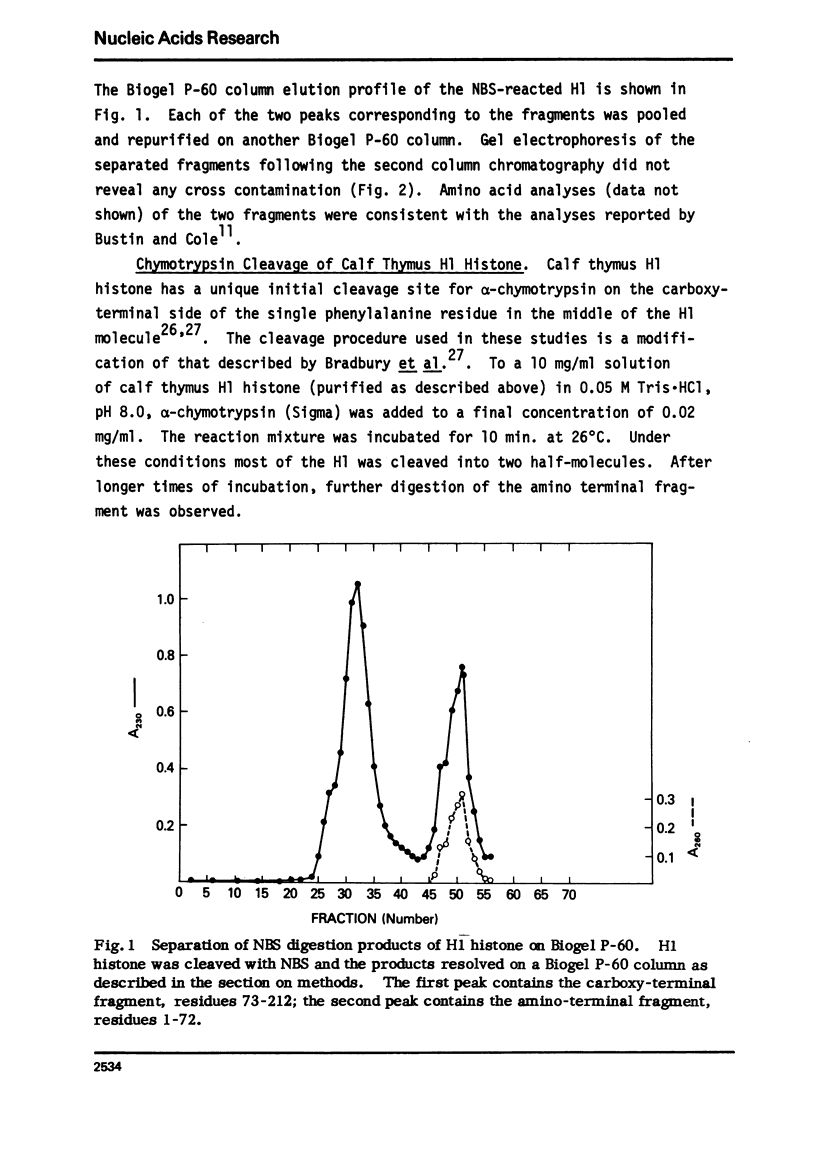
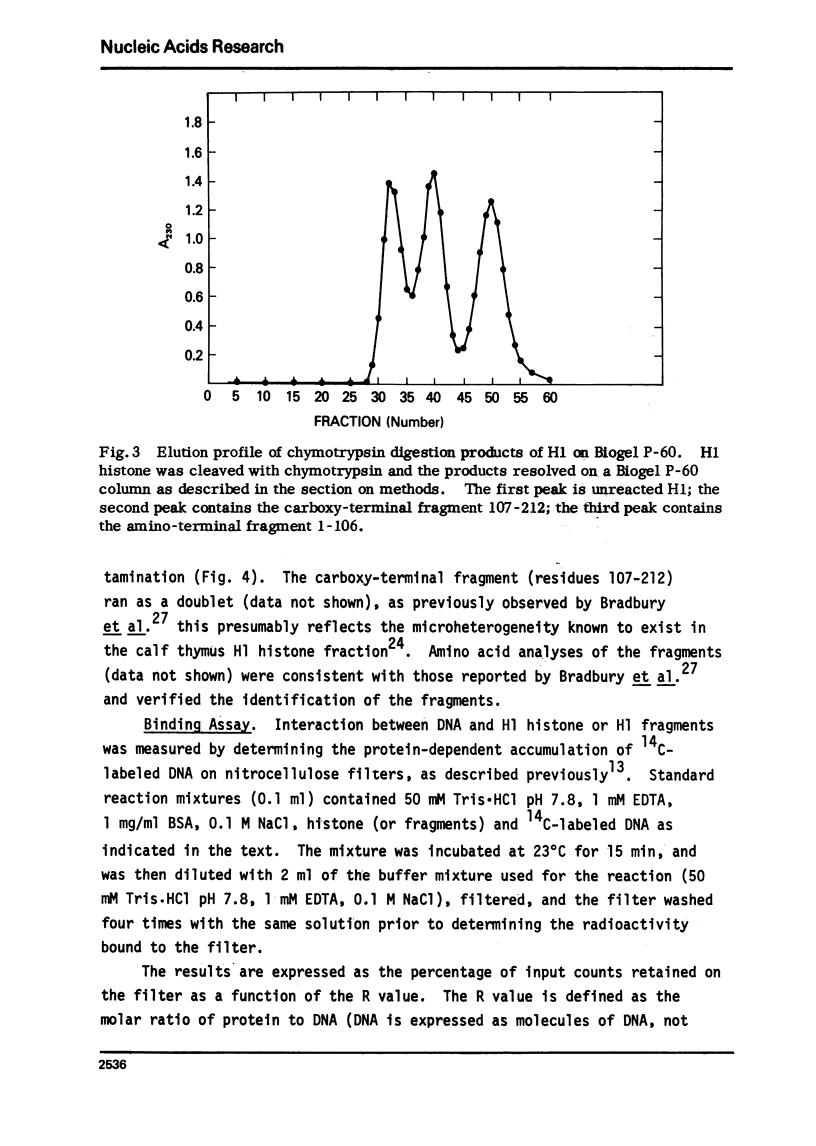
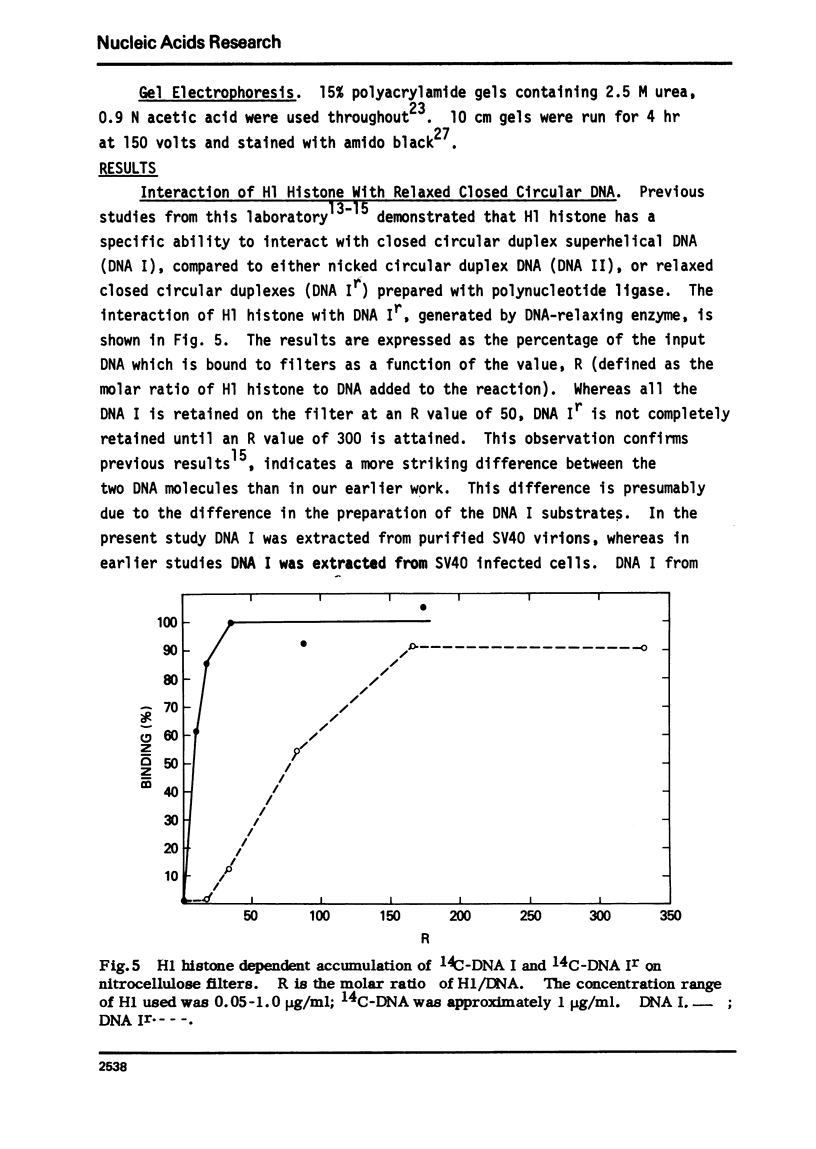
Abstract
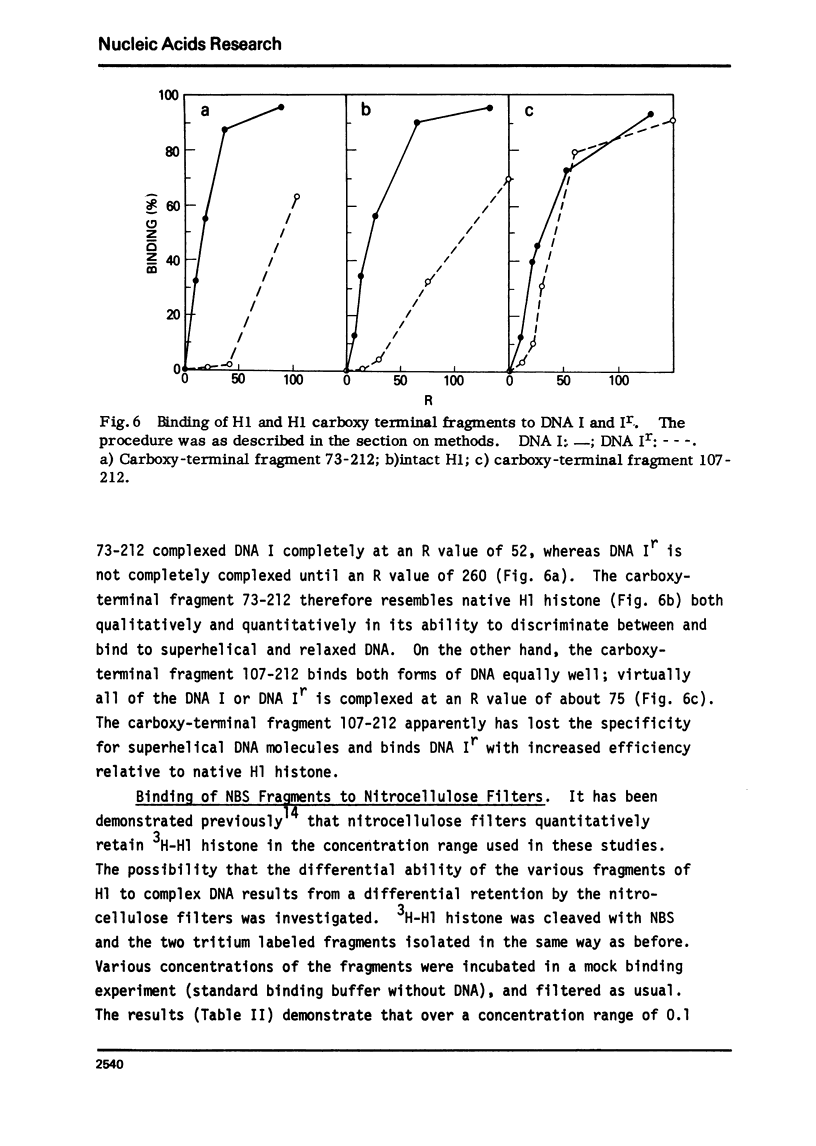
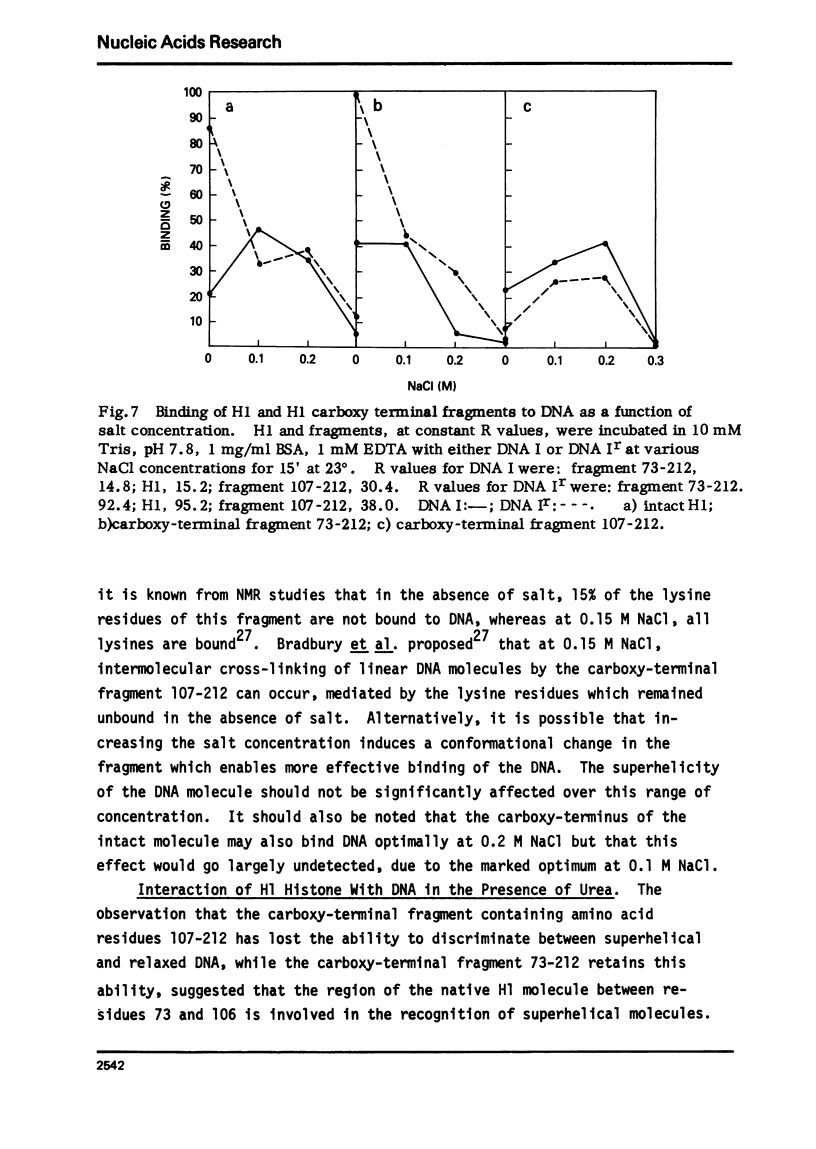
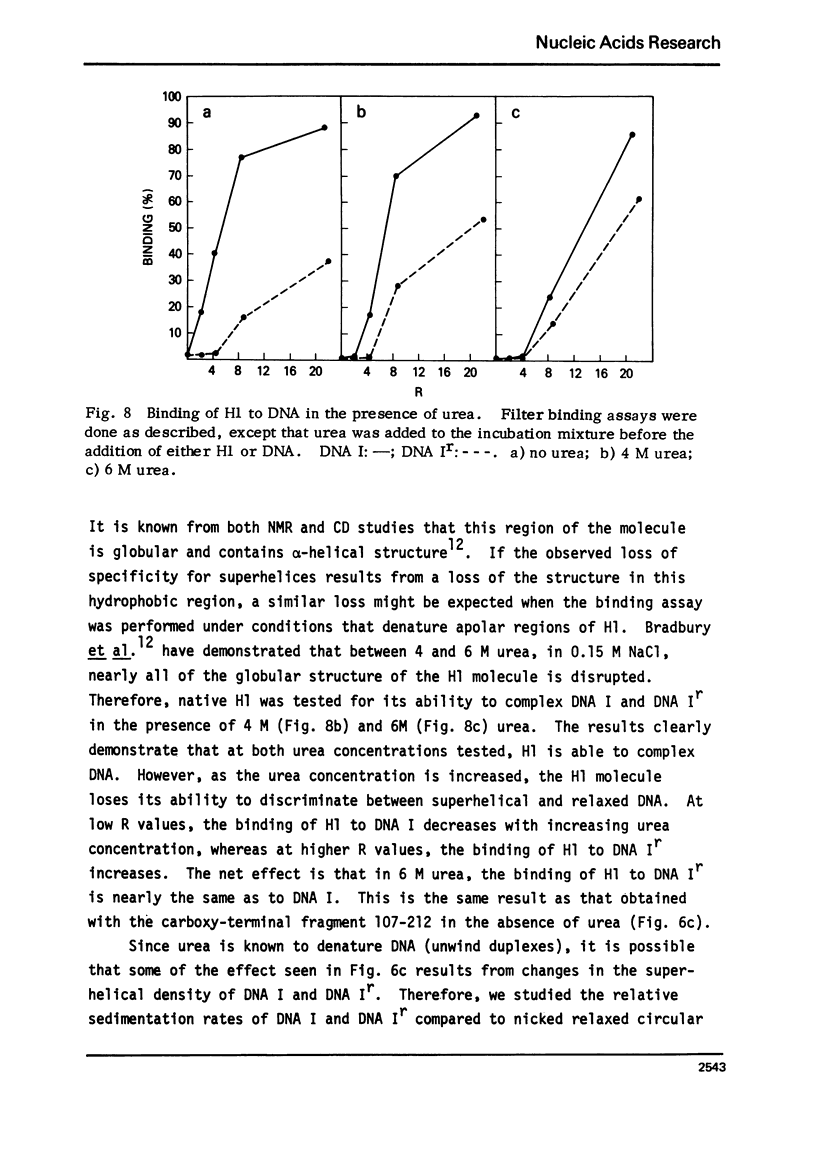
The very lysine rich histone, H1, isolated from a variety of sources interacts preferentially with superhelical DNA compared to relaxed DNA duplexes. The nature of this specific interaction has been investigated by studying the ability of various purified fragments of H1 histone from calf thymus to recognize and bind superhelical DNA. The data suggest that the globular region of the H1 histone molecule (amino acid residues 72-106) is involved in the recognition of superhelical DNA. Thus, the H1 histone carboxy-terminal fragment, 72-212, resembles native H1 histone both quantitatively and qualitatively in its ability to discriminate between and bind to superhelical and relaxed DNA while the H1 histone carboxy-terminal fragment, residues 106-212, has lost this specificity, binding superhelical and relaxed DNA equally well. Furthermore, under conditions in which the globular region of the intact H1 histone has been unfolded, the molecule loses its ability to discriminate between superhelical and relaxed DNA, and binds both forms of DNA equally.