Abstract
After a conditioning period, seed dormancy in obligate root parasitic plants is released by a chemical stimulus secreted by the roots of host plants. Using Phelipanche ramosa as the model, experiments conducted in this study showed that seeds require a conditioning period of at least 4 d to be receptive to the synthetic germination stimulant GR24. A cDNA-AFLP procedure on seeds revealed 58 transcript-derived fragments (TDFs) whose expression pattern changed upon GR24 treatment. Among the isolated TDFs, two up-regulated sequences corresponded to an abscisic acid (ABA) catabolic gene, PrCYP707A1, encoding an ABA 8'-hydroxylase. Using the rapid amplification of cDNA ends method, two full-length cDNAs, PrCYP707A1 and PrCYP707A2, were isolated from seeds. Both genes were always expressed at low levels during conditioning during which an initial decline in ABA levels was recorded. GR24 application after conditioning triggered a strong up-regulation of PrCYP707A1 during the first 18h, followed by an 8-fold decrease in ABA levels detectable 3 d after treatment. In situ hybridization experiments on GR24-treated seeds revealed a specific PrCYP707A1 mRNA accumulation in the cells located between the embryo and the micropyle. Abz-E2B, a specific inhibitor of CYP707A enzymes, significantly impeded seed germination, proving to be a non-competitive antagonist of GR24 with reversible inhibitory activity. These results demonstrate that P. ramosa seed dormancy release relies on ABA catabolism mediated by the GR24-dependent activation of PrCYP707A1. In addition, in situ hybridization corroborates the putative location of cells receptive to the germination stimulants in seeds.
Abbreviations:
- ABA
abscisic acid
- Abz
abscinazole
- AEC
adenylate energy charge
- AFLP
amplified fragment length polymorphism
- RACE
rapid amplification of cDNA ends
- SL
strigolactone
- TDF
transcript-derived fragment
Key words: ABA, CYP707A inhibitor, parasitic plant, Phelipanche ramosa, seed germination, strigolactone
Introduction
Broomrape species (Orobanche spp. and Phelipanche spp.) are obligate root parasitic plants devoid of chlorophyll that exclusively depend on their hosts for their nutritional needs. Although most broomrape species develop in natural ecosystems with no significant damage on their host plants, some are harmful parasitic weeds in important crops. These pests include Orobanche cumana on sunflower, Orobanche crenata and Orobanche foetida on legumes, and Phelipanche ramosa and Phelipanche aegyptiaca on tomato (Parker, 2009). They all have an extraordinary capacity for dissemination because each individual plant may produce up to 500 000 extremely small seeds (between 200 µm and 400 µm) containing an acotyledoneous reduced embryo (Joel et al., 2011). The weedy life cycle of broomrapes is well-described with regard to its major host plants (Joel et al., 2007). Seed germination is induced by chemical signals exuded in the rhizosphere by host roots and leads to the emergence of a radicle that attaches to the host root surface. Most germination stimulants identified thus far belong to the strigolactone (SL) family (Yoneyama et al., 2010), although dehydrocostus lactone, polyphenols, and isothiocyanates may be involved in the germination of O. cumana (Joel et al., 2011), O. foetida (Evidente et al., 2010), and P. ramosa (Auger et al., 2012), respectively. Whatever the nature of the germination stimulant, several preparatory processes generally take place during a conditioning phase before the response to germination stimulants is possible (Joel et al., 1995). Seed hydration (Joel et al., 2007) and major metabolic pathways are initiated during seed conditioning, which thus displays a characteristic pattern of respiration, protein synthesis, and the utilization of reducing sugars (Bar-Nun and Mayer, 1993, 2002). However, some broomrape species may not require this conditioning phase (Plakhine et al., 2009).
SLs are a novel class of plant hormones involved in controlling shoot branching inhibition (Gomez-Roldan et al., 2008; Umehara et al., 2008). Several studies have investigated the SL signalling pathway in plants as well as the relationships between SLs and other phytohormones during the control of plant architecture. SLs interact with auxin and cytokinins (CK) in bud outgrowth control (Crawford et al., 2010; Dun et al., 2012) or during adventitious root initiation (Rasmussen et al., 2012). In addition, cross-talk can occur between SLs, auxin, and ethylene in the control of root hair elongation (Kapulnik et al., 2011). Finally, an ABC transporter has been shown to be a component of SL transport functioning as a cellular exporter (Kretzschmar et al., 2012). By contrast, although the key role of SLs as germination stimulants has been known for several decades (Cook et al., 1966), almost nothing is known about the early molecular events governing the germination of root parasitic plants in response to SLs, nor about how SLs interact with parasite phytohormones during this process. The isolation of SL receptors has remained elusive and their subcellular location is still unknown despite several structure–activity studies (Zwanenburg et al., 2009). To date, the only systematic analysis of hormonal patterns in parasitic plant seeds was performed in Striga hermonthica, a chlorophyllous parasitic plant, and suggests that gibberellins (GA), abscisic acid (ABA), and CK are involved in seed germination (Toh et al., 2011). In addition, several studies, mainly using pharmacological approaches, have shed light on the hormonal control of the seed response to germination stimulants. The application of GA biosynthesis inhibitors (e.g. uniconazole) during seed conditioning inhibits the subsequent germination of P. ramosa, P. aegyptiaca, and O. minor in response to the germination stimulants, GR24 and strigol (Zehhar et al., 2002; Song et al., 2005; Uematsu et al., 2007) suggesting that broomrape seed germination takes place under the control of GA. Nevertheless, it has been demonstrated that compounds such as uniconazole are also potent inhibitors of ABA catabolism (Saito et al., 2006). Thus, the inhibitory effect of uniconazole leading to persistent seed dormancy may also be due to enhanced levels of ABA in treated seed. Accordingly, exogenous ABA application inhibits P. ramosa seed germination (Zehhar et al., 2002). In addition, 7 d conditioned seeds of O. minor show a massive reduction in ABA levels together with a high release in the medium before responding positively to germination stimulants (Chae et al., 2004). These results suggest that broomrape seeds require a sufficiently low ABA content to germinate upon the application of germination stimulation. Ethylene has also been considered as a potential regulator of broomrape germination since the application of ethylene synthesis inhibitors during seed conditioning inhibits the subsequent germination of P. ramosa seeds in the presence of GR24 (Zehhar et al., 2002).
The molecular response of seeds to SLs has not been well investigated due to the lack of genomic resources in parasitic plants. However, the Parasitic Plant Genome Project (PPGP) has made progress recently and ESTs from key developmental stages of S. hermonthica and P. aegyptiaca have been identified (Westwood et al., 2011). Here, the study starting from a genome-wide expression profiling (cDNA-AFLP) on P. ramosa—a closely related species to P. aegyptiaca—demonstrates the relationships between ABA catabolism and the expression of an ABA catabolism gene, PrCYP707A1, during the initiation of seed germination. The results indicate that PrCYP707A1 may be a major molecular component of the seed response to SLs in a root parasitic plant.
Materials and methods
Plant material and chemical treatments
P. ramosa L. Pomel seeds were collected in 2011 from mature flowering spikes growing on winter oilseed rape (Brassica napus L.) in Saint-Martin-de-Fraigneau, France, and stored at 25 °C in darkness. P. ramosa seeds were surface-sterilized for 5min with 12% sodium hypochlorite and thoroughly rinsed three times with sterile distilled water. Seeds were then suspended in 1 mM Na/K phosphate buffer (pH 7.5) with a ratio of 10mg seeds ml– 1. Seeds were then placed in the dark at 21 °C during the conditioning period. Unless otherwise mentioned, the conditioning period was 7 d. The conditioned seeds were stimulated by adding the synthetic SL GR24 at a final concentration of 10– 9 M in 0.1% acetone. GR24 treatments were carried out at 21 °C in the dark. Corresponding control seeds were treated with 0.1% acetone. After these treatments, seeds were collected by filtration onto a 100 µm nylon mesh, blotted on absorbent paper and weighed. Seeds were then frozen in liquid nitrogen and stored at –80 °C before RNA, ABA or adenylate extraction. ABA 8'-hydroxylase (CYP707A) inhibitors abscinazole-E1 (Abz-E1) and abscinazole-E2B (Abz-E2B) (Okazaki et al., 2011, 2012) were solubilized in acetone and used for germination assays at various concentrations in 0.1% acetone.
Imbibition and adenylate energy charge determination
Seed imbibition was determined as described by Joel et al. (2011). Adenylate Energy Charge (AEC=ATP+0.5 ADP/AMP+ADP+ATP) was determined by quantifying adenine nucleotides extracted from 100mg of seeds, essentially as described by Borisjuk et al. (2007). AMP, ADP, and ATP were separated by high-performance liquid chromatography (HPLC) on an IonPac AS11 column (Dionex Corp., Sunnyvale, CA, USA) and quantification was done using a standard curve of known concentrations.
