Abstract
The proteasome is the most complex protease known, with a molecular mass of approximately 3 MDa and 33 distinct subunits. Recent studies reported the discovery of four chaperones that promote the assembly of a 19-subunit subcomplex of the proteasome known as the regulatory particle, or RP. These and other findings define a new and highly unusual macromolecular assembly pathway. The RP mediates substrate selection by the proteasome and injects substrates into the core particle (CP) to be degraded. A heterohexameric ring of ATPases, the Rpt proteins, is critical for RP function. These ATPases abut the CP and their C-terminal tails help to stabilize the RP-CP interface. ATPase heterodimers bound to the chaperone proteins are early intermediates in assembly of the ATPase ring. The four chaperones have the common feature of binding the C-domains of Rpt proteins, apparently a remarkable example of convergent evolution; each chaperone binds a specific Rpt subunit. The C-domains are distinct from the C-terminal tails but proximal to them. Some but probably not all of the RP chaperones appear to compete with CP for binding of the Rpt proteins, as a result of the proximity of the tails to the C-domain. This competition may underlie the release mechanism for these chaperones. Genetic studies in yeast point to the importance of the interaction between the CP and the Rpt tails in assembly, and a recent biochemical study in mammals suggests that RP assembly takes place on pre-assembled CP. These results do not exclude a parallel, CP-independent pathway of assembly. Ongoing work should soon clarify the roles of both the CP and the four chaperones in RP assembly.
Keywords: proteasome, assembly, regulatory particle, AAA protein, chaperone
ATP-dependent proteases are the primary mediators of selective protein degradation in cells [1–3]. They differ from other proteases in that they actively unfold their substrates before hydrolyzing them. Among other things, this property allows them to select substrates on the basis of specific degradation signals, such as ubiquitin, rather than the accessibility of potential cleavage sites. The eukaryotic proteasome, like most ATP-dependent proteases, has a peptide-hydrolyzing core particle (CP) and an ATP-hydrolyzing regulatory particle (RP) (Figure 1a). Although the ability of ATP-dependent proteases to unfold substrates is not fully understood, it is generally thought that unfolding is achieved by pulling the substrate through a translocation channel. The channel is formed at the center of a hexameric ring of ATPases (Figure 2a). This review is concerned primarily with how the ATPase ring is assembled. The central channel is narrow, so that a substrate can only pass through after unfolding, which is forced by an ATP-dependent pulling action exerted by specific loop elements lining the channel. The channel leads to an internal chamber in the enzyme that houses its proteolytic sites. Sequestration of the proteolytic sites in this way isolates these sites from the cytosol, minimizing nonspecific protein degradation.
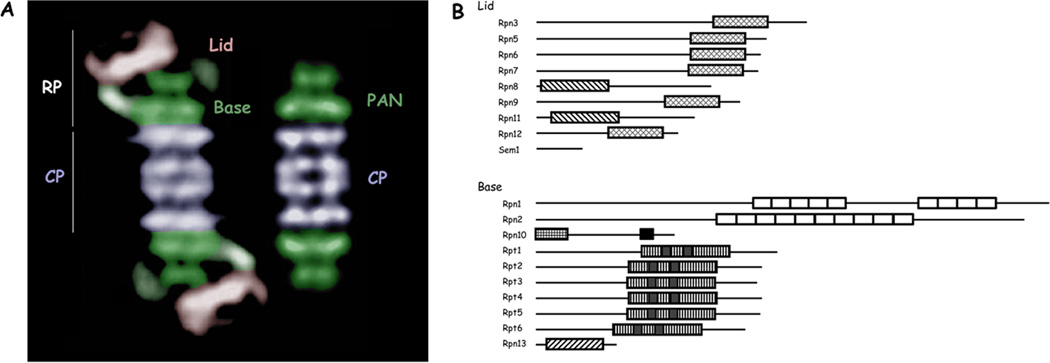
Figure 1. Structure and composition of the proteasome.
(A) Electron micrographs of the eukaryotic proteasome and the archaeal PAN complex, colored to highlight subcomplexes. The line of demarcation between the lid and base subcomplexes is a rough estimate. The lid and base together comprise the RP. The base is rendered in green. The ATPases are colored in dark green to illustrate the close match of this part of the base to the homomeric PAN complex. The distal green ring apparently corresponds to the CC-OB domain of PAN [6]. Electron micrographs are from [4] with permission.
(B) Subunits of the proteasome RP. Major structural domains are indicated approximately to scale: PCI domains in Rpn3, 5, 6, 7, 9, and 12; MPN domains in Rpn8 and Rpn11; alpha helical repeats in Rpn1 and Rpn2, which together form a toroid in each subunit; in Rpn10 an N-terminal VWA domain and a C-terminal UIM domain mediating ubiquitin recognition; the ATPase domains of Rpt1–6; and a ubiquitin-binding PH domain in Rpn13. Modified from [7] with permission.
