Abstract
Bacteria can coordinate group behavior using chemical signals in a process called quorum sensing (QS). The QS system in the opportunistic pathogen Pseudomonas aeruginosa is largely governed by the LasR receptor and its cognate chemical signal, N-(3-oxo)-dodecanoyl l-homoserine lactone (OdDHL). LasR also appears to share this signal with an orphan LuxRtype receptor in P. aeruginosa, termed QscR, which represses LasR activity. Non-native molecules that modulate QscR would represent valuable tools to study the role of this novel QS repressor protein in P. aeruginosa. We performed a critical analysis of previously identified, non-native N-acylated l-homoserine lactone (AHL) activators and inhibitors of QscR to determine a set of structure–activity relationships (SARs). Based on these SAR data, we designed, synthesized, and screened several second-generation libraries of AHLs for new ligands that could target QscR. These studies revealed the most active AHL agonists and antagonists of QscR reported to date, with activities ranging from nanomolar to low micromolar in a QscR bacterial reporter strain. Several of these AHLs were highly selective for QscR over LasR and other LuxR-type receptors. A small subset of the new QscR activators, however, were also found to inhibit LasR; this demonstrates the exciting potential for the synergistic modulation of these integral P. aeruginosa QS receptors by using a single synthetic compound.
Keywords: acylated homoserine lactone, biological activity, Pseudomonas aeruginosa, quorum sensing, screening
Introduction
Small molecule signals provide bacteria with a chemical language that can direct individual cells to behave as a unified group. Bacteria frequently use this signaling mechanism, termed quorum sensing (QS), to establish relationships with a eukaryotic host.[1–3] While these bacteria–host associations can be symbiotic, as in the case of the marine symbiont Vibrio fischeri, in which QS was first characterized,[4, 5] bacteria often use QS to coordinate group processes that can intensify their pathogenic response and result in host infection. Gram-negative bacteria primarily use diffusible N-acylated l-homoserine lactones (AHLs) for QS (Scheme 1).[6–8] Binding of the AHL signals to their cognate receptors, or LuxR-type proteins,[9] controls the expression of genes responsible for myriad group behaviors, including virulence factor and antibiotic production, swarming, and biofilm formation.[10,11]
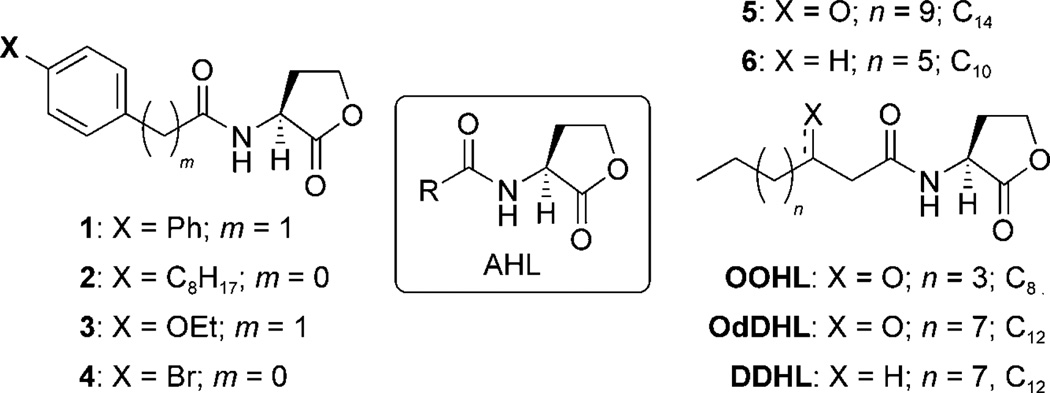
Scheme 1.
General structure of AHLs (center) and control compounds used in this study. The number of acyl tail carbons in selected control AHLs is indicated for clarity.
QS has attracted considerable interest as a new antivirulence target in humans,[12, 13] and several methods to attenuate QS in bacterial pathogens have been developed.[14, 15] Significant recent work has focused on the modulation of QS signaling pathways in Gram-negative bacteria by intercepting native AHL–LuxR-type receptor binding using non-native molecules.[16–18] The rational design and screening of focused libraries has been effectively implemented to identify potent modulators of QS receptor proteins by using cell-based reporter gene systems.[19, 20] Our laboratory has contributed to this area, and we have recently focused on the rational design of libraries of AHL analogues.[16, 21–27] These libraries total over 140 non-native AHLs that largely vary in the composition of the acyl tail. We have evaluated these compounds in reporter strains for a range of LuxR-type receptors, including TraR from Agrobacterium tumefaciens, LuxR from V. fischeri, and LasR from Pseudomonas aeruginosa. Through these studies, we have identified several potent non-native AHLs capable of controlling receptor activity at nanomolar to low micromolar concentrations and numerous interesting structure–activity relationships (SARs).[16, 23]
We now seek to develop new AHL libraries with improved activity over their parent compounds, and more notably, improved receptor selectivity. Such selectivity would be extremely valuable in experiments in which selective modulation of LuxR-type proteins would be desirable. For example, bacteria commonly live in multispecies communities, and selectively silencing QS in a single species could provide a significant advantage to neighboring species.[28] Alternatively, single species often use several AHLs to control QS circuits composed of multiple LuxR-type receptors[29] and the ability to target a specific receptor in these bacteria could provide a chemical method to study and delineate such complex signaling networks. This latter challenge in individual bacterial species has become one of our main research foci, and is the motivation for the current study.
