Abstract
LPS-induced TNFα factor (LITAF) is a multiple functional molecule whose sequence is identical to small integral membrane protein of the lysosome/late endosome (SIMPLE). LITAF was initially identified as a transcription factor that activates transcription of proinflammatory cytokine in macrophages in response to LPS. Mutations of the LITAF gene are associated with a genetic disease, called Charcot-Marie-Tooth syndrome. Recently we have reported that mRNA levels of LITAF and tumor necrosis factor superfamily member 15 (TNFSF15) are upregulated by AMPK. The present study further assesses their biological functions. Thus, we show that AICAR, a pharmacological activator of AMPK, increases the abundance of LITAF and TNFSF15 in the LNCaP and C4-2 prostate cancer cells, which is abrogated by shRNA or dominant negative mutant of AMPK α1 subunit. Our data further demonstrate that AMPK activation upregulates the transcription of LITAF. Intriguingly, silencing LITAF by shRNA enhances proliferation, anchorage-independent growth of these cancer cells, and tumor growth in xenograft model. In addition, our study reveals that LITAF mediates the effect of AMPK by binding to a specific sequence in the promoter region. Furthermore, we show that TNFSF15 remarkably inhibits the growth of prostate cancer cells and bovine aortic endothelial cells in vitro with a more potent effect toward the latter. In conjuncture, intratumor injection of TNFSF15 significantly reduces the size of tumors and number of blood vessels and induces changes characteristic of tumor cell differentiation. Therefore, our studies for the first time establish the regulatory axis of AMPK-LITAF-TNFSF15. They also suggest that LITAF may function as a tumor suppressor.
Keywords: AMPK, LITAF, TNFSF15, p53, tumor suppressor, tumorigenesis
Introduction
Lipopolysaccharide-induced tumor necrosis factor alpha factor (LITAF) was initially characterized as a transcription factor that activates transcription of cytokines, such as TNFα, IL6, sTNF-RII, and CXCL in macrophages in response to lipopolysaccharide (LPS) (Myokai et al., 1999). It was also described as a p53-inducible gene (PIG7) (Polyak et al., 1997). Upon stimulation, LITAF translocates to the nucleus and binds to a specific sequence of the TNFα promoter to cooperate with other transcription factors such as STAT6(B) to upregulate the transcription (Tang et al., 2005). Therefore, LITAF plays an important role in inflammatory response. Interestingly, the sequence of LITAF is identical to the Small Integral Membrane Protein of the Lysosome/late Endosome (SIMPLE), whose mutations are associated with a genetic disease called Charcot-Marie-Tooth disease (CMT), characterized by demyelinating disorders of peripheral nervous system (Niemann et al., 2006). It is not clear whether the mutations disable the transcriptional activity of LITAF or other functions.
Tumor necrosis factor superfamily member 15 (TNFSF15) was first identified as an inhibitor of vascular endothelial cell growth factor, thus also named VEGI (Sethi et al., 2009). This TNF subfamily contains three isoforms resulting from different splicing; the first one consists of 174 amino acids, which shares 20 to 30% homology to other TNF family members and, two additional isoforms comprise 192 and 251 amino acids, respectively (Sethi et al., 2009). All the three isoforms are identical in the carboxyterminal 151 amino acids. TNFSF15 has been shown to potently inhibit the growth of vascular endothelial cells (Yue et al., 1999; Zhai et al., 1999a) as well as tumor cells (Haridas et al., 1999). Several tumor xenograft studies have revealed that TNFSF15 suppresses tumor growth (Hou et al., 2005; Pan et al., 2004; Zhai et al., 1999b). In addition, the longer form, VEGI-251, also called TNF-like ligand 1 (TL1A), has been implicated in autoimmunity and inflammation-induced immunity (Bayry, 2010; Sethi et al., 2009).
AMP-activated protein kinase (AMPK) is a highly conserved serine/threonine protein kinase, consisting of three subunits, a catalytic subunit (α) and two regulatory subunits (β and γ) (Hardie, 2007). In mammals, each subunit of AMPK contains two to three isoforms (α1, α2; β1, β2; γ1, γ2, γ3). AMPK serves as a fuel gauge in maintaining energy homeostasis. Thus, it is activated under metabolic stress that increases intracellular levels of AMP, which servers as an allosteric activator by binding to the γ regulatory subunit. In addition, AMPK is phosphorylated and activated by upstream kinases, such as the tumor suppressor LKB1/Stk11 (Hardie, 2007; Luo et al.,2010). Under physiological conditions, AMPK is activated by hormones and cytokines, including leptin (e.g. in skeletal muscle) and adiponectin secreted from adipocytes, interleukin 6 (IL6), and ciliary neurotrophic factor (CNTF) (Zhang et al., 2009). In addition, AMPK can be activated by a variety of pharmacological agents. The prototypical activator is 5-aminoimidazole-4-carboxamide 1-D-ribonucleoside (AICAR), a cell permeable agent that is phosphorylated after entering the cell and converted to ZMP, an AMP analogue. Importantly, two clinically used anti-diabetic drugs, metformin and thiazolidinediones, have been known to activate AMPK (Zhang et al., 2009).
Recently, we have demonstrated that inhibition of AMPK exacerbates the malignant behavior of prostate cancer cells (Zhou et al., 2009). In exploring the mechanism underlying the inhibitory effects of AMPK, we have identified several interesting targets, among which are LITAF and TNFSF15. In the present study, we aim to explore the relationship of these molecules with AMPK and assess their biological effects on tumor cell growth and tumor development. Our results show that AMPK activation by AICAR upregulates LITAF, which in turn increases TNFSF15 expression. All these regulations occur at the level of transcription. We further show that knockdown of LITAF expression by shRNA enhances cell proliferation, anchorage-independent growth of prostate cancer cells and tumor development in xenograft model. Finally, we show that TNFSF15 inhibits the growth of prostate cancer cells in vitro and in vivo, with a more potent effect on endothelial cell growth and tumor angiogenesis.