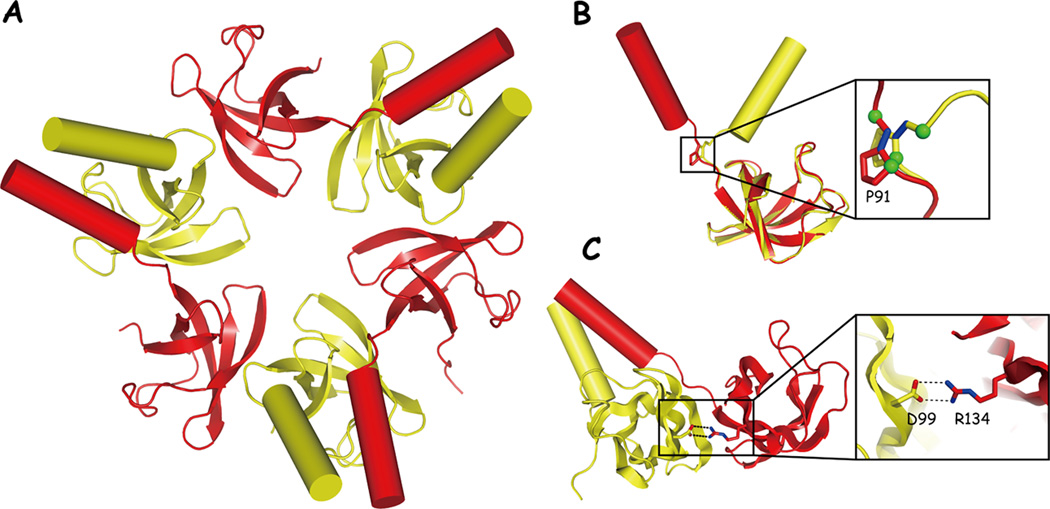
Figure 2. Structure of the CC-OB domain from PAN.
(A) The CC-OB hexamer; view of the face most distal to the CP. C-type subunits are shown in yellow, t-type in red; they alternate around the ring to form a trimer of dimers. Alpha helices are represented as cylinders. The β-sheet elements correspond to the OB fold domains. The open space at center is thought to be the entry port for substrate translocation.
(B) Detail of the two forms of PAN, exhibiting either a cis or trans main-chain configuration at Pro91. Trans segment shown in red, cis shown in yellow.
(C) Detail of a key salt bridge in PAN, which is found in intradimer but not interdimer contacts.
Figure adapted from [6] with permission.
The eukaryotic proteasome (hereafter simply the proteasome) represents a major elaboration of the architecture of ATP-dependent proteases [1]. The PAN complex of archaea is smaller than the proteasome but is remarkably similar in certain respects [4] (Figure 1a), and can be regarded as a close approximation of its evolutionary precursor. PAN’s CP is a 28-subunit structure assembled from 2 gene products, the α and β subunits, which are arranged into four stacked seven-membered rings. Thus, the four rings form a barrel-like a7β7β7α7 structure [5, 6]. The archael RP is a 6-subunit ring structure formed from the product of one gene, the AAA ATPase PAN. In the holocomplex, the symmetry axes of the CP and RP are approximately aligned. A curiosity of the structure, which is also seen in the proteasome, is the alignment of a seven-fold symmetric complex (the CP) with a six-fold symmetric complex (the RP). The utility of this symmetry mismatch, if any, has not been explained.
As we move from the PAN complex to the proteasome, two basic architectural differences are seen. First, 13 additional subunits are present in the proteasome [1] (Figure 1b). Our functional understanding of these “new” subunits is limited, but it seems that many or all of them serve to assist, directly or indirectly, in the recognition and processing of ubiquitin-protein conjugates. For example, subunits Rpn10 and Rpn13 are ubiquitin receptors, and Rpn11 is a deubiquitinating enzyme [1]. Rpn1 is a receptor for the ubiquitin-like proteins Rad23, Dsk2, and Ddi1, which are themselves ubiquitin receptors [1]. Thus, Rpn1 serves indirectly in the delivery of substrates to the proteasome. The 19-subunit RP of yeast is readily fragmented into base and lid subassemblies [7, 8], which are illustrated in Figure 1. Whether these are assembly intermediates is a question we will return to. Several other complexes in the cell are related to the lid in composition and may have evolved from it [7].
The second major architectural difference between PAN and the proteasome is that the homomeric rings of the PAN complex have evolved to be heteromeric in the proteasome. For example, the CP is specified by 14 gene products in eukaryotes, rather than two [2]. Despite this increase in complexity, the eukaryotic CP remains an a7β7β7α7 structure [9]. Each of the seven α subunits takes a specific place within the α ring, and similarly each β subunit is uniquely positioned with the β ring [2, 9]. Also, the ATPase ring, a homomer in PAN, is formed from six distinct gene products in the proteasome [1] (Figure 1b). What may have been gained through these diversifications of subunits is a problem for future study.
Discovery of chaperones for proteasome assembly
A remarkable series of papers from the past year has suggested that diversification of the ATPase ring was a complex evolutionary process involving a cohort of chaperone proteins [10–15](reviewed in [16]). The chaperones seem to have evolved to facilitate proper assembly of the ring, and possibly the proper placement of individual ATPases around the ring. These “RP-chaperones” are known in yeast as Hsm3, Nas2, Nas6, and Rpn14. Their mammalian cognates are S5b, p27, gankyrin/p28, and PAAF1, respectively. The RP chaperones are not generalized chaperones, such as Hsp70 and Hsp90, but seem to be dedicated to proteasome assembly. In yeast, no other function has been convincingly described for these proteins, though surprisingly in mammals there are two cases of functions that seem unrelated to proteasome assembly: p27 has been implicated in the control of insulin production [17–19] whereas gankyrin/p28 is thought to be an oncoprotein, functioning via the promotion of Rb and p53 degradation [20, 21].
Many of the RP chaperones have previously been suggested to be proteasome subunits. In retrospect, this might reflect that the samples in which they were first observed contained some proteasome subassemblies: Hsm3, Rpn14 and Nas6 can all bind to free RP [12, 15], whereas they bind the RP-CP complex (whole proteasome) weakly or not at all. So far no proteins other than the RP chaperones are known to have this property.