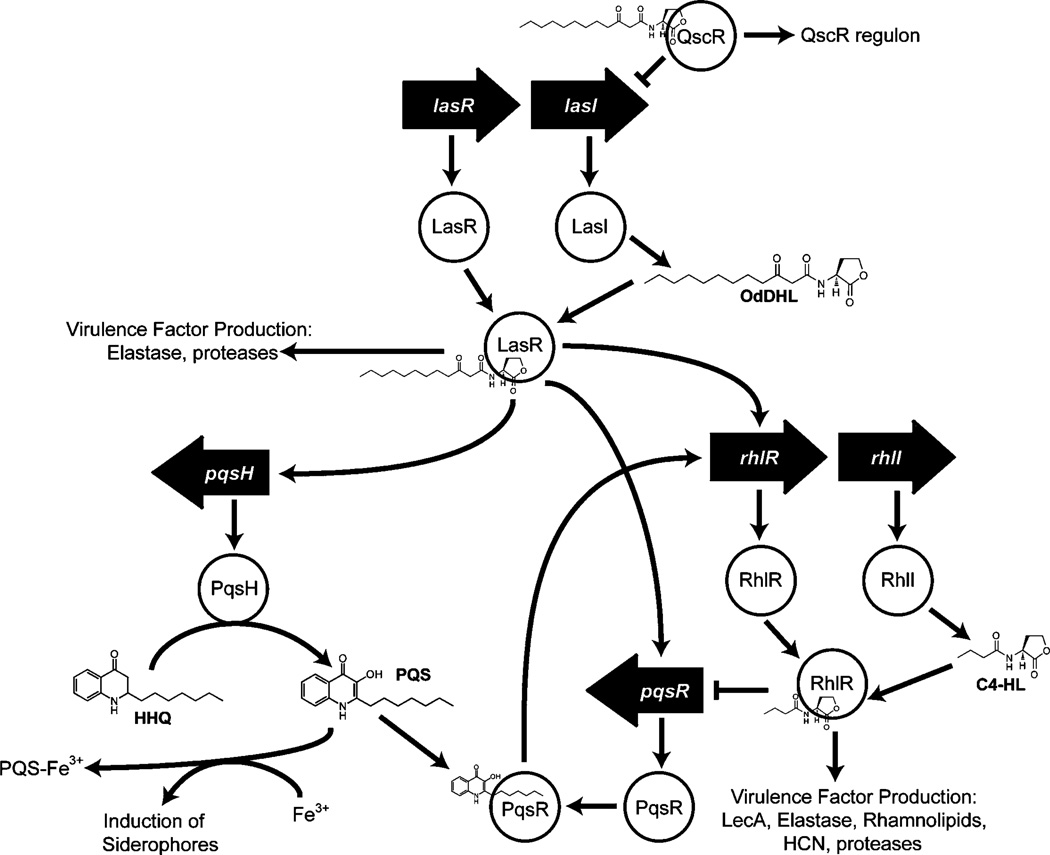
The opportunistic pathogen P. aeruginosa provides a clinically relevant model system with such a multipartite QS circuit (Scheme 2). This organism harbors two main AHL signals, N-(3-oxo)-dodecanoyl HL (OdDHL) and N-butanoyl HL (C4-HL), and at least three LuxR-type receptors (LasR, RhlR, and QscR) that govern a range of phenotypes at high cell densities, including the production of virulence factors, rhamnolipids, and biofilms.[30–35] Each of these receptors represents an attractive target for interception by using non-native AHL mimics.[20] At the center of the QS circuit, LasR uses OdDHL to control multiple signaling pathways, including the RhlR–C4-HL circuit. The QS control repressor (QscR) protein is an orphan receptor that lacks an associated synthase enzyme, and appears to utilize LasR’s signal, OdDHL, for activation. Such orphan receptors or “solos” have emerged as relatively common components in LuxR-type QS systems.[36, 37] However, in contrast to LasR, QscR is a negative regulator of QS-controlled phenotypes. QscR represses both the Las and Rhl systems, in addition to controlling its own independent regulon.[38] P. aeruginosa QscR mutants are hypervirulent, and in turn, QscR overproducing strains are avirulent.[39] As such, the repressor QscR holds a pivotal position in the P. aeruginosa QS system, and synthetic modulators of QscR are of significant interest. For example, selective activation of this LuxR-type receptor in P. aeruginosa could attenuate virulence factor production in this organism. Further, the development of chemical methods to target LasR over QscR (or vice versa) despite their use of an identical native AHL (OdDHL), would help to clarify their roles in controlling P. aeruginosa virulence and could contribute to novel antivirulence approaches.
Scheme 2.
Simplified schematic of the QS signaling circuit in P. aeruginosa. The LuxI-type synthases, LasI and RhlI, generate the AHL ligands OdDHL and C4-HL, respectively. LasR is the dominant system, and together with the RhlR system controls a host of QS controlled genes. QscR negatively regulates lasI in addition to controlling its own regulon.[40, 41] The Las system also regulates production of the Pseudomonas quinolone signal (PQS), which is involved in siderophore production and other processes.[42]
Herein, we report our discovery of a set of new and potent non-native AHLs capable of strongly modulating QscR in P. aeruginosa. These compounds were designed through several cycles of design, screening, and SAR analyses. In an earlier study, we screened a library of ~100 unbiased AHL mimics for QscR inhibition and activation.[26] We delineated key SARs from this previous study for QscR modulation, and synthesized three second-generation AHL libraries that were designed to target QscR. Cell-based screens of these libraries revealed some of the most potent agonists and antagonists of QscR reported to date. Several of these compounds were selective for QscR over LasR and other LuxR-type receptors. Moreover, a set of these compounds were capable of both activating QscR and inhibiting LasR, thereby demonstrating, to our knowledge, for the first time, the potential for synergistic QS control through the modulation of two LuxR-type receptors with one nonnative AHL.
Results and Discussion
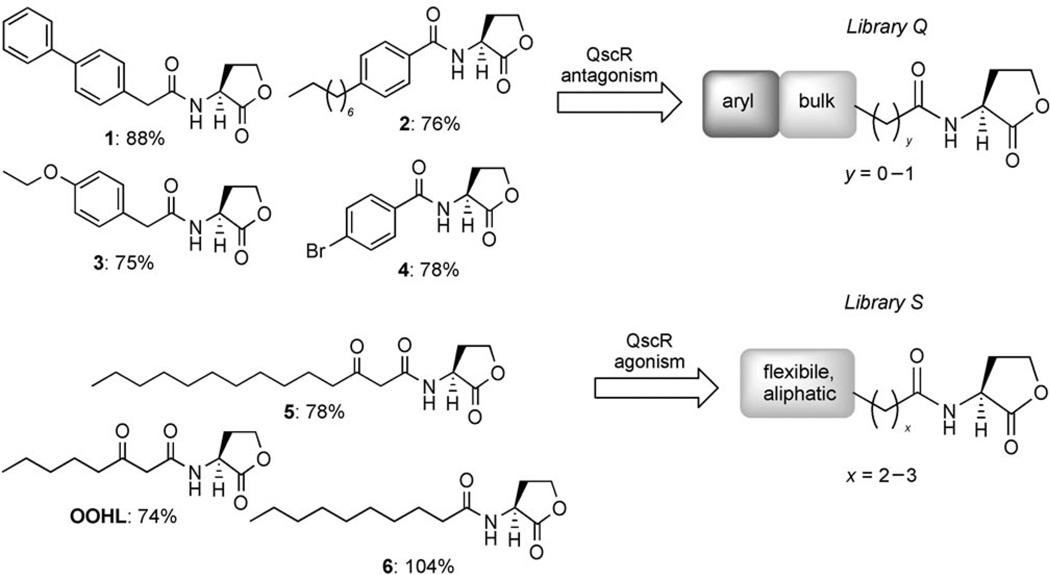
In our preliminary studies of QscR, we evaluated a library of ~100 non-native AHL derivatives for QscR inhibition and activation in an E. coli reporter strain (see the Experimental Section for all strain details).[26] The most active AHL antagonists uncovered in this library were shown to inhibit binding of QscR to its target DNA sequence, possibly by destabilizing the protein complex upon out-competing OdDHL (assuming the ligands target the same site). We scrutinized the structures of these initial AHL leads to determine SARs for both QscR antagonism and agonism (Scheme 3). This analysis framed the design of 29 new synthetic AHLs that comprise libraries Q, R, and S.