Results
AMPK upregulates protein levels of LITAF
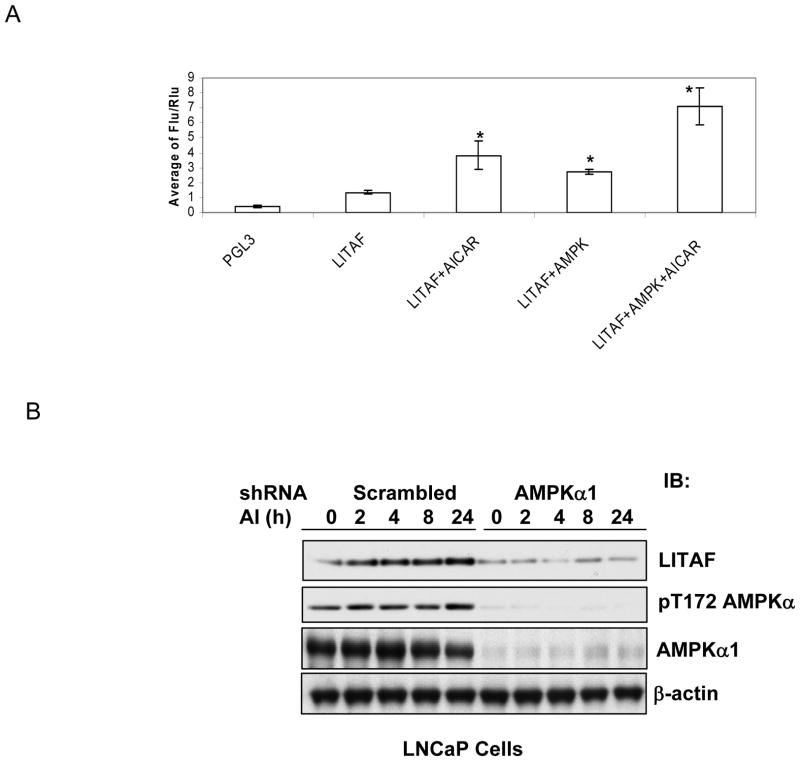
Previously we have shown that the expression of LITAF mRNA is reduced in prostate cancer cells containing the dominant negative mutant of AMPKα1 subunit or shRNA, whereas it is upregulated by AICAR treatment (Zhou et al., 2009). In the present study, we assessed if AMPK plays a role in regulating the promoter activity of LITAF. Toward this end, we cloned 500 bp 5′ upstream of the transcriptional initiation site of the LITAF gene and inserted to a luciferase reporter plasmid. We then transfected LNCaP cells with this plasmid together with a plasmid encoding Renilla luciferase in the presence or absence of the plasmid for wild type α1 subunit of AMPK. After 48 hours, the cells were treated with AICAR for 8 hours. Fire fly luciferase activity was assayed and normalized with Renilla luciferase activity. As shown in Figure 1A, AICAR enhanced the luciferase activity by about two folds, while transfection of AMPK α1 subunit caused a 1.5-fold increase, which was further increased by the treatment with AICAR. To ascertain if the change holds true at protein levels, we treated a prostate cancer cell line, LNCaP cells, with AICAR for different periods of time and examined the expression of LITAF. As shown in Fig 1B, the treatment progressively increased the abundance of LITAF, whereas the induction was diminished in the cells expressing shRNA. A similar result was obtained with the dominant negative mutant of α1 subunit (Data not shown).
Figure 1. AMPK-dependent induction of LITAF by AICAR.
A. Promoter activity assay. LNCaP cells were transfected with a promoter-less fire fly luciferase reporter plasmid (PGL3) or the plasmid with insertion of LITAF promoter region and Renilla luciferase plasmid in the presence or absence of a plasmid encoding wild type AMPK α1 subunit. Two days later, the cells were treated with AICAR for 8 hours and luciferase assay was conducted as described in Materials and Methods. The luciferase activity was normalized with Renilla lucicerase activity and expressed as ratios (Means ± SD, n=3). “*” denotes P<0.01 as compared to basal LITAF promoter activity. B. LNCaP cells stably infected with α1 shRNA or scrambled shRNA retrovirus were treated with AICAR (AI, 1 mM) for indicated times. Cell lysates were blotted with antibodies, as indicated.