RP chaperones bind C domains of the proteasomal ATPases
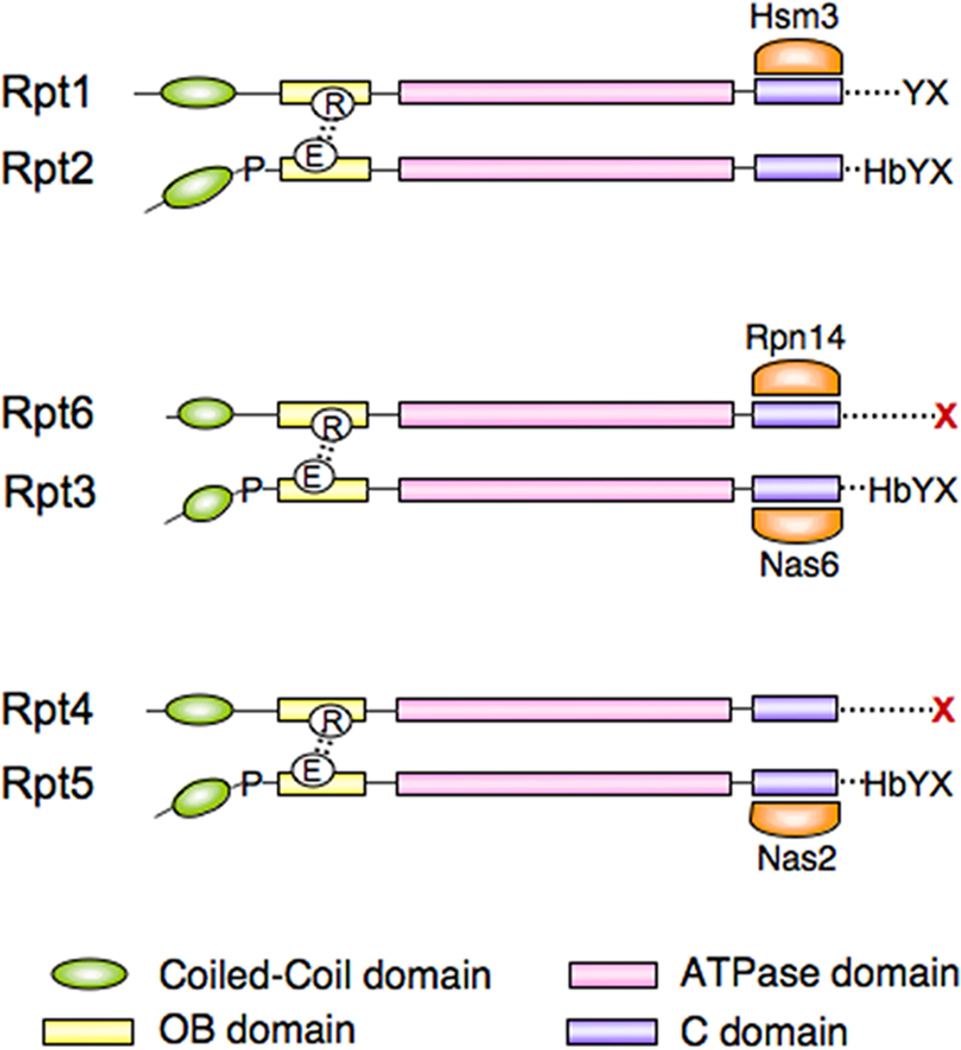
A common feature of the RP chaperones is that each binds the C-domain of a proteasomal ATPase [10, 12, 15, 22]. These ATPases are known in yeast as Rpt1-Rpt6. Specifically, Hsm3 binds the C-domain of Rpt1, whereas Nas2 binds that of Rpt5, Nas6 that of Rpt3, and Rpn14 that of Rpt6 (Figure 3). The C-domain (see Figure 3) is a four-helix bundle that is characteristic of AAA-type ATPases [23], a family to which the proteasomal ATPases (Rpt proteins) belong. The inner surface of the bundle forms a part of the ATPase active site [6, 23]. The outer surface is exposed, and provides the chaperone binding site [24]. Given that all four of the RP chaperones bind Rpt C-domains, it is striking that they are unrelated evolutionarily: Hsm3 is composed primarily of Armadillo/HEAT repeats, Nas6 of ankyrin repeats, Rpn14 of WD40 repeats, and Nas2 contains a PDZ domain. Each of these structural elements is presumably responsible for binding a given C-domain. Notably, no RP chaperone has been found to bind two Rpt proteins, and therefore they do not appear to perform a scaffolding function in assembly. Thus, it is unlikely that the chaperones provide spatial information for assembly in this sense.
Figure 3. Schematic representation of the Rpt proteins and their chaperones.
Structural domains shown are indicated at bottom. Pro91 and the key salt bridge are represented towards the left, and the HbYX motif at right. C-termini that have been implicated in RP assembly are indicated in red. Note that in the Rpt proteins the cognate residue of Asp99 is in each case a glutamic acid (E). Chaperones are highlighted in orange and aligned with their specific Rpt recognition sites.
Protruding from the C-domain is a flexible segment known as the C-terminal tail [25] (Figure 3). The six Rpt tails extend towards the CP and apparently participate in stabilization of the CP-RP association [26–28]. These six tails insert into well-defined pockets formed between the seven α subunits [27–31]. Interestingly, mutations in several tail elements lead to clear assembly-defective phenotypes in yeast [14]. These data suggested that the C-terminal tails help to promote RP assembly, presumably by inserting into the CP pockets. The assembly defects arising from Rpt tail mutations are strong and typically more severe than those seen when individual RP chaperones are deleted [10–15].