Germination assays
Seeds were conditioned by suspending around 100 sterilized seeds in 1mM Na/K phosphate buffer (pH 7.5) and distributing them in a 96-well plate (Cell Culture Multiwell Plate Cellstar; Greiner Bio-One, Frickenhausen, Germany), and then stored for 7 d at 21 °C in the dark. Abz-E1, Abz-E2B and/or GR24 solutions were added to each well and volumes were adjusted to 100 µl with sterile distilled water to 0.2% acetone (final concentration). A 0.2% acetone solution was used as a negative control. Subsequently, plates were incubated for 3 d at 21 °C in the dark and germinated seeds were counted under a stereo microscope (Olympus SZX10; Olympus Europa GmbH, Hamburg, Germany). Seeds were considered as germinated when the radicle protruded out of the seed coat. Each germination assay was repeated at least three times.
For the ABA catabolism inhibitors (Abz-E1 and Abz-E2B) and GR24, IC50 and EC50 ±standard errors (SE), respectively, were determined from the dose–response curves [g=f(c), where g is the germination percentage as a function (f) of (c) concentration of the compound tested] and modelled with a four parameter logistic curve on at least three independent dilution ranges. Data were computed with SigmaPlot® V.10.0 (Systat Software Inc., San Jose, CA, USA). An analysis of variance was performed on the results using SigmaPlot version 10.0 (P <0.05; Student–Newman–Keuls test, SNK).
For seed viability tests following the addition of Abz-E1 and Abz-E2B, treated seeds were washed three times with 100 µl of 1mM Na/K phosphate buffer (pH 7.5) after an initial count. Then, 100 µl of 10– 9 GR24 in 0.1% acetone were applied to the washed seeds. Plates were incubated as mentioned above prior to the determination of germination percentage.
cDNA-AFLP analysis
Total RNA was extracted from 100mg of 7 d conditioned seeds (control) and conditioned seeds treated with GR24 for 2h and 6h, using the RNeasy Plant Mini Kit (Qiagen, Courtaboeuf, France). Extracts were treated with DNase I (0.02U µl–1, New England Biolabs, Ipswich, MA, USA). The integrity of total RNA was checked by electrophoresis on a 2% (w/v) agarose gel and RNA was quantified spectrophotometrically (A260/280; NanoDrop Spectrophotometer ND-1000, Labtech International Ltd, Rigmer, UK). Starting from 2 µg of total RNA, the AFLP-based transcript profiling (cDNA-AFLP) was performed as described by Vuylsteke et al. (2007). All 32 possible primer combinations were performed. Selective [γ-33P] ATP-labelled amplification products were separated on 8% polyacrylamide gels with the Model S2001 apparatus (Life Technologies, Paisley, UK). Dried gels were exposed to Biomax film (Sigma Aldrich, St Louis, MO, USA).
Sequence analysis of TDFs
The GR24-regulated TDFs were recovered by PCR under the same conditions used for the pre-amplification. Purified PCR products were sequenced (Eurofins MWG Operon, Ebersberg, Germany) and a similarity search was done with BLASTN and BLASTX sequence alignments against the nucleotide and protein sequences in the available databases from the Parasitic Plant Genome Project (PPGP, http://ppgp.huck.psu.edu/) and The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/). Functional categorization of TDFs was done using the Blast2Go program (www.Blast2GO.de).
Cloning of PrCYP707A cDNAs
Total RNA isolated from 6h GR24-treated conditioned seeds underwent a reverse transcription procedure. cDNAs were synthesized from 0.5 µg of total RNA using the Superscript II Reverse Transcriptase kit (Invitrogen, Carlsbad, CA, USA). Degenerate and specific primers corresponding to highly conserved regions between P. aegyptiaca and S. hermonthica CYP707A sequences were designed. After denaturation at 94 °C for 5min, amplification consisted of 35 cycles of 45 s at 94 °C, 45 s at 55 °C, and 30 s, 45 s or 90 s at 72 °C. A final step of elongation was done at 72 °C for 5min. The amplified DNA fragments were purified and cloned into a pGEM-T Easy vector (Promega, Madison, WI, USA). Recombinant plasmid DNAs were sequenced. Based on these partial CYP707A sequences, new primers were generated for rapid amplification of cDNA ends (RACE) of each fragment using the Generacer kit (Invitrogen). RACE products corresponding to different CYP707A-encoding genes were cloned and sequenced. To amplify the PrCYP707A full-length cDNAs, specific primer pairs were designed. Sequence data from this article can be found in the GenBank/EMBL databanks under accession numbers JQ838174 (PrCYP707A1) and JQ838175 (PrCYP707A2).
Real-time RT-PCR analysis
Total RNA was extracted from 200mg of seeds and DNase I-treated using the same procedure as for the cDNA-AFLP analysis. cDNA was synthesised from 0.5 µg of total RNA using the Superscript II Reverse Transcriptase (Invitrogen) following the manufacturer’s instructions. Then, 5ng of cDNA was used in 25 µl reactions containing 0.3 µM gene-specific primers and 6.25 µl platinum SYBR Green qPCR SuperMix with ROX (Invitrogen). PCR reactions for three biological replicates were performed each in triplicate with a 7300 real-time PCR system according to the manufacturer's protocol (Applied Biosystems, Carlsbad, CA, USA). Fold change in RNA expression was estimated using threshold cycles. The amplicon of the constitutive elongation factor PrEF1α1 (forward, 5'-AGTGCTCAGTGGTGGCTCAAC-3' and reverse, 5'-CTGGAGCAACAACCTTAATCTTC-3'), which showed low cycle threshold (Ct) variation (standard deviation <1 Ct), was used as an internal control to normalize all the data (Péron et al., 2012). A control experiment without cDNA was included for each PCR mix. The following gene-specific primers were used: PrCYP707A1 (forward, 5'-GCCCGCTCTCAAAAGCTAAA-3' and reverse, 5'-TTGTAACAGATTTGGGCTTTTGG-3') and PrCYP707A2 (forward, 5'-TCCTCTTCCCCAAAATGGTTT-3' and reverse, 5'-TTTGGTTTTGGACACATGTTACTCTT-3'). An analysis of variance was performed on the results from qPCR analyses using SigmaPlot version 10.0. Means of three independent RNA isolations were tested at P <0.05 (SNK test).
PrCYP707A1 in situ hybridization
Digoxigenin (DIG)-labelled RNA probes were prepared using an in vitro transcription kit (Riboprobe Combination Systems, Promega, Madison, USA) according to the manufacturer’s instructions. The riboprobes were synthesized from the full-length PrCYP707A1 clone. Antisense and sense probes were transcribed from SP6 or T7 RNA polymerase promoters after linearisation of the vector with ApaI or NdeI, respectively. Samples of conditioned untreated or GR24-treated (6h) seeds were prepared and in situ hybridization experiments were performed as previously described (Péron et al., 2012).
ABA quantification
ABA concentrations in seeds were determined according to Müller and Munné-Bosch (2011) with minor modifications. First, 200mg of seeds were ground in liquid nitrogen and extracted in acetonitrile:water:acetic acid (49.5:49.5:1, by vol.). D6-ABA, [(S)-5-[2H 6](1-hydroxy-2,6,6 -trimethyl-4-oxocyclohex-2-en-1-yl)-3-methyl-(2Z,4E)-pentadienoic acid)] was used as the internal standard and added in all samples (5×10-9 mol) and in non-labelled ABA standard calibration solutions (5×10–9 to 10–5 mol l–1). Analysis were performed on a liquid chromatograph Agilent 1200 series system (Agilent Technologies Inc., Santa Clara, CA, USA) coupled to a LTQ OrbitrapMS (Thermo Fisher Scientific, Waltham, MA, USA). A Hypersil GOLD column (100×2.1mm, 1.9 µm) equipped with a guard column (Phenomenex, Le Pecq, France) was used. Gradient elution was done with water:0.1% acetic acid (solvent A) and acetonitrile (solvent B). The gradient profile was linear and applied as follows: (t (min), % A): (0, 100%), (1, 100%), (10, 0%), (15, 0%), (18, 100%), (20, 100%). The flow rate was maintained constant at 0.5ml min–1. ABA and D6-ABA were ionized in an Atmospheric Pressure Ionization (API) source operated in the negative electrospray mode. Ion characterization was realized at a resolution better than 30 000 (FWHM). A mass accuracy better than 10 ppm was assured for parent ion (ABA: m/z, 263.1277856; rt=6.94±0.01min; D6-ABA: m/z, 269.166543; rt=6.92±0.02min). An analysis of variance was performed on the results from ABA quantification using SigmaPlot version 10.0. Means of six independent metabolite extractions were tested at P <0.05 (SNK test).