LITAF inhibits malignancy of tumor cells
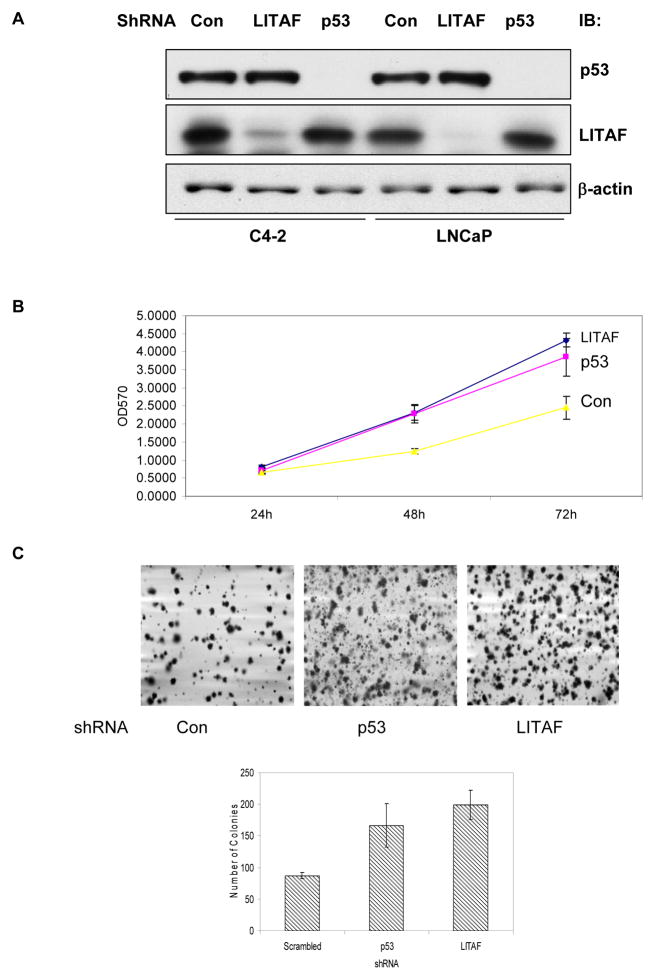
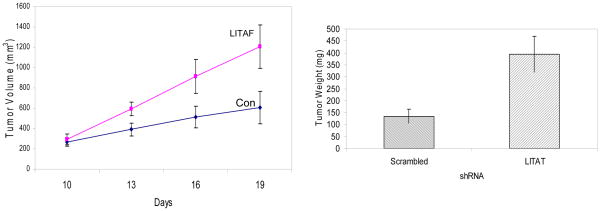
To assess the biological effects of LITAF, we decided to knock down LITAF expression by shRNA. Out of 4 shRNA retroviral plasmids, one was found to yield more than 80% of silencing effect. Thus, the retrovirus for this clone was packaged in HEK293T cells and used to infect into LNCaP and C4-2 cells (Figure 2A). We also included p53 shRNA as a positive control, as previous studies identified LITAF as one of p53-induced genes. However, our current data revealed that knocking down p53 did not affect LITAF expression, or its response to AICAR (data not shown). We then compared behaviors of prostate cancer cells with or without silencing LITAF using MTT assay and found that silencing LITAF significantly accelerated cell proliferation (Figure 2B). Furthermore, it enhanced anchorage-independent growth of prostate cancer cells on soft agars (Figure 2C). All these effects were as great as those by knocking down p53. We then inoculated these cells into athymic nude mice and assessed tumor development. As shown in Figure 3, our results revealed that prostate cancer cells containing LITAF shRNA form tumors at a significantly higher rate than the cells without knockdown.
Figure 2. LITAF inhibits malignant phenotype of prostate cancer cells.
A. C4-2 and LNCaP cells were stably infected with shRNA for LITAF, p53 and scrambled control (Con) and cell extracts were blotted with antibodies, as indicated. B. Proliferation of LNCaP cells carrying LITAF, p53 and scrambled (Con) shRNA was assayed by MTT method. C. Anchorage-independent growth of LNCaP cells with different shRNA on soft-agar. One representative of triplicates was shown. The graph represents means ± STDV (Student’s t test, P<0.05).
Figure 3. LITAF inhibits tumor growth in xenograft model.
LNCaP cells bearing LITAF or scrambled (Con) shRNA were inoculated subcutaneously on the flank of athymic nude mice. Ten days after inoculation, tumors were measured every three days and volumes calculated (Left). At the end of experiment, tumor tissues were removed and weighed. The graph represents average volumes (means ± SE, n=10). Statistic test was conducted using two-way ANOVA method (P<0.05). The bars show differences in average weight of tumors in two groups (mean ± SE, Student’s t test, P<0.01)
LITAF mediates AMPK activation to upregulate TNFSF15
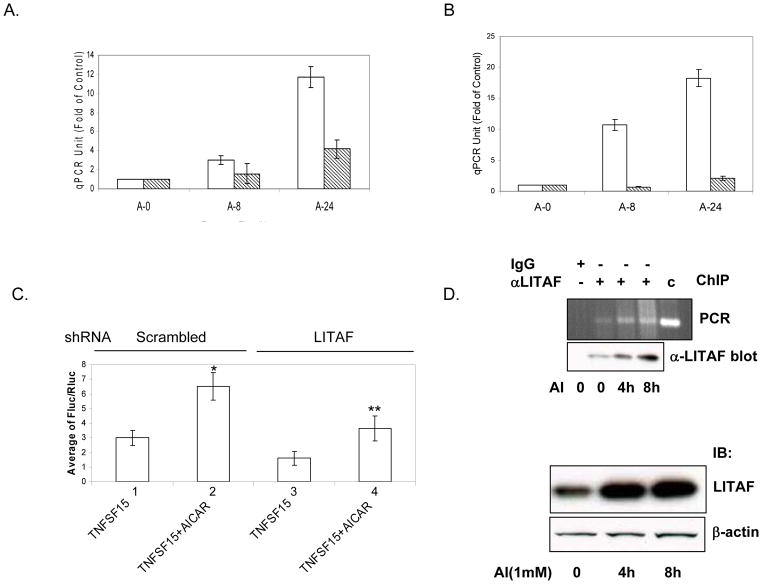
Previous studies have shown that LITAF responds to LPS to induce expression of proinflammatory cyokines such as TNFα. We have reported that AMPK activation increases expression of TNFSF15 in prostate cancer cells where TNFα is barely detectable (Zhou et al., 2009). In this study, we were curious to ask if AICAR-induced expression of TNFSF15 was mediated by LITAF. Toward this end, we treated LNCaP and C4-2 cells containing LITAF shRNA or scrambled shRNA with AICAR and analyzed mRNA abundance by quantitative PCR. The data revealed that AICAR increased levels of TNFSF15 mRNA in control cells, while the induction was markedly reduced in cells where LITAF was silenced (Figure 4A & 4B). To examine if the change of mRNA is attributed to the regulation of transcription, we isolated 1000 bp 5′ upstream of the transcriptional initiation site of TNFSF15 and placed it to upstream sequence of luciferase reporter. We then transfected the plasmid to LNcaP cells carrying scrambled or LITAF shRNA and analyzed the luciferase activity. As shown in Figure 4C, AICAR significantly stimulated the luciferase activity in control cells, whereas the stimulation was diminished by LITAF shRNA. As a putative LITAF-binding element (CTCCC) is present in the promoter region of TNFSF15 (Tang et al., 2003), we asked if LITAF directly binds to this sequence. In the ChIP assay, we found that the sequence surrounding the binding element was specifically pulled down by LITAF antibody, which was increased by the treatment of LNCaP cells with AICAR.
