlings using Trizol (Invitrogen) and DNase treated prior to cDNA synthesis using Superscript II reverse transcriptase (Applied Biosystems). qRT-PCR was carried out using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich). Three biological replicates were performed. Gene-specific primers for qRT-PCR were KAN1 (5'-GATCCAGCATTCAAAATCAGG-3' and 5'-TTTCTCGTGCCAATCTGGTCT-3'), KAN2 (5'-TTTGCAT- GGGAAGTTAATCG-3' and 5'-TTGTTCCCGAGATGCTTGAT-3'), ETT (5'-GGAAAGCCTGATATCCCTGTC-3' and 5'-ACCATCCG- AACAAGTGTTGA-3'), REV (5'-ATATTCGATGAATCGGGTCGTA-3' and 5'-ATAACTCACATGTCTTCCCATCG-3') and ACTIN2 (5'-GCACCCTGTTCTTCTTACCG-3' and 5'-AACCCTCGTAGATTGG- CACA-3'). qRT-PCR data was analysed using MJ Opticon Monitor Data Analysis software. Gene transcript accumulation was normalized to ACTIN2. Statistical analysis was carried out using ANOVA and values ≦0.01 were considered significant.
In situ hybridization
RNA in situ hybridizations were performed as previously described using a DIG-labelled antisense probe generated from a linearizing plasmid carrying the 5' region of FIL (Long et al., 1996; Siegfried et al., 1999).
Histology
Tissue for sectioning was fixed in 2.5% glutaraldehyde, dehydrated in an ethanol series to 100% and embedded in Technovit 7100 (Heraeus). Resin-embedded samples were sectioned to 10 μm, and stained with 0.1% toluidine blue. For analysis of leaf morphology, three leaves of each genotype were analysed.
Results
edd1 is a weak mutant allele of a nuclear gene encoding an organelle-localized glycyl-tRNA synthetase
In a genetic screen for modifiers of as1 a mutant that produced radialized leaves was identified. To identify the new enhancer of as1 a mapping population was generated by crossing as1 plants carrying the enhancer in Ler to Col-0. Analysis of 1050 chromosomes from double mutants located the enhancer to a 44-kb region on chromosome 3 between 17.756 and 17.803kb. Candidate genes in this region were sequenced and revealed a G to A base pair change in the coding sequence of the gene At3g48110. This gene encodes a glycyl-tRNA synthetase (GlyRS). Aminoacyl-tRNA synthetases catalyse the addition of amino acids to their cognate tRNAs (Dang, 1986). At3g48110 has been previously named EDD1 (Uwer et al., 1998). The new mutant allele of this gene was therefore named edd1-3. Unless otherwise stated, this allele will be referred to here as edd1.
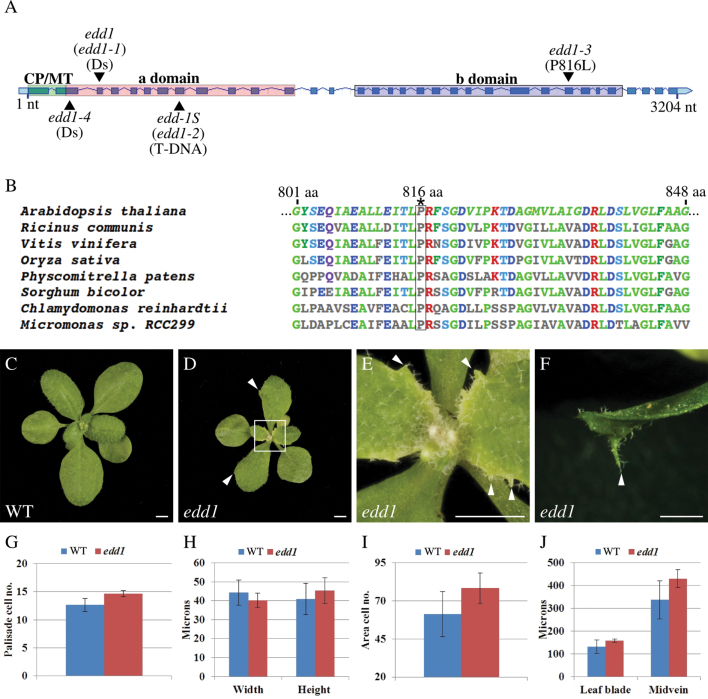
EDD1 is composed of 33 exons and encodes a 117-kDa protein of 1068 amino acids (Fig. 1A). EDD1 has an N-terminal transit peptide sequence that targets both plastids and mitochondria, and this protein is likely to function in both organelles (Duchene et al., 2001). EDD1 is further divided into an N-terminal ‘a domain’ and a C-terminal ‘b domain’ (Uwer et al., 1998). These two domains share homology with the two subunit proteins of GlyRS from Escherichia coli. The ‘a domain’ shares 59% identity with the α subunit protein of GlyRS, which is involved in ATP-dependent formation of the enzyme-bound aminoacyl-adenylate, and the ‘b domain’ shares 36% identity with the β subunit of GlyRS, which is involved in binding the tRNA (Nagel et al., 1984; Toth and Schimmel, 1990; Uwer et al., 1998). The mutation in edd1-3 resulted in a proline to leucine substitution in a highly conserved amino acid (816) in the ‘b domain’ of the protein (Fig. 1A, 1B). Since the previously reported null insertion mutant alleles, edd1-1 and edd1-S (designated here as edd1-2), are homozygous embryo lethal (Uwer et al., 1998; Berg et al., 2005), it is proposed that the mutation in edd1-3 is a partial loss-of-function allele, which encodes a protein that retains some activity.
Fig. 1.
The edd1 mutation is a partial loss-of-function allele of EDD1 that disrupts patterning in marginal and distal regions of leaves. (A) The EDD1 gene, with exons displayed as blue boxes, interconnected by introns displayed as lines. Regions encoding an N-terminal chloroplast and mitochondrial (CP/MT) transit peptide (green), the ‘a domain’ (red), and the ‘b domain’ (blue) are shaded. The sites of the edd1-3 point mutation and other insertion mutant alleles are marked. (B) Alignment of selected region of glycyl-tRNA synthetase genes from different species, showing that the edd1-3 point mutation causes a proline to leucine substitution in a highly conserved amino acid, marked with an *. (C) Rosette of wild type. (D, E) Rosette of edd1 (D) and in close-up (E), showing that leaves are pointed and have more prominent serrations (arrowheads). Regions of pale green tissue are visible (E). (F) edd1 cauline leaves produce an abaxial midvein protrusion (arrowhead). (G–J) edd1 does not alter cells of the leaf lamina but alters the region of the midvein. Comparison of wild-type and edd1 cell number (G), cell size (H), number of cells in transverse sections that include midvein and leaf blade (I), and distance between adaxial and abaxial leaf surfaces in the region of the leaf blade and midvein (J). Bars, 2mm (A–D). Error bars (G–J) are standard deviations from three replicates.