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- AKINRIMISI E. O., BONNER J., TSO P. O. BINDING OF BASIC PROTEINS TO DNA. J Mol Biol. 1965 Jan;11:128–136. doi: 10.1016/s0022-2836(65)80178-4. [DOI] [PubMed] [Google Scholar]
- Balhorn R., Chalkley R., Granner D. Lysine-rich histone phosphorylation. A positive correlation with cell replication. Biochemistry. 1972 Mar 14;11(6):1094–1098. doi: 10.1021/bi00756a023. [DOI] [PubMed] [Google Scholar]
- Beard P., Morrow J. F., Berg P. Cleavage of circular, superhelical simian virus 40 DNA to a linear duplex by S1 nuclease. J Virol. 1973 Dec;12(6):1303–1313. doi: 10.1128/jvi.12.6.1303-1313.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury E. M., Cary P. D., Chapman G. E., Crane-Robinson C., Danby S. E., Rattle H. W., Boublik M., Palau J., Aviles F. J. Studies on the role and mode of operation of the very-lysine-rich histone H1 (F1) in eukaryote chromatin. The conformation of histone H1. Eur J Biochem. 1975 Apr 1;52(3):605–613. doi: 10.1111/j.1432-1033.1975.tb04032.x. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Chapman G. E., Danby S. E., Hartman P. G., Riches P. L. Studies on the role and mode of operation of the very-lysine-rich histone H1 (F1) in eukaryote chromatin. The properties of the N-terminal and C-terminal halves of histone H1. Eur J Biochem. 1975 Sep 15;57(2):521–528. doi: 10.1111/j.1432-1033.1975.tb02327.x. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Inglis R. J., Matthews H. R. Control of cell division by very lysine rich histone (F1) phosphorylation. Nature. 1974 Feb 1;247(5439):257–261. doi: 10.1038/247257a0. [DOI] [PubMed] [Google Scholar]
- Bustin M., Cole R. D. Bisection of a lysine-rich histone by N-bromosuccinimide. J Biol Chem. 1969 Oct 10;244(19):5291–5294. [PubMed] [Google Scholar]
- Böhm E. L., Strickland W. N., Strickland M., Thwaits B. H., van der Westhuizen D. R., von Holt C. Purification of the five main calf thymus histone fractions by gel exclusion chromatography. FEBS Lett. 1973 Aug 15;34(2):217–221. doi: 10.1016/0014-5793(73)80797-5. [DOI] [PubMed] [Google Scholar]
- Böttger M., Scherneck S., Fenske H. A sedimentation study of the interaction of superhelical SV40 DNA with H1 histone. Nucleic Acids Res. 1976 Feb;3(2):419–429. doi: 10.1093/nar/3.