Figure 4. LITAF mediates the effect of AMPK on TNFSF15 expression.
A. & B. C4-2 and LNCaP cells bearing LITAF shRNA or scrambled shRNA were treated with or without AICAR (1 mM) for 8 and 24 hours (A-0, A-8, and A-24), respectively. Total RNA was isolated and reverse transcription performed, followed by quantitative PCR. Each sample was assayed in triplicate and normalized as opposed to GAPDH. Units are expressed as fold of control without AICAR treatment. Bars represent mean ± SD (n=6, two-way ANOVA analysis shows P<0.01). C. LNCaP cells carrying scrambled and LITAF shRNA were transfected with the plasmids containing TNFSF15 promoter-driven fire fly luciferase reporter and Renilla luciferase, and treated with or without AICAR for 8 hours. Luciferase activity was assayed as for Figure 1A. Bars represents means ± SD (n = 4). “*” denotes P<0.01 (2 vs 1); “**”, P<0.05 (4 vs 2). D. ChIP assay. LNCaP cells were treated with or without AICAR (1 mM) for indicated times and chromatin immunoprecipitation was performed using control IgG or anti-LITAF antibody as described in Materials and Methods. The immunoprecipitates were analyzed by PCR and resolved on agarose gel electrphoresis. Geneomic DNA was amplified using specific primers as a positive control (C). Aliquots of ChIP and cell extracts were examined by Western blot with antibodies against LITAF and actin.
TNFSF15 inhibits angiogenesis and tumorigenesis
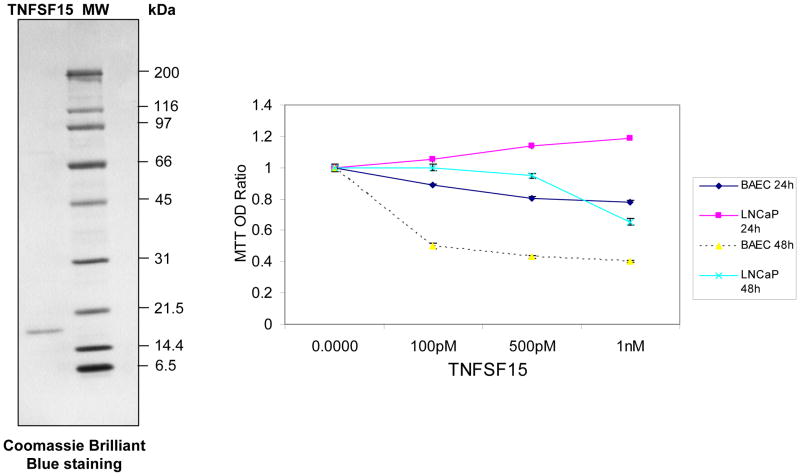
To assess if TNFSF15 plays a role in tumor development, we cloned cDNA encoding the longest version of TNFSF15, VEGI-251, from LNCaP cells by reverse transcription coupled PCR and then expressed the carboxyterminal 151 amino acids, which are identical among all three isoforms (Figure 5). We then examined if the carboxyterminal piece of TNFSF15 exerted an inhibitory effect on the growth of LNCaP cells and bovine aortic endothelial cells (BAEC). In this experiment, the cells were treated with different doses of TNFSF15 for 24 and 48 hours and assayed by MTT. At 24 h, OD570 of LNCaP cells exhibited a nearly linear increase with increasing doses of TNFSF15, reaching a maximal 20% at 1 nM (P<0.01), while at 48 h, the OD progressively decreased with increasing doses (P<0.01. The reason for the increase of MTT readings at 24 hours is not clear. It possibly reflects increased biogenesis of mitochondria within this specific time frame. In contrast, BAEC cells were markedly suppressed and appeared to be more sensitive to TNSF15 treatment at both 24 and 48 hour points (P<0.01). Two-way ANOVA analysis indicates that differences between different groups (cell types and times) are significant (P<0.05).
Figure 5. TNFSF15 inhibits cell proliferation in vitro.
TNFSF15 was expressed as a fusion protein aminoterminally tagged by maltose-binding protein, purified with amylose agarose and cleaved by Factor X. The purified protein (300 ng) was separated onto SDS-PAGE and stained by Coomassie Brilliant Blue (Left). Recombinant TNFSF15 was added to LNCaP and BAEC cells at different concentrations and cell culture continued for 24 and 48 hours, respectively. The effects were evaluated by MTT assays and expressed as ratio of untreated cells cultured in parallel. The curves represent means ± SD (n=6). Statistic analysis was performed using One-way and Two-way ANOVA methods, respectively (P<0.05).
To test if TNFSF15 inhibits tumor growth in vivo, we inoculated LNCaP cells into nude mice and allowed tumor grow for 10 days. When tumors were palpable, intratumor injection of TNFSF15 was performed every two days for two weeks. In the end, mice were sacrificed and tumors removed and weighed. Our data revealed that the average weight of tumors from TNFSF15-admininstered mice was half of that injected with PBS vehicle (Figure 6A). In general, tumors contained fewer visible blood vessels. To evaluate tumor vascularization, the tumor tissues were sectioned for examining histopathological morphology and immunohistochemical reactivity to antibody for Von Willebrand Factor. As shown in Figure 6B, the tumor tissues of the control animals displayed uniform and anaplastic structure without evidence of differentiation. In contrast, experimental animals exhibited multiple lobular structure surrounded and separated by connective tissue. This structure apparently manifests a likelihood of differentiated tumor. In addition, the number of blood vessels formed in the tumor tissue of the control animals was significantly more than that of the experimental animals.
