Abstract
The RecA/RAD51 family of rice (Oryza sativa) consists of at least 13 members. However, the functions of most of these members are unknown. Here the functional characterization of one member of this family, RAD51C, is reported. Knockout (KO) of RAD51C resulted in both female and male sterility in rice. Transferring RAD51C to the RAD51C-KO line restored fertility. Cytological analyses showed that the sterility of RAD51C-KO plants was associated with abnormal early meiotic processes in both megasporocytes and pollen mother cells (PMCs). PMCs had an absence of normal pachytene chromosomes and had abnormal chromosome fragments. The RAD51C-KO line showed no obvious difference from wild-type plants in mitosis in the anther wall cells, which was consistent with the observation that the RAD51C-KO line did not have obviously abnormal morphology during vegetative development. However, the RAD51C-KO line was sensitive to different DNA-damaging agents. These results suggest that RAD51C is essential for reproductive development by regulating meiosis as well as for DNA damage repair in somatic cells.
Key words: DNA damage repair, mitosis, Oryza sativa, reproductive development, synapsis
Introduction
During their lifetimes, all organisms suffer DNA damage, which includes double-strand breaks (DSBs), caused by intercellular events or environmental insults. DSBs can be generated spontaneously during DNA replication in dividing cells. Many endogenous and exogenous DNA-damaging agents, including ionizing radiation, DNA-methylating reagents, oxygen free radicals, and DNA cross-linking reagents, can also cause DSBs. DSB repair is important for genome stability, and eukaryotes are equipped with two major DSB repair pathways: homologous recombination (HR) and non-homologous end-joining (Li and Heyer, 2008; Weterings and Chen, 2008). The former pathway, which repairs DNA with homologous sequences, is a more accurate mechanism, while the latter pathway is an error-prone mechanism. Different organisms use one of the two major repair pathways predominantly to different extents (Bleuyard et al., 2006; Wyman and Kanaar, 2006; Agmon et al., 2009). HR is also important for cell division and generation of genetic diversity.
Many genes are involved in HR. Among these genes, Escherichia coli RecA and its eukaryotic homologue RAD51 have been extensively studied (San Filippo et al., 2008). Sequence and phylogenetic analyses suggest that the two eukaryotic RecA homologues, RAD51 and DMC1, may be generated by the duplication of an ancestral gene derived from the ancestor of eukaryote RecA/RAD51-like genes (Lin et al., 2006). RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3, frequently referred to paralogues of RAD51, are presumably generated from another ancestral gene derived from the ancestor of eukaryote RecA/RAD51-like genes, but they have relatively divergent functions (Lin et al., 2006; San Filippo et al., 2008). In addition, there are four RecA genes which are more similar to eubacterial RecA-like genes than the eukaryotic RAD51-like genes in rice and Arabidopsis (Lin et al., 2006). Proteins encoded by RecA-like and RAD51-like genes share a highly conserved central RecA/RAD51 domain; they are suggested to be evolutionarily related and thus are classified into the RecA/RAD51 family (Lin et al., 2006). The RecA/RAD51 family of eukaryotes includes RAD51, RAD51B, RAD51C, RAD51D, DMC1, XRCC2, XRCC3, and RecA, but not every species has all these members (Lin et al., 2006). Genes of the RecA/RAD51 family play a vital role in HR, HR-dependent DNA repair, and/or other DNA repair processes in both somatic and meiotic cells (Couteau et al., 1999; Osakabe et al., 2002; Bleuyard and White, 2004; Bleuyard et al., 2005; Li et al., 2005; Hamant et al., 2006; Li and Ma, 2006).
Yeast is an excellent model organism for understanding meiosis. Cytological and molecular genetic studies support the idea that some members of the RecA/RAD51 family play an essential role in yeast meiosis, and it is similar in higher eukaryotes. The widely accepted model for recombination in meiosis is the DSB repair model (San Filippo et al., 2008). In yeast meiotic processes, homologous chromosome recombination is initiated by the generation of DSBs. DSBs are catalysed by the conserved topoisomerase-like enzyme SPO11, and the situation is similar in rice (Keeney, 2001; Yu et al., 2010). The RAD51 and RAD51 paralogues with the mediation of many other proteins bind to the single-stranded DNA generated from a DSB; then the nucleoprotein filament searches and attaches to the intact homologue to form a D-loop, which is triple-stranded DNA in which the two strands of a double-stranded DNA molecule are separated by a third strand of DNA (Li and Heyer, 2008). In mammals, some members of the RecA/RAD51 family are essential for the viability, and RAD51 or RAD51C deficiency can result in embryonic lethality (Sonoda et al., 1998; Kuznetsov et al., 2007). Recently, a viable mouse model with a hypomorphic allele of RAD51C was used to investigate the role of RAD51C in meiotic recombination; the results revealed that RAD51C is associated with resolution of the Holliday junction, which is a mobile junction between four strands of DNA, in mice (Kuznetsov et al., 2007).
In plants, the mutants of some RecA/RAD51 family members are sterile but do not show severe irregularities in vegetative growth (Couteau et al., 1999; Bleuyard and White, 2004; Li et al., 2004, 2005; Abe et al., 2005; Osakabe et al., 2005). The Arabidopsis RecA/RAD51 family consists of 11 members (Lin et al., 2006). Some mutants of Arabidopsis RecA/RAD51 family members (RAD51, RAD51C, XRCC3, and DMC1) cause abnormalities in meiosis, including chromosome fragmentation and defective homologue pairing and synapsis (Couteau et al., 1999; Osakabe et al., 2002; Bleuyard and White, 2004; Bleuyard et al., 2005; Li et al., 2005). Knockdown of rice DMC1 leads to abnormal bivalent formation and unequal chromosome segregation in meiosis (Deng and Wang, 2007). Maize (Zea mays) RAD51 is associated with chromosome synapsis and segregation in meiosis (Franklin et al., 2003; Pawlowski et al., 2003).
Some RecA/RAD51 family members are required for efficient HR and/or various types of DNA repair in somatic cells. Mammalian RAD51C, RAD51D, XRCC2, and XRCC3 play roles in DNA damage repair in somatic cells (Kurumizaka et al., 2001, 2002; Sigurdsson et al., 2001; Brenneman et al., 2002; French et al., 2002; Godthelp et al., 2002). RAD51C is needed for Holliday junction resolvase activity in human cells (Liu et al., 2004). In Arabidopsis, some RecA/RAD51 family members, including RAD51, RAD51B, RAD51C, RAD51D, XRCC3, and DMC1, are required for DNA repair, and mutants of these members are hypersensitive to different DNA-damaging agents (Couteau et al., 1999; Osakabe et al., 2002; Bleuyard and White, 2004; Bleuyard et al., 2005; Li et al., 2005; Osakabe et al., 2005; Durrant et al., 2007). Two RAD51 proteins showed ability to bind to single- and double-stranded DNA and strand annealing and strand exchange activities in vitro in physcomitrella moss (Physcomitrella patens; Ayora et al., 2002).
Although RecA/RAD51 family members in various species have been reported to be involved in meiosis and DNA damage repair in somatic cells, not all of them are involved in both biological activities, and the functions of these family members have differentiated during evolution (Lin et al., 2006). For example, disruption of Arabidopsis RAD51B, RAD51D, or XRCC2 does not influence meiosis (Bleuyard et al., 2005; Durrant et al., 2007), but yeast DMC1 only specifically functions in meiosis (Bishop et al., 1992). Mutants of Drosophila XRCC3 (spn-B) and RAD51C (spn-D) do not have defects in DNA repair in somatic cells (Abdu et al., 2003). Mammalian RAD51D is involved in DSBs and is specifically linked with telomere protection in both meiotic and somatic cells (Tarsounas et al., 2004). Rice RAD51 has also been reported to have ATPase activity that is stimulated by non-specific single-stranded DNA in vitro (Rajanikant et al., 2008). Thus it will be possible to understand the RecA/RAD51 family in a species only after elucidating the molecular functions of all members in this family.