Results
GR24 response, imbibition, and energy metabolism of seeds during conditioning
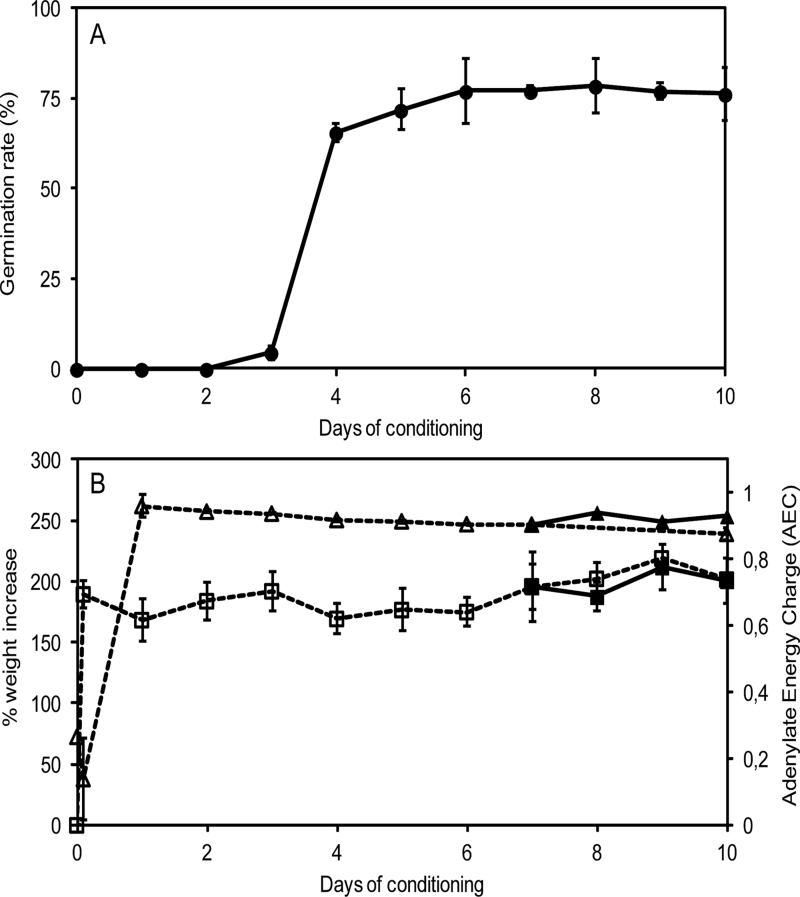
Prior to germination, broomrape seeds require a conditioning period with moist conditions and suitable temperatures to be receptive to germination stimulants (Joel et al., 1995). To evaluate the effect of conditioning period length on the GR24-triggered germination, conditioning periods ranging from 1–10 d were tested before seeds were treated with GR24 (Fig. 1A). Conditioning periods of 4 d or longer led to a statistically equivalent optimal germination response to GR24, ranging from 66±3 to 78±8% (ANOVA, P=0.287), whereas shorter conditioning periods from 0–3 d hampered seed germination. When conditioning was sufficiently long, seed germination, corresponding to the radicle protrusion out of the seed coat, was synchronous and occurred 3 d after adding GR24.
Fig. 1.
Characterization of P. ramosa seed conditioning and germination. (A) Effect of the length of the conditioning period on seed germination. Germination percentages of seeds conditioned for different periods were determined 3 d after the addition of 1nM GR24. Means are values ± SE. (B) Imbibition (squares) and adenylate energy charge (triangles) time-course during conditioning (open symbols and dotted line) and germination (filled symbols and solid line). Seeds were stimulated with 1nM GR24 after a 7 d conditioning period. Means are values ± SE.
Imbibition and AEC of seeds were determined during conditioning (Fig. 1B). P. ramosa seeds were fully imbibed after 1h of soaking (189±5% weight increase) and seed fresh weight remained constant over the next 10 d, in both untreated seeds and seeds treated with GR24 after the 7 d conditioning period. Along with rapid imbibition, a rapid and transient decrease in AEC was observed during the first hour of conditioning. AEC then increased and reached a maximum of 0.9 after 1 d of conditioning and remained constant for the next 9 d, for both GR24-treated seeds and the controls. These results indicate that, as of the first day of the conditioning period, seeds were fully hydrated and not limited in terms of energy metabolism, but unable to respond to GR24 before 4 d of conditioning. A standard 7 d conditioning period was chosen arbitrarily for the subsequent experiments.
Transcriptomic response of P. ramosa seeds to GR24
To investigate the early molecular response of P. ramosa seeds to GR24, the transcriptomic profiles of GR24-treated seeds (2h and 6h treatments) and non-GR24-treated seeds (control) were compared using cDNA-amplified fragment length polymorphism (Vuylsteke et al., 2007). Both GR24-treatment triggered seed germination 3 d later. The 32 primer combinations produced some 2500 TDFs, of which 58 showed an apparent differential expression between samples, including 43 up-regulated genes and 15 down-regulated genes when compared with control levels. Among the 58 sequenced TDFs, 44 showed a significant BLAST hit to sequences found in public databases (Table 1), with 12 TDFs corresponding to unknown proteins. Functional categorization was done using the Blast2GO program. Analysing the 32 annotated sequences according to biological function revealed that, nearly one-third of the sequences (12) encode proteins involved in ‘stress responses’, followed by sequences encoding proteins involved in metabolic processes (9), nucleotide binding (4), oxidation-reduction processes (2), translation (1), transport (1), and, finally, unknown (3). Among the annotated TDFs, two (TDF30 and 37) corresponded to sequences encoding an ABA 8'-hydroxylase (cytochrome P450 CYP707A) that catalyses the C8'-hydroxylation of ABA to 8'-hydroxy-ABA and phaseic acid (Nambara and Marion-Poll, 2005). Compared with the control, both TDFs showed a strong up-regulation in seeds after 2h and 6h of GR24 treatment. Because CYP707A proteins belong to an enzyme family involved in the control of ABA levels during seed dormancy maintenance and breaking (Kushiro et al., 2004; Saito et al., 2004; Okamoto et al., 2006), TDF30 and 37 were selected for further analysis.
Table 1.