Figure 6. TNFSF15 inhibits tumor growth in xenograft model.
A. LNCaP cells were injected into nude mice as described for Figure 3. Ten days after inoculation, intratumor injection of TNFSF15 was performed (10μg/ml, 100 μl for each injection) every two days for two weeks. In the end, mice were sacrificed and tumors removed and weighed. The bars represent average weights ± SE (n=10, Student test, P=0.012953). B. Tumor tissues isolated from A were snap-frozen and sectioned. Histopathological morphology of tumors and immunohistochemical staining with antibodies against Von Willebrand Factor for visualizing blood vessels were examined. Two representative views from tumors injected with TNFSF15 and PBS are presented. Tumor tissues of the control animals exhibited uniform and anaplastic structure without evidence of differentiation, while experimental animals exhibited multiple lobular structure surrounded and separated by connective tissue. This structure apparently indicates the likelihood of differentiated tumor. In addition, the number of blood vessels formed in the tumor tissues of the control animals was significantly more than that of the experimental animals.
Discussion
Recently, we have shown that inhibition of endogenous AMPK by expression of the dominant negative mutant of AMPK α1 subunit or its shRNA augments malignant behaviors of prostate cancer cells, whereas activation of AMPK by AICAR or introducing LKB1 to cancer cells containing loss-of-functional mutations causes changes in an opposite direction. The effects of LKB1/AMPK might be mediated by concerted actions of various downstream targets (Zhou et al., 2009). Among them, we have identified two novel targets, LITAF and TNFSF15. In the present study, we have for the first time demonstrated that activation of AMPK stimulates expression of LITAF, which in turns upregulates TNFSF15, and that knockdown of LITAF accelerates prostate cancer cell proliferation and tumor development. Our data also show that TNFSF15 remarkably inhibits the growth of prostate cancer cells and angiogenesis and induces morphological transition from malignant to benign tumors.
AMPK has been shown to play important roles in mediating the tumor suppressive function of LKB1. To understand the underlying mechanisms, our exciting but challenging task is to identify new targets downstream of AMPK in addition to those well characterized, such as mTOR, FASN, p53, p27 and FOX3a (Luo et al.2010). In this study, we have found that AMPK regulates the transcription of LITAF, which then binds to a specific sequence in the promoter region of TNFSF15 and regulates its transcription. Currently, we do not know how AMPK regulates the transcription of LITAF. It is possible that AMPK indirectly exerts such an effect through regulation of an unidentified transcription factor. However, our data do not support that this occurs through p53. Therefore, it will be our interest to identify the transcription factor that mediates this effect.
Existing data support that LITAF is a transcription factor involved in regulating production of proinflammatory cytokines in response to LPS (Tang et al., 2005; Tang et al., 2006). An additional function of LITAF relates to sorting and degradation of plasma membrane proteins by lysosome and late endosome (Niemann et al., 2006). Mutations of LITAF/SIMPLE have been suggested to alter this second function, which may play a role in the pathogenesis of CMT1C (Beauvais et al., 2006; Bennett et al., 2004; Latour et al., 2006). Thus far, no direct evidence indicates a role of LITAF in tumorigenesis. However, several recent studies suggest that there is such a link. For instance, homozygous deletion the LITAF gene and promoter hypermethylation are reported in some types of lymphomas (Mestre-Escorihuela et al., 2007). Secondly, LITAF was found to associate with WW domain oxidoreductase (WWOX) (Ludes-Meyers et al., 2004). The latter has been found altered in multiple types of cancer and its ectopic expression inhibits xenograft tumor growth of breast cancer cells (Chang et al.2010). Therefore, it will be interesting to assess the direct effect of LITAF on tumorigenesis and elucidate the underlying mechanisms.
Although LITAF is known to upregulate transcription of TNFα, little information has been documented on its role in regulating other TNF superfamily members. A possible link between LITAF and TL1A has been suggested by the findings that their expressions are increased in macrophages of inflammatory bowel diseases such as Crohn’s disease and ulcerative colitis (Picornell et al., 2007; Stucchi et al., 2006; Young and Tovey, 2006). Interestingly, one study has so far reported that bacterially expressed chicken LITAF or LITAF expressed, secreted and purified from COS7 cells, stimulates transcription of TNFSF15 in chicken macrophages (Hong et al., 2006). This report is coincidently in line with our findings where we placed LITAF as an upstream modulator of TNFSF15 in response to AMPK activation.
A clinical investigation has indicated that high levels of TNFSF15 are associated with increased survival rate of breast cancer patients (Parr et al., 2006). Numerous studies have reported that exogenous application of different isoforms of recombinant TNFSF15 to culture media or to animals inhibits tumor cell growth and tumor development and angiogenesis (Haridas et al., 1999; Hou et al., 2005; Pan et al., 2004; Yue et al., 1999; Zhai et al., 1999a; Zhai et al., 1999b). Our present study demonstrates that bovine aorta endothelial cells are more sensitive to TNFSF15 than LNCaP cells, which is in accord with previous reports that TNFSF15 exerts anti-tumor effects mainly through inhibition of angiogenesis (Sethi et al., 2009). Consistently, our animal study showed that the treatment of xenograft tumors with TNFSF15 not only inhibits angiogenesis, but also induces morphological changes characteristic of benign tumors.
