Abstract
The sfr3 mutation causes freezing sensitivity in Arabidopsis thaliana. Mapping, sequencing, and transgenic complementation showed sfr3 to be a missense mutation in ACC1, an essential gene encoding homomeric (multifunctional) acetyl-CoA carboxylase. Cuticle permeability was compromised in the sfr3 mutant when plants were grown in the cold but not in the warm. Wax deposition on the inflorescence stem of cold-grown sfr3 plants was inhibited and the long-chain components of their leaf cuticular wax were reduced compared with wild-type plants. Thus, freezing sensitivity of sfr3 appears, from these results, to be due to cuticular deficiencies that develop during cold acclimation. These observations demonstrated the essential role of the cuticle in tolerance to freezing and drought.
Key words: acetyl-CoA carboxylase, Arabidopsis, cuticle, freezing tolerance, wax
Introduction
Freezing tolerance in plants is generally acknowledged to be a complex trait, with many mechanisms contributing to the survival of the hardy plant. Comparisons of hardy and non-hardy plants have yielded a bewildering array of differences at physiological, biochemical, and molecular genetic levels. Freezing tolerance can be treated as a developmental trait, as hardy species are freezing sensitive in the summer and develop their freezing tolerance only after exposure to cold, a process known as cold acclimation (Guy, 1990). However, unlike morphological development, the development of freezing tolerance does not reveal qualitatively different stages or components; thus, it has been difficult to distinguish what, if anything, is contributed by each of the many changes during cold acclimation.
Several complementary approaches have been used in an attempt to overcome this problem. The investigation of genes with altered expression during cold acclimation (reviewed by Thomashow, 1999, 2010) has proved to be an extremely powerful tool, leading to identification of the C-repeat (CRT)-binding factor/dehydration-responsive element (DRE)-binding protein 1 (CBF/DREB1) regulon and a growing understanding of cold acclimation signalling pathways (Stockinger et al., 2001; Thomashow et al., 2001; Seki et al., 2003; Vogel et al., 2005; Doherty et al., 2009). A reporter-based screen was used to identify a large set of mutants specifically altered in their signalling response to cold (and osmotic stress) (Ishitani et al., 1997; Gong et al., 2002), and through this, a number of novel components of the CBF/DREB1 signal transduction pathway were identified (reviewed by Xiong et al., 2002). Through similar approaches, ICE1, an upstream regulator of CBF/DREB1, was isolated and characterized (Chinnusamy et al., 2003; Miura et al., 2007) and mutants that affect CBF/DREB1-independent regulatory pathways were identified (Zhu et al., 2004; Medina et al., 2005). These studies have not, however, led to an understanding of the role of individual genes in freezing tolerance, except where the effect has been to control the expression of other genes (Xin and Browse, 2000).
Individual genes with clear effects on freezing tolerance can be selected by their mutant phenotypes. Several groups have isolated such mutants and determined the contribution of individual genes or molecular processes to freezing tolerance. Xin and Browse (1998) and Xin et al. (2007) isolated mutants that increased tolerance to freezing in the absence of cold acclimation. The mutations may be in components of the signalling pathway(s) responsible for cold acclimation or in genes that make self-sufficient individual contributions to protection against freezing. Mutations that decrease freezing tolerance are likely to be in genes required for freezing tolerance in the wild-type. The frs1 mutation was found to be an allele of ABA3 (Llorente et al., 2000) and confirms the importance of abscisic acid in freezing tolerance.
The Arabidopsis sensitive to freezing (sfr) mutants (Warren et al., 1996) are impaired in their freezing tolerance following cold acclimation but have near normal (wild-type) levels of freezing tolerance prior to cold acclimation. The corresponding SFR genes could be components of the cold-induced gene response whose individual contributions to freezing tolerance are not currently understood. Alternatively, they may be genes essential for freezing tolerance that are not transcriptionally regulated by cold. It has been shown that the sfr6 mutant is unable to induce genes with CRT/DRE elements in their promoters (Knight et al., 1999; Boyce et al., 2003). The SFR6 gene was identified as a protein of unknown function that acts post-translationally on CBF function as a key component of the cold acclimation process (Knight et al., 2009). SFR2 encodes a constitutively expressed chloroplast targeted β-glycosidase (Thorlby et al., 2004; Fourrier et al., 2008) that is involved in galactolipid remodelling of the outer chloroplast envelope (Moellering et al., 2010).
The sfr3 mutation causes the greatest freezing sensitivity in young incompletely expanded leaves. Expression analysis (Knight et al., 1999) indicates that it is not compromised in its ability to induce genes associated with cold stress signalling pathways. Plants with this mutation are indistinguishable from wild-type plants at normal growth temperatures but display two pleiotropic effects at 4 °C; in the short term (during cold acclimation), mutant plants are deficient in anthocyanin, and in the long term (8 weeks and above), senescence of older leaves is accelerated (McKown et al., 1996). On current knowledge, neither of these phenotypes would be expected to directly cause freezing sensitivity. Therefore, elucidating the function of the wild-type SFR3 gene will give new insights into the mechanism by which plant cells achieve freezing tolerance. In this paper, we identified SFR3 and investigated its role in freezing tolerance.
Materials and methods
Plant growth, and freeze and drought testing
Plants were grown as previously described (Thorlby et al., 2004). To screen individuals for freezing tolerance, seedlings were grown for 4/5 weeks at 18–20 °C with a 9h photoperiod at 250 µM m–2 s–1, and then subjected to 11 d of cold acclimation at 4 °C, with an 8h photoperiod at 220 µM m–2 s–1. They were placed in a freezer with an air temperature minimum of –6.0 °C for 16h and then returned to their pre-acclimation growth conditions. Injury was assessed after 5 d. Cold treatments were carried out for the time period stated under the same conditions as for cold acclimation. For drought testing, plants were grown at 18–20 °C as above, with or without a period of cold treatment. Water was withheld and the plants monitored until they wilted. Plants were then watered and recovery assessed.
Mapping of SFR3
From the mapping cross of sfr3/sfr3 Col (Columbia) × SFR3 +/SFR3 + Ler (Landsberg erecta), a total of 557 new F2 zygotes were screened for crossovers in this region. Among those zygotes showing crossovers, the SFR3 genotypes were determined by freeze testing the F3 progeny of each zygote. Details of the markers developed in this work are available on request. DNA samples were isolated from F2 plants (Thorlby et al., 1999), and analysed for cleaved amplified polymorphic sequence (CAPS) marker genotypes (Konieczny and Ausubel, 1993) and simple sequence length polymorphism marker genotypes (Bell and Ecker, 1994).
Subcloning for complementation
In silico analysis of the genomic sequence indicated that gene At1g36160 was contained on a 15kb SpeI fragment on bacterial artificial chromosome (BAC) F12D14. A genomic fragment was cloned from this BAC into a modified version of the vector pCAMBIA3300 (CAMBIA, Canberra, Australia: http://www.cambia.org) that had an SpeI site introduced into the multiple cloning site.