The C-terminal tails of the Rpts play several critical roles in the proteasome. They not only participate in RP assembly, but in the mature proteasome they mediate opening the substrate translocation channel, or “gate,” into the CP [27, 29]. This function is conserved between PAN and the eukaryotic Rpt proteins [27]. PAN terminates in the residues LYR. Substitution of these residues in PAN, inspection of the conserved C-termini of the Rpt proteins, and mutagenesis of the RPT genes in yeast has suggested that the gate-opening motif is HbYX—a hydrophobic amino acid followed by tyrosine and an unspecified C-terminal residue [27]. Although the identity of the last residue seems unimportant, the carboxylate of the main chain may be critical for recognition [25, 28]. The structural basis of gate opening by HbYX peptides has been studied in some detail by high-resolution electron microscopy [30] and crystallography [28], but will not be described here. It should be noted that not all Rpt C-termini match the HbYX consensus (Figure 3). Those C-termini that have the strongest effect on gating are of the HbYX type [27], whereas the two C-termini that appear to be most critical for ring assembly are notably not of the HbYX type [14] (Figure 3).
Because the Rpt C-termini dock at the CP, the dramatic effects on assembly seen when they are mutated suggests that formation of the ATPase ring is in part templated on pre-existing CP [14]. In strong support of this idea, numerous mutants with a primary defect in CP assembly show prominent secondary effects on RP assembly [32]. On the basis of these findings, one might expect that the RP chaperones regulate assembly in part by binding to the C-terminal tails of the Rpt proteins, thus regulating their interaction with the CP. However, direct binding assays indicate that the chaperone binding sites in the C-domains do not extend to the Rpt C-termini [15]. (This has not been tested for Nas2, which interestingly has a PDZ domain.) Nonetheless, some of the RP chaperones may regulate the engagement of the tails within the CP pockets through competition. Because the tails are close to the C-domains, chaperone binding could sterically interfere with CP recognition of the tails. This model has been assessed mainly by studying the requirements for chaperone dissociation upon assembly of the proteasome [14, 15].
Determinants of RP chaperone dissociation upon proteasome maturation
The deletion of one residue from the C-terminal tail of Rpt6 leads to a defect in the release of Rpn14, the binding partner of Rpt6, from mature proteasomes [14]. The same single-residue deletion (rpt6-Δ1) results in a strong assembly defect, indicating that it perturbs docking of the tail [14]. These findings are consistent with the competition model described above, and suggest in particular that docking of the Rpt6 tail helps trigger dissociation of chaperone. RP chaperones Hsm3 and Nas6 were released correctly under these conditions, indicating that the dissociation defect is restricted to the cognate chaperone of Rpt6. Similar findings were reported for Rpt3 and its partner Nas6 [14]. One potential caveat of the rpt6-Δ1 experiment is that failure to release Rpn14 could underlie the assembly defect of the tail mutant. However this does not appear to be the case because deletion of the RPN14 gene in the genetic background of the rpt6-Δ1 mutant does not revert the phenotype [14].
Interestingly, extension of the Rpt6 tail, rather than truncation, likewise leads to a failure to expel Rpn14 (but not Hsm3 or Nas6) [14]. The insertion of only a single amino acid, several residues upstream from the C-terminus, is sufficient. This mutation does not cause an assembly defect and therefore the retention of Rpn14 cannot be explained by a failure to dock the extended C-terminal tail [14]; most likely, Rpn14 retention reflects ineffective competition by the CP. A final experiment in support of the model is that Nas6, Hsm3, and Rpn14 can be stripped from purified base by the addition of purified CP [15].
An interesting implication of the model above is that the competition between the RP chaperones and the CP for binding to the Rpt proteins could explain in part how the chaperones promote proteasome assembly. However, that has not been tested; the experiments address the release step but not other aspects of the assembly pathway. Nas2 release is apparently triggered by other events [12], and this may be true of Hsm3 as well (S. Park, unpublished data). Thus, most likely not all RP-chaperones are released through this mechanism.
Dimeric ATPase complexes are early intermediates in assembly
An important insight into assembly of the RP, suggested in part by many groups, but especially clear from Kaneko et al. [13], is that Rpts first form heterodimers on the way to ring formation. Moreover, RP chaperones can be found complexed with these heterodimers. The dimeric pairs are Rpt1/Rpt2, Rpt3/Rpt6, and Rpt4/Rpt5 (Figure 3). These pairings are notably at odds with longstanding ideas as to the topology of subunits within the ring [33], which are therefore almost certainly incorrect.
A second reason to prefer a new ordering of Rpts derives from recent structural insights into PAN [6, 34]. In addition to its ATPase domain, PAN has an OB domain and, near the N-terminus, a coiled-coil domain [6, 34]. The crystal structure of a hexameric fragment of PAN containing the OB domain and the coiled-coil domain revealed that this segment of PAN exists as a trimer of dimers (Figure 2). That is, the hexamer shows three-fold rather than six-fold symmetry. Proline-91, which is located between the OB and coiled-coil domains, plays a key role in this asymmetry [6, 34]. In each dimeric pair, one subunit exhibits a cis configuration of the main chain at Pro91, whereas trans-Pro91 is found in the other subunit. Cis-trans isomerization is required to prevent steric occlusion between neighboring coiled coils. Consequently, cis and trans proline residues strictly alternate around the hexameric ring of PAN (Figure 2).