The rice genome contains at least 13 RecA/RAD51 family genes based on publications (Lin et al., 2006; Rajanikant et al., 2008). However, only one of these, DMC1, has been functionally analysed in vivo (Deng and Wang, 2007). In this study, the rice RAD51C mutant was characterized. The results show that RAD51C is indispensable for meiosis in both female and male gametocytes, and it also plays a role in DNA damage repair in somatic cells. These results deepen our understanding of meiosis in rice.
Materials and methods
Rice materials and genotoxic treatment
Rice (Oryza sativa) RAD51C T-DNA insertion line 4D-50016, which had the genetic background of japonica (O. sativa ssp. japonica) variety Dongjin, was kindly provided by Professor Gynheung An (Jeong et al., 2006). The genotype of this line was confirmed by PCR amplification using RAD51C-specific primers 39630-m-F and 39630-m-R, and the T-DNA primer RB1 (Supplementary Table S1 available at JXB online). Rice japonica varieties Dongjin and Zhonghua 11 were used in crosses with the homozygous 4D-50016 line.
The genotoxic treatments were performed according to reported procedures (Chang et al., 2009). In brief, surface-sterilized seeds of heterozygous 4D-50016 plants were germinated and grown on half-strength Murashige and Skoog medium supplemented with 0.3% phytagel for 12 d. After genotype assays, the leaf lengths of the RAD51C-knockout (KO) plants and wild-type siblings segregated from the 4D-50016 line were measured, and the plants were transferred to a standard rice culture solution supplemented with different concentrations of mitomycin C (MMC; Sangon, Shanghai, China) or methylmethane sulphonate (MMS; Sigma-Aldrich, St. Louis, MO. USA) for 20 d (Yoshida et al., 1976). After treatment, the lengths of leaves were remeasured. The length difference of each leaf before and after treatment was noted and all the length differences of the leaves within a plant were summed as leaf growth. The averaged leaf growth from eight plants within a treatment was used as the index of treatment sensitivity (Chang et al., 2009). For analysing the sensitivity of rice plants to ultraviolet (UV)-C irradiation, rice plants at the tillering stage were exposed to UV light for 6h at an irradiance of ~0.29 J m−2 s−1 (Chang et al., 2009).
Database searches and phylogenetic analysis
To identify the members of the rice RecA/RAD51 family, the sequences of Arabidopsis RecA/RAD51 family genes were used as queries to search different databases by BLAST analysis (Altschul et al., 1997). The databases searched were the Rice Genome Annotation Project (RGAP, http://rice.plantbiology.msu.edu/) and GenBank (http://www.ncbi.nlm.nih.gov/genbank/). The positions of known motifs of the RecA/RAD51 proteins were determined by Motif Scanning (http://hits.isb-sib.ch). The Molecular Evolutionary Genetic Analysis program (version 3.1, Nei and Kumar, 2000) was used to generate phylogenetic trees by using nthe Neighbor–Joining method (bootstrap, 10 000 replicates).
Phenotypic analyses
To estimate the viability of pollen and anther of RAD51C-KO plants, mature flowers were dissected and stained using I2–KI solution on a glass slide (Alexander, 1969). The stained pollen grains were observed using a light microscope under bright-field conditions. The anther and the developmental progression of embryo sacs were observed by WECLSM (whole-mount eosin B-staining confocal laser scanning microscope) under a Leica SP2 laser scanning confocal microscope (Zeng et al., 2007). Young panicles at the meiosis stage were fixed with Carnoy’s fixative to observe the meiotic chromosome behaviour (Chang et al., 2009).
Gene expression analysis
Total RNA from rice leaves and flowers was used for gene expression analysis. The RAD51C-specific primers rad51c-rt-f and rad51c-rt-r were used to detect the expression of RAD51C (Supplementary Table S1 at JXB online). Primers rad51c-2f and rad51c-2r were used to detect the different transcripts of RAD51C in different tissues (Supplementary Table S1). The expression level of the rice actin gene was used as reference for the mRNA level using gene-specific primers actin-F and actin-R (Supplementary Table S1). The assays were repeated at least twice. When similar results compared with the control were obtained in repeated experiments, only the result in one repetition is presented.
Genetic complementation of the RAD51C-KO mutant
An 8.9kb fragment containing the RAD51C gene and its native promoter was obtained by digesting the Nipponbare bacterial artificial chromosome clone OSJNBa0004A24 (kindly provided by Professor Rod A. Wing of the University of Arizona) using restriction enzymes PstI and SacI (Supplementary Fig. S1 ay JXB online). The DNA fragment was cloned into a pCAMBIA2301 vector. The seeds of heterozygous 4D-50016 T-DNA insertion plants were used to induce the calli. After genotype determination, the homozygous 4D-50016 calli were selected for subculture. The pCAMBIA2301-RAD51C construct and the empty vector pCAMBIA2301 were separately transformed into the selected calli by Agrobacterium-mediated transformation as previously described (Lin and Zhang, 2005). Positive plants were confirmed by PCR amplification using T-DNA and the rice primer pair RB1 and 39630-m-R, and vector and rice primer pair 2301-f and rad51-com-r (Supplementary Table S1).
Statistical analysis
The significant differences between control and RAD51C-KO plants were analysed by the pair-wise t-test installed in the Microsoft Office Excel program.
Results
RAD51C is a single copy gene in the rice genome
Arabidopsis RAD51 family sequences were used to search different databases. This search identified 13 rice RecA/RAD51 family members (Supplementary Table S2 at JXB online), which included RAD51B, RAD51C, RAD51D, XRCC2, XRCC3, DMC1a, DMC1b, RecA1, RecA2, RecA3, and RecA4 named by Lin et al. (2006), and RAD51A1 and RAD51A2 named by Rajanikant et al. (2008). At least six members of this family, RAD51A2, RAD51C, RAD51D, DMC1b, XRCC2, and RecA2, have 2–3 alternatively spliced transcripts based on the database searches (Supplementary Table S2). According to motif and domain prediction of the deduced protein sequences, all the members, except XRCC2, encode a RecA/RAD51 domain (Supplemental Fig. S2). The RecA/RAD51 domain has two conserved consensus motifs, Walker A (also known as the Walker loop or P-loop) and Walker B, which are crucial for nucleotide binding and ATP hydrolysis (Walker et al., 1982; Hanson and Whiteheart, 2005). The consensus sequences are GXXXXGKT/S (‘X’ indicating any amino acid) for Walker A and hhhhDE (‘h’ indicating a hydrophobic amino acid) for Walker B (Walker et al., 1982; Hanson and Whiteheart, 2005). All genes of this family except XRCC2 (which putatively encodes a protein harbouring only Walker A) encode both Walker A and Walker B motifs.
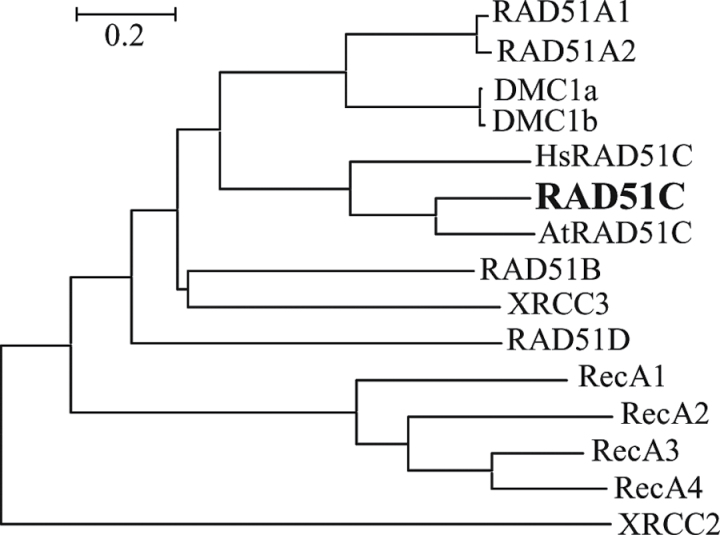
Rice RAD51C (LOC_Os01g39630) was chosen for further functional characterization. Comparative sequence analysis showed that RAD51C is a single copy gene. Phylogenetic analysis revealed that the protein encoded by RAD51C is more closely related to Arabidopsis (AtRAD51C) and human (HsRAD51C) RAD51C than to other rice RecA/RAD51 family proteins (Fig. 1). Rice RAD51C (accession no. BAG87648) has 61.8% and 39.1% sequence identity and 80.8% and 58.9% sequence similarity to AtRAD51C and HsRAD51C, respectively, and has only 5.9–32.0% identity and 10.6–48.9% similarity to other rice RecA/RAD51 family proteins.