Plant transformation and selection
Plasmids were transferred to Agrobacterium tumefaciens GV3101/pM90 (Koncz and Schell, 1986) by electroporation (Cangelosi et al., 1991). Plants were transformed by the floral dip method (Clough and Bent, 1998). Primary transformants were selected for Basta-resistance (the marker in the pCAMBIA vectors) by repeated spraying of the seedlings with a 250mg l–1 of a solution of the herbicide Challenge 60 (AgrEvo Ltd, King’s Lynn, UK) until the growth differential was clear.
Visualization of defects in leaf cuticle using toluidine blue staining
The method of Tanaka et al. (2004) was used. Seeds were grown on Petri dishes of 0.5× MS medium solidified with 0.7% plant culture-tested agar (Sigma). Plants were stratified at 4 °C for 3 d, transferred to a growth chamber at 22 °C with continuous light for 3/4 weeks, and then transferred to a 4 °C growth room for periods of up to 10 d. To stain plants, aqueous 0.05% (w/v) toluidine blue filtered through a 0.2 µm syringe filter (Sartorius, Hannover, Germany) was poured into the culture plate until the plants were submerged and they were incubated for 2min. The staining solution was poured off and the plants were washed by submerging them twice in distilled water. Stained plants were viewed by light microscopy.
Measurement of chlorophyll leaching
To measure chlorophyll leaching, the aerial parts of 4-week-old plants, grown as described for freeze testing, were weighed and immersed in tubes containing 30ml of 80% ethanol. Approximately 0.5g of material was used per assay. Tubes were incubated in the dark with gentle agitation on a rotary shaker, aliquots taken at the indicated times and the absorbance measured at 654nm. Chlorophyll content was calculated according to the formula A 654 × extraction volume (ml) × 1/39.8×1/weight of tissue sample (g) = mg chlorophyll g–1 fresh weight of tissue (Wintermans and de Mots, 1965).
Scanning electron microscopy
Stems from wild-type and sfr3 plants were collected, allowed to air dry, mounted on stubs, and coated with gold particles using a Polaron E5100 coating unit. They were examined using a Hitachi S3000N scanning electron microscope at an accelerating voltage of 20kV.
Analysis of waxes
Detached leaves were immersed with gentle swirling in chloroform for 30 s, carefully removed, and the extracts dried under nitrogen. Metabolite analysis of the extracts was by the gas chromatography/mass spectrometry method of Fiehn et al. (2000) but using a greater range of authentic standards for lipophilic metabolite identification. Each analysis was performed with six biological replicates.
Results
Isolation and identification of the SFR3 gene and sfr3 mutation
The sfr3 mutation was isolated from the pedigreed ethyl methyl sulfonate mutant set of James and Dooner (1990) as a freezing-sensitive mutant (Warren et al., 1996), previously mapped close to the centromere region on chromosome I between markers AIG1 and T27K12 (Thorlby et al., 1999). It had a clear freezing-sensitive phenotype (Fig. 1A).
Fig. 1.
The sfr3 phenotype. (A) Comparison of wild-type (left) and sfr3 (right) plants after a freezing test. Cold acclimated plants were frozen at –6.0 °C for 16h and returned to pre-acclimation growth conditions for recovery. The picture was taken after 4 days recovery. All leaves of the wild-type appeared healthy and green, as did the older, fully expanded leaves of sfr3 plants. The younger leaves of sfr3 plants showed severe damage and were discoloured and collapsed. (B) Wild-type (left) and sfr3 (right) plants after water was withheld for 25 days from plants maintained at 4 °C. Damage was apparent across the whole rosette but was most severe in young leaves (none fully expanded).
The gene was fine mapped to a 220 kb region of the Arabidopsis genome between genes At1g36150 and At1g36410 (Supplementary Fig. S1 at JXB online). This interval was examined for gene annotations. As expected, due to the proximity of the chromosome I centromere, this interval had a low gene density and a high frequency of pseudogenes and transposon genes. Genes with homology to transposable elements and genes annotated as pseudogenes are unlikely to encode a non-redundant function and were not initially selected for analysis. The remaining genes in the 220kb interval were analysed by sequencing and/or by the isolation and analysis of knockout mutants, when available.
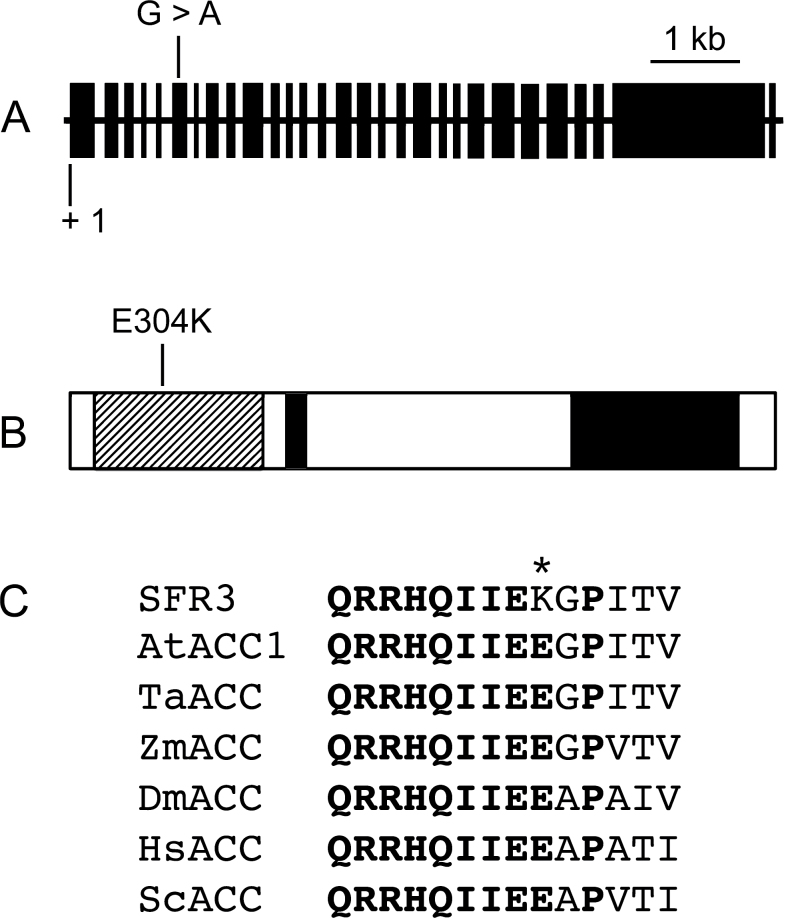
A total of nine genes were sequenced using DNA amplified from the sfr3 mutant and compared with their database versions. The gene At1g36160 was different from the database but the rest were identical. The sequence change in At1g36160, G→A atnt 910 in the coding sequence (nt 1541 in the genomic DNA), created a missense mutation (Fig. 2A) and a restriction site difference; a BseRI site found in the wild-type was not present in sfr3. We exploited this to develop a CAPS marker to distinguish between the wild-type and sfr3, and we used this marker to confirm segregation of the mutant phenotype with the sequence change.