Conservation of proteasomal ATPase features between archaea and eukaryotes
Key characteristics of the OB and coiled-coil domains of PAN are conserved in the eukaryotic proteasome [6, 34]. Most interestingly, structural features of PAN seem to be reflected in sequence features of the Rpt proteins. For example, one would predict that half of the Rpt proteins should be cis (c-type) subunits and half trans (t-type). Although the three-dimensional structure of the Rpt ring is not known, it is indeed the case that half of the Rpts have a strictly conserved proline at the position corresponding to Pro91 of PAN [6, 34] (Figure 3). These are currently thought to be the three c-type subunits, since only proline has a propensity to adopt the cis configuration.
Within each PAN dimer is a salt bridge linking Asp99 and Arg134, both located within the OB domain [6, 34] (Figures 2 and 3). Interdimer interfaces do not contain this salt bridge; the responsible residues are obviously present, but a tilt in the OB domain removes them from proximity. In PAN, the Asp99 that forms the salt bridge is on a c-type subunit, whereas the Arg134 that forms the bridge is on a t-type subunit. Strikingly, the cognate residues of Asp99 are present only on c-type Rpt subunits and the cognate residues of Arg134 are only present in the t-type Rpt subunits (Figure 3). These residues are strictly conserved in the Rpt proteins. Interestingly, the c-type subunits also carry all of the canonical HbYX motifs (Figure 3).
Isomerization at Pro91 and the formation of the OB-OB salt bridge are potentially important for assembly. The salt bridge may stabilize the dimers while formation of a proper dimer might require the participation of a prolyl isomerase. It is unclear at present whether RP chaperone binding stabilizes any of the dimers or any particular conformation of the dimers. Interestingly, the isolated dimeric pairs are lacking in ATPase activity [35]. Thus, either subunit communication around the ring is critical for ATPase activity, or the ATPase domains of the assembly intermediates are not in a mature configuration. Whether the presence of RP chaperones influences ATPase activity has not been studied. Interaction with the CP is not required to activate the ATPase sites, since PA700, the bovine form of the RP, hydrolyzes ATP [36, 37].
In summary, the existence of Rpt-Rpt dimers in eukaryotes as observed in the recent assembly studies is consistent with the structural data from archaea discussed above. Importantly, each assembly module consists of one c-type and one t-type subunit. Based on these and other data, a new model of the order of Rpts within the ring has been proposed [38, 39], though a direct demonstration of the order remains to be accomplished.
Although the view of the Rpt ring as a trimer of dimers probably holds for both the assembly intermediates and for the OB and CC domains of the Rpt’s, it is not clear that it applies to the ATPase domains of the assembled complex. That is, the ring of ATPase domains may be even more asymmetric than the CC-OB ring in the holoenzyme. An interesting scenario for the proteasome holoenzyme is that the ATPase domains do not form a closed ring, because their insertion into only six of the seven alpha cavities of the CP might create a gap between two specific subunits (the identities of which are not yet clear). This is interesting because communication between neighboring subunits within a ring is a well-known feature of AAA family of ATPases [23]. A failure of the Rpt ring to close would thus have interesting implications for the communication between ATPase domains. Essentially, four subunits would be flanked on either side by other ATPases, while one would be flanked only counterclockwise, and another only clockwise. However, the degree to which the ATPase ring is “deformed” to fit the seven-fold symmetry of the CP is unclear. It is even unclear whether all of the C-terminal tails of the ATPases insert into the CP at once. Possibly only a subset are engaged at any given time, and if this is true then the influence of the CP on the detailed positioning of the ATPase domains about the ring could be less than expected.
Possible scaffolds for assembly
Where is the spatial information for ring assembly? Much more work is required in this area, but presumably the dimeric interactions in the CC-OB regions of the Rpts are important, as suggested by studies of PAN [6, 34]. Lateral interactions within the ATPase domains are likely as well, although structural information is lacking because this domain of PAN crystallized as a monomer [6]. It is interesting to consider what components extrinsic to the ring may potentially play a role in its formation, thus “scaffolding” assembly. The dependence of ring or base assembly on the C-termini of Rpt4 and Rpt6 suggests that the CP assists in ring assembly [14], and consistently several CP mutants have been shown to compromise Rpt ring assembly [32]. So far no biochemical intermediates have been isolated that would unequivocally indicate CP-templated assembly, but such species may be nonabundant or labile, or may be involved only at a late stage of assembly, such as completion of the ring (see below). Spatial information supporting ring assembly may also be provided by Rpn1 and Rpn2, as suggested previously [14, 15]. For example, Rpt4 and Rpt6 interact specifically with Rpn2 [40, 41]. The toroidal components of Rpn1 and Rpn2 may predispose them to promote ring assembly [42–44]. It remains unclear whether these toroids are axial [38, 43, 44]. A scaffolding role for Rpn1 and Rpn2 would be most easily imagined if the toroids were axial, but it would not require this positioning.
