Summary
The double-positive (DP) to single-positive (SP) transition during T cell development is initiated by down-regulation of the E-protein transcription factors HEB and E2A. Here, we have demonstrated that in addition to regulating the onset of this transition, HEB and E2A also play a separate role in CD4+ lineage choice. Deletion of HEB and E2A in DP thymocytes specifically blocked the development of CD4+ lineage T cells. Furthermore, deletion of the E-protein inhibitors Id2 and Id3 allowed CD4+ T cell development but blocked CD8+ lineage development. Analysis of the CD4+ lineage transcriptional regulators ThPOK and Gata3 placed HEB and E2A up-stream of CD4+ lineage specification. These studies identify an important role for E-proteins in the activation of CD4+ lineage differentiation as thymocytes undergo the DP to SP transition.
Introduction
The development of CD4+ and CD8+ lineage αβ T cells in the thymus is carefully regulated by developmental checkpoints which ensure proper T cell receptor (TCR) expression and selection and the appropriate determination of lineage choice (Carpenter and Bosselut, 2010). Of these, the TCR selection checkpoint at the CD4+CD8+ double-positive (DP) stage is critical for ensuring that each αβ T cell proceeding to the subsequent CD4+ or CD8+ single-positive (SP) stage expresses a functional TCR, capable of recognition of peptide presented by major histocompatibility complex class I (MHC I) or class II (MHC II) molecules. In addition to this positive selection step, cells expressing an auto-reactive TCR can undergo negative selection, eliminating them from the mature T cell pool (von Boehmer and Kisielow, 2006).
The DP to SP transition also involves the critical decision to enter either the CD4+ or CD8+ lineage (Singer et al., 2008). First, positively selected DP thymocytes progress through a transitional CD4+CD8lo stage (He et al., 2010), where the lineage choice decision is suggested to occur (Brugnera et al., 2000). Then, depending on the MHC specificity of their TCR, cells will proceed to either the CD4+ or CD8+ lineage, expressing only the CD4+ or CD8+co -receptor, respectively. Precise transcriptional regulation of CD4+ vs. CD8+ lineage choice ensures proper functional divergence and matching of co-receptor and TCR specificity (CD4+ and MHC II restriction or CD8+ and MHC I restriction). How the TCR signal translates into activation of CD4+ vs. CD8+ lineage transcriptional programs is still a topic of great interest and debate.
The E-protein transcription factors HEB and E2A are critical regulators of the DP to SP transition. E-proteins are required at the DP stage to enforce positive selection, maintaining expression of DP-associated genes while repressing expression of SP-associated genes (Jones and Zhuang, 2007). Upon receipt of a TCR-mediated positive selection signal, E-protein activity is down-regulated and cells proceed to the SP stage (Bain et al., 2001; Engel et al., 2001; Pan et al., 2002). We have shown previously that the removal of both HEB and E2A at the DP stage is sufficient for cells to initiate SP development in the absence of a TCR signal, suggesting that the down-regulation of E-proteins in response to a TCR-mediated positive selection signal is the molecular switch controlling DP to SP differentiation (Jones and Zhuang, 2007). Because the majority of cells, both TCR+ and TCR−, proceeding to the SP stage in the absence of HEB and E2A were CD8+, we hypothesized that E-proteins may play additional roles in regulating CD4+ vs. CD8+ lineage choice. Even though E-proteins are down-regulated during the window when cells undergo lineage choice, it was still possible that there were additional functions yet to be described for E-proteins during the DP to SP transition. We therefore asked the following questions: Are E-proteins specifically required for the development of CD4+ cells? If so, when during the CD4+ lineage developmental process are they required? What E-protein targets may be disrupted to cause the absence of CD4+ cells upon removal of both HEB and E2A?