Fig. 2.
Location of the sfr3 mutation in the ACC1 gene. (A) Schematic representation of the ACC1 gene showing the location of the sfr3 point mutation. Solid boxes represent exons. (B) Amino acid change in the conceptual translation product of the ACC1 gene. The hatched box, dotted box, and solid box represent the biotin carboxylase domain, biotin carboxyl carrier domain and carboxyl transferase domain, respectively. (C) Alignment of amino acids in the conserved region of the biotin carboxylase domain of acetyl-CoA carboxylases of various species. Bold letters indicate invariant amino acids while the asterisk identifies the amino acid changed in the sfr3 mutant. AtACC1, Arabidopsis thaliana ACC1 (At1g36160); TaACC, Triticum aestivum cytosolic ACCase (A57710); ZmACC, Zea mays ACCase (T02235); DmACC, Drosophila melanogaster ACCase (CG11198-PB); HsACC, Homo sapiens ACCase (S41121); ScACC, Saccharomyces cerevisiae ACCase (P11029).
At1g36160 encodes a homomeric acetyl-CoA carboxylase (ACCase), ACC1, shown previously to be essential for embryo survival (Baud et al., 2003). Recently, a weak allele of ACC1, glossyhead (gsd1), was identified (Lu et al., 2011) that does not cause embryo death (unlike other ACC1 mutants) but rather confers phenotypic changes associated with perturbation in the biosynthesis of cuticular waxes.
The sfr3 mutation is complemented by ACC1
The ACC1 gene was cloned from BAC F12D14, which contains the ACC1 gene in its entirety, into a plant transformation vector. A region of 4265bp upstream of the translation start codon and 1188bp of the downstream sequence were included with the gene. The incorporation of these regions should give normal control of gene expression in the resulting transgenic lines. No other genes, according to current genome annotations, were present in the cloned fragment.
Out of 24 freeze-tested primary sfr3 transformant plants, 23 displayed freezing tolerance indistinguishable from the wild-type (Supplementary Fig. S2 at JXB online). A number of freezing-tolerant primary transformants that showed complementation with the wild-type gene were selected for further analysis. The plants were left to self-fertilize and set seed. Testing of plants grown from this seed confirmed that the transgene segregated with the freezing-tolerant phenotype, confirming ACC1 as the site of the sfr3 mutation.
In Arabidopsis, there are potentially two genes, located contiguously within a 25kb region near the centre of chromosome 1, encoding the homomeric form of ACCase, ACC1 and ACC2 (At1g36160 and At1g36180) (Yanai et al., 1995). As the gene At1g36180/ACC2 was within the area delineated as containing SFR3, it was sequenced, but it did not contain a sequence change. A T-DNA knockout homozygous for the T-DNA insert within At1g36180/ACC2 was also isolated. This line did not show either a pre- or post-freezing phenotype, suggesting that this second homomeric ACCase is not an essential gene and is not necessary for freezing tolerance. Baud et al. (2003) obtained similar results, demonstrating that no ACCase protein could be detected in acc1 knockout mutant embryos, suggesting that ACC2 is either not expressed or is expressed only at a very low level.
The sfr3 mutation is within a highly conserved motif
The nucleotide change in the sfr3 mutant created a missense mutation that changes aa 304 in the ACC1 protein from a glutamic acid to a lysine (Fig. 2B). This amino acid is part of a highly conserved motif, the carbamoyl-phosphate synthase L chain ATP binding subdomain of the biotin carboxylase domain, which is conserved in plants, animals, and yeast (Fig. 2C). Interestingly, the mutation E86K in gsd1, the only other non-embryo-lethal mutation currently identified in ACC1, also lies in the biotin carboxylase domain (Lu et al., 2011).
The sfr3 mutant is drought sensitive in the cold
As ACC1 has been implicated in the development and maintenance of the plant cuticle (Baud et al., 2004; Lu et al., 2011), the primary barrier to water loss in plants, the drought tolerance of sfr3 plants was examined. The recovery of sfr3 and wild-type plants after water was withheld for a time and then made available was identical when they were grown in the warm (data not shown). Differences between sfr3 and the wild-type plants were apparent, however, if drought testing was carried out on plants that had been grown in the warm for 4 weeks before transfer to the cold for drought testing. Immediately after transfer to the cold, water was withheld. After 25 d without water, sfr3 plants wilted while wild-type plants remained vigorous (Fig. 1B). Wild-type plants did not show signs of water stress for an additional 5 d, whereas, at this time, watering of sfr3 plants did not result in their recovery.
The sfr3 mutation results in a cold-induced increase in cuticle permeability
Several assays have demonstrated cuticular deficiencies in cold-grown sfr3 plants. Toluidine blue stains the epidermis in plants with damaged cuticle but not when the cuticle is intact (Tanaka et al., 2004), and the degree of staining reflects the severity of cuticular damage. Neither wild-type nor sfr3 plants showed epidermal staining following growth in the warm (data not shown). Wild-type plants that had been transferred from the warm and grown for 10 d at 4 °C also showed no staining (Fig. 3E, 3I), but sfr3 plants treated in the same way showed distinct staining (Fig. 3B, 3F, and 3J; C, G, and K; and D, H, and L). The youngest leaves stained strongly, while older leaves stained less intensely or not at all. There was very little staining of the leaf pedicle, even in the most intensely stained plants. The toluidine blue staining procedure was also performed on plants that had been grown at 4 °C for shorter periods. No staining could be detected on plants that had spent less than 6 d in the cold (data not shown).
Fig. 3.
Toluidine blue staining of sfr3 and wild-type seedlings grown in the cold for 10 d. A control wild-type plant before (A) and after (E, I) staining is shown. Three different sfr3 plants are shown before (B–D) and after (F–H and J–L) staining. The images in the lower panel (I–L) are close ups of the images in the panel above to show details of the different staining patterns.