To confirm the new modifier of as1 is due to the mutation in EDD1, a 10.5-kb genomic fragment encompassing EDD1 was tested for complementation of the edd1 as1 mutant. In four independent transgenic lines, the wild-type EDD1 transgene fully suppressed the trumpet and radial leaf phenotypes of edd1 as1 and the phenotype of these transgenic plants was indistinguishable from as1 single mutants. Genetic analysis was also carried out to determine whether the mutation in edd1-3 disrupts EDD1 function. edd1-3 was crossed with a Ds transposon insertion allele (GT_5_108612), designated edd1-4 (Fig. 1A). As reported for edd1-1, homozygous edd1-4 individuals were not identified and siliques of edd1-4/+ plants had 25% aborted seed, indicating this allele is embryo lethal. Plants from a cross of edd1-3 with edd1-4/+ resulted in 54% (n = 120) embryo lethality, and a reciprocal cross of edd1-4/+ with edd1-3 resulted in 53% embryo lethality (n = 99). The lack of complementation demonstrates edd1-3 is a mutant allele of EDD1. Together, complementation and allelism data confirm that the new enhancer of as1 is due to mutation in EDD1.
edd1 disrupts leaf development
To determine whether the new enhancer of as1 affects leaf development independently to as1, the leaf phenotype of the single edd1 mutant was examined. In wild type, rosette leaves were elongate and rounded and have very subtle marginal serrations (Fig. 1C). The edd1 mutant produced pointed leaves with more pronounced marginal serrations but did not produce trumpet-shaped or radial leaves (Fig. 1D, 1E). The edd1 mutant was also characterized by pale green tissue or chlorosis in regions where cells were undergoing proliferation and expansion, including the proximal region of young leaves and immature floral organs (Fig. 1E). The extent of pale tissue was progressively reduced during growth, such that mature organs lacked visible chlorosis. In the majority of plants, at least one cauline leaf had an abaxial outgrowth at the distal tip of the leaf, and this outgrowth appeared to be associated with the midvein (Fig. 1F). The frequency of this phenotype was rare in rosette leaves of plants grown at 22 °C, but increased when plants were grown at 18 °C, where 70% of edd1 plants had at least one rosette leaf with an abaxial outgrowth at the distal tip of the leaf.
To determine whether edd1 disrupts morphology of the leaf, the arrangements of cells in internal tissue layers of wild-type and edd1 leaves were compared in transverse sections of mature leaves. In wild type, the palisade mesophyll comprises a subepidermal layer of tall, narrow, closely packed cells on the adaxial side of the leaf (Fig. S1A). edd1 mutants also had a distinct palisade mesophyll layer (Fig. S1C). Comparison of the palisade mesophyll cell number and size showed no significant difference between the wild type and the edd1 mutant (Fig. 1G, 1H). The cell number in an area of the leaf transverse sections that included the leaf vasculature and leaf lamina was also compared. edd1 had a slight increase in the number of cells in the transverse dimension compared with wild type (Fig. 1I). The distance between the adaxial and abaxial sides of the leaf (thickness) was also used to compare the morphology of edd1 with wild type. There was no difference in the thickness of the leaf in the region of the leaf lamina, whereas the midvein thickness was greater in edd1 compared with wild type (Fig. 1J). The increase in cell number in the edd1 leaf may therefore be associated with a change in development of the midvein. A change in the thickness of the midvein may be associated with the abaxial outgrowth at the distal tip of the leaf. However, the change in leaf shape in edd1 does not appear to be associated with major changes in internal mesophyll morphology.
edd1 as1 mutants produce trumpet-shaped and radial leaves
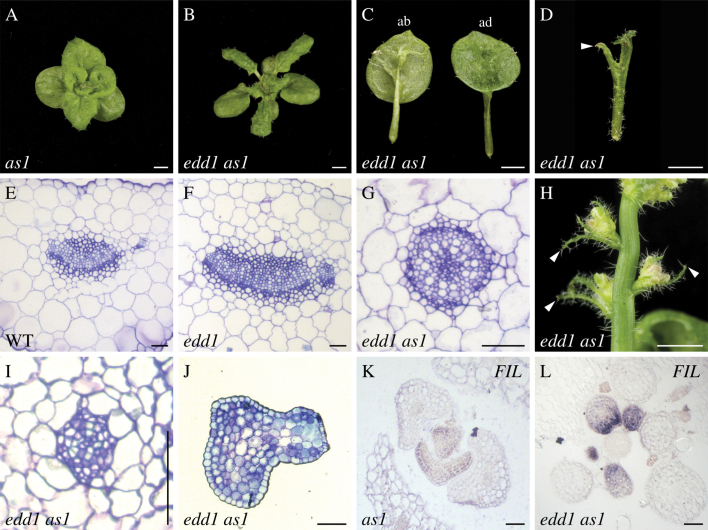
The edd1 mutant had a more severe effect on leaf development in the edd1 as1 double mutant. The as1 single mutant had short, broad, rounded, and slightly epinastic leaves (Fig. 2A). By comparison, the edd1 as1 double mutant had narrower leaves than as1 (Fig. 2B). Rosette and cauline leaves of edd1 as1 were often trumpet-shaped or radial (Fig. 2C, 2D, 2H). The inner side of trumpet-shaped leaves was dark green and resembled adaxial tissue, while the outer side was light green and resembled abaxial tissue, suggesting that trumpet leaves had reduced adaxial fate and were abaxialized (Fig. 2C). In addition, some edd1 as1 leaves produced an abaxial outgrowth at the distal tip of the leaf (Fig. 2D). To further determine whether leaf dorsoventral polarity was disrupted in edd1 as1, internal vascular tissue in transverse sections of leaves from wild type, edd1, and edd1 as1 were compared. In wild type, vasculature was oriented along the dorsoventral axis with xylem tissue adaxial to phloem (Fig. 2E). Vascular patterning in edd1 was similar to wild type, whereas vasculature of trumpet-shaped edd1 as1 leaves was radial with phloem surrounding xylem, indicating these leaves were abaxialized (Fig. 2F, 2G). In transverse sections of more severely affected edd1 as1 radial cauline leaves, the vasculature was disorganized in the proximal region of the leaf and was not clearly evident in the distal region of the leaf, suggesting that radial leaves may have a general reduction in dorsoventral polarity (Fig. 2I, 2J).