Fig. 1.
Phylogenetic relationship of rice RecA/RAD51 family proteins and Arabidopsis (AtRAD51C; accession no. ACA14294) and human (HsRAD51C; AAC39604) RAD51C proteins. For rice RecA/RAD51 family genes encoding multiple proteins, only the sequence of the largest protein is compared with that of other proteins.
Knockout of RAD51C resulted in sterile rice
To study the function of rice RAD51C, a rice T-DNA insertion mutant line 4D-50016 was obtained; this line had a T-DNA inserted into the third exon of RAD51C (POSTECH; http://signal.salk.edu/cgi-bin/RiceGE), with the genetic background of japonica rice variety Dongjin (Supplementary Fig. S1A, B at JXB online). No RAD51C expression was detected in the homozygous 4D-50016 plants (Supplementary Fig. S1C); thus they are referred to as RAD51C-knockout (KO) plants in the following text. Two sequences of alternatively spliced transcripts of rice RAD51C, RAD51C-1 and RAD51C-2, from japonica rice variety Nipponbare were retrieved from GenBank (Supplementary Table S2). RAD51C-1 (GenBank accession no. AK060971) and RAD51C-2 (AK068701) putatively encode proteins consisting of 349 and 309 amino acids, respectively. Comparative analysis of the genomic and cDNA sequences of RAD51C (GenBank accession no. JN394076) in Dongjin identified two alternatively spliced transcripts in both leaf and panicle tissues. The first transcript was RAD51C-1, which had a sequence identical to the cDNA AK060971 from Nipponbare in the coding region (Supplementary Fig. S1A). The second transcript, named RAD51C-3, putatively encoded a protein consisting of 124 amino acids (Supplementary Figs S1A, S3). RAD51C-2 was not detected in the leaf and panicle tissues of Dongjin or in the indica (O. sativa ssp. indica) rice variety Minghui 63. RAD51C-1 and RAD51C-3 were co-expressed in various tissues of Dongjin, including stem, leaf, sheath, pistil, stamen, and spikelet at different developmental stages (including the stages covering meiosis when spikelets were 3–6mm in size), and callus, with RAD51C-1 as the major transcript (Supplementary Fig. S3). Minghui 63 also expressed RAD51C-1 and RAD51C-3. The insertion of T-DNA into RAD51C blocked the expression of both RAD51C-1 and RAD51C-3 in the 4D-50016 line (Supplementary Fig. S1).
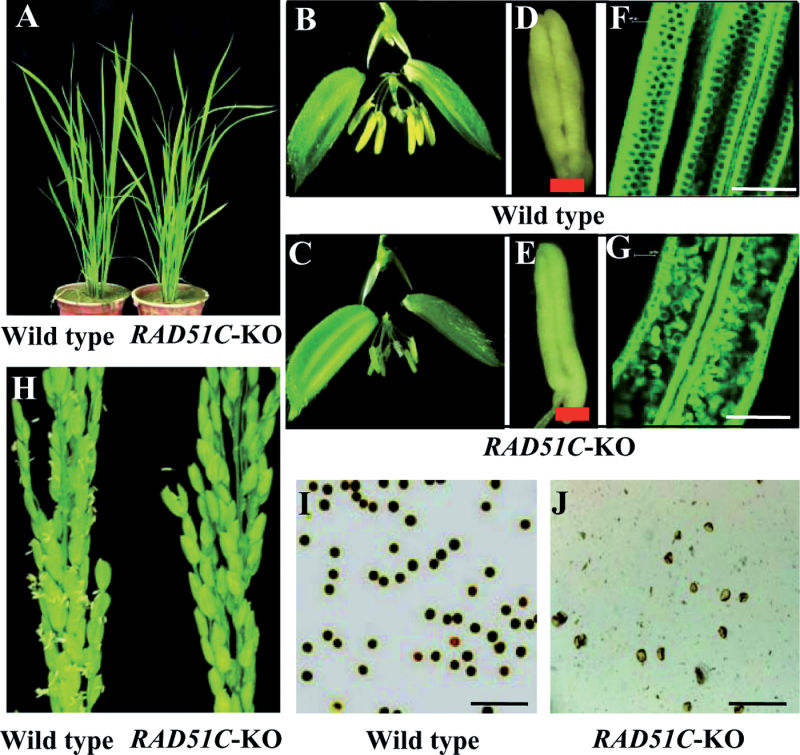
The RAD51C-KO plants did not show any obviously abnormal morphology during vegetative development (Fig. 2A); however, they failed to produce seeds. The general morphology of spikelets of RAD51C-KO plants did not differ from those of wild-type Dongjin except for the anthers (Fig. 2B, 2C). The anthers of wild-type plants were plumper than those of RAD51C-KO plants (Fig. 2D, 2E). The anthers of RAD51C-KO plants were filled with shrivelled pollen, whereas the anthers of wild-type plants were filled with orbicular pollen (Fig. 2F, 2G). Staining pollen with iodium potassium iodide showed that RAD51C-KO plants produced mostly aborted pollen (Fig. 2I, 2J). In addition, RAD51C-KO plants could not release pollen (Fig. 2H). These results suggest that RAD51C-KO plants appear to be male sterile due to abnormal male gametophyte development.
Fig. 2.
Phenotypes of RAD51C-KO plants. Dongjin is the wild type (WT). Bar=150μm. (A) Vegetative development stage. (B and C) Spikelets. (D and E) Anthers. (F and G) Photographs of eosin-B-stained anthers under confocal microscopy. (H) Panicles. (I and J) Iodium potassium iodide-stained pollen.