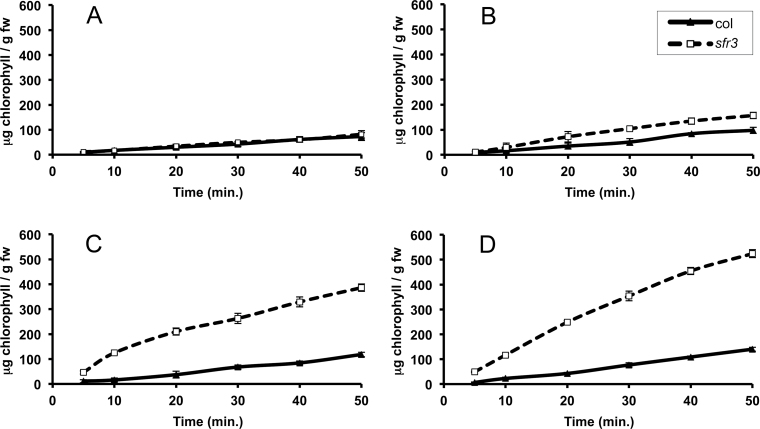
The permeability of the plant cuticle can also be assessed by measuring the release of chlorophyll from leaves immersed in 80% ethanol (Aharoni et al., 2004; Schnurr et al., 2004). No difference in chlorophyll release was detected between wild-type and sfr3 plants grown in warm conditions (Fig. 4A), but after only 24h of growth in the cold, differences were apparent (Fig. 4B). Chlorophyll was leached more rapidly from sfr3 plants, and the amount released increased with longer growth in the cold (Fig. 4C, 4D). Extended growth at low temperature had little effect on chlorophyll leached from wild-type plants.
Fig. 4.
Graphs showing the rate of leaching of chlorophyll into ethanol from sfr3 plants compared with wild-type plants. The results from warm-grown plants (A) and warm-grown plants transferred to the cold for 24h (B), 4 d (C), and 10 d (D) are shown. Experiments were performed in triplicate and results are shown as means ± standard deviation. The total amount of chlorophyll present in the leaves of sfr3 plants, measured by total extraction of chlorophyll in 96% ethanol, was not significantly different from that of wild-type plants (data not shown).
The loss of water by evaporation from wild-type and sfr3 plants was compared (Supplementary Fig. S3 at JXB online). Loss from whole detached rosettes of warm-grown wild-type and sfr3 plants were similar. After 10 d of growth in the cold, wild-type plants lost water at a similar rate to warm-grown plants, but sfr3 plants lost significantly more water (Supplementary Fig. S3).
Epicuticular wax morphology is altered on cold-grown sfr3 inflorescence stems
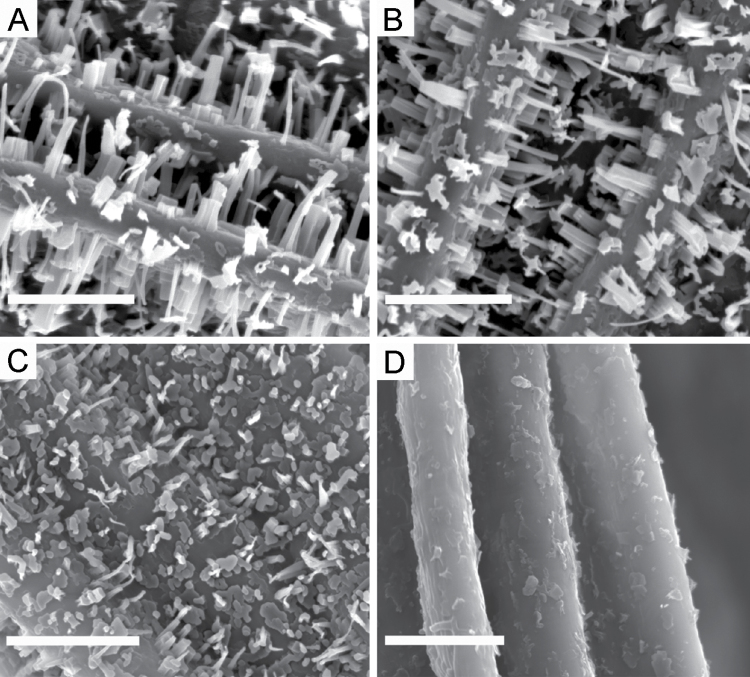
Scanning electron microscopy was used to examine epicuticular wax crystals on wild-type and sfr3 inflorescence stems. These observations were not possible with leaf material due to the lack of visible epicuticular wax crystals on Arabidopsis leaves. Plants that developed an inflorescence stem in the warm (data not shown) or that were transferred to 4 °C for a further 21 d of growth after inflorescence stem development showed a dense pattern of crystals over the surface of the stem for both wild-type (Fig. 5A) and sfr3 (Fig. 5B) plants. Plants that had initiated bolting but had not produced a significant inflorescence stem before transfer to the cold showed a different result; wild-type stems were decorated with a dense pattern of wax crystals (Fig. 5C) similar to those produced in the warm, whereas the short stems that developed on sfr3 plants in the cold were devoid of wax crystals (Fig. 5D). Thus, wax is deposited normally on sfr3 stems that develop in the warm but not in the cold.
Fig. 5.
Scanning electron microscopy images showing wax crystals on the inflorescence stem of wild-type (A, C) and sfr3 (B, D) plants. The surface of wild-type (A) and sfr3 (B) stems that developed in the warm before transfer to 4 °C for 21 d are decorated by densely distributed columnar-shaped epicuticular wax crystals. Wild-type stems that had initiated bolting in the warm before transfer to the cold showed a dense decoration of wax crystals (C). sfr3 stems that were grown in the same way (initiation in the warm and growth in the cold) showed a total lack of visible wax crystals (D). Bars, 10 µm.
Wax composition is altered on sfr3 leaves that develop in the cold
To test whether the cuticular deficiencies of sfr3 were associated with changes in wax composition, a comparative analysis of the major classes of wax components present on leaves was carried out. A previous analysis of foliar fatty acids (McKown et al., 1996) demonstrated no differences between sfr3 and wild-type plants in either the warm or cold. As Arabidopsis shows only limited growth at 4 °C, analysis was performed on plants that had been maintained at this temperature for a relatively long period (4 weeks) following initial growth at 20 °C for 3 weeks. The youngest fully expanded leaves were analysed, as these were likely to have had the highest proportion of their wax load deposited under cold growth conditions.