Scheme 3.
Graphical representation of selected SARs generated from analysis of preliminary QscR “hits” (1–6; left). The activities of these AHLs in the primary screens are indicated as percent relative to the positive control (DDHL; see main text). In previous studies, AHL antagonists 1–4 inhibited LasR by <30%, and only AHL agonist 5 activated LasR significantly (87 %).[23, 24] These initial hits were used as controls in this study. Activity trends originating from these SARs were utilized in part for the design of the second-generation libraries, shown on the right (libraries Q and S).
General ligand design, synthesis, and biological screening
In each of the following sections, we provide a brief outline of our design process for libraries Q, R, and S, followed by a detailed discussion of the effects of these compounds on receptor activity using reporter gene assays. The AHL libraries were synthesized using our previously reported solid-phase synthesis methods[21, 23] (see the Experimental Section) in high purities and in sufficient quantities for numerous biological assays (ca. 20 mg).
Reporter gene assays are standard methods for the screening of small-molecule libraries for LuxR-type receptor modulation, and we utilized established strains for this purpose in the current study. N-Dodecanoyl HL (DDHL), instead of OdDHL, was used as a positive control in the QscR assays (Scheme 1). DDHL has previously been shown to activate QscR at least as well as OdDHL, and is more straightforward to synthesize.[26, 41] Compounds were tested in reporter strains for QscR antagonism (in the presence of the DDHL at its EC50: 20 nm) and QscR agonism (synthetic compound alone). Briefly, ligand screening in the QscR reporter system consisted of incubating compounds in a 96-well plate in the presence of mid-log phase E. coli cells containing a QscR β-galactosidase (β-gal) reporter, followed by Miller assay evaluation (see the Experimental Section).
Designing AHLs for QscR antagonism: library Q
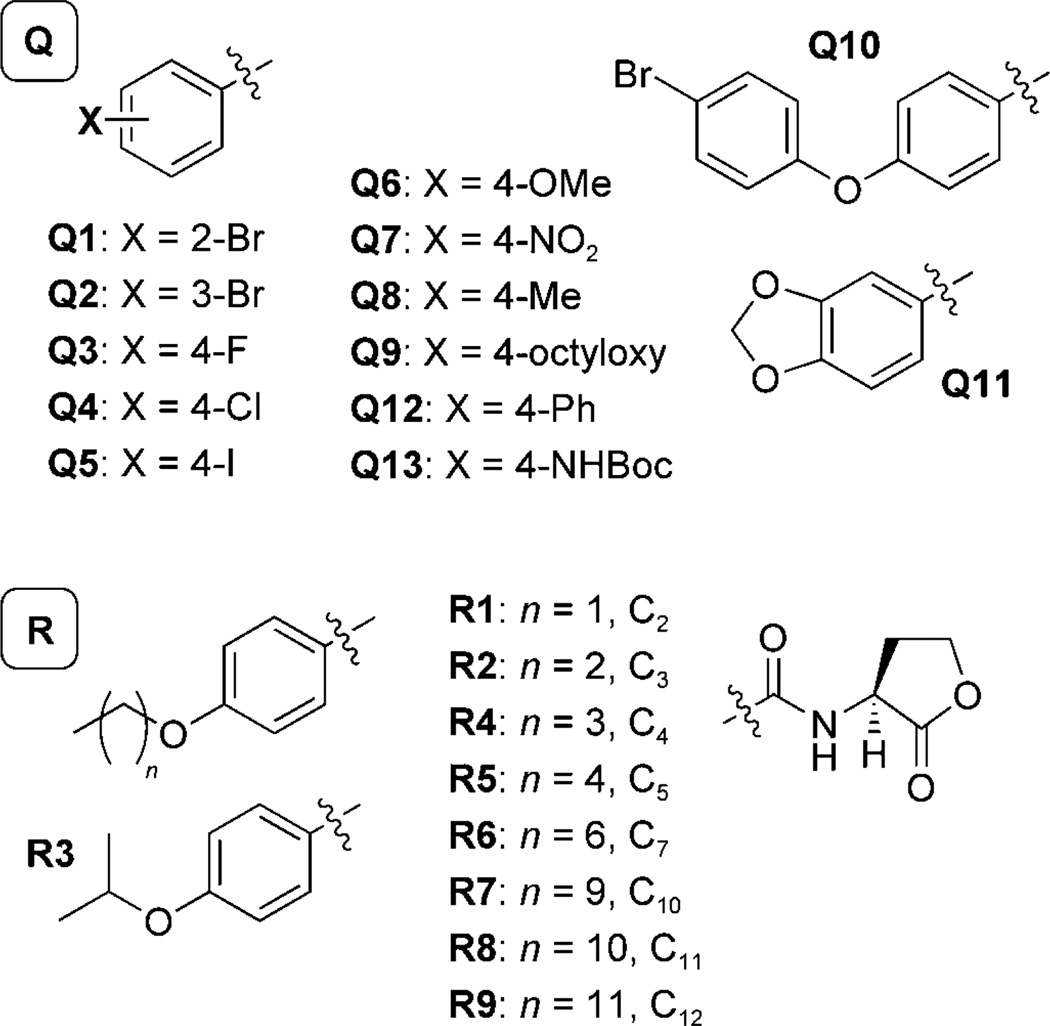
Structural trends from the earlier AHL library that conferred QscR antagonistic activity were used to design library Q. Sterically bulky synthetic AHLs, especially those bearing aromatic groups (e.g., control compounds 1–4; Scheme 3), were observed to be strong antagonists of QscR with EC50 values in the mid- to high-nanomolar range (Table 1). Especially interesting from this set of hits were the N-benzoyl HLs (BnHLs) 2 and 4, which were two of the five most potent antagonists of QscR identified. BnHLs had not shown significant antagonistic activities against other LuxR-type receptors (i.e., LasR, LuxR, or TraR) in our previous studies.[23, 24] In contrast to these receptor homologues, QscR appeared to be antagonized by AHLs with steric bulk/substituents on the acyl chain adjacent to the amide. We incorporated these structural features, along with acyl chain aromatic functionality, into the design of AHL library Q. Compounds Q1 to Q13 (Scheme 4) more thoroughly explore the effect of BnHL functionality on QscR antagonism by positioning electron-donating (OCH3, CH3) and electron-withdrawing groups (Br, F, Cl, I, NO2) around the benzoyl ring (AHLs Q1–Q8). To probe steric tolerance farther away from the amide moiety, BnHLs Q9–Q13 were designed to move and expand steric bulk along the AHL acyl chain.