Fig. 2.
edd1 as1 produces trumpet-shaped and radial leaves. (A) Rosettes of as1. (B) Rosettes of edd1 as1. (C) A trumpet-shaped leaf of edd1 as1, showing the abaxial (left) and adaxial (right) sides of a leaf. (D) A radial leaf of edd1 as1, with minimal lamina and an abaxial midvein protrusion (arrowhead). (E–G) Transverse sections through the midvein vasculature of wild type (E), edd1 (F), and edd1 as1 (G). (H) edd1 as1 radial cauline leaves (arrowhead). (I, J) Transverse sections of moderately (I) and more severely (J) affected radial cauline edd1 as1 leaves. (K, L) in situ analysis of FIL in transverse sections of as1 (K) and edd1 as1 (L) young vegetative shoots. Bars, 2mm (A–D, H) and 50 µm (E–G, I–L).
Expression of the gene FILAMENTOUS FLOWER (FIL) is restricted to the abaxial side in developing leaves (Sawa et al., 1999; Siegfried et al., 1999). To further examine leaf polarity in edd1 as1, the expression pattern of FIL in edd1 as1 leaves was compared with wild type by in situ hybridization. The expression of FIL was restricted to the abaxial side of as1 leaves (Fig. 2K). In edd1 as1 radial leaves, FIL was expressed throughout initiating leaves and, as leaves developed, FIL became more highly expressed on the adaxial side of the leaf than on the abaxial side of the leaf (Fig. 2L). This expression pattern confirmed that edd1 as1 leaves were compromised in adaxial fate. Based on the single mutant phenotype, it is concluded that EDD1 has a role in leaf dorsoventral polarity that requires AS1, whereas EDD1 patterning of leaf margins and the distal tip of the leaf is independent of AS1.
edd1 interacts synergistically with pgy2 in leaf development
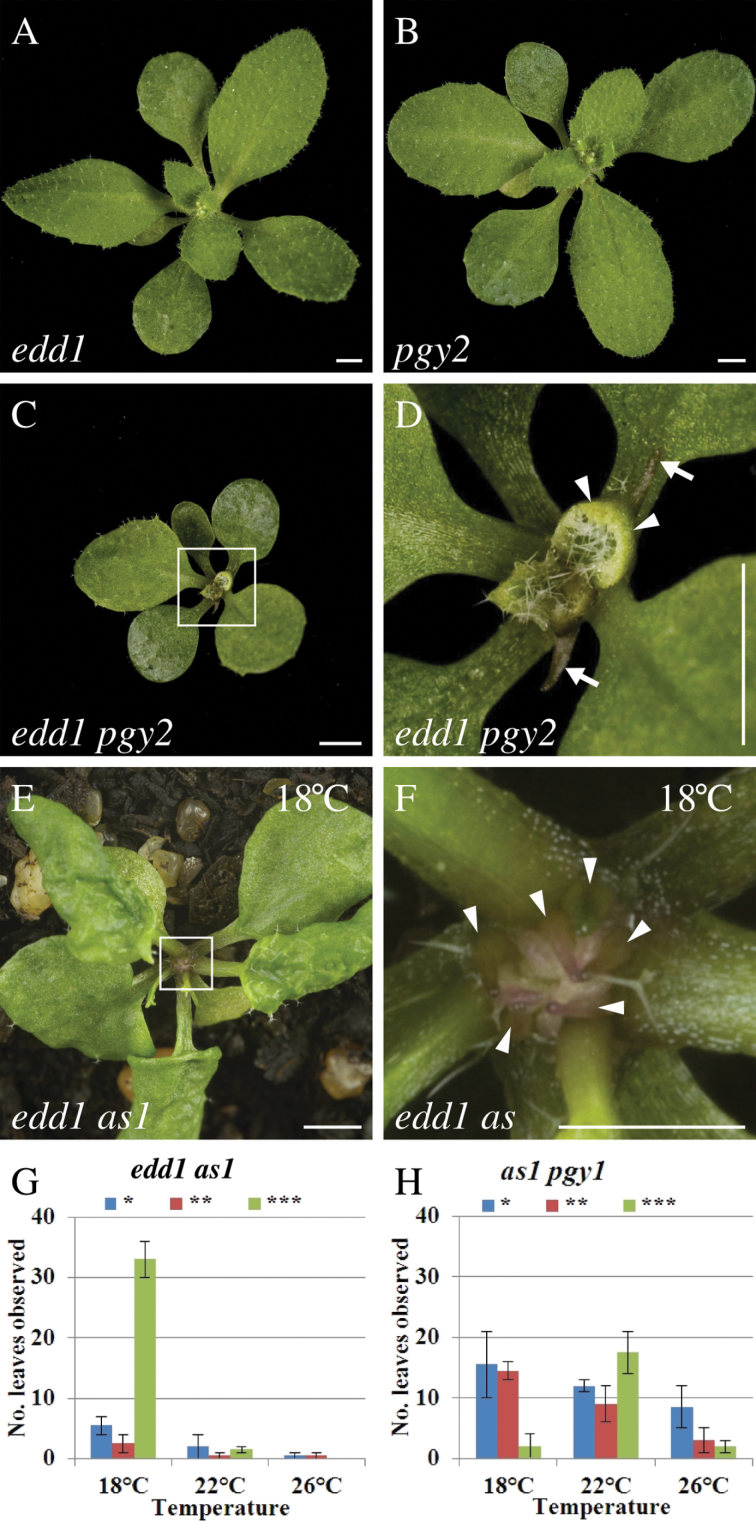
Other enhancers of as1 with leaf shape phenotypes similar to edd1 are mutants in ribosomal proteins (Pinon et al., 2008; Yao et al., 2008; Horiguchi et al., 2011; Szakonyi and Byrne, 2011). Ribosomal proteins act with AS1 to promote adaxial fate. In an as1 background, ribosomal protein mutants have ectopic outgrowths on the adaxial side of the leaf or have radial leaves. Single ribosomal protein mutants have pointed and serrated leaves like edd1, but ribosomal protein mutants also have defects in the internal palisade mesophyll, where cells are less closely aligned than in wild type (Pinon et al., 2008; Yao et al., 2008; Horiguchi et al., 2011; Szakonyi and Byrne, 2011). The edd1 mutant leaf phenotype therefore has some overlap with ribosomal protein mutants including pgy1, which is defective in the ribosomal protein gene RPL10aB, and pgy2, which is defective in the ribosomal protein gene RPL9C (Pinon et al., 2008). To determine whether EDD1 regulates leaf development in a common genetic pathway with ribosomal proteins, edd1 was combined with pgy2 and the leaf phenotype in the double mutant was analysed. Both edd1 and pgy2 single mutants had pointed and serrated leaves (Fig. 3A, 3B). Compared with either single mutant, the double mutant was reduced in size (Fig. 3C). Leaves of the edd1 pgy2 double mutant were pointed and serrated, had pale margins, and had an abaxial outgrowth at the distal tip of the leaf suggesting that these two mutations have additive effects on leaf development (Fig. 3C, 3D).