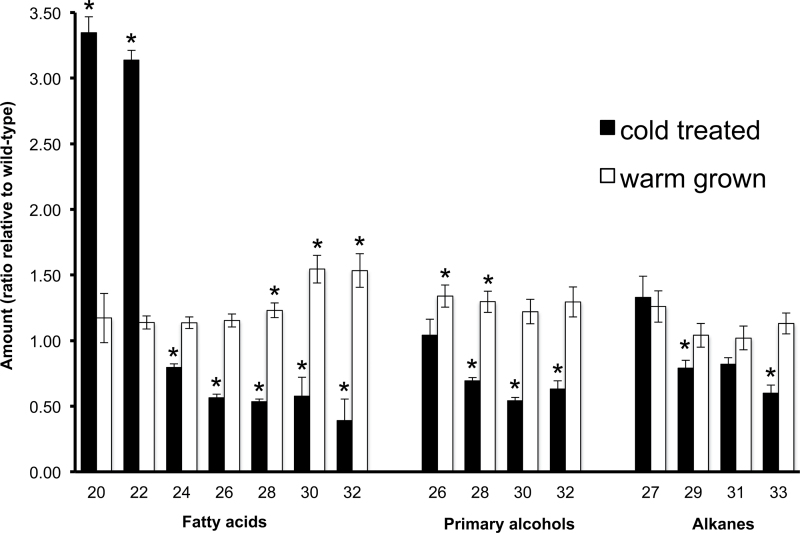
This comparative analysis showed that the leaf wax composition of warm-grown sfr3 plants was similar to that of wild-type plants (Fig. 6). No significant alterations in the amount of alkanes, the largest component of Arabidopsis leaf wax, were found. The relative amounts of primary alcohols and fatty acids were also similar. There was a small increase in the amount of the longer-chain-length varieties of these components on sfr3 leaves, suggesting that the mutant is not deficient in malonyl-CoA or in its ability to produce cuticular wax in the warm. Substantial differences between sfr3 and wild-type wax composition became apparent when plants were grown in the cold (Fig. 6). The amount of alkanes, the major wax constituent, was reduced in sfr3 plants compared with wild-type plants. There was a significant decrease (P <0.05) in the amount of two of the longer alkane components, with C33 reduced to 60% of the amount seen in wild-type plants. There was also a decrease in the amount of longer-chain-length primary alcohol and fatty acid components of the wax, and an increase in the amount of the shorter C20 and C22 fatty acids.
Fig. 6.
Graph showing a comparison of the relative amounts of the major classes of leaf wax components isolated from warm-grown and cold-treated sfr3 plants. The ratio of each wax component relative to the amount on wild-type plants grown under the same conditions (warm or cold) is shown. Chemical classes and chain lengths are labelled on the horizontal axis. Statistically significant differences (Student’s t-test, P <0.05) between wild-type and sfr3 plants treated in the same way are indicated by an asterisk.
Long-term growth in the cold affects flower development
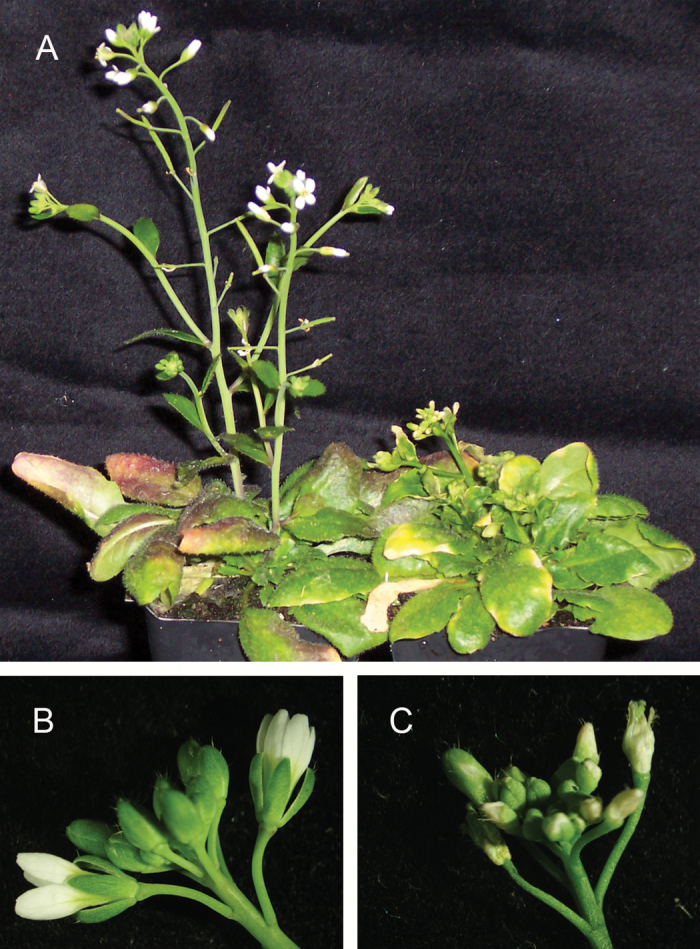
To investigate the effect of longer periods of cold on the growth of sfr3 plants, 4-week-old warm-grown plants were transferred to cold growth conditions and maintained under these conditions for 10 weeks. Previous observations have shown that, after 8 weeks’ growth in the cold, accelerated senescence of older leaves occurs (McKown et al., 1996). After a 10-week cold growth period, accelerated senescence of older leaves was again observed, but in other respects the mutant appeared similar to wild-type plants. Differences were, however, observed in the development of flowers. If plants were transferred to cold growth conditions shortly after the initiation of bolting, the inflorescence stem of the sfr3 mutant was stunted compared with the wild type, and flower development was abnormal (Fig. 7). Shrunken slightly browned structures developed in place of petals, and these senesced without expanding (Fig. 7). Normal siliques developed in wild-type plants, but were not observed in sfr3 mutants.
Fig. 7.
Image showing the effects of long-term growth at 4 °C on inflorescence development in sfr3 plants. Warm-grown 4-week-old plants were transferred to 4 °C growth conditions shortly after the inflorescence stem had initiated and maintained at this temperature for 10 weeks. Wild-type plants (A, left plant) developed normal inflorescence stems supporting flowers and siliques, while sfr3 plants developed only stunted inflorescence stems (A, right plant). Flowers of normal appearance developed on wild-type plants (B), while those on sfr3 plants were abnormal (C). Petals were shrunken and brown and did not expand, and no siliques were observed.
Discussion
A missense mutation in ACC1, a gene encoding a homomeric acetyl-CoA carboxylase (ACCase) is responsible for the freezing sensitivity of sfr3. ACCase catalyses the ATP-dependent carboxylation of acetyl-CoA to form malonyl-CoA, the first and committing step in de novo fatty acid biosynthesis (Nikolau et al., 2003). Malonyl-CoA is also essential for further elongation of fatty acids to long-chain and very-long-chain fatty acid forms.
ACCases are composed of three distinct functional domains. A structural domain carries the biotin prosthetic group (the biotin carboxyl carrier domain). One catalytic domain, the biotin carboxylase domain, catalyses carboxylation of the biotin prosthetic group, and a second, the carboxyltransferase domain, transfers the carboxyl group from carboxy-biotin to acetyl-CoA (Nikolau et al., 2003). Each of these domains can be encoded as a separate polypeptide. This is the case for the heteromeric ACCases found in plastids of most non-graminacea plants, which are responsible for the production of the plastidic malonyl-CoA pool required for de novo fatty acid synthesis. In the second type of ACCase, the homomeric form, all of the functional domains are encoded on a single polypeptide.