List of TDFs modulated in P. ramosa conditioned seeds treated for 2h or 6h with GR24
| TDF no. | Regulationa | Best Arabidopsis hit (Accession no.) | Functional category | E value | |
|---|---|---|---|---|---|
| 2 h | 6 h | ||||
| 1 | + | + | Sulphite reductase (NP_196079) | Oxidation–reduction processes | 2.00E–08 |
| 3 | + | + | 60S ribosomal protein L8-3 (NP_195336) | Translation | 3.00E–12 |
| 5 | + | O | Sucrose synthase 3 (NP_192137) | Carbohydrate metabolic processes | 6.00E–30 |
| 6 | – | – | High mobility group (HMG1/2) domain-containing rotein (NP_565788) | Nucleotide binding | 8.00E–17 |
| 7 | O | – | Ninja-family protein AFP3 (NP_189598) | Nucleotide binding | 7.00E–19 |
| 8 | O | + | Peptidylprolyl isomerase ROF2 (NP_199668) | Response to stress | 6.00E–22 |
| 11 | O | + | Heat shock protein 81.4 (NP_200411) | Response to stress | 3.00E–65 |
| 13 | O | + | PPPDE putative thiol peptidase family protein (NP_187365) | Unknown | 1.00E–24 |
| 14 | O | + | Rossmann-fold NAD(P)-binding domain-containing protein (NP_175552) | Oxidation–reduction processes | 1.00E–16 |
| 15 | + | O | Aldolase-type TIM barrel family protein (AED97862) | Response to stress | 8.00E–15 |
| 16 | + | + | Phosphatidylethanolamine-binding protein (NP_195750) | Unknown | 0.001 |
| 20 | + | O | Trans-cinnamate 4-monooxygenase (NP_180607) | Secondary metabolic processes | 0.006 |
| 26 | + | O | HIPL2 protein (NP_201069) | Carbohydrate metabolic processes | 3.00E–08 |
| 28 | + | + | Methionine synthase 2 (NP_001118564) | Cellular amino acid metabolic processes | 3.00E–21 |
| 30 | + | + | Abscisic acid 8'-hydroxylase 1 (NP_974574 ) | Response to stress | 0.007 |
| 31 | + | O | Acetylornithine deacetylase (NP_001190758) | Protein metabolic processes | 0.091 |
| 32 | – | – | 26S proteasome regulatory subunit 4-A (NP_194633) | Protein metabolic processes | 1 |
| 33 | + | + | RNA recognition motif-containing protein (NP_197436) | Nucleotide binding | 2.00E–23 |
| 36 | O | + | Heat shock 70kDa protein 1 (NP_195870) | Response to stress | 0.073 |
| 37 | + | + | Abscisic acid 8'-hydroxylase 1 (NP_974574) | Response to stress | 0.001 |
| 38 | O | – | BI1-like protein (NP_567466) | Unknown | 3.00E–16 |
| 39 | + | + | Heat shock protein 81-1 (NP_200076) | Response to stress | 3.00E–23 |
| 40 | + | O | Putative aquaporin TIP3-2 (NP_173223) | Transport | 6.00E–50 |
| 41 | + | + | Beta-glucosidase 44 (NP_188436) | Carbohydrate metabolic processes | 5.00E–41 |
| 42 | + | + | 5-methyltetrahydropteroyltriglutamate homocysteine methyltransferase (NP_197294) | Cellular amino acid metabolic processes | 1.00E–16 |
| 44 | O | + | Glutathione S-transferase PM24 (NP_192161) | Response to stress | 6.00E–06 |
| 45 | O | + | Heat shock protein 21 (NP_194497) | Response to stress | 0.02 |
| 48 | + | O | Splicing factor U2af large subunit B (NP_176287) | Nucleotide binding | 1.00E–06 |
| 49 | + | O | Catalase 2 (NP_195235) | Response to stress | 1.00E–48 |
| 50 | + | O | Heat shock protein 70B (NP_173055) | Response to stress | 1.00E–79 |
| 51 | + | O | Phenylalanine ammonia-lyase 3 (NP_001190223) | Response to stress | 6.00E–10 |
| 58 | O | + | Putative xyloglucan glycosyltransferase 8 (NP_180039) | Carbohydrate metabolic processes | 1.00E–26 |
aCompared with the non-treated control sample, (+) corresponds to an up-regulation, (–) a down-regulation, and (O) no change
Molecular cloning of CYP707A homologuesin P. ramosa
Sixty-three and 25 ESTs correspond to putative sequences encoding a cytochrome P450 CYP707A in P. aegyptiaca and S. hermonthica EST libraries (PPGP database), respectively. Using alignment and contig procedures, three full-length cDNA sequences (PaCYP707A1, PaCYP707A2, and PaCYP707A3) were identified in P. aegyptiaca and two full-length (ShCYP707A1 and ShCYP707A2) and one partial (ShCYP707A3) cDNA sequences in S. hermonthica. Based on these sequences, several sets of primers were designed and used in RACE experiments, leading to the identification of two full-length P. ramosa cDNA sequences of 1770 (PrCYP707A1) and 1645bp (PrCYP707A2). Despite several attempts, amplification of a sequence corresponding to PaCYP707A3 was unsuccessful. Possible reasons include that the corresponding gene is not present in P. ramosa or is not expressed at the times tested or exhibits a divergent sequence. The predicted amino-acid sequences of the PrCYP707A1 and PrCYP707A2 genes showed high sequence identity with each other (77.6%), and with AtCYP707A3 (74.1%) and AtCYP707A1 (73.5%), respectively, when compared with the four Arabidopsis CYP707A protein sequences. Sequence comparison did not allow the attribution of Arabidopsis orthologues to both P. ramosa sequences. Both TDF30 and TDF37 corresponded to PrCYP707A1.
Change in PrCYP707A gene expression inP. ramosa seeds after GR24 treatment
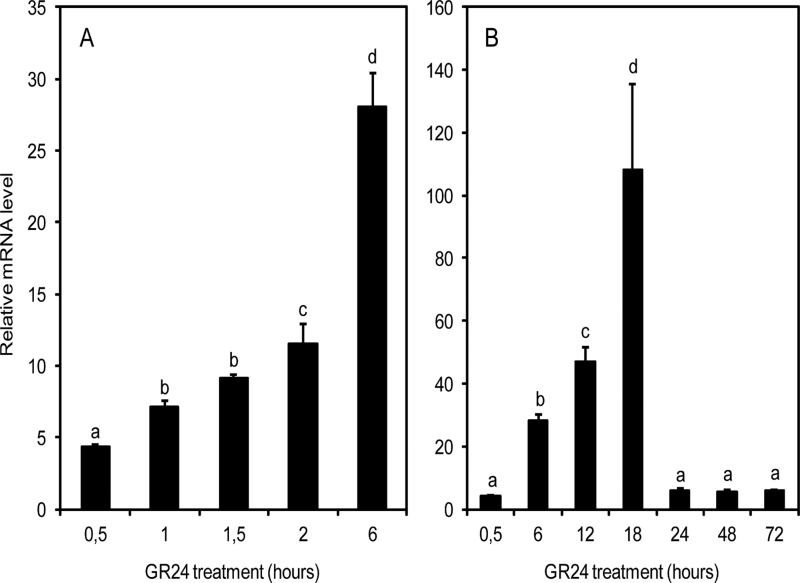
After conditioning, the expression patterns of PrCYP707A1 and PrCYP707A2 were examined in detail during a 3 d GR24 treatment (Fig. 2). While levels of PrCYP707A2 mRNA were low and did not change upon GR24 treatment (data not shown), the expression level of PrCYP707A1 showed a rapid and strong significant increase as of 1h (Fig. 2A) and reached a peak 18h (Fig. 2B) after the addition of GR24. After 24h, PrCYP707A1 mRNA levels dropped, reaching a value similar to that observed the first 30min, and remained stable for the next 48h.
Fig. 2.
Time-course of PrCYP707A1 expression in 7 d conditioned seeds treated with 1nM GR24. (A) Time-course during a GR24 treatment of 6h. (B) Time-course during a GR24 treatment of 3 d. Means are values ±SD (n=6). Means denoted by the same letter do not differ significantly at P <0.05 (SNK test).
PrCYP707A1 is up-regulated by GR24 after a minimum conditioning period
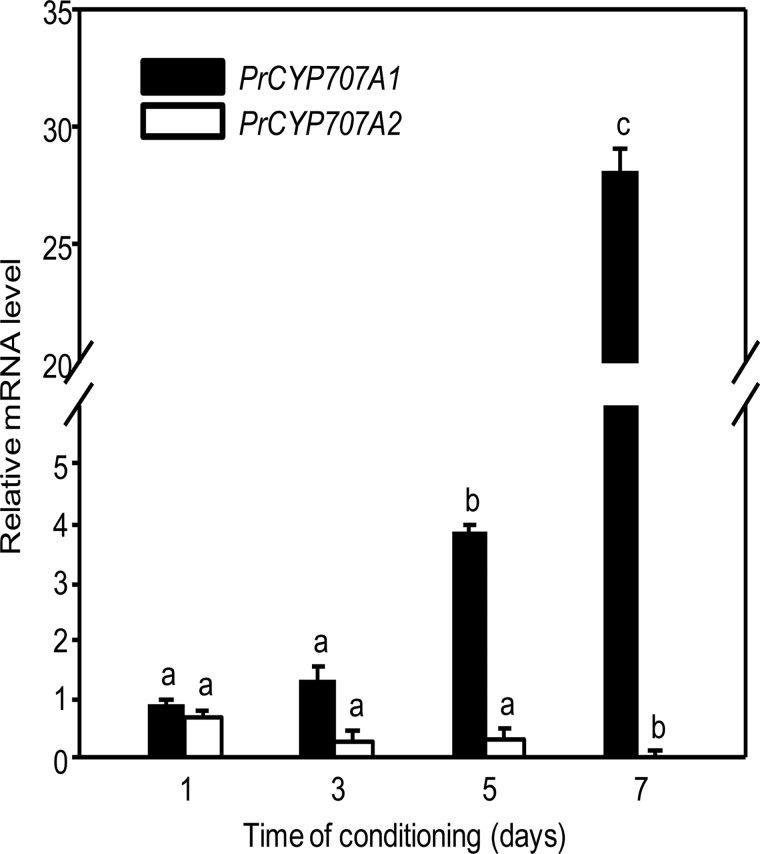
First, the expression patterns of PrCYP707A1 and PrCYP707A2 were examined during conditioning. No change in mRNA accumulation of either gene was recorded (data not shown). Because P. ramosa seeds were receptive to GR24 after a minimum 4 d conditioning period (Fig. 1A), the expression of PrCYP707A1 and PrCYP707A2 were examined in 6h GR24-treated seeds after various conditioning periods (1, 3, 5, and 7 d) (Fig. 3). In seeds that underwent 1 d and 3 d of conditioning, GR24 triggered neither germination nor PrCYP707A1 mRNA accumulation. By contrast, PrCYP707A1 was significantly up-regulated in GR24-stimulated seeds that were conditioned for 5 d and 7 d, with germination percentages of 72±6% and 77±2%, respectively. Whatever the conditioning period, PrCYP707A2 did not exhibit any major change in its expression level.