Table 1.
IC50 and EC50 values in E. coli reporter strains for the most active non-native AHLs in LasR and QscR.[a]
| Compound | IC50 [µm] | EC50 [µm] | ||
|---|---|---|---|---|
| QscR | LasR | QscR | LasR | |
| OdDHL | – | –[d] | 0.022[c] | 0.01[b] |
| DDHL | – | – | 0.005[c] | 0.04[b] |
| OOHL | – | 0.11[b] | 0.011[c] | –[d] |
| 1 | 0.030[c] | – | – | – |
| 2 | 0.13[c] | – | – | – |
| 3 | 0.16[c] | – | – | – |
| 4 | 0.18[c] | – | – | – |
| 5 | – | – | 0.056[c] | 0.01[b] |
| 6 | – | 0.25[b] | 0.006[c] | >200[b] |
| Q4 | 0.56[e] | – | – | – |
| Q5 | 0.11[e] | – | – | – |
| Q9 | 0.011 | – | – | – |
| Q10 | – | – | 1.1 | – |
| Q12 | 0.14[e] | – | – | – |
| Q13 | 0.22[e] | – | – | – |
| R1 | 1.9[e] | – | – | >200 |
| R4 | – | – | 2.0 | – |
| R5 | – | – | 0.89 | – |
| R6 | 0.057 | 1.0 | – | – |
| S1 | – | – | – | – |
| S2 | – | – | 0.10 | – |
| S3 | – | – | 0.079 | >200 |
| S6 | – | – | – | – |
| S7 | 0.020[e] | 10 | 0.078 | – |
For strain and assay information, see the Experimental Section. Determined by testing AHLs over a range of concentrations (10−2–105 nm). All assays were performed in triplicate; italicized compounds are controls.
Values reported previously, see ref. [24].
Values reported previously, see ref. [26].
Not determined.
Antagonism dose–response curve up-turned at higher concentrations; see the Supporting Information.
Scheme 4.
Structures of the AHL library Q (Q1–Q13, top) and AHL library R (R1–R9, bottom) designed to antagonize QscR. All library members contained the l-HL head-group (right) and varied in their acyl group substitution. The carbon number is indicated in the library R acyl groups for clarity.
Assay results: library Q
Over two-thirds of the AHLs in library Q inhibited QscR by at least 50%, and five AHLs inhibited QscR by >67% at 5 µm in the presence of 20 nm DDHL (250:1; see the Supporting Information for all primary screening data). Key trends in antagonistic activity and new SAR are outlined below. The most active compounds (>67% β-gal inhibition) from these primary assays were further evaluated by using dose–response experiments in the E. coli QscR reporter strain, and IC50 values were calculated and compared to those for the controls (Table 1).
Compounds Q1–Q8 revealed several interesting trends in QscR antagonistic activity caused by altering the electronics of the BnHL benzoyl group. The inhibitory activities of bromo-substituted BnHLs Q1, Q2, and 4 increased as the substituent was moved further from the amide bond (ortho < meta < para), and inhibitory activity of the para-substituted BnHLs increased with the size of the halogen (Q3 < Q4 < 4 < Q5). These SAR trends closely mirror those observed for N-phenylacetanoyl HL (PHL) antagonists for other LuxR homologues in our previous studies.[22–25] Interestingly, the BnHL bearing the electron-withdrawing para-NO2 group (Q7) decreased QscR activity to the same level as the electron-donating para-CH3 BnHL Q8 (48±4 vs. 49±3% inhibition, respectively), providing support that steric bulk, as opposed to electronic nature, on an aryl group para to the amide group is favorable for QscR antagonism. Despite the nanomolar potency of the bi-aryl AHL control antagonist 1 (Table 1), size appeared to not be the exclusive feature required for QscR antagonistic activity; for example, the bulkier BnHLs Q10 and Q11 were less potent antagonists (<45 %). Incorporating both structural flexibility and steric bulk, as exemplified by Q9 and Q13, appears to be beneficial for QscR antagonism.
Several of the Q library antagonists exhibited weak agonism at high micromolar concentrations (Table 1), a trend that we have previously interpreted as characteristic of partial agonism at these very high concentrations.[23] However, only Q10 showed appreciable agonistic activity (Table 1, 30 %), highlighting the effectiveness of using our SARs for antagonist design. The most active inhibitor from this family of antagonists was Q9 (IC50=11 nm, Table 1). Compound Q9 blends the structural elements of control antagonists 2 and 3 by converting the benzyl-ether phenylacetic AHL 3 into a BnHL like 2. This compound was tenfold more active than its parent compounds and is one of the most active synthetic antagonists reported, to our knowledge, for QscR to date. Notably, Q9 can competitively inhibit QscR by 50% at a twofold lower concentration than DDHL.
Designing AHLs for QscR antagonism: library R
The high antagonistic activity observed for Q9 against QscR motivated us to design a second-generation library (library R) to explore the structural features of this compound critical for activity. Library R contained Q9 analogues with acyl tails of varying lengths and branching (Scheme 4). We subjected this library to the analogous QscR antagonism and agonism screens as for library Q, and determined IC50 values for the four most potent compounds (Table 1).
Assay results: library R
In general, library R provided weaker QscR antagonists relative to lead AHL Q9. The most potent antagonists in library R were AHLs R1 and R6, which exhibited IC50 values of 1.9 µm and 57 nm, respectively (Table 1). Their higher activities relative to other library R members potentially could be attributed to their optimal acyl group size; the acyl chain of R1 extends eight atoms from the amide bond, as does the QscR agonist OOHL, while the acyl chain of R6 extends 13 atoms from the amide bond, and is the closest in length to the agonist OdDHL (Table 1). As such, these antagonists could fulfill a steric requirement in a QscR ligand binding site that makes them strong competitors with natural AHL ligands. Compounds R4 and R5 were actually modest QscR agonists, as opposed to antagonists, with low micromolar and submicromolar EC50 values (Table 1).