In the end, we would emphasize that our present study has for the first time demonstrated that AMPK upregulates LITAF, which in turn increases the expression of TNFSF15. The regulation occurs at the transcriptional level. Collectively, our data suggest that LITAF and TNFSF15 may function as tumor suppressors. Therefore, we propose a model illustrated in Figure 7. LITAF serves as one of downstream targets of AMPK to inhibit cancer cell growth, possibly by multiple mechanisms, one of which is TNFSF15 that potently inhibits angiogenesis. As for other functions of LITAF, such as those related to lysomal/late endosome trafficking and degradation of membrane proteins, it is worthwhile to explore if they participate in tumorigenesis. Overall, our work points to a new direction in investigating the correlation of tumor progression and prognosis with expression levels and activity of AMPK, LITAF and TNFSF15 and the mechanism by which they are involved in tumorigenesis. Likewise, our data also suggest that targeting AMPK pathway would represent a promising approach for prevention and treatment of cancer, as it acts on both tumor cells and angiogenesis.
Figure 7. Model of LITAF regulation of tumorigenesis.
LITAF mediates the inhibitory effects of AMPK on tumorigenesis by directly regulating TNFSF15, which in turn inhibits angiogenesis, and other unidentified targets.
Materials and Methods
Materials
AICAR was purchased from Toronto Research Chemicals Inc (North York, ON, Canada). Antibodies for total and phospho-T172 of AMPK alpha subunit were from Cell Signaling Technology, Inc. (Danvers, MA). Antibody for p53 was from Millipore (Billerica, MA). Monoclonal antibodies for β-actin and flag epitope (M2) were purchased from Sigma (Saint Louis, MO). Monoclonal antibody for LITAF was purchased from BD Biosciences (San Jose, CA).
Stable cell lines
Retroviral plasmids for LITAF shRNA and scrambled shRNA were purchased from Open Biosystems (Huntsville, AL). A set of four plasmids (Catalog number: RHS4529-NM_001136472) were transfected into HEK293T cells to examine a clone that produces the most efficient silencing effect and one (V2HS_247509) was selected. ShRNA for p53 was in pSuperRetro vector, a gift from Dr. Zhixiong Jim Xiao. Retrovirus was packaged in HEK293T cells and the virus supernatant was infected into two prostate cancer cell lines, LNCaP and C4-2. Two days after infection, the cells were selected with puromcyine and maintained in RPMI1640 medium supplemented with 10% FBS. The stable cell lines for the dominant negative mutant of human AMPK α1 catalytic subunit (D139A) and α1 shRNA were described previously (Zhou et al., 2009).
Immunoblot
Equal amounts of cell extracts (25 μg) were separated onto SDS-PAGE and electrophoretically transferred to PVDF membranes (Millipore). The membranes were sequentially blotted with the first and second antibodies, and developed by the enhanced chemiluminescence (ECL) method, according to the protocol provided by manufactures.
Soft Agar Assay for Colony Formation
Assays on anchorage independent growth were carried out on soft agar, as described previously (Zhou et al., 2009).
Expression and purification of TNFSF15
Total RNA was prepared from LNCaP cells and cDNA synthesized. TNFSF15 was amplified by PCR using the primers based on the sequence of VEGI-251 (forward: 5′ ATGGCCGAGGATCTGGGACTGAGC 3′, reverse: 5′ CTCTCCTCCTATAGTAAGAAGGCTCC 3′) and subcloned into pCR-BluntII-TOPO (Invitrogen, Carlsbad, CA). After sequencing validation, the cDNA segment encoding the carboxyterminal 151 amino acids was amplified by PCR using the primers (forward: 5′ agctggatccGTTGTGAGACAAACTCCCACACAGC 3′, reverse: 5′ agctgtcgacCTCTCCTCCTATAGTAAGAAGGCTCC 3′). The PCR product was subcloned into pMal-C2 plasmid at the BamHI and SalI sites. The plasmid was transformed into JM109 cells and the recombinant TNFSF15 was induced by IPTG, purified using Amylose beads, cleaved with Factor X, and purified further to remove Factor X, according to the manufacture’s protocol (New England BioLabs, Ipswich, MA).
MTT assay
Cells (5×103) were seeded into 96-well plates and cell viability was measured at different days using MTT kit according to the manufacture’s protocol (Invitrogen).
Quantitative Real-time PCR Analysis
the expression of mRNA were examined by real-time PCR with the ABI 7300 Real-time PCR System (Applied Biosystems, Foster City, CA), using SYBRGREEN PCR Master Mix 2× reagent in 20 μl reaction volume. The primers for RT-PCR were designed as described previously (Zhou et al., 2009). Each sample was amplified in triplicate and normalized to Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression. Results were evaluated by the comparative threshold cycle value method (2−ΔΔCt) for relative quantification of gene expression.
Xenograft model
Stable LNCaP cells (2×106) bearing LITAF shRNA or scrambled shRNA were resuspended in 100 μl phenol red-free RPMI medium and mixed with 100 μl matrigel (BD Biosciences, Bedford, MA) for each injection. The cell suspensions were then injected subcutaneously into the flank of nude mice (Charles River Laboratories, Wilmington, MA). Each group consisted of 10 mice and two spots were inoculated for each mouse. Ten days after inoculation, tumors were measured with calipers and tumor volume was calculated using the formula W2xL/2 (W, shorter diameter; L, longer diameter) (mm3).
For TNFSF15 administration, when tumors grew palpable, TNFSF15 prepared in PBS (100μl, 10 μg/ml) or PBS as vehicle control was injected inside tumors every 2 days. After two weeks injection, mice were euthanized and tumors removed and weighed.
Immunohistochemical staining procedures
Tumor tissues were snap-frozen and sectioned immediately or stored at −80°C. The frozen sections were fixed with pre-cooled acetone/methanol (1:1) fixative for 10 min at RT, endogenous peroxidase activity was quenched with 3% H2O2 in methanol for 10 min. The slides were washed in Tris-buffered saline (TBS) three times 10 min for each, blocked with 5% calf serum in TBS at RT for 1 h. They were then briefly rinsed with TBS and incubated with rabbit polyclonal antibodies against human Von Willebrand Factor (Catalog number, ab6994, Abcam, Cambridge, MA) in 1:200 dilution with 5% calf serum-TBS at 4°C overnight. Non-immunized rabbit IgG (Catalog number, I-1000, Vector Laboratories, Burlingame, CA) was used as a negative control. The specimens were washed in TBS three times with gentle agitation and incubated with HRP-conjugated anti-rabbit IgG antibodies (Catalog number, NEFB 12001EA, PerkinElmer, Covina, CA) at RT for 1 h. After washing, the DAB substrate (Catalog number, SK-4100, Vector Laboratories) was applied and signals were developed following the manufacture’s instruction. The specimens were counterstained with hematoxylin and dehydrated and mounted.