Fig. 3.
Genetic interactions between edd1 and pgy1 are additive. (A) Rosette of edd1. (B) Rosette of pgy2. (C, D) Rosette of edd1 pgy2 (C) and in close-up (D), showing leaves with pale margins (arrowheads), prominent serrations and an abaxial midvein protrusion (arrows). (E, F) edd1 as1 grown at 18 °C had trumpet-shaped and radial leaves (arrowhead) (square in E is enlarged in F). (G, H) Phenotype severity of as1 pgy1 and edd1 as1 grown at 18, 22, and 26 °C. The leaf phenotype severity was scored, with *, **, and *** for mild trumpet shape, severe trumpet shape, and radial leaf shape, respectively. Scores are means for leaves of 20 individual plants. Bars, 2mm (A–E) and 1mm (F).
The severity of the edd1 as1 leaf phenotype was found to be temperature sensitive. Therefore the severity of the edd1 as1 and as1 pgy1 leaf phenotypes were examined at 18, 22, and 26 °C. The edd1 as1 adaxial defect was most prominent when plants were grown at 18 °C, where one-third of leaves were trumpet-shaped or radial compared to a low level of defective leaves when plants were grown at 22 and 26 °C (Fig. 3E–G). At all temperatures, the severity of the leaf defect was greatest in late rosette and cauline leaves. By contrast, the severity of the as1 pgy1 leaf phenotype was not increased by growth at 18 °C and instead the frequency of abaxialized leaves in as1 pgy1 mutants was decreased at 18 °C (Fig. 3H). These leaf phenotypes and the genetic interactions indicate that EDD1 and the ribosomal PGY genes act independently in leaf development.
edd1 enhances rev and kan1 kan2 phenotypes
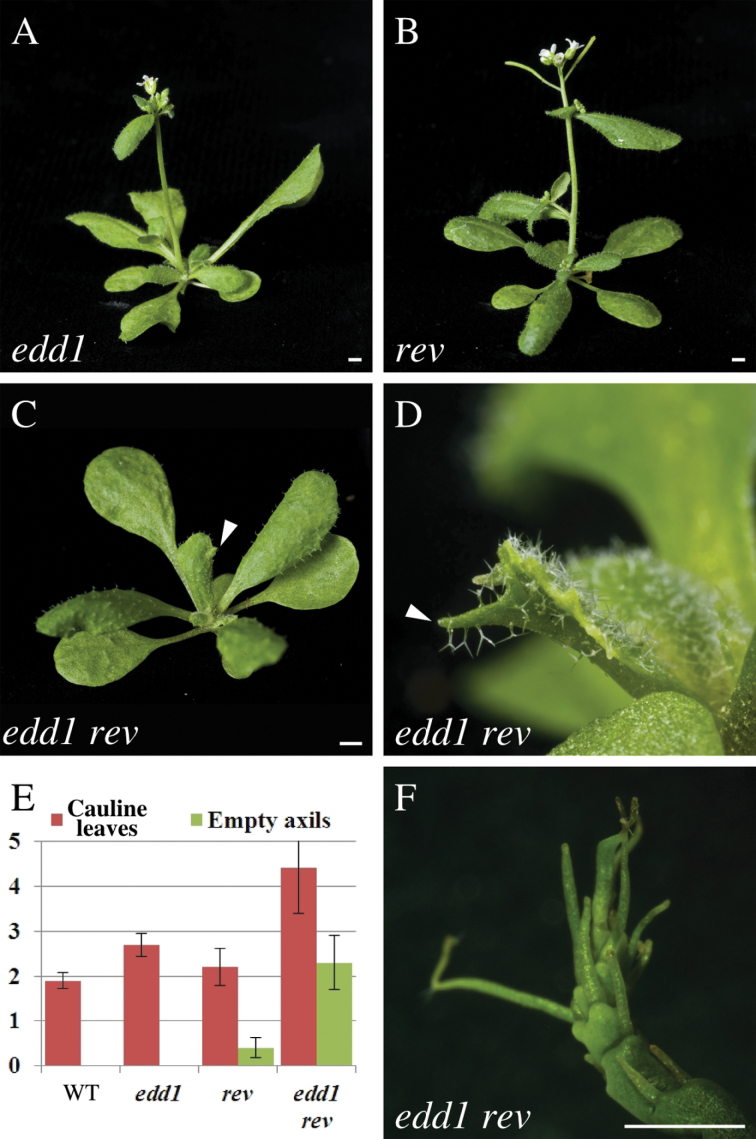
To establish whether EDD1 interacts with other genes involved in leaf dorsoventral polarity, edd1 was combined with rev and with kan1 kan2, and the leaf phenotypes were compared to the single mutants. rev mutants have long and narrow leaves and a reduced number of lateral branches (Talbert et al., 1995; Zhong and Ye, 1999; Otsuga et al., 2001) (Fig. 4A, 4B). The edd1 rev double mutant had phenotypes of both single mutants. Leaves of the double mutant were long, narrow, and serrated, had pale margins, and often had an abaxial midvein protrusion (Fig. 4C, 4D). The edd1 rev double mutant had an increased number of leaves relative to rev and many of these leaves were not associated with an axillary branch (Fig. 4E). The edd1 rev mutant also had an abnormal pin-like inflorescence, which produced a small number of flowers as well as radial outgrowths that were interpreted to be rudimentary flowers (Fig. 4F). The double mutant phenotypes indicate that EDD1 and the HD-ZIPIII gene REV have additive interactions in leaf development and synergistic interactions in the inflorescence and suggest that these genes act independently in organ development.
Fig. 4.
edd1 enhanced rev inflorescence phenotypes. (A) edd1. (B) rev. (C, D) edd1 rev showed an additive leaf phenotype including long leaves that had serrations (arrowhead) (C) and abaxial midvein protrusion (D). (E) edd1 enhanced the number of cauline leaves and the number of leaves without branches compared to rev. (F) edd1 rev produced disorganized floral meristems with radial organs. Bars, 2mm (A–C, F).