Comparing QscR antagonistic activities for compounds R1 through R5 revealed decreasing activity with increasing alkyl chain length (two to five carbons; see the Supporting Information for all primary data). However, this trend halted abruptly with R6, which contained a seven-carbon alkyl chain and was the most potent antagonist. Inhibitory activity followed another stepwise decrease as the alkyl chain lengthened from R7– R9. Such a specific preference for length has been previously observed for AHL antagonists of TraR and LuxR, which are most strongly antagonized by AHLs with seven or ten atoms in the acyl tail, respectively.[24]
Designing AHLs for QscR agonism: library S
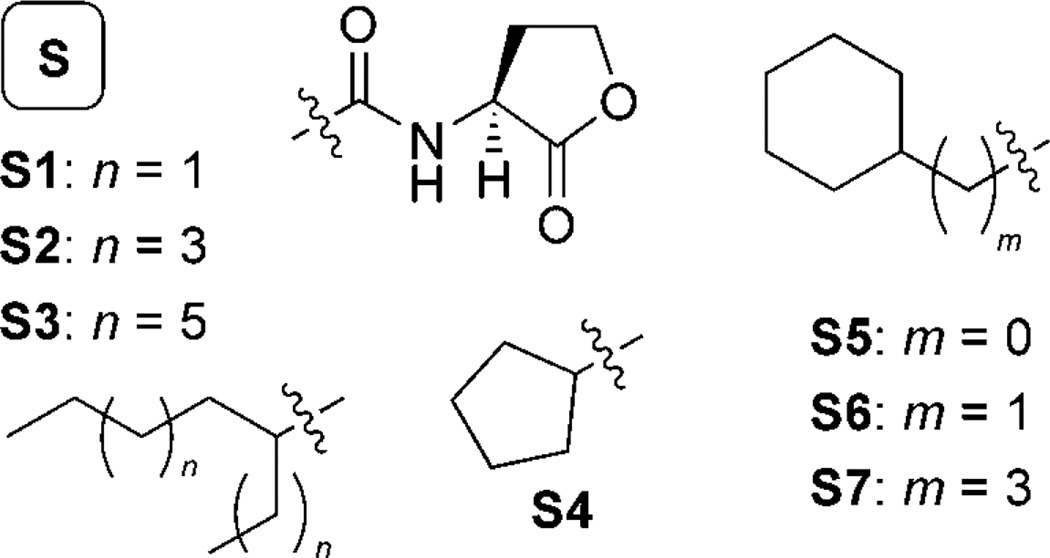
Turning next to agonist design, we utilized trends in QscR agonistic activity derived from our initial AHL libraries to design library S (Scheme 5). Several of our initial agonist hits were natural AHLs (5, 6, and OOHL; Scheme 3) and could strongly activate QscR (>50%; 5 µm). The other most potent agonists had lipophilic alkyl chains, but also contained sterically bulky substituents alpha to the amide group. We, therefore, incorporated these two structural features into library S. Compounds S1–S3 contained α-branched aliphatic acyl groups of varying lengths, while S4–S7 were designed to test the effects of moving the branched substituent in a stepwise fashion down the acyl chain away from the amide. The acyl chain of each member of library S consisted solely of sp3-hybridized carbons, imparting an element of flexibility into these AHLs that further distinguishes them from the members of libraries Q and R.
Scheme 5.
Structures of the AHL library S (S1–S7) designed to agonize QscR. All library members contained the l-HL head-group (top, center) and varied in their acyl group substitution.
Assay results: library S
Screening of library S in the E. coli QscR reporter strain revealed three new agonists of QscR (AHLs S2, S3, and S7). Interestingly, butyl-cyclohexyl HL S7 was also found to be a weak antagonist of QscR. However, no other library S member displayed appreciable antagonistic activity against QscR, and further demonstrated the utility of our SAR analyses in developing agonist-specific AHL libraries for QscR.
We performed dose–response analyses on agonists S2, S3, and S7 to determine their EC50 values (Table 1). While potent, all three were ~tenfold less active than the native AHL controls (OdDHL, DDHL, OOHL, and 6). The most active QscR agonists were the branched-alkyl HLs S3 and S7, both with EC50 values below 80 nm. As was demonstrated by library R, QscR appears to prefer AHL ligands with significantly more acyl chain steric bulk than other LuxR homologues. In library S, this trend is best shown by branched AHLs S1–S3. Agonistic activity decreased as the lengths of the branched alkyl tails decreased; S2 was a less potent agonist than S3 (Table 1), while the still shorter S1 showed minimal agonistic activity in QscR (7 %; see the Supporting Information).
Ligand selectivity for QscR
The library design and screening studies detailed above revealed a new set of potent QscR modulators. As introduced above, non-native ligands with selectivity for either LasR or QscR, as well as ligands that have activity in both receptors (i.e., tandem QscR activation and LasR inhibition (QS inhibition), or tandem QscR inhibition and LasR activation (QS activation)) would be especially powerful tools for the study of QS in P. aeruginosa (Figure 1). Indeed, the signaling crosstalk and interplay between the LasR and QscR receptors provides a unique opportunity to apply a chemical approach for synergistic modulation of QS activity and virulence factor production with a single compound. Therefore, we sought to examine the selectivity of libraries Q–S for QscR over LasR, and we screened these libraries in analogous bacterial reporter gene assays for LasR antagonism and agonism (see the Experimental Section for strain information). An analysis of the LasR screening data is provided below (see the Supporting Information for full primary LasR screening data).