Here, we utilized our Tcf12f/fTcfe2af/fCd4cre double conditional deletion model for the removal of Tcf12, the gene encoding HEB, and Tcfe2a (also known as Tcf3), the gene encoding E2A, at the DP stage of thymocyte development. Through the use of this model and other powerful genetic systems, we have shown that E-proteins are required for the development of CD4+T cells, and MHC II-restricted thymocytes can instead enter the CD8+ lineage in the absence of HEB and E2A. Perturbation of E-protein down-regulation by removal of the E-protein inhibitors Id2 and Id3 further supporteda role for E-proteins in directing CD4+ development, at the expense of CD8+ development. Analysis of CD4+ lineage factors and potential E-protein targets placed E-proteins up-stream of CD4+ lineage specification. We propose that proper regulation of the timing or amount of E-protein down-regulation during the DP to SP transition is critical to allow for CD4+ lineage development. In addition to revealing a role for E-proteins in CD4+ T cell development, this work identifies E-proteins as transcriptional regulators controlling both positive selection and lineage choice, serving to further elucidate the enigmatic relationship between these two events.
Results
CD4+ lineage T cells fail to develop in the absence of HEB and E2A
Initial analysis of the Tcf12f/fTcfe2af/fCd4cre+mice demonstrated a severe defect in the CD4+ T cell compartment in both the thymus and periphery (Jones and Zhuang, 2007). Because this defect could be due to either a failure of CD4+ T cells to develop or enhanced negative selection of CD4 SP thymocytes, we first determined if inhibition of negative selection could rescue the CD4+ population. Two approaches were taken to analyze the effect of eliminating mediators of negative selection. First, Tcf12f/fTcfe2af/fCd4cre+mice were crossed with Bim-deficient mice (Bcl2l11−/−), a model defective in cell death of negatively selected thymocytes (Bouillet et al., 2002). Elimination of Bim, however, failed to significantly rescue the CD4 SP defect observed in Tcf12f/fTcfe2af/fCd4cre+mice (Figure 1A). Second, Tcf12f/fTcfe2af/fCd4cre+ mice were crossed with MHC I and II-double deficient mice. Even though CD8+ T cells can develop in the absence of a TCR upon removal of HEB and E2A (Jones and Zhuang, 2007), we reasoned that an MHC-mediated TCR signal would still be required to induce negative selection among the population of thymocytes successfully expressing a TCR on their cell surface. Therefore, because the CD4 SP thymocytes in Tcf12f/fTcfe2af/fCd4cre+mice are TCR+, removal of both MHC I and II was an additional test to determine if blocking the negative selection signal could rescue this population. However, similar to the Bim-deficiency, there was no rescue of CD4 SP cells upon removal of MHC I and II, through use of β2 microglobulin- (B2m) and Abb-targeted mice, respectively (Figure 1B). In fact, the CD4 SP defect was more severe in Tcf12f/fTcfe2af/fAbb−/−B2m+/−Cd4cre+ and Tcf12f/fTcfe2af/fAbb−/−B2m−/−Cd4cre+ mice than in Tcf12f/fTcfe2af/fCd4cre+ mice. This result confirmed that the development of the CD4+TCR+T cells observed in Tcf12f/fTcfe2af/fCd4cre+ mice requires MHC II and TCR-mediated positive selection, indicating that these cells represent a small population of MHC II-restricted thymocytes likely receiving a positive selection signal prior to complete loss of residual HEB and E2A protein.
Figure 1. Elimination of negative selection mediators fails to rescue the CD4+ T cell defect.
(A) Tcf12f/fTcfe2af/fCd4cre+ mice were crossed onto a Bim-deficient background (Bcl2l11−/−). Representative staining of thymus from indicated genotypes. Total thymocyte cell number means ± SD (x108) for 1–2 month old mice were 1.9 ± 0.97 for control (n=5); 2.4 ± 0.55 for Bcl2l11−/− (n=4); 1.5 ± 0.30 for cre+ (n=5); 2.3 ± 1.3 for cre+ Bcl2l11−/− (n=7). Percentages in each quadrant are displayed. Percent means ± SD for the CD4 SP (CD4+CD8−) gate were 18 ± 4.9 for control (n=5); 21 ± 8.5 for Bcl2l11−/− (n=5); 1.7 ± 0.37 for cre+ (n=6); 4.5 ± 3.1 for cre+ Bcl2l11−/− (n=7). (B) Tcf12f/fTcfe2af/fCd4cre+ mice were crossed onto MHC class I- (B2m−/−) and/or class II- (Abb−/−) deficient backgrounds. Representative staining of thymus from indicated genotypes. Total thymocyte cell number means ± SD (x108) for 1.5–4 month old mice were 1.9 ± 1.2 for control (n=3); 1.3 ± 0.48 for cre+ (n=4); 0.67 ± 0.32 for Abb−/−cre+ (n=4); 1.2 ± 0.76 for Abb−/−B2m−/−cre+ (n=5). Percentages in each quadrant are displayed. Percent means ± SD for the CD4 SP (CD4+CD8−) gate were 10 ± 1.9 for control (n=3); 1.5 ± 0.61 for cre+ (n=4); 0.60 ± 0.088 for Abb−/−cre+ (n=4); 0.30 ± 0.14 for Abb−/−B2m−/−cre+ (n=5). CD4 SP percent values for Abb−/−cre+ and Abb−/−B2m−/−cre+ are significantly different from cre+ (p=0.023 and p=0.003, respectively). Abb−/−cre+ animals used for these statistics are either B2m+/− or B2m+/+. See also Figure S1.