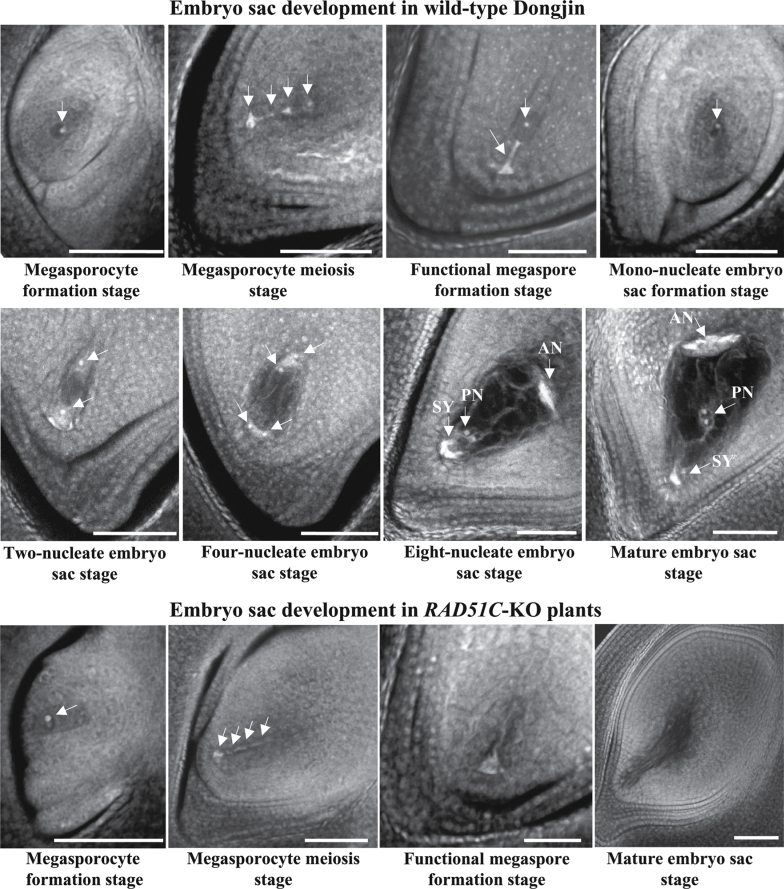
To ascertain whether RAD51C also influences female organ development, embryo sac formation in RAD51C-KO plants was examined. Compared with the processes of embryo sac development in wild-type Dongjin, RAD51C-KO plants were also able to form a megasporocyte in each embryo sac (Fig. 3). However, abnormalities were observed from the megasporocyte meiosis stage to the mature embryo sac stage in RAD51C-KO plants. The four megaspores formed via two meiotic divisions of the megasporocyte showed aberrant alignment in the tetrad of megaspores, and no functional megaspores were formed. This resulted in degenerated embryo sacs at the mature embryo sac stage of wild-type plants. These results suggest that RAD51C-KO plants may also be female sterile due to abnormal meiosis.
Fig. 3.
Development of the embryo sac in RAD51C-KO and wild-type (Dongjin) plants. Arrows indicate the megasporocyte or megaspore. AN, antipodals; PN, polar nucleus; SY, synerids. Bar=50μm.
The inference that RAD51C-KO plants are both male and female sterile is supported by the analysis of reciprocal crosses between RAD51C-KO plants and wild-type Dongjin. No hybrid seed was obtained for 508 and 293 spikelets in three independent plants using RAD51C-KO plants as paternal and maternal recipients in crosses with Dongjin, respectively (Table 1). In contrast, reciprocal crosses between wild-type siblings segregated from the RAD51C-KO line and Dongjin showed that 42% and 60% of spikelets were fertile (Table 1). These results suggest that RAD51C-KO plants are completely female and male sterile.
Table 1.
Reciprocal crosses between RAD51C-KO and wild-type (Dongjin) plants
| Cross combination | No. of seeds/ no. of spikelets | Seed setting rate (%) |
|---|---|---|
| Dongjin (♀)×wild-type siblings segregated from the RAD51C-KO line (♂) | 42/99 | 42 |
| Wild-type siblings segregated from the RAD51C-KO line (♀)×Dongjin (♂) | 57/94 | 60 |
| Dongjin (♀)×RAD51C-KO line (♂) | 0/115 | 0 |
| 0/176 | 0 | |
| 0/217 | 0 | |
| RAD51C-KO line (♀)× Dongjin (♂) | 0/83 | 0 |
| 0/124 | 0 | |
| 0/86 | 0 |
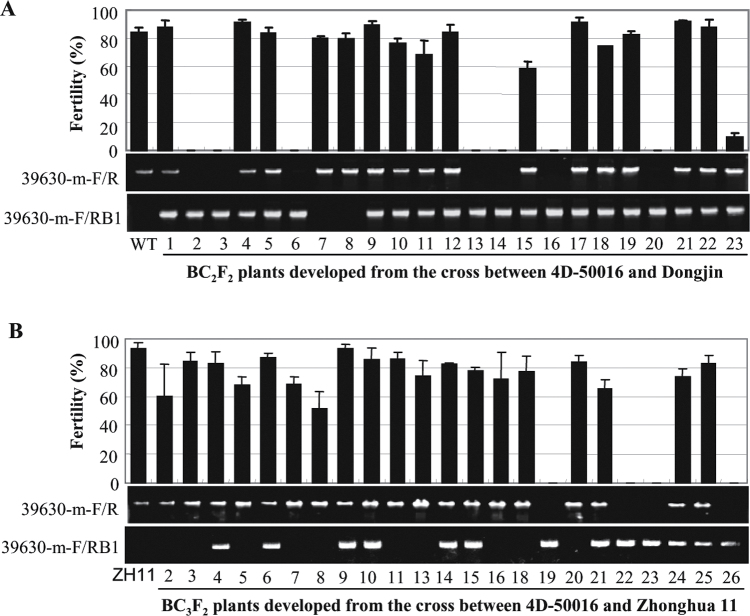
To determine whether the sterile phenotype of RAD51C-KO plants was due to knockout of RAD51C, the T2 generation from three T1 heterozygous T-DNA insertion (4D-50016) plants was analysed. A total of 265 T2 plants were examined for their fertility. Among these plants, 66 plants with homozygous T-DNA insertion in RAD51C were sterile, whereas 141 plants with heterozygous T-DNA insertion in RAD51C and 58 wild-type siblings segregated from the 4D-50016 line were fertile. In order to exclude the possibility that any somatic change caused by tissue culture influenced the fertility of RAD51C-KO plants, a heterozygous 4D-50016 plant was crossed with wild-type Dongjin. An F1 hybrid was backcrossed (BC) with Dongjin twice, selecting for the presence of the insertion, and the progeny were selfed to produce a BC2F2 population consisting of 23 plants. All RAD51C-KO plants (2, 3, 6, 13, 14, 16, and 20) were sterile, but the plants with a heterozygous T-DNA insertion (1, 4, 5, 9–12, 15, 17–19, and 21–23) or without the T-DNA insertion (7 and 8) were fertile (Fig. 4A). A BC3F2 population consisting of 23 plants was also developed from crossing a heterozygous 4D-50016 plant with japonica rice variety Zhonghua 11. Consistent with the results from the BC2F2 population, the sterile phenotype was only associated with RAD51C-KO plants (19, 22, 23, and 26) in the BC3F2 population (Fig. 4B). These results suggest that knockout of RAD51C is associated with sterility in rice.
Fig. 4.
Genotype and fertility of BC2F2 and BC3F2 populations segregated for T-DNA inserted in the RAD51C gene. 39630-m-F and 39630-m-R are RAD51C-specific PCR primers, and RB1 is a T-DNA-specific primer. Plants showing only the band amplified by 39630-m-F and RB1 were RAD51C-KO plants with a homozygous T-DNA insertion. Plants showing only the band amplified by 39630-m-F and 39630-m-R were negative plants without a T-DNA insertion. Plants showing both PCR amplification bands were heterozygous plants with heterozygous T-DNA insertion. The bar represents the mean (three panicles) ±SD. (A) Plant fertility of a BC2F2 population developed from the cross between the RAD51C-T-DNA insertion line (4D-50016) and wild-type (WT) Dongjin. (B) Plant fertility of a BC3F2 population developed from the cross between the RAD51C-T-DNA insertion line and rice variety Zhonghua 11 (ZH11).