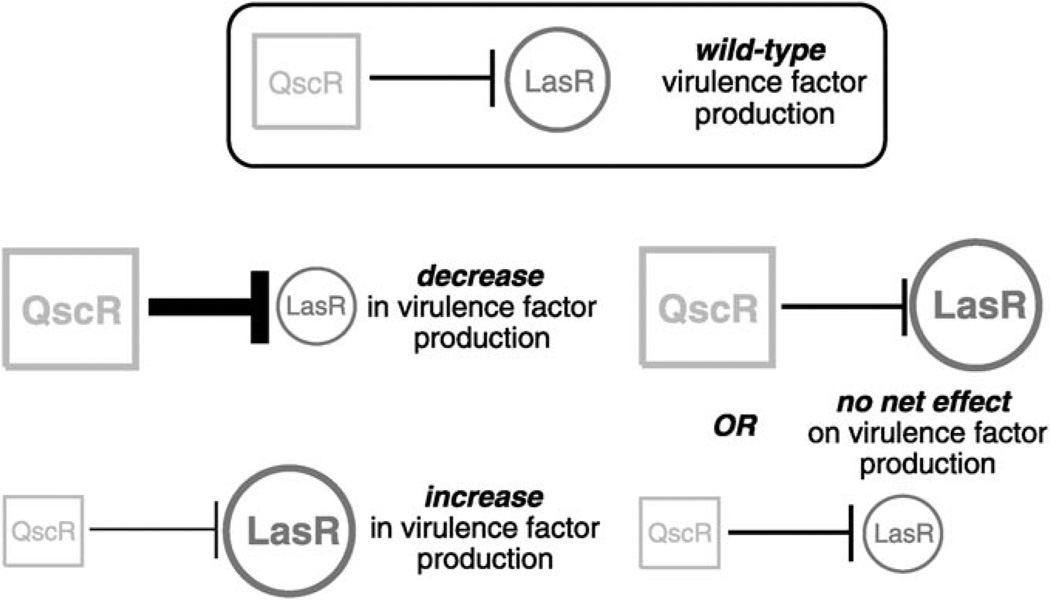
Figure 1.
Schematic of the potential effects of modulating QscR and LasR receptor activities in P. aeruginosa. A) Normal receptor function: QscR negatively regulates LasR and provides a limit on virulence factor production. B) Increased QscR activity decreases LasR activity. C) Decreased QscR activity increases LasR activity. D) Simultaneous increase or decrease in QscR and LasR activities might show no net change in virulence factor production.
Interestingly, no AHLs in libraries Q–S were found to be agonists of both QscR and LasR. This was somewhat surprising since these receptors are both strongly activated by the same native ligand, OdDHL. However, our screens revealed that numerous library members (Q1–Q4, Q6, Q7, Q12, Q13, R2, R3, R6, R7, S1, S4, S5, and S7) were at least weak antagonists of both receptors (antagonism activity >15% with <10% agonism in primary screens). These findings provide some insight into a mode of action for these ligands in QscR and LasR, by suggesting that the mechanism of inhibition between the two receptors is similar, while the requirements for agonism are different.
The LasR primary screening studies of libraries Q–S yielded several other activity trends that were directly relevant to our interest in the selective and synergistic modulation of multiple LuxR-type receptors in P. aeruginosa. Each trend is discussed in turn. First, we were excited to observe that many of the most active synthetic modulators of QscR reported herein were also selective for QscR over LasR in reporter strains. Notably, the BnHLs Q5 and Q9 antagonized QscR by at least 94% at 5 µm, yet displayed no activity in LasR (≤1% agonism or antagonism). In addition, antagonists Q8, Q10, Q11, R8, and S6 inhibited QscR by 25–60% and were also inactive in LasR. BnHL Q10 was the only QscR agonist identified in this study that lacked LasR antagonistic activity. In turn, the only compound that selectively modulated LasR was R9, which weakly antagonized LasR by 22% and displayed minimal QscR activity (only 2% activation).
Second, in terms of synergistic modulation trends, the lipophilic AHLs S3 and S2 were found to be both potent QscR agonists (EC50 values=~100 nm) and weak LasR inhibitors (32 and 16%, respectively). AHLs R4 and R5, which were relatively moderate agonists of QscR (<50% at 5 µm), were also weak inhibitors of LasR (<15% at 10 µm). Only one compound identified in this study had the opposite activity: R1 strongly antagonized QscR (IC50=1.9 µm) and simultaneously was a weak agonist of LasR (37% at 10 µm). Despite their relatively low activities compared to other ligands uncovered in this study, the discovery of these five ligands is significant. Their simultaneous activities in LasR and QscR define them as a set of lead chemical scaffolds on which new generations of ligands could be developed that modulate multiple receptors in P. aeruginosa with improved potencies. Such studies are currently underway in our laboratory.
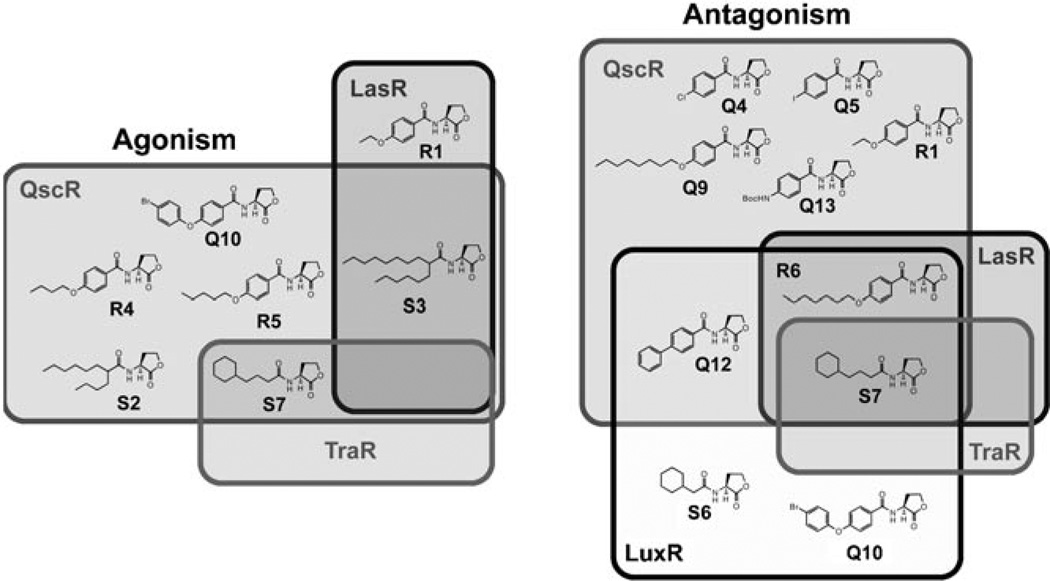
To expand the analysis of these libraries beyond P. aeruginosa and examine their receptor selectivity further, we also screened libraries Q–S for agonistic and antagonistic activity in LuxR (in V. fischeri) and TraR (in A. tumefaciens). These experiments complement previous comparative studies reported by our laboratory to probe the activities of AHLs between different Gram-negative bacteria.[16, 23, 25] Generally, our new QscR ligands displayed limited activities in these other two species; this further highlights the selectivity of these compounds for P. aeruginosa, and more specifically, for QscR (Figure 2; see the Supporting Information for full screening data for LuxR and TraR). One compound, however, did not fit this trend. AHL S7 was found to be a potent (>50%) antagonist of all four receptors (QscR, LasR, TraR, and LuxR). Intriguingly, it also agonized QscR (EC50=0.02 µm; see above) and very weakly agonized TraR (8% at high concentrations). The activity of S7 in multiple LuxR-type receptors demonstrates the potential that this compound holds as broader-spectrum modulator of QS in Gram-negative bacteria. Previous studies in our laboratory have revealed additional ligands of this activity class (e.g., 4-Br phenyl-propionoyl HL);[16, 23] however, AHL S7 represents the first wholly aliphatic variant that we have uncovered with such multireceptor activity.