Homomeric ACCase is responsible for the production of the independent cytosolic pool of malonyl-CoA. This pool is separate from the plastid pool because membranes are impermeable to acyl-CoAs (Jacobson and Stumpf, 1972). The cytosolic pool of the malonyl-CoA pool produced by homomeric ACCase is required for a wide range of reactions. These include the elongation of plastid-synthesized C16 and C18 fatty acids to long-chain and very-long-chain fatty acids for seed storage triacylglycerols, waxes, or sphingolipids. Malonyl-CoA is also required for the synthesis of various secondary metabolites, including flavonoids, and the malonation of amino acids and glycosides (Roesler et al., 1994). Mutation of ACC1 probably reduces (or eliminates) the cytosolic malonyl-CoA pool and thus affects those pathways that utilize it.
A number of plants carrying mutations in ACC1 have been characterized and the gene has been shown to be essential for the production of very-long-chain fatty acids. The gurke and pasticcino3 mutants of Arabidopsis have mutations in ACC1 (Baud et al., 2003, 2004) and develop abnormal embryos with altered cotyledon primordia. The embryos eventually die, leading to embryo lethality. The sfr3 mutant is thus unlikely to represent a complete loss of function. More recently, a weak mutant allele of ACC1, gsd1, has been identified (Lu et al., 2011). This mutant does not confer an embryo-lethal phenotype but produces plants with glossy inflorescence stems, post-genital fusion in floral organs, and reduced fertility. Characterization of gsd1 has demonstrated the essential role of ACC1 in the biosynthesis of cuticular waxes and the production of a functioning cuticle.
Several studies have established the ubiquitous expression of the Arabidopsis ACC1 gene (Roesler et al., 1994; Baud et al., 2003; Lu et al., 2011). This is not surprising, considering the wide range of reactions requiring malonyl-CoA. Transcript levels are highest in flowers and the silique walls of developing embryos, but the gene is also expressed in roots, stems, leaves, and developing seeds. A high proportion of very-long-chain fatty acids are incorporated into triacylglycerols deposited in seeds during maturation, but they are also essential for cuticle and wax synthesis (Baud et al., 2004). Evidence from Nikolau and Wurtele (1998) using Northern blotting and in situ hybridization showed that ACC1 mRNA accumulated in epidermal cells involved in epicuticular wax deposition. Similarly, studies in pea demonstrated that the homomeric form of ACCase is concentrated in the epidermal layer of leaves (Alban et al., 1994). More recently, Arabidopsis microarray data (Suh et al., 2005) and GUS constructs (Lu et al., 2011) have confirmed the preferential expression of ACC1 in the epidermis of the upper section of inflorescence stems actively involved in cuticle deposition. This is consistent with the major destination of cytosolic malonyl-CoA in the developing plant being for the production of waxes for cuticle deposition. Indeed, even within the mutated embryos of lethal acc1 alleles, there was evidence of severely altered cuticle formation (Baud et al., 2004).
The broad lipid profiling studies carried out with gsd1 have confirmed the essential role of ACC1 in generating malonyl-CoA for wax synthesis (Lu et al., 2011). The gsd1 mutant exhibits a large reduction in total wax that is due mainly to the decrease in the production of the very-long-chain alkane, secondary alcohol, and ketone precursors (Lu et al., 2011). Synthesis of cutin, the other major cuticle component, is enhanced in gsd1, probably because a reduced malonyl-CoA pool leads to the build-up of shorter acyl chains, which are shunted into the shorter-chain-length cutin (Lu et al., 2011). Direct comparison of gsd1 and sfr3 wax composition is complicated by the cold-induced nature of the sfr3 phenotype (see below). A substantial portion of the wax load on sfr3 plants transferred to the cold is likely to be deposited before transfer, when plants are in the warm and the leaves are growing more actively. However, a comparative analysis (Fig. 6), using leaves from plants growing in the cold for relatively long periods, showed similar trends. Little difference was observed between sfr3 and wild-type plants grown in the warm, but clear differences were present after transfer to (and further growth in) the cold. The longer-chain-length components of alkanes, primary alcohols, and fatty acids were all reduced compared with wild-type plants (Fig. 6), as might be expected if the malonyl-CoA pool required for their elongation is limited. The shorter C20 and C22 chain-length fatty acid components of the wax were increased in the cold, probably as a result of insufficient malonyl-CoA being present for further elongation.
The results presented here showed that the sfr3 mutant cannot be distinguished from the wild-type when grown in the warm. Indeed, it has been shown previously that the ability of non-acclimated (warm-grown) wild-type plants to tolerate only mild freezing temperatures is not further compromised in the sfr3 mutant (Warren et al., 1996). Cold acclimation, however, has a clear detrimental affect on the ability of sfr3 to develop the pronounced freezing tolerance associated with this process in wild-type. A number of assays clearly demonstrated that sfr3 plants had cold-induced cuticular deficiencies that were not detectable in the warm. Evidence from chlorophyll efflux experiments (Fig. 4) suggested that there was some damage to the effectiveness of the cuticle within 24h of transfer to the cold, and that damage increased with prolonged incubation. The results of the toluidine blue assay, where no staining was visible until 6 d after transfer to the cold, also suggested a gradual degradation of the effectiveness of the cuticle. This was further supported by the images of the rich decoration of wax crystals on the inflorescence stems of sfr3 plants that developed in the warm and their absence on those that developed in the cold (Fig. 5), as well as by the comparative analysis of leaf cuticular waxes (above).
These observations suggest that, in the sfr3 mutant, ACCase functions normally in the warm or that sufficient ACCase activity is present to carry out all required metabolic functions. In contrast, following growth in the cold, the activity of ACCase becomes limiting and leads to clear deficiencies. This suggests that the sfr3 mutation results in a cold-sensitive allele of ACC1. This would explain the normal growth of sfr3 plants in the warm, while other mutations of ACC1 have been shown to be embryo lethal (Baud et al., 2004) or, in the case of gsd1 (Lu et al., 2011), to show clear phenotypic differences. Real-time PCR analysis (Supplementary Fig. S4 at JXB online) showed that the transcript level of ACC1 did not differ between wild-type and sfr3 plants in either warm or cold growth conditions. Thus, cold sensitivity is unlikely to be a product of transcript instability.
It is possible that sfr3, like gsd1, is a weak allele of ACC1 and (unlike gsd1) is able to meet the metabolic requirement for cytosolic malonyl-CoA in the warm but not to meet an increased demand necessary for growth in the cold. If the sfr3 phenotype were a result of an inability to respond to conditions requiring an increased malonyl-CoA supply, it might be expected that sfr3 would differ from wild-type plants when the plants were subjected to drought stress in the warm as this treatment has been shown to lead to a substantial increase in the deposition of waxes (Kosma et al., 2009) and presumably the demand for malonyl-CoA. However, sfr3 plants only showed a drought-induced phenotype following growth in the cold. Likewise, gsd1, a weak allele of ACC1, which shows a clear phenotype in the warm, appears to have a less severe stem wax phenotype (based on comparison with electron microscopy images presented by Lu et al., 2011) than cold-grown sfr3 plants. If sfr3 is simply a weaker mutant allele of ACC1 than gsd1, one might expect a less severe phenotype than is present with gsd1 unless the increased malonyl-CoA demand in the cold is substantial.