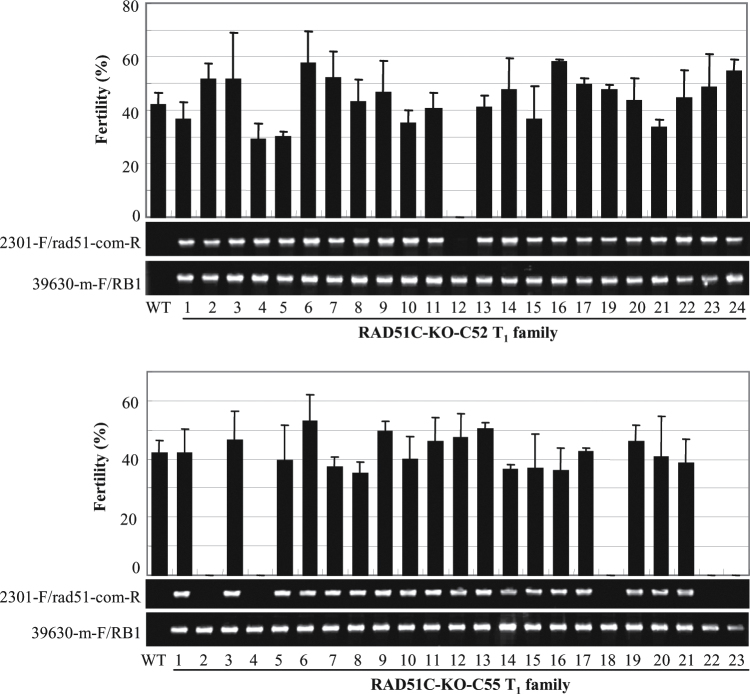
The above inference was further confirmed by genetic complementation analysis of RAD51C-KO plants. RAD51C regulated by its native promoter and empty vector (control) were transformed into RAD51C-KO calli. Sixty independent positive plants transformed with RAD51C, named RAD51C-KO-C, and eight independent positive plants transformed with empty vector, named RAD51C-KO-V, were obtained. The fertility of 48 of the 60 T0 RAD51C-KO-C plants was restored, but all eight RAD51C-KO-V control plants were still sterile (Supplementary Table S3 at JXB online). Plants in two T1 families from two fertile T0 plants (RAD51C-KO-C52 and RAD51C-KO-C55) were further analysed individually for their fertility and the existence of the RAD51C transgene. All the T1 plants carrying the RAD51C transgene were fertile, whereas the negative siblings segregated from the RAD51C-KO-C plants were sterile (Fig. 5). All these results suggest that RAD51C is essential for rice fertility.
Fig. 5.
The fertility of rice plants (RAD51C-KO-C) in two T1 families was associated with the existence of the RAD51C transgene in a RAD51C-KO background. The bar represents the mean (three panicles) ±SD. The vector-specific primer 2301-F and RAD51C promoter-specific primer rad51-com-R were used to examine the existence of the RAD51C transgene. The RAD51C-specific primer 39630-m-F and T-DNA-specific primer RB1 were used to examine the existence of a T-DNA insertion in RAD51C. The RAD51C-KO plant, which was the recipient of the RAD51C transgene, had the genetic background of Dongjin (wild type, WT).
Knockout of RAD51C disrupted meiosis but not mitosis
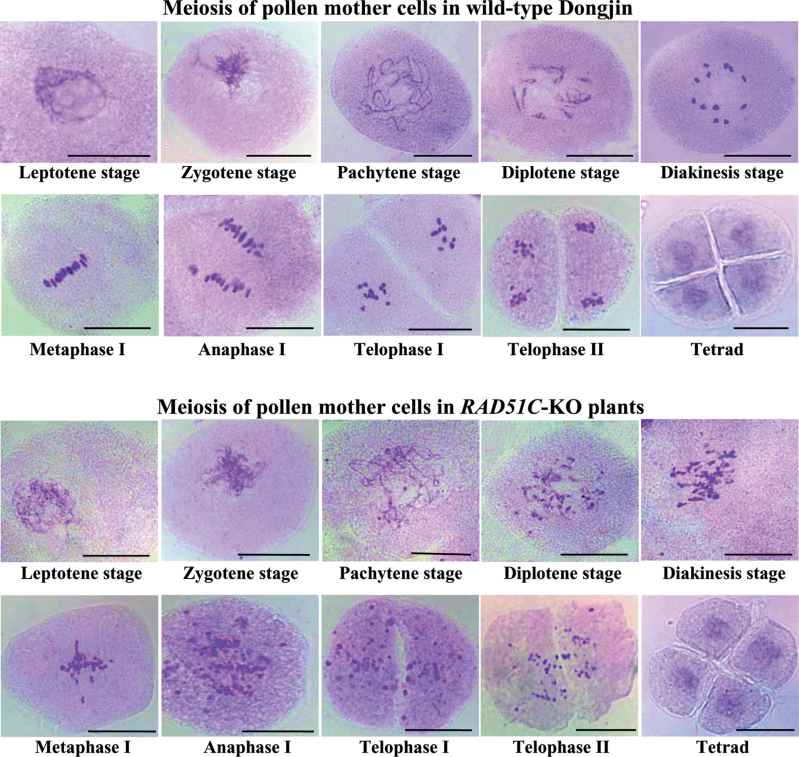
The fact that RAD51C-KO plants had abnormal megasporocyte meiosis (Fig. 3) led to examination of the processes of male gametophyte development. The first meiosis of pollen mother cells (PMCs) begins with chromosome condensation and formation of a thin thread-like structure at the leptotene stage (Ma, 2005). The PMCs in RAD51C-KO and wild-type (Dongjin) plants showed no obvious differences at the leptotene stage and subsequent zygotene stage (Fig. 6). However, abnormalities were observed from the pachytene stage in RAD51C-KO plants compared with wild-type plants. The PMCs of RAD51C-KO plants did not form a complete synaptonemal complex at the pachytene stage and exhibited visible abnormal chromosome fragments from the diplotene stage to telophase II, the end of the second meiosis. Because of these chromosome fragments, the numbers of bivalents in the PMCs of RAD51C-KO plants were difficult to distinguish at the diakinesis stage, at which stage 12 bivalents were clearly observed in the PMCs of wild-type plants. In the metaphase I stage, most of the chromosome fragments were aligned on the division plane of PMCs; however, some fragments were dispersed in the cytoplasm in RAD51C-KO plants. The chromosome fragments of PMCs of RAD51C-KO plants again became obvious at anaphase I and telophase I stages. However, despite these abnormalities in the first meiosis, the PMCs of RAD51C-KO plants could enter into the second meiosis to form tetrads. All these results suggested that RAD51C plays an important role in meiosis.
Fig. 6.
Cytological analysis of meiotic processes of pollen mother cells in RAD51C-KO and wild-type plants. Bar=25μm.
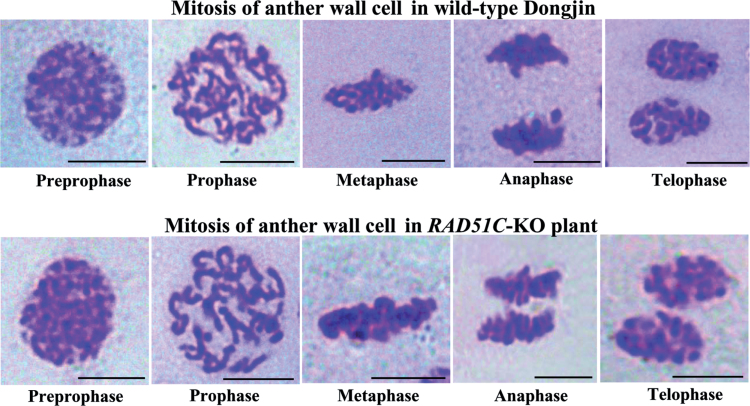
RAD51C was constitutively expressed in different rice tissues (Supplementary Fig. S3 at JXB online). To ascertain whether rice RAD51C also functioned in mitosis, the mitotic processes of anther wall cells in RAD51C-KO plants were examined. The mitotic processes, the pre-prophase, prophase, metaphase, anaphase, and telophase stages, showed no obvious difference from those in the cells of wild-type Dongjin (Fig. 7). In the prophase stage, the chromosomes are normal; no visible chromosome fragments, as seen in meiosis, were observed in the anther wall cells of RAD51C-KO plants. This cytological result is consistent with the observation that RAD51C-KO plants did not have obviously abnormal morphology during vegetative development (Fig. 2A). Thus, RAD51C may not be essential for somatic growth.
Fig. 7.