Figure 2.
Venn diagrams showing the receptor selectivity of the most active agonists and antagonists identified in this study in QscR, LasR, TraR, and LuxR. Overlapping regions indicate ligands with notable activity in two or more receptors (see main text). Left: active agonists; right: active antagonists. The QscR agonists and antagonists were the most selective ligands identified in this study. See the Supporting Information for primary screening and dose–response characterization data for compounds in A. tumefaciens and V. fischeri.
Conclusions
We have designed and synthesized three second-generation AHL libraries and evaluated their activity as non-native modulators of QscR in P. aeruginosa. These studies have yielded several highly active and QscR-selective agonists and antagonists, with activities ranging from nanomolar to low micromolar in a QscR bacterial reporter strain. Generally, the most active QscR antagonists had N-benzoyl acyl groups (i.e., BnHLs, such as the nanomolar inhibitor Q9), and the QscR agonists had branched lipophilic acyl groups, establishing that synthetic AHLs with sterically bulky acyl chains are effective non-native ligands for QscR. This preference for size was exemplified by the nanomolar agonistic activities of branched aliphatic AHLs, such as S3.
The QscR-selective AHLs reported herein represent new chemical tools for controlling QS in P. aeruginosa. These compounds could be especially useful for further elucidating the roles of QscR in QS-controlled phenotypic responses, and potentially even in interspecies signal sensing. This latter pathway is being invoked for QscR with increasing frequency.[38, 40, 43] In addition, these new ligands provide a route toward further probing of the complex relationship between QscR and LasR in P. aeruginosa. The application of non-native AHLs with synergistic activities against multiple QS receptors in P. aeruginosa, such as the initial leads S2, S3, and S7, could permit virulence attenuation by using a single compound. To probe these and related research questions, we are currently developing new assays in our laboratory that allow for the quantitative analysis of the effects of these compounds on receptor specific, phenotypic QS responses in P. aeruginosa. The selective regulation of one QS circuit within one organism, and also within mixed bacterial communities, by using receptor/species selective chemical probes represents a novel approach toward the modulation of QS-controlled phenotypes, and could have broad utility. The results presented herein suggest that such chemical probes will be accessible.
Experimental Section
Chemistry
AHL libraries Q–S and the control compounds 1–6 were prepared by microwave-assisted solid-phase synthesis using previously described methods.[21, 23] Purities and isolated yields for these AHLs were 91–99% and 15–90%, respectively. Compounds were submitted to bacteriological assays following resin cleavage and an aqueous work-up without further purification. Full characterization data for the active compounds, as well as details of the instrumentation and analytical methods used in this work can be found in the Supporting Information.
Stock solutions of synthetic compounds were prepared in DMSO and stored at room temperature in sealed vials. Compounds were transferred into solvent resistant polypropylene or polystyrene 96- well multititer plates when appropriate for small-molecule screening, with the amount of DMSO never exceeding 2% (by volume).
Bacteriology
The four bacterial reporter strains utilized in this study were E. coli DH5α harboring the QscR expression vector pJN105Q and a plasmid-born PA1897-lacZ expression vector (pJL101),[41] E. coli DH5α harboring the LasR expression vector pJN105L and a plasmid-born PlasI–lacZ fusion (pSC11),[41] V. fischeri ES114 (ΔluxI),[44] and A. tumefaciens WCF47 (ΔtraI) harboring a plasmid-born PtraI–lacZ fusion (pCF372).[45] All bacteria were grown in a standard laboratory incubator with shaking (200 rpm). Absorbance and luminescence measurements were obtained by using a PerkinElmer Wallac 2100 EnVision multilabel plate reader with Wallac Manager v 1.03 software. All bacteriological assays were performed in triplicate.
Reporter gene assay protocols
Primary assays for QscR, LasR, LuxR, and TraR activity were performed as reported,[23, 26] with the following adaptations. In the case of the QscR reporter, DDHL was used in place of OdDHL as the native ligand for the system. For all antagonism assays, the concentration of native ligand utilized was approximately equal to its EC50 value in each bacterial reporter strain. For QscR antagonism assays, synthetic ligand (5 µm) was screened against DDHL (20 nm). For LasR, TraR, and LuxR antagonism assays, synthetic compound (10 µm) was screened in the presence of OOHL (100 nm), OdDHL (10 nm), or N-(3-oxo-hexanoyl) HL (3 µm; OHHL), respectively. For QscR agonism assays, synthetic compound (5 µm) was screened against DDHL (5 µm). For TraR, LasR, and LuxR agonism assays, synthetic compound (10 µm) was screened alongside the natural ligand (10 µm) for the system (see the Supporting Information for full protocols and primary assay data). Dose response reporter gene assays were performed according to the protocols outlined above by using varying concentrations of compound. The IC50 and EC50 values were calculated with GraphPad Prism software (v. 4.0) by using a sigmoidal curve fit.
Supplementary Material
Acknowledgements
Financial support for this work was provided by the NIH (AI063326), Greater Milwaukee Foundation Shaw Scientist Program, Burroughs Welcome Fund, Camille & Henry Dreyfus Foundation, Research Corporation, and Johnson & Johnson. We gratefully acknowledge Professors Peter Greenberg (University of Washington), Stephen Winans (Cornell University), and Edward Ruby (UW–Madison) for donations of bacterial strains and advice on their manipulation.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201000708.