The failure of Bim- or MHC I and II-deficiency to rescue CD4 SP cells in the thymus, which is also reflected in analysis of the peripheral CD4+ T cell compartment in the spleen of these animals (Figure S1A and S1B), suggested that enhanced negative selection is not the major factor driving the CD4+ T cell phenotype in Tcf12f/fTcfe2af/fCd4cre+mice. The absence of CD4+ T cells upon removal of MHC II demonstrated that CD4+T cells fail to develop independently of TCR signaling in the absence of HEB and E2A. This result is specific to the CD4+ lineage as CD8+ T cells, both TCR− and TCR+, continue to be produced in the absence of both MHC I and II-mediated selection (Figure S1C). Together, these findings indicate that the reduced CD4+ compartment in Tcf12f/fTcfe2af/fCd4cre+mice results from a failure to generate CD4+ lineage T cells.
Removal of E-proteins drives CD4+ lineage-destined cells to the CD8+ lineage
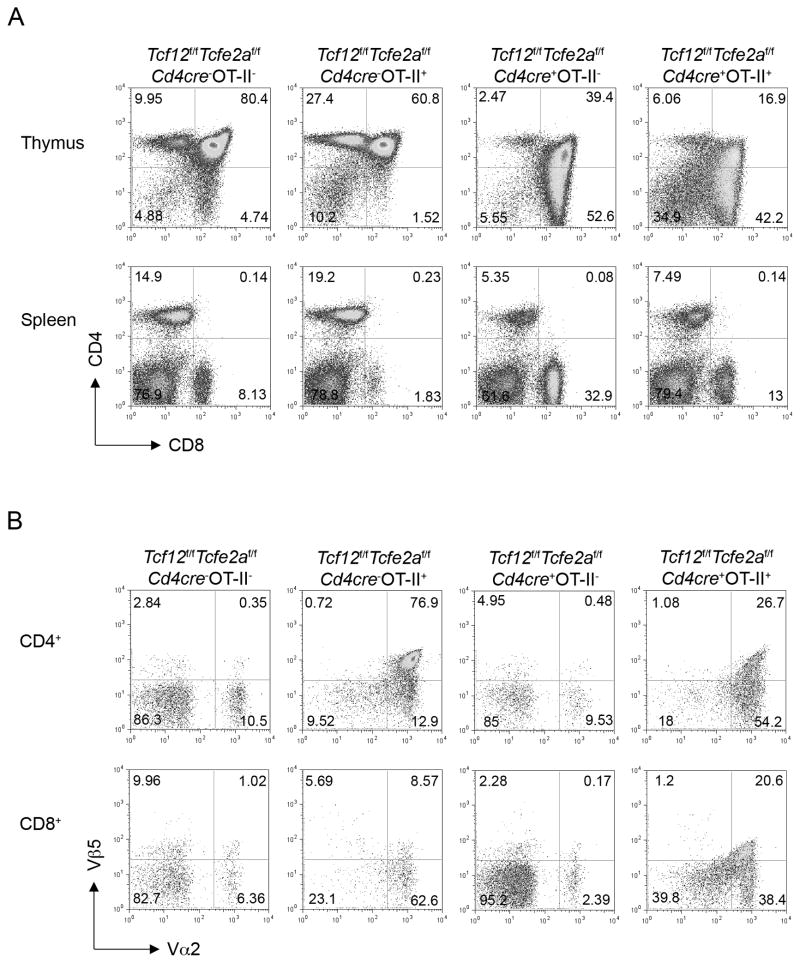
If CD4+ lineage T cells fail to develop in the absence of HEB and E2A, are MHC II-restricted thymocytes blocked in their development or can they instead proceed by entering the CD8+ lineage? To address this question, we next crossed the Tcf12f/fTcfe2af/fCd4cre+mice onto an OT-II TCR transgenic background. OT-II transgenic mice express a MHC II-restricted TCR, composed of a Vβ5 and a Vα2 chain, which drives primarily CD4+T cell production (Barnden et al., 1998). However, analysis of Tcf12f/fTcfe2af/fCd4cre+OT-II+ mice revealed an increased ratio of CD8+:CD4+ in both the thymus and spleen, similar to that of Tcf12f/fTcfe2af/fCd4cre+ mice alone (Figure 2A). To confirm that CD8+ T cells expressing the components of the OT-II transgenic TCR were being generated in Tcf12f/fTcfe2af/fCd4cre+OT-II+mice, Vβ5 and Vα2 expressing populations among splenic T cells and thymocytes were analyzed. Indeed, a large population of CD8+T cells co-stained positive for Vβ5 and Vα2 in Tcf12f/fTcfe2af/fCd4cre+OT-II+ mice, and the percent of CD8+ T cells within the Vβ5+Vα2+ compartment was significantly higher in Tcf12f/fTcfe2af/fCd4cre+OT-II+ mice when compared to Tcf12f/fTcfe2af/fCd4cre−OT-II+ mice (Figure 2B and S2A), revealing that developing thymocytes that would normally be directed to the CD4+ lineage, as seen in the Tcf12f/fTcfe2af/fCd4cre−OT-II+ control, could instead become CD8+ T cells in the absence of HEB and E2A.