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman G. E., Hartman P. G., Bradbury E. M. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The isolation of the globular and non-globular regions of the histone H1 molecule. Eur J Biochem. 1976 Jan 2;61(1):69–75. doi: 10.1111/j.1432-1033.1976.tb09998.x. [DOI] [PubMed] [Google Scholar]
- DeLange R. J., Smith E. L. Histones: structure and function. Annu Rev Biochem. 1971;40:279–314. doi: 10.1146/annurev.bi.40.070171.001431. [DOI] [PubMed] [Google Scholar]
- Eason R., Vinograd J. Superhelix density heterogeneity of intracellular simian virus 40 deoxyribonucleic acid. J Virol. 1971 Jan;7(1):1–7. doi: 10.1128/jvi.7.1.1-7.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Jones G. M., Rall S. C., Cole R. D. Extension of the amino acid sequence of a lysine-rich histone. J Biol Chem. 1974 Apr 25;249(8):2548–2553. [PubMed] [Google Scholar]
- Keller W. Characterization of purified DNA-relaxing enzyme from human tissue culture cells. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2550–2554. doi: 10.1073/pnas.72.7.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkade J. M., Jr, Cole R. D. The resolution of four lysine-rich histones derived from calf thymus. J Biol Chem. 1966 Dec 25;241(24):5790–5797. [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Lake R. S., Salzman N. P. Occurrence and properties of a chromatin-associated F1-histone phosphokinase in mitotic Chinese hamster cells. Biochemistry. 1972 Dec 5;11(25):4817–4826. doi: 10.1021/bi00775a027. [DOI] [PubMed] [Google Scholar]
- Ohlenbusch H. H., Olivera B. M., Tuan D., Davidson N. Selective dissociation of histones from calf thymus nucleoprotein. J Mol Biol. 1967 Apr 28;25(2):299–315. doi: 10.1016/0022-2836(67)90143-x. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Panyim S., Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969 Mar;130(1):337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- Rall S. C., Cole R. D. Amino acid sequence and sequence variability of the amino-terminal regions of lysine-rich histones. J Biol Chem. 1971 Dec 10;246(23):7175–7190. [PubMed] [Google Scholar]
- Sebring E. D., Kelly T. J., Jr, Thoren M. M., Salzman N. P. Structure of replicating simian virus 40 deoxyribonucleic acid molecules. J Virol. 1971 Oct;8(4):478–490. doi: 10.1128/jvi.8.4.478-490.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H. T., Smith C. A., Sharp P. A., Vinograd J. Sequence heterogeneity in closed simian virus 40 deoxyribonucleic acid. J Virol. 1972 Feb;9(2):317–325. doi: 10.1128/jvi.9.2.317-325.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. A model for particulate structure in chromatin. Nucleic Acids Res. 1974 Nov;1(11):1579–1586. doi: 10.1093/nar/1.11.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T., Singer M. F. Interaction of f1 histone with superhelical DNA. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2597–2600. doi: 10.1073/pnas.72.7.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T., Singer M. F. The effect of superhelicity on the interaction of histone f1 with closed circular duplex DNA. J Biol Chem. 1976 Apr 25;251(8):2334–2338. [PubMed] [Google Scholar]
- Vogel T., Singer M. The interaction of histones with simian virus 40 supercoiled circular deoxyribonucleic acid in vitro. J Biol Chem. 1975 Jan 25;250(2):796–798. [PubMed] [Google Scholar]
- Woodworth-Gutai M., Lebowitz J. Introduction of interrupted secondary structure in supercoiled DNA as a function of superhelix density: consideration of hairpin structures in superhelical DNA. J Virol. 1976 Apr;18(1):195–204. doi: 10.1128/jvi.18.1.195-204.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]