Fig. 3.
Expression of PrCYP707A1 and PrCYP707A2 in seeds conditioned for 1, 3, 5 or 7 d and then treated for 6h with 1nM GR24. Means are values ± SD (n=6). For each gene, means denoted by the same letter do not differ significantly at P <0.05 (SNK test).
GR24 induces PrCYP707A1 mRNA accumulation in cells close to the micropyle
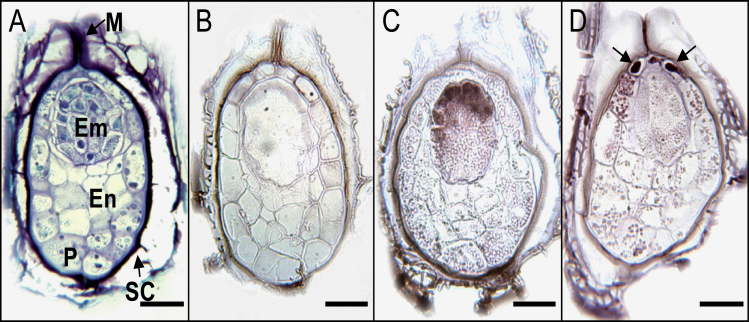
Seeds strongly accumulated PrCYP707A1 transcripts during the first 18h of GR24 treatment (Fig. 2). In situ hybridization experiments were performed on longitudinal sections of 6h GR24-treated seeds to localize this accumulation spatially (Fig. 4). To allow better visualization and identification of the different parts of 6h GR24-treated seeds (cf. Joel et al., 2011), a seed section was stained with toluidine blue (Fig. 4A). Positive hybridization with the specific antisense probe indicated PrCYP707A1 transcript accumulation (Fig. 4C, 4D), whereas no signal was observed after hybridization with the sense probe (Fig. 4B). In 7 d conditioned seeds, transcripts accumulated mainly in the embryo cells facing the micropyle (Fig. 4C). By contrast, PrCYP707A1 mRNA accumulated markedly in the cells near the micropyle in 6h GR24-treated seeds, whereas no staining was detected in the embryo cells (Fig. 4D). According to Joel et al. (2011), these stained cells may correspond to perisperm tissue. These results indicate that GR24 induced a change in the spatial localization of PrCYP707A1 expression in seeds.
Fig. 4.
In situ localization of PrCYP707A1 transcripts in longitudinal sections of P. ramosa seeds. (A) Section stained with toluidine blue for visualization of seed anatomy: M, micropyle; Em, embryo; En, endosperm; P, perisperm; SC, seed coat (Joel et al., 2011). Seeds were conditioned for 7 d (A, B, C) and treated for 6h with 1nM GR24 (D). Sections were hybridized with the sense probe as a negative control (B) and with the antisense probe (C, D). Positive hybridization signals are indicated by brown-violet staining (arrows) using a digoxigenin-labelled RNA immunodetection system. Bars, 50 µm.
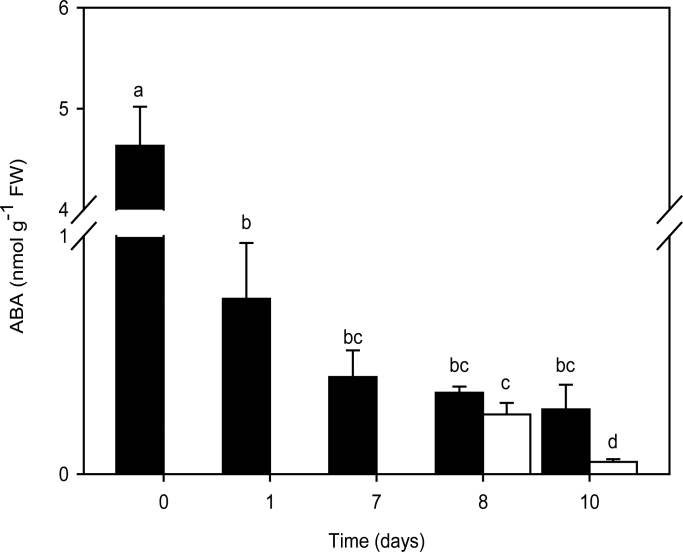
Change in seed ABA levels during conditioning and GR24 treatment
ABA levels in seeds were determined during the 7 d conditioning period and the following 3 d GR24 treatment. A 6.3-fold decrease in ABA content occurred during the first day of conditioning (Fig. 5). Although no significant decrease was observed in seeds during the next 6 d of conditioning, a second, 8-fold drop in ABA content was observed specifically in 3 d GR24-treated seeds compared with 10 d conditioned seeds. Interestingly, this decline in ABA levels in GR24-treated seeds followed the up-regulation of PrCYP707A1 occurring during the first 18h of GR24 treatment (Fig. 2).
Fig. 5.
Change in ABA levels during seed conditioning and GR24-triggered germination over time. Seeds without GR24 treatment (black bars) and with 1nM GR24 added on day 7 of conditioning (white bars). Means are values ± SE. Means denoted by the same letter do not differ significantly at P <0.05 (SNK test).
Abz-E1 and Abz-E2B inhibit GR24-triggered seed germination of P. ramosa
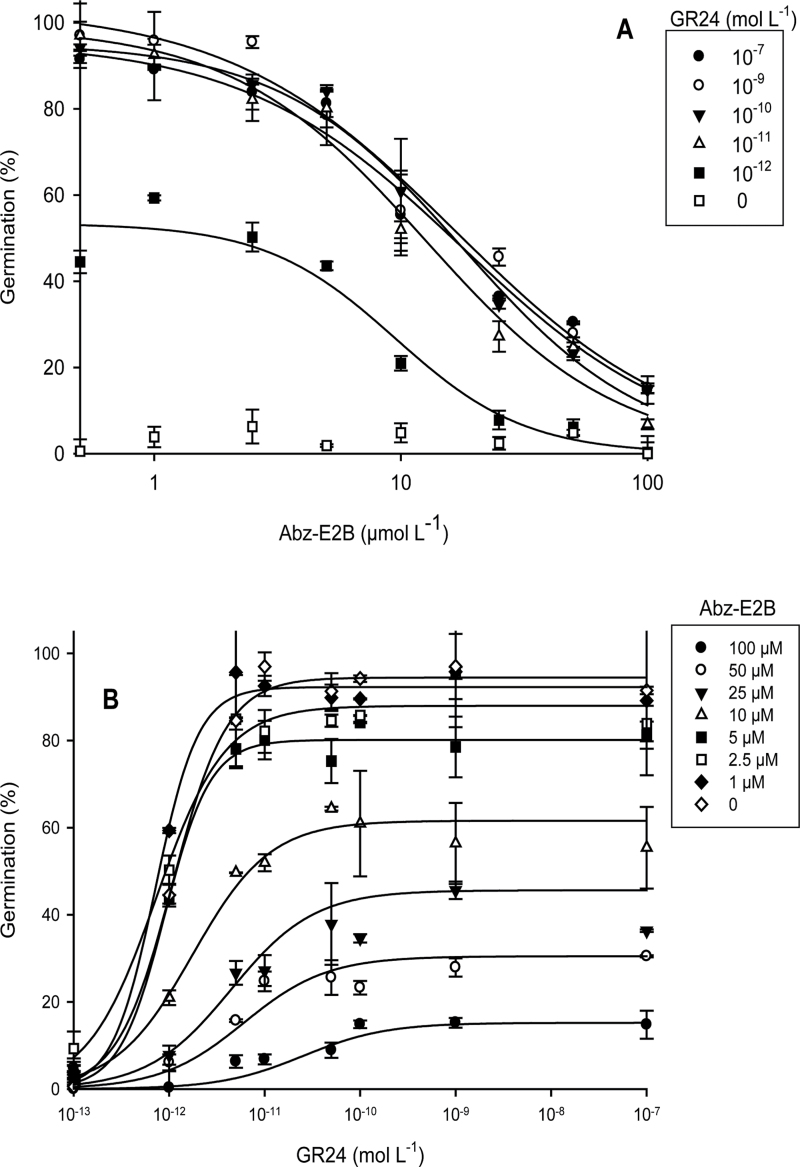
The preponderant role of ABA catabolism in the GR24-dependent germination of P. ramosa seeds was confirmed by using two CYP707A inhibitors, Abz-E1 and Abz-E2B (Okazaki et al., 2011, 2012). When 7 d conditioned seeds were treated for 3 d with 10–9 M GR24 together with Abz-E1 or Abz-E2B at various concentrations, seed germination was inhibited in a concentration-dependent manner. Inhibition was maximal at 100 µM, 84±5% and 90±3% for Abz-E1 or Abz-E2B, respectively, with IC50 reaching 30±16 µM and 17±9 µM, respectively (data not shown). The most effective inhibitor, Abz-E2B was used for further experiments. Using GR24 concentrations ranging from 10-12 to 10-7 M with Abz-E2B concentrations ranging from 10–6 to 10–4 M, no GR24 concentration was able to overcome Abz-E2B inhibition (Fig. 6A). IC50 values ranged insignificantly from 8.3 µM and 17 µM according to GR24 concentration (ANOVA, P=0.877). Germination inhibition was total with 100 µM Abz-E2B and 10–12 M GR24. Increasing concentrations of Abz-E2B did not significantly modify the EC50 of GR24 (values ranging from 0.9nM to 11.3nM; ANOVA, P=0.181) (Fig. 6B). This indicates that Abz-E2B did not interfere with the perception of the germination stimulant. Significant reduction in the maximum germination percentage was observed with Abz-E2B concentrations of 5 µM or higher (ANOVA, P <0.001). To ensure that the Abz-E2B inhibition of germination was not due to seed mortality, treated seeds were subsequently washed with distilled water and treated once again with GR24 in the absence of the inhibitor. Once washed, the seeds showed a germination rate similar to that of untreated seeds (data not shown). Taken together, these results demonstrate that, with regard to germination, Abz-E2B acts as a non-competitive antagonist of GR24 with reversible inhibitory activity.