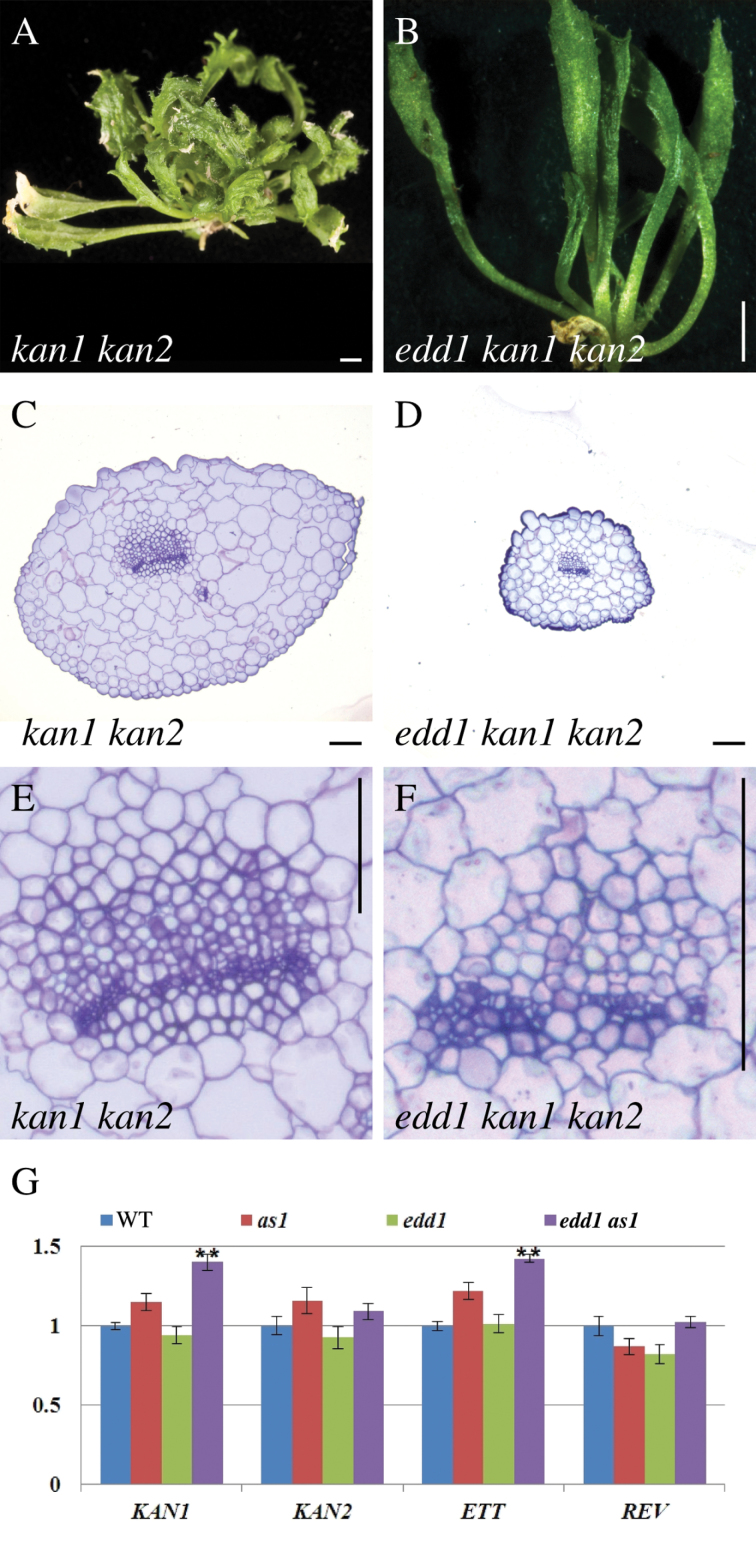
The kan1 kan2 double mutant has small leaves with ectopic leaf lamina protrusions from the abaxial side of the leaf (Eshed et al., 2001) (Fig. 5A). In the edd1 kan1 kan2 triple mutant, leaves were smaller and narrower than kan1 kan2 leaves and had a significant reduction in abaxial ectopic outgrowths (Fig. 5B). To further analyse the effect of edd1 on kan1 kan2 leaf dorsoventral polarity, the morphology of edd1 kan1 kan2 and kan1 kan2 leaves in transverse sections was examined. Leaves in the triple mutants were more rounded than in kan1 kan2, consistent with reduced leaf lamina (Fig. 5C, 5D). Although kan1 kan2 leaves had reduced lamina, the leaf vasculature retained dorsoventral polarity (Fig. 5E). In transverse section, edd1 kan1 kan2 leaf vasculature was less organized than in kan1 kan2 but vasculature retained dorsoventral polarity (Fig. 5F). Together, the edd1 mutation suppressed the kan1 kan2 ectopic abaxial outgrowths and enhanced the leaf lamina expansion defect of kan1 kan2 but otherwise did not affect leaf dorsoventral polarity.
Fig. 5.
edd1 alters the kan1 kan2 leaf phenotype. (A) Rosette of kan1 kan2. (B) Rosette of edd1 kan1 kan2. (C) Transverse section of kan1 kan2 leaf. (D) Transverse section of edd1 kan1 kan2 leaf. (E) Close-up of kan1 kan2 leaf midvein vasculature. (F) Close-up of edd1 kan1 kan2 leaf midvein vasculature. (G) qRT-PCR expression analysis of KAN1, KAN2, ETT, and REV in wild type, as1, edd1, and edd1 as1 (**, P < 0.01). Bars, 2mm (A, B) and 50 µm (C–F).