The cuticular deficiencies associated with cold-treated sfr3 (Figs 3 and 4) have been found in a number of mutants with deficient cuticular membranes (Chen et al., 2003; Aharoni et al., 2004; Schnurr et al., 2004), as well as the gsd1 allele of ACC1 (Lu et al., 2011). Many of these mutants display developmental defects that are not present in sfr3 plants. Fusion of leaf surfaces and floral organs is often associated with a defective cuticle, and it has been suggested that this is due to the lack of cuticle allowing the transmission of signals that would not normally occur (Nawrath, 2006). The lack of such abnormalities observed on sfr3 plants used in freezing tolerance experiments is likely to be a product of the cold-induced nature of the deficiencies that are not present during the normal growth and development of the plant. The abnormal flower development and infertility found following long-term growth in the cold (Fig. 7) suggested that this is the case and provides further evidence of the cold-sensitive nature of the mutation.
This work clearly demonstrates the critical role of the plant cuticle in resisting freezing and drought stress. The cuticle becomes compromised during cold acclimation (when plants are held at 4 °C), and plants are then sensitive to freezing in subsequent freezing assays. The primary function of the cuticle is to control water loss from leaves and other aerial parts of the plant (Riederer and Schreiber, 2001). The clear lack of tolerance to drought in cold-treated sfr3 plants (Fig. 1B) suggests that the cold-treated mutant has an inability to control the loss of water. This alone may be sufficient to explain the freezing sensitivity of the sfr3 mutant. The mutant may not be able to resist increased dehydration stress associated with recovery from freezing.
Evidence from other genes involved in cuticular wax deposition has indicated that wax deposition is associated with regions undergoing elongation (Xia et al., 1996) and the biosynthetic flux into waxes is tightly coordinated with surface area expansion (Nawrath, 2006), i.e. wax deposition occurs as the leaf expands. Leaves of the sfr3 mutant that have developed in the warm would be expected to have normal cuticle development and be less sensitive to a freezing challenge. However, leaves that develop further in the cold, after transfer to cold acclimation conditions, would have a compromised cuticle and be more sensitive to freezing. The greater sensitivity of younger leaves of sfr3 plants, which mature in the cold, to freezing damage (Fig. 1A) is indicative of this, as is the more intense staining seen in younger leaves in the toluidine blue staining assay (Fig. 3). As the more mature leaves, which developed in the warm but have also experienced cold, are also damaged in freezing, and particularly in drought stress, it is clear that there is a need for continued wax deposition after leaves have matured. This may be associated with the repair of damaged cuticle or increased deposition occurring after transfer to the cold. Previous studies have suggested that alterations in stomatal density (Aharoni et al., 2004) affect the ability of cuticle mutants to resist water loss. However, this does not appear to be a factor in sfr3 plants, as no differences were detected between the stomatal density of wild-type and sfr3 plants that were either grown in the warm or cold acclimated for 10 d (Supplementary Table S1 at JXB online).
The sfr3 mutant provides a tool for further investigations of the role of the plant cuticle in plant development and resistance to both biotic and abiotic stresses. The ability to compromise the function of a previously effective cuticle by incubation of intact plants in the cold will facilitate further studies of cuticle function. The ability to inactivate (or at least reduce the activity of) ACC1 using the same treatment will allow investigation of the role of this enzyme in plant metabolism and gene expression.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Mapping of the sfr3 gene.
Fig. S2. Complementation of the sfr3 mutation with the wild-type ACC1 gene.
Fig. S3. Water loss from excised rosettes of sfr3 and wild-type plants.
Fig. S4. Expression levels of SFR3 in wild-type and sfr3 plants in both cold and warm growth conditions.
Table S1. Stomatal density in wild-type and sfr3 plants.
Glossary
Abbreviations:
- ACCase
acetyl-CoA carboxylase;
- BAC
bacterial artificial chromosome;
- CAPS
cleaved amplified polymorphic sequence;
- CRT
C-repeat;
- CBF/DREB1
CRT-binding factor/DRE-binding protein 1;
- DRE
dehydration-responsive element
Footnotes
© 2012 The Author(s).
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/2.0/uk/) which permits unrestricted noncommercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16:2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alban C, Baldet P, Douce R. Localization and characterization of two structurally different forms of acetyl-CoA carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxypropionate herbicides. Biochemical Journal. 1994;300:557–565. doi: 10.1042/bj3000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Bellec Y, Miquel M, Bellini C, Caboche M, Lepiniec L, Faure JD, Rochat C. gurke and pasticcino3 mutants affected in embryo development are impaired in acetyl-CoA carboxylase. EMBO Reports. 2004;5:515–520. doi: 10.1038/sj.embor.7400124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuilleme S, Miquel M, Caboche M, Lepiniec L, Rochat C. Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. The Plant Journal. 2003;33:75–86. doi: 10.1046/j.1365-313x.2003.016010.x. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Boyce JM, Knight H, Deyholos M, Openshaw MR, Galbraith DW, Warren G, Knight MR. The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. The Plant Journal. 2003; 34:395–406. doi: 10.1046/j.1365-313x.2003.01734.x. [DOI] [PubMed] [Google Scholar]
- Cangelosi GA, Best EA, Martinetti G, Nester EW. Genetic analysis of Agrobacterium. Methods in Enzymology. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- Chen X, Goodwin M, Boroff VL, Liu X, Jenks MA. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane synthesis. Plant Cell. 2003;15:1170–1185. doi: 10.1105/tpc.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong XH, Agarwal M, Zhu JK. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes & Development. 2003;17:1043–1054. doi: 10.1101/gad.1077503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AS. Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Doherty CJ, Van Buskirk HA, Myers SJ, Thomashow MF. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009;21:972–984. doi: 10.1105/tpc.108.063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. Metabolite profiling for plant functional genomics. Nature Biotechnology. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- Fourrier N, Bedard J, Lopez-Juez E, Barbrook A, Bowyer J, Jarvis P, Warren G, Thorlby G. A role for SENSITIVE to FREEZING2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. The Plant Journal. 2008;55:734–745. doi: 10.1111/j.1365-313X.2008.03549.x. [DOI] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK. RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proceedings of the National Academy of Sciences, USA. 2002;99:11507–11512. doi: 10.1073/pnas.172399299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL. Cold acclimation and freezing stress tolerance: role of protein metabolism. Annual Review of Plant Physiology and Plant Molecular Biology. 1990;41:187–223. [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson BS, Stumpf PK. Fat metabolism in higher plants: LV. Acetate uptake and accumulation by class I, class II chloroplasts from Spinacia oleracea. Archives of Biochemistry and Biophysics. 1972;153:656–663. doi: 10.1016/0003-9861(72)90384-0. [DOI] [PubMed] [Google Scholar]