Fig. 6.
Abz-E2B inhibition of P. ramosa seed germination. (A) Abz-E2B IC50 curve of the concentration response of P. ramosa seed germination at different GR24 concentrations. (B) GR24 EC50 curve of the concentration response of P. ramosa seed germination at different Abz-E2B concentrations. All data are reported as means ± SE.
Discussion
Germination of obligate root parasitic plants is stimulated by the perception of secondary metabolites released from the roots of a potential host plant (Estabrook and Yoder, 1998). Among the germination stimulants identified so far, SLs have been the most extensively studied (Yoneyama et al., 2010). In addition to their capacity to induce broomrape seed germination at nanomolar concentrations, SLs are host recognition signals for symbiotic arbuscular mycorrhizal fungi (Besserer et al., 2006) and constitute new plant hormones that inhibit shoot branching (Gomez-Roldan et al., 2008; Umehara et al., 2008). Although much progress has been made on the signalling activities of SLs in non-parasitic plants, the signalling pathway triggered by SLs leading to broomrape seed germination remains unclear. Here, early transcriptome modifications of P. ramosa seeds triggered by a synthetic SL, the germination stimulant GR24, were studied using an AFLP-based transcript profiling procedure (Vuylsteke et al., 2007). The overall results from the cDNA-AFLP experiments indicate that GR24 does not induce massive modification of the transcriptome because, among the 2500 TDFs visualized on gels, only 58 showed significant differential expression between the control and the 2h and 6h GR24-treated samples. Two of the most distinct TDFs, in terms of expression pattern and biologically significant association with germination, corresponded to a CYP707A gene encoding an ABA 8'-hydroxylase (Table 1). Interestingly, an AtCYP707A gene has been already shown to be up-regulated in Arabidopsis seedlings treated with GR24 (Mashiguchi et al., 2009). Based on both TDF sequences, a search in the Parasitic Plant Genome Project databank uncovered three distinct CYP707A sequences in P. aegyptiaca and S. hermonthica. RACE-PCR strategies revealed two CYP707A full-length cDNAs, named PrCYP707A1 and PrCYP707A2.
ABA is known to play a major role in seed dormancy and germination (Koornneef et al., 2002). Its hormonal action is controlled by a fine-tuned balance between biosynthesis and catabolism (Nambara and Marion-Poll, 2005). Seed dormancy maintenance involves ABA synthesis (Finkelstein et al., 2008) whereas a decrease in ABA content triggered by after-ripening, stratification, and other dormancy-releasing mechanisms promote the germination process in dormant seeds (Gubler et al., 2005). Thus, dormancy release relies mainly on ABA catabolism by specific ABA 8'-hydroxylases encoded by the cytochrome P450 CYP707A family (Kushiro et al., 2004; Saito et al., 2004; Okamoto et al., 2006). ABA 8'-hydrolases catalyse ABA hydroxylation and produce 8'-hydroxy ABA which is then spontaneously isomerized to phaseic acid (Nambara and Marion-Poll, 2005). CYP707A-related sequences have been characterized in many plant species. For instance, four genes encoding CYP707A activity have been identified in Arabidopsis (Kushiro et al., 2004; Saito et al., 2004) and two in barley (Millar et al., 2006). Among the four Arabidopsis sequences, AtCYP707A2 is up-regulated in association with a rapid decrease in ABA content during seed imbibition. Seeds of a cyp707a2 mutant were hyperdormant and accumulated 6-fold higher ABA levels than the wild type (Kushiro et al., 2004; Millar et al., 2006), whereas constitutive expression of AtCYP707A1 in Arabidopsis results in decreased ABA levels in seeds along with dramatically reduced dormancy (Millar et al., 2006). Similarly, ABA content in barley is higher in embryos of after-ripened dormant seeds than of non-dormant seeds in association with higher HvCYP707A1 expression levels in non-dormant compared to dormant seeds (Millar et al., 2006). Altogether, these results highlight the major role of CYP707A genes in regulating the ABA level during seed dormancy and release.
In root parasitic plants, the control of ABA levels is also thought to be involved in the seed germination process (Zehhar et al., 2002; Chae et al., 2004). In the present study, a strong decrease in ABA levels in P. ramosa seeds occurred during the first day of conditioning (Fig. 5). This decrease was maintained to a lesser extent for the next 6 d of conditioning. Interestingly, neither gene, PrCYP707A1 nor PrCYP707A2, exhibited any change in their expression levels during the 7 d conditioning period (data not shown). Based on these results, the decrease in ABA levels in conditioned P. ramosa seeds does not seem to be associated with ABA catabolism, but rather with ABA release in the medium as previously demonstrated in the 1 d conditioned seeds of O. minor (Chae et al., 2004).
The present study shows that GR24 treatment, following conditioning, induced a second decrease in the ABA levels in seeds (Fig. 5). These results suggest that GR24 promotes a sufficient and necessary reduction in ABA content for P. ramosa seed germination. Accordingly, when exogenous ABA is applied during conditioning and GR24 treatments, germination of P. ramosa seeds is inhibited (Zehhar et al., 2002). Here, the second ABA reduction in response to GR24 was associated with the strong and rapid up-regulation of PrCYP707A1 that started as early as 1h after the addition of GR24 and persisted with increasing intensity for 18h (Fig. 2). By contrast, PrCYP707A2 did not show any change in its expression during the 72h GR24 treatment. These findings indicate that conditioned P. ramosa seeds can germinate only after endogenous ABA content reaches a sufficiently low level through CYP707A-dependent catabolism triggered by GR24. The important role of CYP707A genes in the regulation of ABA levels and dormancy release in response to environmental cues has already been investigated in non-parasitic plant: exogenous nitrate and light, two seed dormancy releasing stimuli in Arabidopsis, induce an up-regulation of AtCYP707A2 (Seo et al., 2006; Matakiadis et al., 2009). A PrCYP707A1-dependent release of dormancy in P. ramosa seeds appears to rely on the perception of another environmental cue: the exogenous germination stimulant produced by the host plant. This primordial role of CYP707A1 in P. ramosa seed germination was confirmed by the application of specific inhibitors of ABA 8'-hydroxylase, Abz-E1 and Abz-E2B (Okazaki et al., 2011, 2012) on conditioned seeds together with GR24 (Fig. 6). These inhibitors prevented seed germination. Abz-E1 and Abz-E2B correspond to structural analogues of uniconazole, which has been shown to inhibit the germination of P. ramosa, O. aegyptiaca, and O. minor seeds when applied during conditioning (Zehhar et al., 2002; Song et al., 2005; Uematsu et al., 2007). This study now provides evidence that uniconazole inhibited broomrape seed germination by inhibiting both gibberellin synthesis and ABA catabolism.