Genetic interactions between edd1 and the leaf polarity genes, rev and kan1 kan2, indicated that EDD1 may influence leaf adaxial fate via a pathway partly involving these genes. To further test this possibility, transcript levels of KAN1, KAN2, ETT, and REV in the edd1 as1 mutant were compared with wild type and the two single mutants by qRT-PCR (Fig. 5G). Transcript levels of abaxial fate genes, KAN1, KAN2 and ETT, showed no significant changes in the edd1 single mutant compared to wild type and as1. In the edd1 as1 double mutant, KAN2 showed no significant changes compared to wild type and as1, whereas KAN1 and ETT were upregulated in the double mutant. REV transcript levels showed no change in either edd1 as1 or in the single mutants compared to wild type. Expression of dorsoventral polarity genes in edd1 as1 suggests either that the edd1 as1 abaxialized phenotype is due to upregulation of KAN1 and ETT or that upregulation of these genes is a consequence of reduced adaxial fate in edd1 as1 leaves. EDD1 may therefore not act as a canonical dorsoventral polarity gene.
edd1 and scabra3 act synergistically in organ patterning
EDD1 is targeted to plastids and mitochondria, and potentially the adaxial defect in edd1 as1 mutants is due to reduced translation in one or both of these organelles. Some other nuclear-encoded genes encoding plastid-localized proteins required for nonphotosynthetic functions of chloroplasts also disrupt leaf development. One of these, SCABRA3 (SCA3), was used to test whether disruption of plastid transcription affects leaf adaxial fate. SCA3 encodes the protein RpoTp, a nuclear-encoded, plastid-targeted RNA polymerase (Hricova et al., 2006). SCA3 transcribes a subset of plastid genes including genes coding for translational machinery, such as ribosomal proteins and the plastid-encoded DNA polymerase (Hajdukiewicz et al., 1997). Mutations in SCA3 result in reduced plant growth and abnormal leaf morphology, including pale and serrate margins, similar to edd1 (Hricova et al., 2006) (Fig. 6A, 6B). Given the similarity between the leaf phenotypes of sca3 and edd1, the role of SCA3 in leaf development and adaxial fate was examined by analysis of edd1 sca3 and sca3 as1 double mutants. The edd1 sca3 double mutant was greatly reduced in size, and leaves had large regions of pale green and white tissue (Fig. 6C). Most leaves had an abaxial outgrowth at the distal tip of the leaf, which occurred at a higher frequency than in edd1 (Fig. 6D). These abaxial protrusions were not present in sca3. edd1 sca3 mutants also had additional defects in shoot development. The inflorescence formed a pale and translucent pin-like structure with rudimentary growths, which was interpreted to be floral meristems that have failed to develop organs (Fig. 6E). The enhanced phenotype of the edd1 sca3 is consistent with both EDD1 and SCA3 involvement in organ development, and this may reflect the role of both genes in plastid function. To determine whether plastid function is required for adaxial fate, leaf development in the sca3 as1 double mutant was examined. Like edd1 as1, the sca3 as1 double mutant produced trumpet-shaped leaves (Fig. 6F). Together, these results indicate that organelles, particularly plastids, are involved in organ patterning.
Fig. 6.
sca3 interacts with edd1 and as1 in leaf development. (A) Rosette of edd1. (B) Rosette of sca3. (C, D). Leaves of edd1 sca3 showing chlorosis and abaxial midvein protrusion (arrowheads). (E) edd1 sca3 floral meristems were surrounded by radial organs (arrowheads). (F) sca3 as1 produced trumpet-shaped leaves (arrowheads). Bars, 2mm (A–F).
Discussion
EDD1 is essential for plant viability and null mutations in this gene are embryo lethal, which has precluded analysis of EDD1 function in plant development (Uwer et al., 1998). However, edd1-3 described here is a partial loss-of-function mutation that has uncovered a role for EDD1 in leaf morphogenesis. edd1 as1 leaves had trumpet-shaped and radial leaves with reduced adaxial fate. Consistent with EDD1 function in adaxial fate the abaxial genes KAN1 and ETT were upregulated in edd1 as1 plants. edd1 as1 did not alter leaf dorsoventral polarity in the rev mutant, although it is possible that interaction with this class III HD-ZIP gene is masked by redundancy with PHB and PHV. Furthermore edd1 suppressed abaxial outgrowths of kan1 kan2 but reduced the leaf lamina expansion of kan1 kan2. The edd1 kan1 kan2 leaves are reminiscent of kan1 kan2 leaves that have further reduced abaxial fate through loss of KAN3 or YABBY gene function (Eshed et al., 2004). Therefore EDD1 may influence adaxial as well as abaxial fate through the class III HD-ZIP and KANADI pathway. The edd1 leaf has a shape similar to pgy ribosomal protein mutants. However, genetic interactions and temperature sensitivity of edd1 as1 and as1 pgy mutants suggest that EDD1 functions independent of ribosomes in leaf dorsoventral polarity.
Single edd1 mutants had leaves with pronounced marginal serrations. Leaf serrations are formed at regular intervals along the leaf margin at localized points of auxin maxima, which are established by the auxin efflux transporter PINFORMED1 (PIN1) and the transcription factor CUPSHAPED COTYLEDON2 (CUC2) (Kawamura et al., 2010; Bilsborough et al., 2011; Hasson et al., 2011; Byrne, 2012). The more prominent serrations in edd1 may reflect a disruption in auxin gradients mediated by PIN1 and CUC2. edd1 also produced an abaxial outgrowth at the distal tip of the leaf and these outgrowths appeared to be extensions of the midvein. Although the nature and morphogenesis of these outgrowths is unclear, they may reflect a lack of coordination between development of the vasculature and leaf lamina. Similar outgrowths develop on the abaxial side of leaves that are mutant for the two related zinc-finger transcription factor genes JAGGED (JAG) and NUBBIN (NUB) (Dinneny et al., 2004, 2006; Ohno et al., 2004). In jag nub double mutants, these outgrowths have epidermal cells similar to vasculature, consistent with the possibility that they represent vascular growth without lamina growth. JAG and NUB promote growth of lateral organs. During leaf development, JAG expression becomes restricted to the distal region of the leaf, whereas NUB is expressed on the abaxial side of the leaf (Dinneny et al., 2004, 2006; Ohno et al., 2004). Possibly the abaxial outgrowth at the distal tip of the leaf in edd1 is associated with misregulation of JAG and NUB. The leaf distal tip phenotypes edd1 suggests that EDD1 is differentially required along the proximodistal axis of the leaf.