References
- 1.Camilli A, Bassler BL. Science. 2006;311:1113. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler BL, Losick R. Cell. 2006;125:237. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Platt TG, Fuqua C. Trends Microbiol. 2010;18:383. doi: 10.1016/j.tim.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nealson KH, Platt T, Hastings JW. J. Bacteriol. 1970;104:313. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nealson KH, Hastings JW. Microbiol. Rev. 1979;43:496. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazdunski AM, Ventre I, Sturgis JN. Nat. Rev. Microbiol. 2004;2:581. doi: 10.1038/nrmicro924. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua C, Greenberg EP. Nat. Rev. Mol. Cell Biol. 2002;3:685. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 8.Welch M, Mikkelsen H, Swatton JE, Smith D, Thomas GL, Glansdorp FG, Spring DR. Mol. Biosyst. 2005;1:196. doi: 10.1039/b505796p. [DOI] [PubMed] [Google Scholar]
- 9.Schuster M, Greenberg EP. In: Chemical Communication among Bacteria. Winans SC, Bassler BL, editors. ASM Press; Washington, DC: 2008. p. 133. [Google Scholar]
- 10.de Kievit TR, Iglewski BH. Methods Enzymol. 1999;310:117. doi: 10.1016/s0076-6879(99)10010-7. [DOI] [PubMed] [Google Scholar]
- 11.de Kievit TR, Iglewski BH. Infect. Immun. 2000;68:4839. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanley S, Hung D. Biochemistry. 2009;48:8776. doi: 10.1021/bi9009083. [DOI] [PubMed] [Google Scholar]
- 13.Rasko DA, Sperandio V. Nat. Rev. Drug. Discovery. 2010;9:117. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 14.Pacheco AR, Sperandio V. Curr. Opin. Microbiol. 2009;12:192. doi: 10.1016/j.mib.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufmann GF, Park J, Janda KD. Expert Opin. Biol. Ther. 2008;8:719. doi: 10.1517/14712598.8.6.719. [DOI] [PubMed] [Google Scholar]
- 16.Geske GD, O’Neill JC, Blackwell HE. Chem. Soc. Rev. 2008;37:1432. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni N, Li M, Wang J, Wang B. Med. Res. Rev. 2009;29:65. doi: 10.1002/med.20145. [DOI] [PubMed] [Google Scholar]
- 18.Suga H, Smith KM. Curr. Opin. Chem. Biol. 2003;7:586. doi: 10.1016/j.cbpa.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Janssens JCA, De Keersmaecker SCJ, De Vos DE, Vanderleyden J. Curr. Med. Chem. 2008;15:2144. doi: 10.2174/092986708785747580. [DOI] [PubMed] [Google Scholar]
- 20.Mattmann ME, Blackwell HE. J. Org. Chem. 2010;75:6737. doi: 10.1021/jo101237e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. J. Am. Chem. Soc. 2005;127:12762. doi: 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]
- 22.Geske GD, O’Neill JC, Blackwell HE. ACS Chem. Biol. 2007;2:315. doi: 10.1021/cb700036x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geske GD, O’Neill JC, Miller DM, Mattmann ME, Blackwell HE. J. Am. Chem. Soc. 2007;129:13613. doi: 10.1021/ja074135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geske GD, O’Neill JC, Miller DM, Wezeman RJ, Mattmann ME, Lin Q, Blackwell HE. ChemBioChem. 2008;9:389. doi: 10.1002/cbic.200700551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geske GD, Mattmann ME, Blackwell HE. Bioorg. Med. Chem. Lett. 2008;18:5978. doi: 10.1016/j.bmcl.2008.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mattmann ME, Geske GD, Worzalla GA, Chandler JR, Sappington KJ, Greenberg EP, Blackwell HE. Bioorg. Med. Chem. Lett. 2008;18:3072. doi: 10.1016/j.bmcl.2007.11.095. [DOI] [PubMed] [Google Scholar]
- 27.Praneenararat T, Geske GD, Blackwell HE. Org. Lett. 2009;11:4600. doi: 10.1021/ol901871y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowery CA, Dickerson TJ, Janda KD. Chem. Soc. Rev. 2008;37:1337. doi: 10.1039/b702781h. [DOI] [PubMed] [Google Scholar]
- 29.Boyer M, Wisniewski-Dyé F. FEMS Microbiol. Ecol. 2009;70:1. doi: 10.1111/j.1574-6941.2009.00745.x. [DOI] [PubMed] [Google Scholar]
- 30.Wagner V, Li L, Isabella V, Iglewski BH. Anal. Bioanal. Chem. 2007;387:469. doi: 10.1007/s00216-006-0964-6. [DOI] [PubMed] [Google Scholar]
- 31.Venturi V. FEMS Microbiol. Rev. 2006;30:274. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 32.Xu H, Lin W, Xia H, Xu S, Li Y, Yao H, Bai F, Zhang X, Bai Y, Saris P, Qiao M. FEMS Microbiol. Lett. 2005;253:103. doi: 10.1016/j.femsle.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 33.Ren D, Zuo R, Wood T. Appl. Microbiol. Biotechnol. 2005;66:689. doi: 10.1007/s00253-004-1691-6. [DOI] [PubMed] [Google Scholar]
- 34.Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, Venturi V, Zennaro E, Leoni L. Mol. Microbiol. 2007;66:1557. doi: 10.1111/j.1365-2958.2007.06029.x. [DOI] [PubMed] [Google Scholar]
- 35.Wade DS, Calfree MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. J. Bacteriol. 2005;187:4372. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramoni S, Venturi V. Microbiology-Sgm. 2009;155:1377. doi: 10.1099/mic.0.026849-0. [DOI] [PubMed] [Google Scholar]
- 37.Patankar AV, Gonzalez JE. FEMS Microbiol. Rev. 2009;33:739. doi: 10.1111/j.1574-6976.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- 38.Fuqua C. J. Bacteriol. 2006;188:3169. doi: 10.1128/JB.188.9.3169-3171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chugani SA, Whiteley M, Lee KM, D’Argenio D, Manoil C, Greenberg EP. Proc. Natl. Acad. Sci. USA. 2001;98:2752. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. J. Bacteriol. 2006;188:3365. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JH, Lequette Y, Greenberg EP. Mol. Microbiol. 2006;59:602. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 42.Dubern J-F, Diggle SP. Mol. Biosyst. 2008;4:882. doi: 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- 43.Ledgham F, Ventre I, Soscia C, Foglino M, Sturgis JN, Lazdunski A. Mol. Microbiol. 2003;48:199. doi: 10.1046/j.1365-2958.2003.03423.x. [DOI] [PubMed] [Google Scholar]
- 44.Lupp C, Urbanowski M, Greenberg EP, Ruby EG. Mol. Microbiol. 2003;50:319. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhu J, Beaber JW, More MI, Fuqua C, Eberhard A, Winans SC. J. Bacteriol. 1998;180:5398. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.