Figure 2. Deletion of E-proteins on a TCR transgenic background generates CD8+ T cells expressing components of a MHC II-restricted TCR.
Tcf12f/fTcfe2af/fCd4cre+ mice were crossed onto an OT-II TCR transgenic background. (A) Representative staining of thymus and spleen from indicated genotypes. Percentages in each quadrant are displayed. Percent means ± SD for the CD4 SP (CD4+CD8−) thymocyte gate were 11 ± 0.74 for control (n=5); 28 ± 2.1 for OT-II+ (n=5); 3.3 ± 1.8 for cre+ (n=4); 5.2 ± 2.1 for cre+OT-II+ (n=7). Percent means ± SD for the splenocyte CD4+ T (CD4+CD8−) and CD8+ T (CD4−CD8+) gates, respectively, were 18 ± 5.7 (n=5) and 8.7 ± 2.3 (n=7) for control; 17 ± 4.6 (n=4) and 2.2 ± 0.63 (n=5) for OT-II+; 4.8 ± 0.92 (n=4) and 29 ± 9.2 (n=5) for cre+; 7.6 ± 2.2 (n=6) and 21 ± 11 (n=8) for cre+OT-II+. (B) Representative staining of spleen from indicated genotypes. Plots are pre-gated on CD4+ or CD8+ cells, as labeled. Percentages in each quadrant are displayed. Percent means ± SD for the splenocyte Vα2+Vβ5+ gate within the CD8+ T cell compartment were 1.4 ± 0.32 (n=5) for control; 14 ± 5.2 (n=3) for OT-II+; 0.20 ± 0.044 (n=3) for cre+; 35 ± 17 (n=6) for cre+OT-II+. See also Figure S2.
To confirm that the CD8+Vβ5+Vα2+ cells developing in the Tcf12f/fTcfe2af/fCd4cre+OT-II+ mice could be activated and behave like CD8+ lineage T cells, we isolated and stimulated splenocytes in vitro. Even though these CD8+ cells expressed the components of a MHC II-restricted TCR, they were able to produce IFN-γ, up-regulate the activation marker CD69, and proliferate upon stimulation (Figure S2 B-D). Together, these results suggest that the removal of HEB and E2A blocks the ability of thymocytes to become CD4+ lineage T cells, and the resulting E-protein-deficient cells undergo the DP to SP transition to become CD8+ lineage T cells, independent of TCR expression and specificity. E-proteins are therefore required first, for ensuring that T cells express a functional TCR and second, for ensuring that cells enter the appropriate lineage to match their TCR specificity.