EDD1 encodes a glycyl-tRNA synthetase that is localized to plastids and mitochondria (Uwer et al., 1998; Duchene et al., 2001). A reasonable prediction is that translation in these organelles is disrupted in the edd1 mutant. Other mutants disrupting the nonphotosynthetic functions of chloroplasts or mitochondrial function demonstrate both organelles are required during leaf development. Weak or unstable mutations in a number of genes required for chloroplast function have defects in development of the leaf palisade cell layer. The tomato gene DEFECTIVE CHLOROPLASTS AND LEAVES (DCL) and the closely related Arabidopsis gene AtDCL are nuclear genes encoding plastid-localized proteins involved in ribosomal rRNA processing (Bellaoui et al., 2003; Bellaoui and Gruissem, 2004). In unstable dcl mutants, plastid organization is aberrant and palisade cells fail to expand in mutant sectors of the leaf (Keddie et al., 1996). The Antirrhinum gene DIFFERENTIATION AND GREEENING (DAG), which encodes a protein of unknown function, and the tobacco gene VARIEGATED AND DISTORTED LEAF (VDL), which encodes a predicted plastid RNA helicase, are required for the development of mesophyll cell chloroplasts and for development of the leaf palisade (Chatterjee et al., 1996; Wang et al., 2000). Another factor demonstrating a link between chloroplast function and leaf development is CRUMPLED LEAF (CRL) (Asano et al., 2004). CRL is a novel protein that localizes to the outer envelope of plastids. In the crl mutant, plastids are enlarged and planes of cell division are aberrant in multiple tissues. In addition, mutation of the plastid-encoded ribosomal protein gene rpl36 in tobacco results in slender hyponastic leaves (Fleischmann et al., 2011). Leaf morphology also depends on mitochondrial function. For example, mutations in AtFtsH4, a nuclear-encoded mitochondrial ATP-dependent metalloproteases, have asymmetric leaves with irregular serrations and irregular arrangement of cells in the palisade (Gibala et al., 2009). These mutants have abnormal mitochondria but also have abnormal chloroplasts, so it is unclear whether the leaf defects are a primary or secondary consequence of changes in mitochondrial function.
The correlation between nonphotosynthetic plastid function and leaf development has prompted the suggestion that a plastid-derived signal promotes palisade cell division and expansion (Chatterjee et al., 1996; Keddie et al., 1996). Many plastid-localized proteins are nuclear encoded, and retrograde signalling, from the chloroplast to the nucleus, controls expression of nuclear genes encoding plastid-localized proteins involved in photosynthesis (Nott et al., 2006). Such retrograde signalling may also control the transition from cell proliferation to cell expansion (Andriankaja et al., 2012). Although significant alteration in cell number or size in the leaf blade of edd1 was not detected, it may be possible that small changes in palisade cell division and/or expansion in edd1 and sca3 lead to disruption of the palisade and that, in combination with as1, this results in reduced adaxial fate. One reason for the adaxial palisade mesophyll possibly being more sensitive to signalling from the chloroplast might relate to the duration of cell divisions in this tissue, which is prolonged relative to that of adjacent epidermal cells and spongy mesophyll cell layers (Donnelly et al., 1999). In this case, EDD1 and SCA3 would be acting relatively late in leaf patterning and subsequent to the initial establishment of adaxial fate. Alternatively, the role of plastids in adaxial fate may involve intracellular signalling to determine adaxial fate that is independent of retrograde signalling. EDD1 is likely to be essential for general cellular function. However, some aspects of leaf development have a greater requirement for EDD1 function. Although the mechanism by which EDD1 affects development remains speculative, it is clear that organelle function is critical for leaf morphogenesis.
Supplementary material
Supplementary data are available at JXB online.
Supplementary Fig. S1. Transverse sections of wild-type and edd1 leaves
Supplementary Material
Acknowledgements
The authors thank Catherine Kidner for comments on the manuscript, Andrew Davis for plant photography, Adrian Turner for qRT-PCR advice, José Micol for providing seed, and John Bowman for providing the FIL in situ clone. This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) and strategic funding from the University of Sydney to MEB.
References
- Andriankaja M, Dhondt S, De Bodt S, et al. Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Developmental Cell. 2012;22:64–78. doi: 10.1016/j.devcel.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Asano T, Yoshioka Y, Kurei S, Sakamoto W, Machida Y. A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis . The Plant Journal. 2004;38:448–459. doi: 10.1111/j.1365-313X.2004.02057.x. [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Gruissem W. Altered expression of the Arabidopsis ortholog of DCL affects normal plant development. Planta. 2004;219:819–826. doi: 10.1007/s00425-004-1295-5. [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Keddie JS, Gruissem W. DCL is a plant-specific protein required for plastid ribosomal RNA processing and embryo development. Plant Molecular Biology. 2003;53:531–543. doi: 10.1023/B:PLAN.0000019061.79773.06. [DOI] [PubMed] [Google Scholar]
- Berg M, Rogers R, Muralla R, Meinke D. Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis . The Plant Journal. 2005;44:866–878. doi: 10.1111/j.1365-313X.2005.02580.x. [DOI] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proceedings of the National Academy of Sciences, USA. 2011;108:3424–3429. doi: 10.1073/pnas.1015162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME. Making leaves. Current Opinion in Plant Biology. 2012;15:24–30. doi: 10.1016/j.pbi.2011.10.009. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis . Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis . Development. 2002;129:1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Sparvoli S, Edmunds C, Garosi P, Findlay K, Martin C. DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus . EMBO Journal. 1996;15:4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta . Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. Multienzyme complex of aminoacyl-tRNA synthetases: an essence of being eukaryotic. The Biochemical Journal. 1986;239:249–255. doi: 10.1042/bj2390249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Weigel D, Yanofsky MF. NUBBIN and JAGGED define stamen and carpel shape in Arabidopsis . Development. 2006;133:1645–1655. doi: 10.1242/dev.02335. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Yadegari R, Fischer RL, Yanofsky MF, Weigel D. The role of JAGGED in shaping lateral organs. Development. 2004;131:1101–1110. doi: 10.1242/dev.00949. [DOI] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis . Developmental Biology. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- Duchene AM, Peeters N, Dietrich A, Cosset A, Small ID, Wintz H. Overlapping destinations for two dual targeted glycyl-tRNA synthetases in Arabidopsis thaliana and Phaseolus vulgaris . Journal of Biological Chemistry. 2001;276:15275–15283. doi: 10.1074/jbc.M011525200. [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Current Biology. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. Establishment of polarity in lateral organs of plants. Current Biology. 2001;11:1251–1260. doi: 10.1016/s0960-9822(01)00392-x. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Izhaki A, Baum SF, Floyd SK, Bowman JL. Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development. 2004;131:2997–3006. doi: 10.1242/dev.01186. [DOI] [PubMed] [Google Scholar]
- Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schottler MA, Bock R. Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. The Plant Cell. 2011;23:3137–3155. doi: 10.1105/tpc.111.088906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Current Biology. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Gibala M, Kicia M, Sakamoto W, Gola EM, Kubrakiewicz J, Smakowska E, Janska H. The lack of mitochondrial AtFtsH4 protease alters Arabidopsis leaf morphology at the late stage of rosette development under short-day photoperiod. The Plant Journal. 2009;59:685–699. doi: 10.1111/j.1365-313X.2009.03907.x. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO Journal. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. The Plant Cell. 2011;23:54–68. doi: 10.1105/tpc.110.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Molla-Morales A, Perez-Perez JM, Kojima K, Robles P, Ponce MR, Micol JL, Tsukaya H. Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. The Plant Journal. 2011;65:724–736. doi: 10.1111/j.1365-313X.2010.04457.x. [DOI] [PubMed] [Google Scholar]
- Hricova A, Quesada V, Micol JL. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis . Plant Physiology. 2006;141:942–956. doi: 10.1104/pp.106.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pi L, Liang W, Xu B, Wang H, Cai R, Huang H. The proteolytic function of the Arabidopsis 26S proteasome is required for specifying leaf adaxial identity. The Plant Cell. 2006;18:2479–2492. doi: 10.1105/tpc.106.045013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, Semiarti E, Machida Y, Machida C. Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. The Plant Journal. 2007;51:173–184. doi: 10.1111/j.1365-313X.2007.03132.x. [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant and Cell Physiology. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- Kawamura E, Horiguchi G, Tsukaya H. Mechanisms of leaf tooth formation in Arabidopsis . The Plant Journal. 2010;62:429–441. doi: 10.1111/j.1365-313X.2010.04156.x. [DOI] [PubMed] [Google Scholar]
- Keddie JS, Carroll B, Jones JD, Gruissem W. The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO Journal. 1996;15:4208–4217. [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Arreola A, Gallagher TL, Gasser CS. ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis . Development. 2012;139:1105–1109. doi: 10.1242/dev.067918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. KANADI regulates organ polarity in Arabidopsis . Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–84. doi: 10.1038/nature02366. [DOI] [PubMed] [Google Scholar]
- Kojima S, Iwasaki M, Takahashi H, Imai T, Matsumura Y, Fleury D, Van Lijsebettens M, Machida Y, Machida C. ASYMMETRIC LEAVES2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana . Plant and Cell Physiology. 2011;52:1259–1273. doi: 10.1093/pcp/pcr083. [DOI] [PubMed] [Google Scholar]
- Li H, Xu L, Wang H, Yuan Z, Cao X, Yang Z, Zhang D, Xu Y, Huang H. The Putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. The Plant Cell. 2005;17:2157–2171. doi: 10.1105/tpc.105.033449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. The Plant Cell. 2003;15:2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis . Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411:709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- McHale NA, Koning RE. PHANTASTICA regulates development of the adaxial mesophyll in Nicotiana leaves. The Plant Cell. 2004;16:1251–1262. doi: 10.1105/tpc.019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel GM, Cumberledge S, Johnson MS, Petrella E, Weber BH. The β subunit of E. coli glycyl-tRNA synthetase plays a major role in tRNA recognition. Nucleic Acids Research. 1984;12:4377–4384. doi: 10.1093/nar/12.10.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu Review of Plant Biology. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Ohno CK, Reddy GV, Heisler MG, Meyerowitz EM. The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development. 2004;131:1111–1122. doi: 10.1242/dev.00991. [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. REVOLUTA regulates meristem initiation at lateral positions. The Plant Journal. 2001;25:223–236. doi: 10.1046/j.1365-313x.2001.00959.x. [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. The Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development. 2008;135:1315–1324. doi: 10.1242/dev.016469. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. The Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa S, Ito T, Shimura Y, Okada K. FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems. The Plant Cell. 1999;11:69–86. doi: 10.1105/tpc.11.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. Seed and molecular resources for Arabidopsis . Plant Physiology. 2000;124:1477–1480. doi: 10.1104/pp.124.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. Members of the YABBY gene family specify abaxial cell fate in Arabidopsis . Development. 1999;126:4117–4128. doi: 10.1242/dev.126.18.4117. [DOI] [PubMed] [Google Scholar]
- Szakonyi D, Byrne ME. Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana . The Plant Journal. 2011;65:269–281. doi: 10.1111/j.1365-313X.2010.04422.x. [DOI] [PubMed] [Google Scholar]
- Talbert PB, Adler HT, Parks DW, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana . Development. 1995;121:2723–2735. doi: 10.1242/dev.121.9.2723. [DOI] [PubMed] [Google Scholar]
- Tattersall AD, Turner L, Knox MR, Ambrose MJ, Ellis TH, Hofer JM. The mutant crispa reveals multiple roles for PHANTASTICA in pea compound leaf development. The Plant Cell. 2005;17:1046–1060. doi: 10.1105/tpc.104.029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth MJ, Schimmel P. A mutation in the small (α) subunit of glycyl-tRNA synthetase affects amino acid activation and subunit association parameters. Journal of Biological Chemistry. 1990;265:1005–1009. [PubMed] [Google Scholar]
- Ueno Y, Ishikawa T, Watanabe K, Terakura S, Iwakawa H, Okada K, Machida C, Machida Y. Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis . The Plant Cell. 2007;19:445–457. doi: 10.1105/tpc.106.042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwer U, Willmitzer L, Altmann T. Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. The Plant Cell. 1998;10:1277–1294. doi: 10.1105/tpc.10.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R, Hudson A. phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus . Development. 1995;121:2143–2154. [Google Scholar]
- Waites R, Selvadurai HR, Oliver IR, Hudson A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum . Cell. 1998;93:779–789. doi: 10.1016/s0092-8674(00)81439-7. [DOI] [PubMed] [Google Scholar]
- Wang Y, Duby G, Purnelle B, Boutry M. Tobacco VDL gene encodes a plastid DEAD box RNA helicase and is involved in chloroplast differentiation and plant morphogenesis. The Plant Cell. 2000;12:2129–2142. doi: 10.1105/tpc.12.11.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lin WC, Huang T, Poethig RS, Springer PS, Kerstetter RA. KANADI1 regulates adaxial-abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2 . Proceedings of the National Academy Sciences, USA. 2008;105:16393–16398. doi: 10.1073/pnas.0803997105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Huang H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development. 2003;130:4097–4107. doi: 10.1242/dev.00622. [DOI] [PubMed] [Google Scholar]
- Yang L, Huang W, Wang H, Cai R, Xu Y, Huang H. Characterizations of a hypomorphic argonaute1 mutant reveal novel AGO1 functions in Arabidopsis lateral organ development. Plant Molecular Biology. 2006;61:63–78. doi: 10.1007/s11103-005-5992-7. [DOI] [PubMed] [Google Scholar]
- Yao Y, Ling Q, Wang H, Huang H. Ribosomal proteins promote leaf adaxial identity. Development. 2008;135:1325–1334. doi: 10.1242/dev.017913. [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. The Plant Cell. 1999;11:2139–2152. doi: 10.1105/tpc.11.11.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant and Cell Physiology. 2004;45:369–385. doi: 10.1093/pcp/pch051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.