Identification of early assembly intermediates via pulse-labeling
Radiochemical pulse-labeling studies indicate that RP assembly is a very rapid process [45, 46] (S. Park, unpublished data), posing a significant impediment to understanding the pathway. Many putative assembly intermediates have been described, but it is critical to show in each case that the intermediate is transient–otherwise it cannot be excluded that the complex is simply a fragment of the proteasome. This criterion has so far been satisfied only for one defined precursor, a complex of Rpt1, Rpt2, Rpn1, and Hsm3 that we have referred to as BP1 [14, 15]. If the proteasome dissociates to some extent during cell lysis, or during purification, the resulting subcomplexes will not disappear in a pulse-chase experiment, at least not any faster than the proteasome itself. However, given the complexity of the assembly pathway, it would be a daunting task to describe precursor-product relationships by this method. Depending on how BP1 is isolated, Rpt5 is sometimes found associated [14, 15]. This is probably a physiological interaction, based on current models of the topology of the ring [38, 39]. However, in our view the Rpt5-containing form of BP1 is not likely to represent a productive or major assembly intermediate, because Rpt1 incorporation into the nascent RP seems to involve a Rpt4-Rpt5 heterodimer [11–13, 35].
In mammalian cells, pulse-chase analysis has been used to show, quite clearly, that new labeled RP assembles onto pre-existing unlabeled CP, consistent with a relatively rapid pace of RP assembly [45, 46]. In addition, a small complex containing Rpt3 and Rpt6 labels very rapidly, arguing that this species is a true assembly intermediate [45]. This complex is expected to contain gankyrin/Nas6 and PAAF1/Rpn14 as well, but the RP-chaperones are not pulse-labeled efficiently, consistent with their being recycled after each round of assembly [14].
In one experiment recently reported by the Hendil group, a pulse-labeled sample was subjected to gel filtration, the proteasome-containing fraction was further purified by immunprecipitation with an antibody to the CP, and the immunoprecipitate was analyzed by two-dimensional IEF/SDS-PAGE [45]. Interestingly, label was found only in the proteins Rpn2, Rpn10, Rpn11, Rpn13, and Txnl1 (thioredoxin like-1, a novel component of the mammalian RP [47, 48]). It was thus proposed that a complex containing only these proteins, deposited on the CP, forms an early assembly intermediate. In this model, the ATPases are added to the “growing” RP in later stages, in a process intimately involving the CP. However, the exact composition of the recovered complex is not certain because unlabeled RP subunits could be present, just as unlabeled CP subunits are. Also, in the two-dimensional gels some RP subunits are poorly visualized [45]. Nonetheless, this labeling-based study is interesting because, in contrast to other studies carried out in mammalian cells (see below), it is most consistent with a CP-templating model. Although, this study does not indicate how the CP might participate in assembly, it suggests that assembly does indeed occur on the CP. Of particular interest is the indication that Rpn2 may play a major role in linking the nascent RP to CP. This finding is consistent with an earlier proposition linking Rpn2 to assembly [14, 15] and with evidence that Rpn2 directly binds to the CP [43].
In vitro RP assembly
A different model for the RP assembly intermediates has emerged from studies by the DeMartino group. This analysis initiated with the purification and identification of three distinct subassemblies of the RP as isolated from lysates of bovine red blood cells [35]. One complex, PS-1, contained Rpt3, Rpt6, Rpn2, Rpn10, Rpn13, Uch37, and all of the lid subunits. PAAF1 (the chaperone for Rpt6) was present though it is unclear whether it was stoichiometrically represented. Gankryin/Nas6 (the chaperone for Rpt3) was notably unidentified. Disregarding the chaperones, PS-1 interestingly contains essentially all of the key RP components of the complex described by the Hendil group [45], but these components are assembled into a larger structure in which all lid subunits are present as well as one of the dimeric ATPase assemblies. It is unclear why the complexes differ between these two studies, which complex is most relevant physiologically, and whether the two complexes represent different stages of RP assembly. One caveat is that bovine red blood cells are enucleate and have little protein synthetic activity, suggesting that they might not carry out de novo proteasome assembly. Therefore, the complexes isolated may not fully mimic physiological intermediates in the assembly of new proteasome complexes. On the other hand, using bovine cells had the advantage that it allowed for RP assembly to be accomplished for the first time in vitro with purified components, as described below.
In addition to PS-1, complementary complexes were identified containing on the one hand Rpt1, Rpt2, Rpn1, and S5b/Hsm3 (PS-2; equivalent to BP1, as described above), and on the other Rpt4, Rpt5, and p27/Nas2 (PS-3) [49]. PS-1, PS-2, and PS-3 contain in sum all subunits of the RP. Moreover, each PS complex contains a distinct pair of Rpt subunits. When these complexes were mixed, the RP reassembled with impressive if not complete efficiency [35]. Addition of CP to the reaction had no influence on the rate of reassembly. This study provides the best evidence to date for a CP-independent pathway of RP assembly. However, we know too little about the in vivo assembly process to judge how well this in vitro reaction mimics the physiological pathway. For example, it is unclear whether the in vitro reaction shows the chaperone-dependence of the in vivo pathway.
Although many features of the assembly pathway are conserved, as described above, it is uncertain whether the base, which seems to be present as such in yeast, can exist in a free form in mammalian cells. Both the identification of PS-1 and the pulse-labeling study in HeLa cells suggest otherwise: that either the lid or components of the lid join to the nascent RP prior to the completion of the base. In yeast however, mutational defects in base assembly characteristically result in the accumulation of free lid (and not a lid-base hybrid species) [11, 12, 14, 15, 50]. A free base complex is readily observed in gently prepared yeast extracts [11, 12, 14], yet even harsh treatments do not release intact base from the mammalian PA700 complex [35]. These data suggest a structural difference between the yeast and mammalian RP, which might be reflected in the assembly pathway. However, the appearance of free lid as a result of base assembly defects was reported in one study in mammalian cells [13]. Further work to clarify this issue both in yeast and mammals would be of interest. If, in mammals, the lid assembles into the nascent proteasome before the ATPase ring is formed, it could provide numerous subunit-subunit interactions with the potential of altering the assembly pathway. For example, a CP-dependent pathway might become CP-independent with early loading of the lid.