Cytological analysis of mitotic processes of anther wall cells in RAD51C-KO and wild-type plants. Bar=5μm.
RAD51C-KO plants were sensitive to DNA-damaging agents
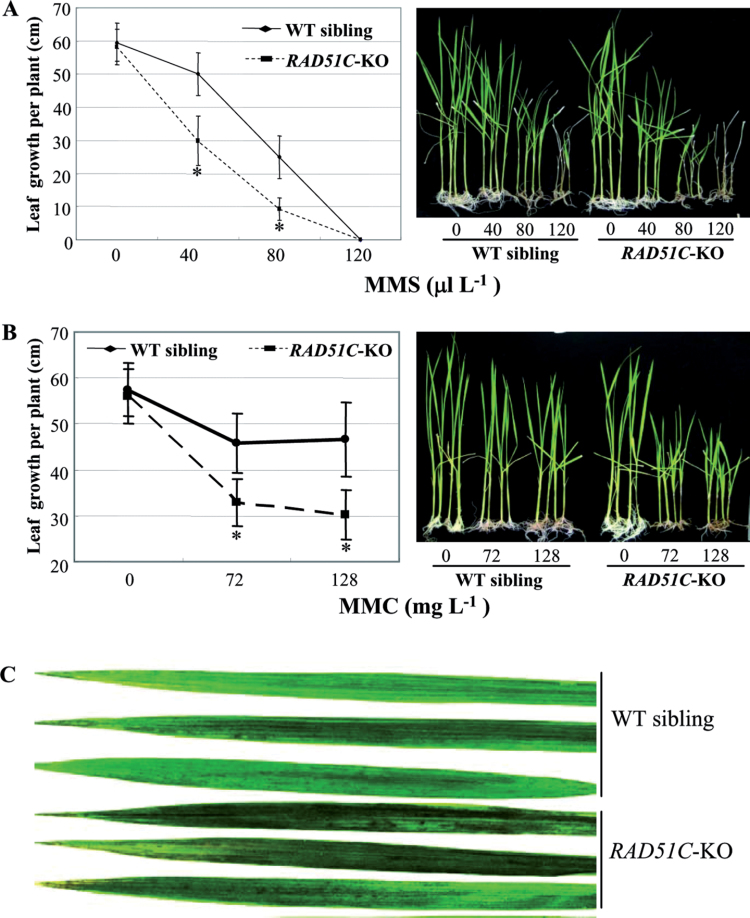
Previous studies have revealed that RAD51 paralogues from species other than rice are required for various types of DNA repair in somatic cells (Kurumizaka et al., 2001, 2002; Sigurdsson et al., 2001; Brenneman et al., 2002; French et al., 2002; Godthelp et al., 2002). To investigate whether RAD51C influenced the cellular response to DNA damage, the sensitivity to the alkylating agent MMS and the cross-linking agent MMC was analysed in RAD51C-KO plants and wild-type siblings (control) segregated from the RAD51C-KO line. An increased concentration of MMS influenced the growth of both RAD51C-KO and control plants; after treatment with 120 µl l−1 MMS, all RAD51C-KO plants died, whereas some of the control plants survived (Fig. 8A). Furthermore, the RAD51C-KO plants grew significantly more slowly (P < 0.0001) than control plants in culture medium supplemented with 40 µl l−1 or 80 µl l−1 MMS. MMC also affected the growth of both RAD51C-KO and control plants (Fig. 8B). However, the growth of the former was significantly slower (P < 0.0005) than that of the latter after treatment with 72mg l−1 or 128mg l−1 MMC. The RAD51C-KO-C plants, which carried the transgene RAD51C in the genetic background of the RAD51C-KO line, were also examined for their responses to MMS and MMC. After MMS and MMC treatments, the RAD51C-KO-C plants showed a similar level of suppressed growth to the wild-type siblings segregated from the RAD51C-KO line (Supplementary Fig. S4 at JXB online).
Fig. 8.
RAD51C-KO plants were hypersensitive to MMS, MMC, and UV-C irradiation. Each data point represents the mean (averaged from eight plants) ±SD. A significant difference between RAD51C-KO plants and wild-type (WT) siblings segregated from the RAD51C-KO line was detected at P < 0.0005 (*). (A) Performance of rice plants after treatment with different concentrations of MMS.(B) Performance of rice plants after treatment with different concentrations of MMC. (C) Rice flag leaves after treatment with UV irradiation.
UV irradiation induces various type of DNA damage (McCready and Marcello, 2003). The RAD51C-KO plants and control wild-type siblings were exposed to UV-C at the tillering stage. Several days after treatment, both types of plants developed necrotic lesions on their leaves (Fig. 8C). However, markedly more lesions were observed in RAD51C-KO plants compared with the control plants. All these results show that the somatic cells of RAD51C-KO plants are more sensitive to DNA-damaging agents than those of control plants, suggesting that RAD51C may play a vital role in DNA repair in somatic cells.
Discussion
Phylogenetic analysis has revealed that rice RAD51C is more closely related to Arabidopsis RAD51C than to other members of the Arabidopsis RecA/RAD51 family (Lin et al., 2006). The present results suggest that rice RAD51C appears to be the functional homologue of Arabidopsis RAD51C in both meiosis of reproductive cells and DNA damage repair of somatic cells. In addition, it has the highest sequence similarity to Arabidopsis RAD51C compared with other rice RecA/RAD51 family proteins.
Rice RAD51C is essential for meiosis
The completely sterile phenotype of RAD51C-KO plants indicates that RAD51C is indispensable for reproductive development. Comparative analysis of the development of PMCs in RAD51C-KO and wild-type plants suggests that RAD51C is required for at least the first meiotic processes of male gametocytes. RAD51C deficiency also resulted in abnormal megaspores that were formed by two meiotic divisions, indicating that RAD51C is also required for the meiosis of female gametocytes. These conclusions are supported by reciprocal crosses between RAD51C-KO and wild-type plants using RAD51C-KO plants as either paternal or maternal recipients, which were unable to produce hybrid seed.
Rice RAD51C is the sequence orthologue of Arabidopsis and mammalian RAD51C genes. Arabidopsis RAD51C is involved in meiosis; its mutant rad51c-1 fails to form synapsis and leads to chromosome fragmentation that results in complete male and female sterility (Bleuyard et al., 2005; Li et al., 2005). Chromosome fragmentation was suppressed in a spo11-1/rad51c-1 double mutant, suggesting that the chromosome fragmentation of the rad51c-1 mutant is related to SPO11-1-generated DSBs (Li et al., 2005). The formation of DSBs initiates meiotic recombination (Grelon et al., 2001). Mouse RAD51C has two distinct functions in meiosis: RAD51-mediated recombination and homologous junction resolution (Kuznetsov et al., 2007). A deficiency of mouse RAD51C leads to early meiotic prophase I arrest in males and precocious separation of sister chromatids at meiotic metaphase II in females. The present results showed that the chromosome behaviour of male gametocytes in RAD51C-KO plants was obviously abnormal from the pachytene stage of the first meiosis. Chromosome fragmentation was observed, which suggests that knockout of RAD51C is leading to broken/unrepaired chromosome. Furthermore, the abnormal pachytene chromosomes suggest that the RAD51C-KO plant may be defective in synapsis and/or in homologue juxtaposition. Since meiotic recombination occurs between the leptotene and zygotene stages (Hamant et al., 2006), these results suggest that rice RAD51C may influence the early meiotic processes prior to the pachytene stage. All these results suggest that rice RAD51C appears to be the functional homologue of Arabidopsis and mammalian RAD51C in meiosis.