Luciferase Assay
The promoter regions for human LITAF (500 bp upstream of the transcription initiation site and human TNFSF15 (1000 bp upstream of the transcription initiation site) were subcloned into pGL3 vector that expresses fire fly luciferase as a reporter (Promega, Madison, WI), respectively. The plasmid was cotransfected with pNull-Rluc plasmid encoding Renilla luciferase into LNCaP cells using Lipofectamine 2000 (Invitrogen). Two days later, the cells were treated with or without AICAR for 8 hours and reporter gene assays conducted using a Dual luciferase assay kit purchased from Promega, as previously described (Xiang et al., 2002).
Chromatin immuoprecipitation (ChIP)
ChIP assay was performed as described by Nelson et al (Nelson et al., 2006). Briefly, LNCaP cells at an 80% confluent density were treated with or without AICAR for indicated times (see Figure legend). The cells were then cross-linked with formaldehyde, quenched and lysed in lysis buffer. Cell lysates were centrifuged. The supernatant was saved for Western blot. Chromatin pellets were resuspended in the lysis buffer, sonicated and centrifuged again. The supernatant was immunoprecipitated with LITAF antibody (BD Biosciences). The immunoprecipitate was assayed by Western blot and PCR to amplify the sequence (258 bp) containing a potential LITAF binding motif (CTCCC) using the primers (up: 5′ GACAGAGGGCTAGGCAGCAC 3′ and down: 5′ GCTCATGTGCTCCTTCCTTCG 3′. The PCR product was analyzed on agarose gel electrophoresis.
Statistic analysis
Difference between paired groups was tested by student’s t test. Trend difference within groups and between groups at different time points or doses was assessed by one-way and two-way ANOVA methods, respectively. Significance was set at P<0.05
Acknowledgments
This work is supported by NIH grants (R01 CA118918 to ZL, R01 DE014079 to SA, CA124490 to CC). We thank Dr. Zhixiong Xiao (BUMC) for providing p53 shRNA pSuperRetro construct as a generous gift. We are also thankful to Miss Siu Wong for her assistance in animal dissection, to Dr. Wen Guo for helping us with confirmatory qPCR analysis and to Dr. Mengwei Zang for her advice on statistic analysis.
Abbreviation
- AMPK
AMP-activated protein kinase
- AICAR
5-aminoimidazole-4-carboxamide 1-D-ribonucleoside
- BAEC
bovine aortic endothelial cells
- CMT1C
Charcot-Marie-Tooth type 1C disease, an autosomal genetic disease with demyelinating periphery nervous system
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- LKB1
liver kinase B, also named serine/threonine kinase 11 (STK11)
- LITAF
lipopolysaccharide-induced tumor necrosis factor alpha factor
- LPS
lipopolysaccharide
- SIMPLE
small integral membrane Protein of the lysosome/late endosome
- TNFSF15
tumor necrosis factor superfamily member 15
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Bayry J. Immunology: TL1A in the inflammatory network in autoimmune diseases. Nat Rev Rheumatol. 2010;6:67–8. doi: 10.1038/nrrheum.2009.263. [DOI] [PubMed] [Google Scholar]
- Beauvais K, Furby A, Latour P. Clinical, electrophysiological and molecular genetic studies in a family with X-linked dominant Charcot-Marie-Tooth neuropathy presenting a novel mutation in GJB1 Promoter and a rare polymorphism in LITAF/SIMPLE. Neuromuscul Disord. 2006;16:14–8. doi: 10.1016/j.nmd.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Bennett CL, Shirk AJ, Huynh HM, Street VA, Nelis E, Van Maldergem L, et al. SIMPLE mutation in demyelinating neuropathy and distribution in sciatic nerve. Ann Neurol. 2004;55:713–20. doi: 10.1002/ana.20094. [DOI] [PubMed] [Google Scholar]
- Chang JY, He RY, Lin HP, Hsu LJ, Lai FJ, Hong Q, et al. Signaling from membrane receptors to tumor suppressor WW domain-containing oxidoreductase. Exp Biol Med (Maywood) 2010;235:796–804. doi: 10.1258/ebm.2010.009351. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–85. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Haridas V, Shrivastava A, Su J, Yu GL, Ni J, Liu D, et al. VEGI, a new member of the TNF family activates nuclear factor-kappa B and c-Jun N-terminal kinase and modulates cell growth. Oncogene. 1999;18:6496–504. doi: 10.1038/sj.onc.1203059. [DOI] [PubMed] [Google Scholar]
- Hong YH, Lillehoj HS, Lee SH, Park DW, Lillehoj EP. Molecular cloning and characterization of chicken lipopolysaccharide-induced TNF-alpha factor (LITAF) Dev Comp Immunol. 2006;30:919–29. doi: 10.1016/j.dci.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Hou W, Medynski D, Wu S, Lin X, Li LY. VEGI-192, a new isoform of TNFSF15, specifically eliminates tumor vascular endothelial cells and suppresses tumor growth. Clin Cancer Res. 2005;11:5595–602. doi: 10.1158/1078-0432.CCR-05-0384. [DOI] [PubMed] [Google Scholar]
- Latour P, Gonnaud PM, Ollagnon E, Chan V, Perelman S, Stojkovic T, et al. SIMPLE mutation analysis in dominant demyelinating Charcot-Marie-Tooth disease: three novel mutations. J Peripher Nerv Syst. 2006;11:148–55. doi: 10.1111/j.1085-9489.2006.00080.x. [DOI] [PubMed] [Google Scholar]