- James DW, Dooner HK. Isolation of EMS-induced mutants in Arabidopsis altered in seed fatty acid composition. Theoretical and Applied Genetics. 1990;80:241–245. doi: 10.1007/BF00224393. [DOI] [PubMed] [Google Scholar]
- Knight H, Mugford SG, Ülker B, Gao D, Thorlby G, Knight MR. Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. The Plant Journal. 2009;58:97–108. doi: 10.1111/j.1365-313X.2008.03763.x. [DOI] [PubMed] [Google Scholar]
- Knight H, Veale EL, Warren GJ, Knight MR. The sfr6 mutation in Arabidopsis suppresses low temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell. 1999;11:875–886. doi: 10.1105/tpc.11.5.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics. 1986;204:383–396. [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. The Plant Journal. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lü S, Joubès J, Jenks MA. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiology. 2009;151:1918–1929.. doi: 10.1104/pp.109.141911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, Oliveros JC, Martinez-Zapater JM, Salinas J. A freezing-sensitive mutant of Arabidopsis, frs1, is a new aba3 allele. Planta. 2000;211:648–655. doi: 10.1007/s004250000340. [DOI] [PubMed] [Google Scholar]
- Lu SY, Zhao HY, Parsons EP, et al.2011The glossyhead1 allele of ACC1 reveals a principal role for multidomain acetyl-coenzyme A carboxylase in the biosynthesis of cuticular waxes by Arabidopsis. Plant Physiology 1571079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKown R, Kuroki G, Warren G. Cold responses of Arabidopsis mutants impaired in freezing tolerance. Journal of Experimental Botany. 1996;47:1919–1925. [Google Scholar]
- Medina J, Rodriguez-Franco M, Penalosa A, Carrascosa MJ, Neuhaus G, Salinas J. Arabidopsis mutants deregulated in RCI2A expression reveal new signaling pathways in abiotic stress responses. The Plant Journal. 2005;42:586–597. doi: 10.1111/j.1365-313X.2005.02400.x. [DOI] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering ER, Muthan B, Benning C. Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science. 2010;330:226–228. doi: 10.1126/science.1191803. [DOI] [PubMed] [Google Scholar]
- Nawrath C. Unraveling the complex network of cuticular structure and function. Current Opinion in Plant Biology. 2006;9:281–287. doi: 10.1016/j.pbi.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Nikolau BJ, Ohlrogge JB, Wurtele ES. Plant biotin-containing carboxylases. Archives of Biochemistry and Biophysics. 2003;15:211–22. doi: 10.1016/s0003-9861(03)00156-5. [DOI] [PubMed] [Google Scholar]
- Nikolau BJ, Wurtele ES. 1998. In situ transgenic studies of the regulation of acetyl-CoA carboxylase gene expression. In: Sanchez J, Cerda-Olmedo E, Martinez-Force E, eds,Advances in plant lipid research Spain: Universidad de Sevilla; 50 53 [Google Scholar]
- Riederer M, Schreiber L. Protecting against water loss: analysis of the barrier properties of plant cuticles. Journal of Experimental Botany. 2001;52:2023–2032. doi: 10.1093/jexbot/52.363.2023. [DOI] [PubMed] [Google Scholar]
- Roesler KR, Shorrosh BS, Ohlrogge JB. Structure and expression of an Arabidopsis acetyl-coenzyme A carboxylase gene. Plant Physiology. 1994;105:611–617. doi: 10.1104/pp.105.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell. 2004;16:629–642. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K. Molecular responses to drought, salinity and frost: common and different paths for plant protection. Current Opinion in Biotechnology. 2003;14:194–199. doi: 10.1016/s0958-1669(03)00030-2. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Mao YP, Regier MK, Triezenberg SJ, Thomashow MF. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Research. 2001;29:1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. Cuticular lipid composition surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiology. 2005;139:1649–1665. doi: 10.1104/pp.105.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. The Plant Journal. 2004;37:139–146. doi: 10.1046/j.1365-313x.2003.01946.x. [DOI] [PubMed] [Google Scholar]
- Thomashow M. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Thomashow MF, Gilmour SJ, Stockinger EJ, Jaglo-Ottosen KR, Zarka DG. Role of the Arabidopsis CBF transcriptional activators in cold acclimation. Plant Physiology. 2001;112:171–175. [Google Scholar]
- Thomashow MF. Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiology. 2010;154:571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlby G, Fourrier N, Warren G. The SENSITIVE TO FREEZING2 gene required for freezing tolerance in Arabidopsis thaliana encodes a β-glucosidase. Plant Cell. 2004;16:2192–2203.. doi: 10.1105/tpc.104.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlby G, Veale E, Butcher K, Warren G. Map positions of SFR genes in relation to other freezing-related genes of Arabidopsis thaliana. The Plant Journal. 1999;17:445–452. doi: 10.1046/j.1365-313x.1999.00395.x. [DOI] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. The Plant Journal. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- Warren G, McKown R, Marin A, Teutonico R. Isolation of mutations affecting the development of freezing tolerance in Arabidopsis thaliana. Plant Physiology. 1996;111:1011–1019. doi: 10.1104/pp.111.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermans JFGM, de Mots A. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochimica et Biophysica Acta. 1965;109:448–453. doi: 10.1016/0926-6585(65)90170-6. [DOI] [PubMed] [Google Scholar]
- Xia Y, Nikolau BJ, Schnable PS. Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell. 1996;8:1291–1304. doi: 10.1105/tpc.8.8.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Browse J. eskimo1 mutants of Arabidopsis are constitutively freezing-tolerant. Proceedings of the National Academy of Sciences, USA. 1998;95:7799–7804. doi: 10.1073/pnas.95.13.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Browse J. Cold comfort farm: the acclimation of plants to freezing temperatures. Plant, Cell & Environment. 2000;23:893–902. [Google Scholar]
- Xin Z, Mandaokar A, Chen Last RL, Browse J. Arabidopsis ESK1 encodes a novel regulator of freezing tolerance. The Plant Journal. 2007;49:786–799. doi: 10.1111/j.1365-313X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Xiong LM, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14:S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N. Genomic organization of 251kDa acetyl-CoA carboxylase genes in Arabidopsis: tandem gene duplication has made two differentially expressed isozymes. Plant Cell Physiology. 1995;36:779–787. doi: 10.1093/oxfordjournals.pcp.a078822. [DOI] [PubMed] [Google Scholar]
- Zhu J, Shi H, Lee BH, Damsz B, Cheng S, Stirm V, Zhu JK, Hasegawa PM, Bressan RA. An Arabidopsis homeodomain transcription factor gene HOS9, mediates cold tolerance through a CBF-independent pathway. Proceedings of the National Academy of Sciences, USA. 2004;101:9873–9878. doi: 10.1073/pnas.0403166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.