RAD51C is also important for somatic DNA repair in rice
In contrast to mammalian RAD51C (Sonoda et al., 1998; Kuznetsov et al., 2007), the present results suggest that rice RAD51C may not be essential for vegetative development under normal growth conditions. Arabidopsis RAD51B, RAD51C, and RAD51D are also not essential for vegetative growth (Li et al., 2005; Osakabe et al., 2005; Durrant et al., 2007). However, like some of its orthologues in other species, rice RAD51C appears to be involved in maintaining DNA stability in somatic cells following exposure to DNA-damaging agents. Human RAD51C plays an important role in DNA repair in somatic cells (Somyajit et al., 2010). Human RAD51C is involved in processing of branch migration and homologous junctions (Liu et al., 2004). Haploinsufficiency of hamster RAD51C causes increased sensitivity to DNA damage (Smeenk et al., 2010). Arabidopsis RAD51C is also involved in DNA damage repair in somatic cells caused by cross-linking reagents (Abe et al., 2005; Bleuyard et al., 2005). Interestingly, atrad51c seedlings are hypersensitive to γ-irradiation (Abe et al., 2005), which is a direct DSB inducer, whereas imbibed atrad51c seeds are not sensitive to γ-irradiation compared with the wild type (Bleuyard et al., 2005). The different responses of the Arabidopsis RAD51C mutant to γ-irradiation at different developmental stages could be due to different DNA repair pathways being involved. Both HR and non-homologous end-joining contribute to DSB repair. HR-mediated repair is associated with DNA replication during synthesis and gap 2 phases of the cell cycle (Richardson et al., 2000). It is suggested that the cells of imbibed seeds are in the gap 1 phase; the role of AtRAD51C in DNA damage repair caused by γ-irradiation may not be notable in the imbibed seeds (Abe et al., 2005; Bleuyard et al., 2005). However, Drosophila RAD51C does not appear to be involved in DSB repair in somatic cells, although it is specifically required during meiosis (Abdu et al., 2003). The present results suggest that rice RAD51C also appears to be the functional homologue of Arabidopsis RAD51C in somatic DNA repair caused by DNA-damaging agent.
The mechanisms of repairing DNA damage caused by the alkylating reagent MMS, the cross-linking reagent MMC, and UV irradiation are different (Chang et al., 2009). MMS methylates DNA on N7-deoxyguanine and N3-deoxyadenine (Vazquez et al., 2008), while MMC causes interstrand DNA cross-linking (Lehoczky et al., 2007). UV irradiation induces direct DNA damage, including strand breakage, thymine dimers, and other photoproducts, or indirect damage through reactive oxygen species (McCready and Marcello, 2003). In yeast, the damage caused by MMS can be repaired by base excision repair, HR, and DNA damage tolerance pathways, together with a functional synthesis-phase checkpoint (Vazquez et al., 2008). In addition, MMS can cause derived DSBs and this DSB repair is reliant on the HR gene RAD51 (Ma et al., 2011). There are two main pathways to eliminate the damage of an interstrand cross-linking agent: nucleotide excision and HR repair (Lehoczky et al., 2007). Nucleotide excision repair, base excision repair, HR-dependent repair, and other DNA repair pathways can repair the damage caused by UV irradiation (Kimura et al., 2004). Furthermore, a recent study reported that UV irradiation is associated with DSB repair (Yang et al., 2008).
Arabidopsis RAD51C can repair the DSBs caused by MMC and another DNA cross-linking reagent cisplatin (Abe et al., 2005; Bleuyard et al., 2005). Hypersensitivity to cross-linking agents is a consistent character of HR in deficient mutants described in plant and vertebrate cell lines (Liu et al., 1998; Takata et al., 2000; Sasaki et al., 2004; Abe et al., 2005). As discussed above, the HR-dependent pathway is also involved in repair of DNA damage caused by other agents. The sensitivity of RAD51C-KO plants to MMS, MMC, and UV-C may be because the plants were defective in HR or in both HR and other DNA repair pathways. Since RAD51C-KO plants may have abnormal homologue juxtaposition and synapsis, which was associated with HR, during meiosis, further study is required to ascertain whether RAD51C contributes to DNA repair in somatic cells by HR.
Supplementary Material
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The RAD51C gene and RAD51C-knockout mutant 4D-50016.
Figure S2. The schematic diagram of the domain structures of rice RecA/RAD51 family proteins.
Figure S3. Expression of transcripts RAD51C-1 and RAD51C-3 in rice variety Dongjin.
Figure S4. Plant responses to methylmethane sulphonate (MMS) and mitomycin C (MMC).
Table S1. PCR primers used for construction of vectors and gene structure and expression analyses.
Table S2. Rice RecA/RAD51 gene family.
Table S3. Genetic complementation of the RAD51C-KO line.
Acknowledgements
This work was supported by grants from the National Program of High Technology Development of China (2012AA10A303) and the National Program of Transgenic Variety Development of China (2011ZX08009-003).
References
- Abdu U, Gonzalez-Reyes A, Ghabrial A, Schupbach T. 2003. The Drosophila spn-D gene encodes a RAD51C-like protein that is required exclusively during meiosis Genetics 165 197– 204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe K, Osakabe K, Nakayama S, Endo M, Tagiri A, Todoriki S, Ichikawa H, Toki S. 2005. Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis Plant Physiology 139 896– 908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agmon N, Pur S, Liefshitz B, Kupiec M. 2009. Analysis of repair mechanism choice during homologous recombination Nucleic Acids Research 37 5081– 5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander MP. 1969. Differential staining of aborted and nonaborted pollen Stain Technology 44 117– 122 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs Nucleic Acids Research 25 3389– 3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayora S, Piruat JI, Luna R, Reiss B, Russo VE, Aguilera A, Alonso JC. 2002. Characterization of two highly similar Rad51 homologs of Physcomitrella patens. Journal of Molecular Biology 316 35– 49 [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. 1992. DMC1: a meiosis-specific yeast homolog of E.coli recA required for recombination, synaptonemal complex formation, and cell cycle progression Cell 69 439– 456 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, Savigny F, White CI. 2005. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair The Plant Journal 41 533– 545 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI. 2006. Recent advances in understanding of the DNA double-strand break repair machinery of plants DNA Repair (Amsterdam) 5 1– 12 [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, White CI. 2004. The Arabidopsis homolog of Xrcc3 plays an essential role in meiosis EMBO Journal 23 439– 449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneman MA, Wagener BM, Miller CA, Allen C, Nickoloff JA. 2002. XRCC3 controls the fidelity of homologous recombination: roles for XRCC3 in late stages of recombination Molecular Cell 10 387– 395 [DOI] [PubMed] [Google Scholar]
- Chang Y, Gong L, Yuan W, Li X, Chen G, Li X, Zhang Q, Wu C. 2009.. Replication protein A (RPA1a) is required for meiotic and somatic DNA repair but is dispensable for DNA replication and homologous recombination in rice Plant Physiology 151 2162– 2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP. 1999. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis The Plant Cell 11 1623– 1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng ZY, Wang T. 2007. OsDMC1 is required for homologous pairing in Oryza sativa. Plant Molecular Biology 65 31– 42 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Wang S, Dong X. 2007. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response Proceedings of the National Academy of Sciences, USA 104 4223– 4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AE, Golubovskaya IN, Bass HW, Cande WZ. 2003. Improper chromosome synapsis is associated with elongated RAD51 structures in the maize desynaptic2 mutant Chromosoma 112 17– 25 [DOI] [PubMed] [Google Scholar]
- French CA, Masson JY, Griffin CS, O’Regan P, West SC, Thacker J. 2002. Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability Journal of Biological Chemistry 277 19322– 19330 [DOI] [PubMed] [Google Scholar]