Conclusion
The study of RP assembly, though chronically neglected, has made great progress in the last year. It has been revealed as a novel and extremely sophisticated process. The complexity of the pathway is obvious, with many assembly factors already implicated. Considering this as well as the technical challenges in studying a very fast biosynthetic process, a detailed understanding will require many years of effort to be achieved. Evidence for both CP-dependent and CP-independent assembly pathways has been presented. It is not excluded that both models are correct, because at present there is no compelling evidence that a single unique assembly pathway exists, even within one organism. In fact, yeast strains lacking all four RP-chaperones, though hypersensitive to stress, are viable [11, 12], so the chaperone-dependent pathway itself is not strictly required. The pathway appears to be robust and tolerant to many types of perturbation, perhaps a desirable feature given the importance of proteasomes in surviving cellular stress. Indeed, as recent work has suggested that proteasome activity is subject to a variety of precise controls [1], the chaperone-mediated assembly may provide another target of regulation.
Acknowledgements
We thank Marion Schmidt (Albert Einstein College of Medicine) and Suzanne Elsasser for providing comments on the manuscript. The work was supported by a grant from US National Institutes of Health (NIH) to D.F. (GM043601), an NIH NRSA postdoctoral fellowship (5F32GM75737-2) to S.P., and a Charles A. King Trust postdoctoral fellowship to S.P.
Reference
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murata S, Yashiroda H, Tanaka K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell. Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 3.Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nat. Chem. Biol. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 5.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 Å resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol. Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, Baumeister W, Fried VA, Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 8.Saeki Y, Toh-e A, Yokosawa H. Rapid isolation and characterization of the yeast proteasome regulatory complex. Biochem. Biophys. Res. Commun. 2000;273:509–515. doi: 10.1006/bbrc.2000.2980. [DOI] [PubMed] [Google Scholar]
- 9.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 10.Le Tallec B, Barrault MB, Guerois R, Carre T, Peyroche A. Hsm3/S5b participates in the assembly pathway of the 19S regulatory particle of the proteasome. Mol. Cell. 2009;33:389–399. doi: 10.1016/j.molcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Funakoshi M, Tomko RJ, Jr, Kobayashi H, Hochstrasser M. Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell. 2009;137:887–899. doi: 10.1016/j.cell.2009.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K. Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell. 2009;137:900–913. doi: 10.1016/j.cell.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko T, Hamazaki J, Iemura S, Sasaki K, Furuyama K, Natsume T, Tanaka K, Murata S. Assembly pathway of the Mammalian proteasome base subcomplex is mediated by multiple specific chaperones. Cell. 2009;137:914–925. doi: 10.1016/j.cell.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D. Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature. 2009;459:861–865. doi: 10.1038/nature08063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besche HC, Peth A, Goldberg AL. Getting to first base in proteasome assembly. Cell. 2009;138:25–28. doi: 10.1016/j.cell.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Volinic JL, Banz C, Yao KM, Thomas MK. Interactions with p300 enhance transcriptional activation by the PDZ-domain coactivator Bridge-1. J. Endocrinol. 2005;187:283–292. doi: 10.1677/joe.1.06305. [DOI] [PubMed] [Google Scholar]
- 18.Volinic JL, Lee JH, Eto K, Kaur V, Thomas MK. Overexpression of the coactivator bridge-1 results in insulin deficiency and diabetes. Mol. Endocrinol. 2006;20:167–182. doi: 10.1210/me.2005-0127. [DOI] [PubMed] [Google Scholar]
- 19.Thomas MK, Tsang SW, Yeung ML, Leung PS, Yao KM. The roles of the PDZ-containing proteins bridge-1 and PDZD2 in the regulation of insulin production and pancreatic beta-cell mass. Curr. Protein Pept. Sci. 2009;10:30–36. doi: 10.2174/138920309787315248. [DOI] [PubMed] [Google Scholar]
- 20.Higashitsuji H, Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo Y, Masuda T, Dawson S, Shimada Y, Mayer RJ, Fujita J. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005;8:75–87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Higashitsuji H, Itoh K, Nagao T, Dawson S, Nonoguchi K, Kido T, Mayer RJ, Arii S, Fujita J. Reduced stability of retinoblastoma protein by gankyrin, an oncogenic ankyrin-repeat protein overexpressed in hepatomas. Nat. Med. 2000;6:96–99. doi: 10.1038/71600. [DOI] [PubMed] [Google Scholar]
- 22.Dawson S, Apcher S, Mee M, Higashitsuji H, Baker R, Uhle S, Dubiel W, Fujita J, Mayer RJ. Gankyrin is an ankyrin-repeat oncoprotein that interacts with CDK4 kinase and the S6 ATPase of the 26 S proteasome. J. Biol. Chem. 2002;277:10893–10902. doi: 10.1074/jbc.M107313200. [DOI] [PubMed] [Google Scholar]
- 23.Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J. Struct. Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura Y, Umehara T, Tanaka A, Horikoshi M, Padmanabhan B, Yokoyama S. Structural basis for the recognition between the regulatory particles Nas6 and Rpt3 of the yeast 26S proteasome. Biochem. Biophys. Res. Commun. 2007;359:503–509. doi: 10.1016/j.bbrc.2007.05.138. [DOI] [PubMed] [Google Scholar]