Removal of the E-protein negative regulators Id2 and Id3 permits CD4+ lineage T cell development
Down-regulation of E-protein activity upon receipt of a TCR-mediated positive selection signal is largely mediated by induction of the E-protein inhibitor Id3 (Bain et al., 2001; Engel et al., 2001). The mammalian Id family consists of four members (Id1-4), which form heterodimers with E-proteins to inhibit E-protein DNA-binding activity (Benezra et al., 1990). Of these Id family members, primarily Id3 and Id2 are expressed during T cell development (Engel et al., 2001; Wikstrom et al., 2008). Because Id3-deficient mice demonstrate only a partial block in DP to SP development (Rivera et al., 2000), we hypothesized that Id2 may be performing a similar function and compensating in the absence of Id3 to down-regulate E-protein activity and allow cells to develop beyond the DP stage. In addition, even though the absence of Id3 does not demonstrate a bias for CD4+ over CD8+ T cell development (Guo et al., 2011; Pan et al., 1999; Rivera et al., 2000), as could be anticipated given our E-protein-deficient phenotype, we wondered if the removal of both Id3 and Id2 would exhibit this effect on lineage choice by more strongly perturbing E-protein down-regulation.
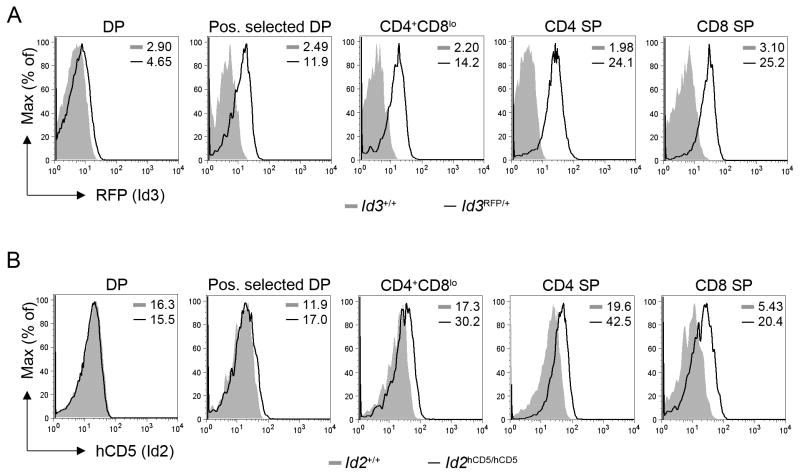
First, we investigated the expression pattern of Id3 and Id2 at the single cell level during DP to SP thymocyte development. Id3 has previously been shown by RT-PCR to be induced in TCR-signaled thymocytes (Bain et al., 2001; Engel et al., 2001), and time course analysis suggests that Id3 is rapidly and transiently expressed in response to this stimulation (Schmitz et al., 2003). In addition, single cell analysis of Id3 expression, using an Id3RFP reporter mouse (McMahon et al., 2008), is consistent with rapid Id3 up-regulation in thymocytes undergoing positive selection (Figure 3A). This result is also consistent with a recent study utilizing an Id3GFP allele (Miyazaki et al., 2011). Id2 has been suggested to be induced during the DP to SP transition as well (Huang et al., 2004; Wang et al., 2006), however with delayed kinetics when compared to Id3 expression (Schmitz et al., 2003). To further investigate the expression pattern of Id2 during thymocyte development, we developed an Id2hCD5 reporter allele by inserting an internal ribosome entry site- (IRES) driven, tailless human-CD5 cDNA cassette into the 3′ untranslated region of the Id2 gene (Figure S3A and S3B). In this model, the coding region of Id2 remains intact, and hCD5 surface expression is used as a reporter of Id2 transcription. Expression of the hCD5 reporter is clearly observed within the Id2-dependent (Yokota et al., 1999) NK cell (NK1.1+) compartment and is also detected within the αβ and γδ T cell lineages (Figure S3C and S3D). In addition, Id2hCD5/hCD5 mice express Id2 at amounts comparable to that of wild-type controls during thymocyte development (Figure S3E). Single cell analysis of hCD5 expression demonstrated slight up-regulation of Id2 expression in positively selected DP thymocytes with expression further increasing as cells transition through the transient CD4+CD8lo compartment and into both the CD4 and CD8 SP stages (Figure 3B and S3 F-H). These results are consistent with previous RT-PCR analysis (Wang et al., 2006) and suggest that, while the kinetics may be delayed (Schmitz et al., 2003), Id2 expression corresponds with that of Id3 during the DP to SP transition.