- Godthelp BC, Wiegant WW, van Duijn-Goedhart A, Scharer OD, van Buul PP, Kanaar R, Zdzienicka MZ. 2002. Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability Nucleic Acids Research 30 2172– 2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G. 2001. AtSPO11-1 is necessary for efficient meiotic recombination in plants EMBO Journal 20 589– 600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O, Ma H, Cande WZ. 2006. Genetics of meiotic prophase I in plants Annual Review of Plant Biology 57 267– 302 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. 2005. AAA+ proteins: have engine, will work Nature Reviews Molecular Cell Biology 6 519– 529 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Park S, et al. 2006. Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice The Plant Journal 45 123– 132 [DOI] [PubMed] [Google Scholar]
- Keeney S. 2001. Mechanism and control of meiotic recombination initiation Current Topics in Developmental Biology 52 1– 53 [DOI] [PubMed] [Google Scholar]
- Kimura S, Tahira Y, Ishibashi T, Mori Y, Mori T, Hashimoto J, Sakaguchi K. 2004. DNA repair in higher plants; photoreactivation is the major DNA repair pathway in non-proliferating cells while excision repair (nucleotide excision repair and base excision repair) is active in proliferating cells Nucleic Acids Research 32 2760– 2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurumizaka H, Ikawa S, Nakada M, Eda K, Kagawa W, Takata M, Takeda S, Yokoyama S, Shibata T. 2001. Homologous-pairing activity of the human DNA-repair proteins Xrcc3·Rad51C Proceedings of the National Academy of Sciences, USA 98 5538– 5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurumizaka H, Ikawa S, Nakada M, Enomoto R, Kagawa W, Kinebuchi T, Yamazoe M, Yokoyama S, Shibata T. 2002. Homologous pairing and ring and filament structure formation activities of the human Xrcc2*Rad51D complex Journal of Biological Chemistry 277 14315– 14320 [DOI] [PubMed] [Google Scholar]
- Kuznetsov S, Pellegrini M, Shuda K, et al. 2007. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. Journal of Cell Biology 176 581– 592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoczky P, McHugh PJ, Chovanec M. 2007. DNA interstrand cross-link repair in Saccharomyces cerevisiae FEMS Microbiology Reviews 31 109– 133 [DOI] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B. 2004. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis Proceedings of the National Academy of Sciences, USA 101 10596– 10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ma H. 2006. Double-stranded DNA breaks and gene functions in recombination and meiosis Cell Research 16 402– 412 [DOI] [PubMed] [Google Scholar]
- Li W, Yang X, Lin ZG, Timofejeva L, Xiao R, Makaroff CA, Ma H. 2005. The AtRAD51C gene is required for normal meiotic chromosome synapsis and doublestranded break repair in Arabidopsis Plant Physiology 138 965– 976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD. 2008. Homologous recombination in DNA repair and DNA damage tolerance Cell Research 18 99– 111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Zhang Q. 2005. Optimising the tissue culture conditions for high efficiency transformation of indica rice Plant Cell Reports 23 540– 547 [DOI] [PubMed] [Google Scholar]
- Lin Z, Kong H, Nei M, Ma H. 2006. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer Proceedings of the National Academy of Sciences, USA 103 10328– 10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tebbs RS, et al. 1998. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages Molecular Cell 1 783– 793 [DOI] [PubMed] [Google Scholar]
- Liu Y, Masson JY, Shah R, O’Regan P, West SC. 2004. RAD51C is required for Holliday junction processing in mammalian cells Science 303 243– 246 [DOI] [PubMed] [Google Scholar]
- Ma H. 2005. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants Annual Review of Plant Biology 56 393– 434 [DOI] [PubMed] [Google Scholar]
- Ma W, Westmoreland JW, Gordenin DA, Resnick MA. Alkylation base damage is converted into repairable double-strand breaks and complex intermediates in G2 cells lacking AP endonuclease. PLoS Genetics. (e1002059) 2011;7 doi: 10.1371/journal.pgen.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready S, Marcello L. 2003. Repair of UV damage in Halobacterium salinarum. Biochemical Society Transactions 31 694– 698 [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. In: Thomas RH, ed. Heredity, Vol. 86 Oxford: Oxford University Press; [Google Scholar]
- Osakabe K, Yoshioka T, Ichikawa H, Toki S. 2002. Molecular cloning and characterization of RAD51-like genes from Arabidopsis thaliana Plant Molecular Biology 50 71– 81 [DOI] [PubMed] [Google Scholar]
- Osakabe K, Abe K, Yamanouchi H, et al. 2005. Arabidopsis Rad51B is important for double-strand DNA breaks repair in somatic cells Plant Molecular Biology 57 819– 833. [DOI] [PubMed] [Google Scholar]
- Pawlowski WP, Golubovskaya IN, Cande WZ. 2003. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition The Plant Cell 15 1807– 1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajanikant C, Melzer M, Rao BJ, Sainis JK. 2008. Homologous recombination properties of OsRad51, a recombinase from rice Plant Molecular Biology 68 479– 491 [DOI] [PubMed] [Google Scholar]
- Richardson C, Jasin M. 2000. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells Molecular and Cellular Biology 20 9068– 9075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annual Review of Biochemistry 77 229– 257 [DOI] [PubMed] [Google Scholar]
- Sasaki MS, Takata M, Sonoda E, Tachibana A, Takeda S. 2004. Recombination repair pathway in the maintenance of chromosomal integrity against DNA interstrand crosslinks Cytogenetic and Genome Research 104 28– 34 [DOI] [PubMed] [Google Scholar]
- Sigurdsson S, Van Komen S, Bussen W, Schild D, Albala JS, Sung P. 2001. Mediator function of the human Rad51B–Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange Genes and Development 15 3308– 3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G, de Groot AJ, Romeijn RJ, van Buul PP, Zdzienicka MZ, Mullenders LH, Pastink A, Godthelp BC. 2010. Rad51C is essential for embryonic development and haploinsufficiency causes increased DNA damage sensitivity and genomic instability. Mutation Research 689 50– 58 [DOI] [PubMed] [Google Scholar]
- Somyajit K, Subramanya S, Nagaraju G. 2010. RAD51C: a novel cancer susceptibility gene is linked to Fanconi anemia and breast cancer Carcinogenesis 31 2031– 2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. 1998. Rad51-deficient vertebrate cells accumulate chromosome breaks prior to cell death EMBO Journal 17 598– 608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Fukushima T, Morrison C, Albala J, Swagemakers MA, Kanaar R, Thompson LH, Takeda S. 2000. The Rad51 paralog Rad51B promotes homologous recombinational repair . Molecular and Cellular Biology 20 6476– 6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Munoz P, Claas A, Smiraldo PG, Pittman DL, Blasco MA, West SC. 2004. Telomere maintenance requires the RAD51D recombination/repair protein Cell 117 337– 347 [DOI] [PubMed] [Google Scholar]
- Vazquez MV, Rojas V, Tercero JA. 2008. Multiple pathways cooperate to facilitate DNA replication fork progression through alkylated DNA DNA Repair (Amsterdam) 7 1693– 1704 [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold EMBO Journal 1 945– 951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E, Chen DJ. 2008. The endless tale of non-homologous end-joining Cell Research 18 114– 124 [DOI] [PubMed] [Google Scholar]
- Wyman C, Kanaar R. 2006. DNA double-strand break repair: all’s well that ends well. Annual Review of Genetics 40 363– 383 [DOI] [PubMed] [Google Scholar]
- Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae . PLoS Genetics. (e1000264) 2008;4 doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. 1976. Laboratory manual for physiological studies of rice 3rd edn Manila, The Philippines: International Rice Research Institute; [Google Scholar]
- Yu H, Wang M, Tang D, Wang K, Chen F, Gong Z, Gu M, Cheng Z. 2010. OsSPO11-1 is essential for both homologous chromosome pairing and crossover formation in rice. Chromosoma 119 625– 636 [DOI] [PubMed] [Google Scholar]
- Zeng YX, Hu CY, Lu YG, Li JQ, Liu XD. 2007. Diversity of abnormal embryo sacs in indica/japonica hybrids in rice demonstrated by confocal microscopy of ovaries Plant Breeding 126 574– 580 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.