To be receptive to germination stimulants, broomrape seeds require a preparatory phase of several days called the conditioning period (Joel et al., 1995). The present study demonstrated that P. ramosa seeds require a minimum of 4 d of conditioning to allow optimal germination in response to GR24 (Fig. 1A). At first glance, this result contradicts a previous study that concluded that P. aegyptiaca, a closely related species, does not need conditioning to respond to germination stimulants (Plakhine et al., 2009). However, non-conditioned P. aegyptiaca seeds stimulated by GR24 only germinate after 7 d, a period that may correspond to a 4 d conditioning period and a 3 d germination process, as reported for P. ramosa seeds here. This study showed that the conditioning period starts with seed imbibition that takes around 1h and optimal AEC (0.9) is reached as of first day of conditioning (Fig. 1B). This rapid imbibition is obtained by water entering the seed through the micropyle which opens after 30min (Joel et al., 2011). Thus, the inability of P. ramosa seeds to respond to GR24 during the first 4 d of conditioning cannot be attributed to a defect in seed hydration nor in energy status of the embryo. An analysis of the expression pattern of PrCYP707A1 in seeds treated for 6h with GR24 demonstrated that PrCYP707A1 expression was not up-regulated until a conditioning period of 5 d or more (Fig. 3). Taken together, these results suggest that P. ramosa seeds do not have physical dormancy and that the minimal conditioning period may correspond to physiological processes resulting in the set-up of the machinery needed for GR24 perception and signalling leading to PrCYP707A1 over-expression.
A recent study on the inheritance of the germination control in P. aegyptiaca seeds suggests that receptors of germination stimulants are located in the living perisperm cells beneath the micropyle (Plakhine et al., 2012). The in situ hybridization experiments support this hypothesis since, upon GR24 treatment, PrCYP707A1 mRNA accumulated rapidly and specifically in similar cells in P. ramosa seeds (Fig. 4). Moreover, such rapid accumulation triggered 1h after GR24 application can be attributed to the location of these cells, which are readily accessible to the germination stimulant entering through the micropyle.
One question that remains is how the germination stimulant activates the signalling pathway leading to rapid transcriptional activation of the PrCYP707A1 gene. Expression of AtCYP707A2 has been shown to be regulated by exogenous nitrate, which releases seed dormancy in Arabidopsis (Matakiadis et al., 2009). The nitrate control of seed dormancy is known to proceed via the production of nitric oxide (NO) (Bethke et al., 2006). In this context, Liu et al. (2009, 2010) demonstrated in Arabidopsis that hydrogen peroxide (H2O2) and NO are involved in the up-regulation of the AtCYP707A2 gene and the subsequent decrease in ABA levels during seed imbibition. Interestingly, the cDNA-AFLP procedure also identified two TDFs (44 and 49) putatively encoding a GST and a catalase in P. ramosa GR24-treated seeds, suggesting that oxidative stress may have occurred upon GR24 stimulation. The possible involvement of NO and H2O2 in SL signalling and PrCYP707A1 activation, as well as the occurrence of oxidative stress during P. ramosa seed germination, are currently under investigation.
The cDNA-AFLP procedure proved to be a powerful tool to identify candidate genes involved in the response of P. ramosa, a non-model plant, to the germination stimulant GR24. In addition to PrCYP707A1, other revealed TDFs may correspond to genes putatively involved in this process (Table 1). For instance, the Arabidopsis genes similar to TDF11-39 and TDF36-50 encode heat shock proteins, HSP90 and HSC70, respectively, known to form a molecular complex that modulates ABA-dependent physiological responses such as stomatal closure and seed germination (Clément et al., 2011). Interestingly, the Arabidopsis proteins peptidylprolyl isomerase ROF1 and ROF2, also called AtFKBP64 and AtFKBP65, corresponding to TDF8, have been shown to bind to HSP90 (Aviezer-Hagai et al., 2007). Similarly, FK506 binding proteins (FKBP) are thought to play a major role in seed germination of sorghum (Sharma and Singh, 2003). Moreover, TDF7 may correspond to a member of a small plant-specific protein family, ABI five binding proteins (AFPs), which interact with the transcription factor ABA-insensitive5 (ABI5), a key regulator of ABA signalling and stress response in Arabidopsis seeds (Garcia et al., 2008). Finally, TDF20 and TDF51 correspond to two sequences encoding a cinnamate 4-hydroxylase (C4H) and a phenyl ammonia lyase (PAL), respectively, involved in phenylpropanoid synthesis. In Arabidopsis, both C4H and PAL2 genes have been shown to be transiently induced in seeds exposed to the germination stimulants karrikins identified in smoke from wildfires (Nelson et al., 2010). Interestingly, karrikins and GR24 signalling require the same F-box protein MAX2 in Arabidopsis germination and shoot branching processes, respectively (Nelson et al., 2011). Thus, all these putative candidate genes deserve further study to investigate their potential implication in P. ramosa seed germination.
In summary, GR24 triggered the dormancy release of P. ramosa seeds by activating a strong and rapid up-regulation of an ABA-catabolic gene PrCYP707A1 that occurs in association with a reduction in ABA levels. However, release from dormancy was shown to require a minimum conditioning period since germination and the activation of PrCYP707A1 expression only occurred 4 d post imbibition. The results on the spatial and temporal expression of PrCYP707A1 corroborate previous studies suggesting that putative receptors of parasitic plant germination stimulants are effective following a conditioning period and are located in the cells between the embryo and the micropyle. How GR24 triggers the ABA decline leading to the P. ramosa seed germination requires further study, as does the possible implication of gibberellins since antagonism between ABA and GA plays a key role in controlling seed germination.
© 2012 The Authors.
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/2.0/uk/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Acknowledgements
This work was supported financially by a PhD fellowship (to M-M. Lechat) and funds from the French Ministry of Education and Research.
References
- Auger B, Pouvreau J-B, Pouponneau K, Yoneyama K, Montiel G, Le Bizec B, Yoneyama K, Delavault P, Delourme R, Simier P. 2012. Germination stimulants of Phelipanche ramosa in the rhizosphere of Brassica napus are derived from the glucosinolate pathway Molecular Plant–Microbe Interactions 25, 993–1004 [DOI] [PubMed] [Google Scholar]
- Aviezer-Hagai K, Skovorodnikova J, Galigniana M, Farchi-Pisanty O, Maayan E, Bocovza S, Efrat Y, von Koskull-Döring P, Ohad N, Breiman A. 2007. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90 Plant Molecular Biology 63 237– 255 [DOI] [PubMed] [Google Scholar]
- Bar-Nun N, Mayer AM. 1993. Preconditioning and germination of Orobanche seeds: respiration and protein synthesis Phytochemistry 34 39– 45 [Google Scholar]
- Bar-Nun N, Mayer AM. 2002. Composition of and changes in storage compounds in Orobanche aegyptiaca seeds during preconditioning Israel Journal of Plant Sciences 50 277– 279 [Google Scholar]
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais J-C, Roux C, Bécard G, Séjalon-Delmas N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biology. 2006;4:e226. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Libourel IGL, Jones RL. 2006. Nitric oxide reduces seed dormancy in Arabidopsis Journal of Experimental Botany 57 517– 526 [DOI] [PubMed] [Google Scholar]
- Borisjuk L, Macherel D, Benamar A, Wobus U, Rolletschek H. 2007. Low oxygen sensing and balancing in plant seeds: a role for nitric oxide New Phytologist 176 813– 823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae SH, Yoneyama K, Takeuchi Y, Joel DM. 2004. Fluridone and norflurazon, carotenoid-biosynthesis inhibitors, promote seed conditioning and germination of the holoparasite Orobanche minor Physiologia Plantarum 120 328– 337 [DOI] [PubMed] [Google Scholar]
- Clément M, Leonhardt N, Droillard M-J, Reiter I, Montillet J-L, Genty B, Laurière C, Nussaume L, Noël LD. 2011. The cytosolic/nuclear HSC70 and HSP90 molecular chaperones are important for stomatal closure and modulate abscisic acid-dependent physiological responses in Arabidopsis Plant Physiology 156 1481– 1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook CE, Whichard LP, Turner B, Wall ME, Egley GH. 1966. Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant Science 154 1189– 1190 [DOI] [PubMed] [Google Scholar]
- Crawford S, Shinohara N, Sieberer T, Williamson L, George G, Hepworth J, Müller D, Domagalska MA, Leyser O. 2010. Strigolactones enhance competition between shoot branches by dampening auxin transport Development 137 2905– 2913 [DOI] [PubMed] [Google Scholar]