- 25.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 26.Kleijnen MF, Roelofs J, Park S, Hathaway NA, Glickman M, King RW, Finley D. Stability of the proteasome can be regulated allosterically through engagement of its proteolytic active sites. Nat. Struct. Mol. Biol. 2007;14:1180–1188. doi: 10.1038/nsmb1335. [DOI] [PubMed] [Google Scholar]
- 27.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stadtmueller BM, Ferrell K, Whitby FG, Heroux A, Robinson H, Myszka DG, Hill CP. Structural models for interactions between the 20S proteasome and its PAN/19S activators. J. Biol. Chem. 2009 Nov 4; doi: 10.1074/jbc.C109.070425. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillette TG, Kumar B, Thompson D, Slaughter CA, Demartino GN. Differential roles of the C-termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26s proteasome. J. Biol. Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 32.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 33.Ferrell K, Wilkinson CR, Dubiel W, Gordon C. Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends. Biochem. Sci. 2000;25:83–88. doi: 10.1016/s0968-0004(99)01529-7. [DOI] [PubMed] [Google Scholar]
- 34.Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, Lupas AN, Zeth K. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol. Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 35.Thompson D, Hakala K, DeMartino GN. Subcomplexes of PA700, the 19 S regulator of the 26 S proteasome, reveal relative roles of AAA subunits in 26 S proteasome assembly and activation and ATPase activity. J. Biol. Chem. 2009;284:24891–24903. doi: 10.1074/jbc.M109.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeMartino GN, Moomaw CR, Zagnitko OP, Proske RJ, Chu-Ping M, Afendis SJ, Swaffield JC, Slaughter CA. PA700, an ATP-dependent activator of the 20 S proteasome, is an ATPase containing multiple members of a nucleotide-binding protein family. J. Biol. Chem. 1994;269:20878–20884. [PubMed] [Google Scholar]
- 37.Hoffman L, Rechsteiner M. Nucleotidase activities of the 26 S proteasome and its regulatory complex. J. Biol. Chem. 1996;271:32538–32545. doi: 10.1074/jbc.271.51.32538. [DOI] [PubMed] [Google Scholar]
- 38.Nickell S, Beck F, Scheres SH, Korinek A, Forster F, Lasker K, Mihalache O, Sun N, Nagy I, Sali A, Plitzko JM, Carazo JM, Mann M, Baumeister W. Insights into the molecular architecture of the 26S proteasome. Proc. Natl. Acad. Sci. U S A. 2009;106:11943–11947. doi: 10.1073/pnas.0905081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forster F, Lasker K, Beck F, Nickell S, Sali A, Baumeister W. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem. Biophys. Res. Commun. 2009;388:228–233. doi: 10.1016/j.bbrc.2009.07.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartmann-Petersen R, Tanaka K, Hendil KB. Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys. 2001;386:89–94. doi: 10.1006/abbi.2000.2178. [DOI] [PubMed] [Google Scholar]
- 41.Richmond C, Gorbea C, Rechsteiner M. Specific interactions between ATPase subunits of the 26 S protease. J. Biol. Chem. 1997;272:13403–13411. doi: 10.1074/jbc.272.20.13403. [DOI] [PubMed] [Google Scholar]
- 42.Kajava AV. What curves alpha-solenoids? Evidence for an alpha-helical toroid structure of Rpn1 and Rpn2 proteins of the 26 S proteasome. J. Biol. Chem. 2002;277:49791–49798. doi: 10.1074/jbc.M204982200. [DOI] [PubMed] [Google Scholar]
- 43.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat. Struct. Mol. Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Effantin G, Rosenzweig R, Glickman MH, Steven AC. Electron microscopic evidence in support of alpha-solenoid models of proteasomal subunits Rpn1 and Rpn2. J. Mol. Biol. 2009;386:1204–1211. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hendil KB, Kriegenburg F, Tanaka K, Murata S, Lauridsen AM, Johnsen AH, Hartmann-Petersen R. The 20S proteasome as an assembly platform for the 19S regulatory complex. J. Mol. Biol. 2009;394:320–328. doi: 10.1016/j.jmb.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y, Fruh K, Ahn K, Peterson PA. In vivo assembly of the proteasomal complexes, implications for antigen processing. J. Biol. Chem. 1995;270:27687–27694. doi: 10.1074/jbc.270.46.27687. [DOI] [PubMed] [Google Scholar]
- 47.Andersen KM, Madsen L, Prag S, Johnsen AH, Semple CA, Hendil KB, Hartmann-Petersen R. Thioredoxin Txnl1/TRP32 is a redox-active cofactor of the 26 S proteasome. J. Biol. Chem. 2009;284:15246–15254. doi: 10.1074/jbc.M900016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiseman RL, Chin KT, Haynes CM, Stanhill A, Xu CF, Roguev A, Krogan NJ, Neubert TA, Ron D. Thioredoxin-related Protein 32 is an arsenite-regulated Thiol Reductase of the proteasome 19 S particle. J. Biol. Chem. 2009;284:15233–15245. doi: 10.1074/jbc.M109.002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeMartino GN, Proske RJ, Moomaw CR, Strong AA, Song X, Hisamatsu H, Tanaka K, Slaughter CA. Identification, purification, and characterization of a PA700-dependent activator of the proteasome. J. Biol. Chem. 1996;271:3112–3118. doi: 10.1074/jbc.271.6.3112. [DOI] [PubMed] [Google Scholar]
- 50.Isono E, Nishihara K, Saeki Y, Yashiroda H, Kamata N, Ge L, Ueda T, Kikuchi Y, Tanaka K, Nakano A, Toh-e A. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]