- Ludes-Meyers JH, Kil H, Bednarek AK, Drake J, Bedford MT, Aldaz CM. WWOX binds the specific proline-rich ligand PPXY: identification of candidate interacting proteins. Oncogene. 2004;23:5049–55. doi: 10.1038/sj.onc.1207680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Zang M, Guo W. AMPK as a metabolic tumor suppressor: control of metabolism and cell growth. Future Oncol. 2010;6:457–70. doi: 10.2217/fon.09.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, Siebert R, Climent J, Fresquet V, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–80. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- Myokai F, Takashiba S, Lebo R, Amar S. A novel lipopolysaccharide-induced transcription factor regulating tumor necrosis factor alpha gene expression: molecular cloning, sequencing, characterization, and chromosomal assignment. Proc Natl Acad Sci U S A. 1999;96:4518–23. doi: 10.1073/pnas.96.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–85. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Niemann A, Berger P, Suter U. Pathomechanisms of mutant proteins in Charcot-Marie-Tooth disease. Neuromolecular Med. 2006;8:217–42. doi: 10.1385/nmm:8:1-2:217. [DOI] [PubMed] [Google Scholar]
- Pan X, Wang Y, Zhang M, Pan W, Qi ZT, Cao GW. Effects of endostatin-vascular endothelial growth inhibitor chimeric recombinant adenoviruses on antiangiogenesis. World J Gastroenterol. 2004;10:1409–14. doi: 10.3748/wjg.v10.i10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr C, Gan CH, Watkins G, Jiang WG. Reduced vascular endothelial growth inhibitor (VEGI) expression is associated with poor prognosis in breast cancer patients. Angiogenesis. 2006;9:73–81. doi: 10.1007/s10456-006-9033-1. [DOI] [PubMed] [Google Scholar]
- Picornell Y, Mei L, Taylor K, Yang H, Targan SR, Rotter JI. TNFSF15 is an ethnic-specific IBD gene. Inflamm Bowel Dis. 2007;13:1333–8. doi: 10.1002/ibd.20223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–5. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- Sethi G, Sung B, Aggarwal BB. Therapeutic Potential of VEGI/TL1A in Autoimmunity and Cancer. Adv Exp Med Biol. 2009;647:207–15. doi: 10.1007/978-0-387-89520-8_15. [DOI] [PubMed] [Google Scholar]
- Stucchi A, Reed K, O’Brien M, Cerda S, Andrews C, Gower A, et al. A new transcription factor that regulates TNF-alpha gene expression, LITAF, is increased in intestinal tissues from patients with CD and UC. Inflamm Bowel Dis. 2006;12:581–7. doi: 10.1097/01.MIB.0000225338.14356.d5. [DOI] [PubMed] [Google Scholar]
- Tang X, Fenton MJ, Amar S. Identification and functional characterization of a novel binding site on TNF-alpha promoter. Proc Natl Acad Sci U S A. 2003;100:4096–101. doi: 10.1073/pnas.0630562100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Marciano DL, Leeman SE, Amar S. LPS induces the interaction of a transcription factor, LPS-induced TNF-alpha factor, and STAT6(B) with effects on multiple cytokines. Proc Natl Acad Sci U S A. 2005;102:5132–7. doi: 10.1073/pnas.0501159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Metzger D, Leeman S, Amar S. LPS-induced TNF-alpha factor (LITAF)-deficient mice express reduced LPS-induced cytokine: Evidence for LITAF-dependent LPS signaling pathways. Proc Natl Acad Sci U S A. 2006;103:13777–82. doi: 10.1073/pnas.0605988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Zang M, Waelde CA, Wen R, Luo Z. Phosphorylation of 338SSYY341 regulates specific interaction between Raf-1 and MEK1. J Biol Chem. 2002;277:44996–5003. doi: 10.1074/jbc.M203953200. [DOI] [PubMed] [Google Scholar]
- Young HA, Tovey MG. TL1A: a mediator of gut inflammation. Proc Natl Acad Sci U S A. 2006;103:8303–4. doi: 10.1073/pnas.0602655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue TL, Ni J, Romanic AM, Gu JL, Keller P, Wang C, et al. TL1, a novel tumor necrosis factor-like cytokine, induces apoptosis in endothelial cells. Involvement of activation of stress protein kinases (stress-activated protein kinase and p38 mitogen-activated protein kinase) and caspase-3-like protease. J Biol Chem. 1999;274:1479–86. doi: 10.1074/jbc.274.3.1479. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Ni J, Jiang GW, Lu J, Xing L, Lincoln C, et al. VEGI, a novel cytokine of the tumor necrosis factor family, is an angiogenesis inhibitor that suppresses the growth of colon carcinomas in vivo. FASEB J. 1999a;13:181–9. doi: 10.1096/fasebj.13.1.181. [DOI] [PubMed] [Google Scholar]
- Zhai Y, Yu J, Iruela-Arispe L, Huang WQ, Wang Z, Hayes AJ, et al. Inhibition of angiogenesis and breast cancer xenograft tumor growth by VEGI, a novel cytokine of the TNF superfamily. Int J Cancer. 1999b;82:131–6. doi: 10.1002/(sici)1097-0215(19990702)82:1<131::aid-ijc22>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Zhang BB, Zhou G, Li C. AMPK: an emerging drug target for diabetes and the metabolic syndrome. Cell Metab. 2009;9:407–16. doi: 10.1016/j.cmet.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Zhou J, Huang W, Tao R, Ibaragi S, Lan F, Ido Y, et al. Inactivation of AMPK alters gene expression and promotes growth of prostate cancer cells. Oncogene. 2009;28:1993–2002. doi: 10.1038/onc.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]