- Dun EA, de Saint Germain A, Rameau C, Beveridge CA. 2012. Antagonistic action of strigolactone and cytokinin in bud outgrowth control Plant Physiology 158 487– 498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estabrook EM, Yoder JI. 1998. Plant–plant communications: rhizosphere signaling between parasitic angiosperms and their hosts Plant Physiology 116 1– 7 [Google Scholar]
- Evidente A, Cimmino A, Fernández-Aparicio M, Andolfi A, Rubiales D, Motta A. 2010. Polyphenols, including the new peapolyphenols A–C, from pea root exudates stimulate Orobanche foetida seed germination Journal of Agricultural and Food Chemistry 58 2902– 2907 [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. 2008. Molecular aspects of seed dormancy Annual Review of Plant Biology 59 387– 415 [DOI] [PubMed] [Google Scholar]
- Garcia M, Lynch T, Peeters J, Snowden C, Finkelstein R. 2008. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings Plant Molecular Biology 67 643– 658 [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. 2008. Strigolactone inhibition of shoot branching Nature 455 189– 194 [DOI] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV. 2005. Dormancy release, ABA and pre-harvest sprouting Current Opinion in Plant Biology 8 183– 187 [DOI] [PubMed] [Google Scholar]
- Joel DM, Bar H, Mayer AM, Plakhine D, Ziadne H, Westwood JH, Welbaum GE. 2011. Seed ultrastructure and water absorption pathway of the root-parasitic plant Phelipanche aegyptiaca (Orobanchaceae) Annals of Botany 109 181– 195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel DM, Hershenhorn J, Eizenberg H, Aly R, Ejeta G, Rich PJ, Ransom JK, Sauerborn J, Rubiales D. 2007. Biology and management of weedy root parasites Horticultural Reviews 33 267– 349 [Google Scholar]
- Joel DM, Steffens JC, Matthews DE. 1995. Germination of weedy root parasites. In: Kigel J, Galili G, eds. Seed development and germination New York: Marcel Dekker, Inc; 567– 597 [Google Scholar]
- Kapulnik Y, Resnick N, Mayzlish-Gati E, Kaplan Y, Wininger S, Hershenhorn J, Koltai H. 2011. Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis Journal of Experimental Botany 62 2915– 2924 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L N, Hilhorst H. 2002. Seed dormancy and germination Current Opinion in Plant Biology 5 33– 36 [DOI] [PubMed] [Google Scholar]
- Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E. 2012. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching Nature 483 341– 344 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. 2004. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism EMBO Journal 23 1647– 1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shi L, Ye N, Liu R, Jia W, Zhang J. 2009. Nitric oxide-induced rapid decrease of abscisic acid concentration is required in breaking seed dormancy in Arabidopsis New Phytologist 183 1030– 1042 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ye N, Liu R, Chen M, Zhang J. 2010. H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination Journal of Experimental Botany 61 2979– 2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Sasaki E, Shimada Y, Nagae M, Ueno K, Nakano T, Yoneyama K, Suzuki Y, Asami T. 2009. Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis Bioscience, Biotechnology, and Biochemistry 73 2460– 2465 [DOI] [PubMed] [Google Scholar]
- Matakiadis T, Alboresi A, Jikumaru Y, Tatematsu K, Pichon O, Renou J-P, Kamiya Y, Nambara E, Truong H-N. 2009. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy Plant Physiology 149 949– 960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Jacobsen JV, Ross JJ, Helliwell CA, Poole AT, Scofield G, Reid JB, Gubler F. 2006. Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8'-hydroxylase The Plant Journal 45 942– 954 [DOI] [PubMed] [Google Scholar]
- Müller M, Munné-Bosch S. Rapid and sensitive hormonal profiling of complex plant samples by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Plant Methods. 2011;7:37. doi: 10.1186/1746-4811-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism Annual Review of Plant Biology 56 165– 185 [DOI] [PubMed] [Google Scholar]
- Nelson DC, Flematti GR, Riseborough J-A, Ghisalberti EL, Dixon KW, Smith SM. 2010. Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana Proceedings of the National Academy of Sciences, USA 107 7095– 7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM. 2011. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana Proceedings of the National Academy of Sciences, USA 108 8897– 8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E. 2006. CYP707A1 and CYP707A2, which encode abscisic acid 8'-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis Plant Physiology 141 97– 107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki M, Kittikorn M, Ueno K, Mizutani M, Hirai N, Kondo S, Ohnishi T, Todoroki Y. 2012. Abscinazole-E2B, a practical and selective inhibitor of ABA 8'-hydroxylase CYP707A Bioorganic and Medicinal Chemistry 20, 3162–3172 [DOI] [PubMed] [Google Scholar]
- Okazaki M, Nimitkeatkai H, Muramatsu T, Aoyama H, Ueno K, Mizutani M, Hirai N, Kondo S, Ohnishi T, Todoroki Y. 2011. Abscinazole-E1, a novel chemical tool for exploring the role of ABA 8'-hydroxylase CYP707A Bioorganic and Medicinal Chemistry 19 406– 413 [DOI] [PubMed] [Google Scholar]
- Parker C. 2009. Observations on the current status of Orobanche and Striga problems worldwide Pest Management Science 65 453– 459 [DOI] [PubMed] [Google Scholar]
- Péron T, Véronési C, Mortreau E, Pouvreau J-B, Thoiron S, Leduc N, Delavault P, Simier P. 2012. Role of the sucrose synthase encoding PrSus1 gene in the development of the parasitic plant Phelipanche ramosa L. (Pomel) Molecular Plant–Microbe Interactions 25 402– 411 [DOI] [PubMed] [Google Scholar]
- Plakhine D, Tadmor Y, Ziadne H, Joel DM. 2012. Maternal tissue is involved in stimulant reception by seeds of the parasitic plant Orobanche Annals of Botany 109, 979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plakhine D, Ziadna H, Joel DM. 2009. Is seed conditioning essential for Orobanche germination? Pest Management Science 65 492– 496 [DOI] [PubMed] [Google Scholar]
- Rasmussen A, Mason M, De Cuyper C, et al. 2012. Strigolactones suppress adventitious rooting in Arabidopsis and pea Plant Physiology doi:10.1104/pp.111.187104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M. 2004. Arabidopsis CYP707As encode (+)-abscisic acid 8'-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid Plant Physiology 134 1439– 1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Okamoto M, Shinoda S, et al. 2006. A plant growth retardant, uniconazole, is a potent inhibitor of ABA catabolism in Arabidopsis Bioscience, Biotechnology, and Biochemistry 70 1731– 1739 [DOI] [PubMed] [Google Scholar]
- Seo M, Hanada A, Kuwahara A, et al. 2006. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism The Plant Journal 48 354– 366 [DOI] [PubMed] [Google Scholar]
- Sharma AD, Singh P. 2003. Evidence for the role of FK506-inhibitable peptidyl-prolyl cis/trans isomerase activity in seed germination Current Science 84 142– 143 [Google Scholar]
- Song WJ, Zhou WJ, Jin ZL, Cao DD, Joel DM, Takeuchi Y, Yoneyama K. 2005. Germination response of Orobanche seeds subjected to conditioning temperature, water potential and growth regulator treatments Weed Research 45 467– 476 [Google Scholar]
- Toh S, Kamiya Y, Kawakami N, Nambara E, McCourt P, Tsuchiya Y. 2011. Thermoinhibition uncovers a role for strigolactones in Arabidopsis seed germination Plant and Cell Physiology 53 107– 117 [DOI] [PubMed] [Google Scholar]
- Uematsu K, Nakajima M, Yamaguchi I, Yoneyama K, Fukui Y. 2007. Role of cAMP in gibberellin promotion of seed germination in Orobanche minor Smith Journal of Plant Growth Regulation 26 245– 254 [Google Scholar]
- Umehara M, Hanada A, Yoshida S, et al. 2008. Inhibition of shoot branching by new terpenoid plant hormones Nature 455 195– 200 [DOI] [PubMed] [Google Scholar]
- Vuylsteke M, Peleman JD, van Eijk MJT. 2007. AFLP-based transcript profiling (cDNA-AFLP) for genome-wide expression analysis Nature Protocols 2 1399– 1413 [DOI] [PubMed] [Google Scholar]
- Westwood JH, dePamphilis CW, Das M, Fernandez-Aparicio M, Honaas LA, Timko MP, Wickett NJ, Yoder JI. 2011. The Parasitic Plant Genome Project: new tools for understanding the biology of Orobanche and Striga Weed Science doi:10.1614/WS-D-11-00113.1 [Google Scholar]
- Yoneyama K, Awad AA, Xie X, Yoneyama K, Takeuchi Y. 2010. Strigolactones as germination stimulants for root parasitic plants Plant and Cell Physiology 51 1095– 1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehhar N, Ingouff M, Bouya D, Fer A. 2002. Possible involvement of gibberellins and ethylene in Orobanche ramosa germination Weed Research 42 464– 469 [Google Scholar]
- Zwanenburg B, Mwakaboko AS, Reizelman A, Anilkumar G, Sethumadhavan D. 2009. Structure and function of natural and synthetic signalling molecules in parasitic weed germination Pest Management Science 65 478– 491 [DOI] [PubMed] [Google Scholar]