Figure 3. Id3 and Id2 are up-regulated from the DP to SP stage.
(A) Because the Id3RFP reporter allele design results in the disruption of the Id3 gene, we analyzed Id3RFP/+ animals here. Mean values for RFP expression in B6 (Id3+/+) and Id3RFP/+ populations are indicated, and data are representative of analysis of three Id3RFP/+ mice. (B) Mean values for expression of hCD5 in B6 (Id2+/+) and Id2hCD5/hCD5 populations are indicated, and data are representative of analysis of three Id2hCD5/hCD5 mice. (A and B) Histograms are pre-gated on the following thymic populations: DP (CD4+CD8+), positively selected DP (CD4+CD8+TCRβ+CD69+), CD4+CD8lo (CD4+CD8lo), CD4SP (CD4+CD8−), and CD8 SP (CD4−CD8+TCRβ+). See also Figure S3.
Because the expression pattern of Id2 suggested that it may serve redundant function with Id3 to down-regulate E-protein activity, the effect of removing both Id2 and Id3 at the DP stage, by use of a conditional Id2f/fId3f/fCd4cre mouse model, was then investigated. In fact, Id2f/fId3f/fCd4cre+ mice did display the expected defect in DP to SP development, further highlighted by the accumulation of TCRβ+ cells within the DP and CD4+CD8lo gates (Figure 4A). The increased percentage of CD69+ cells detected within the DP gate was also supportive of an accumulation of positively-selected thymocytes due to delayed differentiation into the SP stage.
Figure 4. CD4+ T cells can develop in the absence of Id2 and Id3.
(A)Representative staining of thymus from indicated genotypes. First row graphs display total live thymocytes. Second row graphs are pre-gated on DP or total TCRβ+ thymocytes, as shown. Percentages in each gated population are displayed. Percent means ± SD for the CD4 SP (CD4+CD8−) gate within the TCRβ+ compartment were 59 ± 5.5 for cre− (n=14) and 24 ± 11 for cre+ (n=14), with a significant difference (p<0.001). Percent means ± SD for the CD8 SP (CD4−CD8+) gate within the TCRβ+ compartment were 15 ± 2.0 for cre−(n =14) and 0.87 ± 0.84 for cre+ (n=14), with a significant difference (p<0.001). (B) Cell number in the thymus of 1–2 month old mice (n=16). (C) Number of CD4 SP TCRβ+ thymocytes in 1–2 month old mice (n=13). ***p<0.001. Graphed results are means with error bars representing SD. See also Figure S4.
In addition to the defect in progression of positively-selected cells, there was also a differential effect on the generation of CD4 vs. CD8 SP thymocytes in these Id-deficient animals. CD8 SP development demonstrated a near complete block, whereas CD4 SP development was permissible. The unchanged total thymocyte numbers and the reduction of CD4 SP cell numbers suggested that CD4+ T cell development was limited (Figure 4B and 4C), but analysis of TCRβ+ thymocytes clearly showed that the CD4+ lineage pathway remains viable upon deletion of Id2 and Id3 (Figure 4A). DNA analysis of Id2 and Id3 deletion in Id2f/fId3f/fCd4cre+ thymocytes confirmed efficient deletion at the DP stage (Figure S4A). Complete deletion of Id2 and Id3 appeared to be necessary and sufficient for the observed phenotype because SP thymocyte development in mice that retain one allele of either Id2 or Id3 was comparable to that of cre− controls (Figure S4B). Peripheral analysis of T cells in these mice also demonstrated a reduction in the CD4+ T cell compartment and a nearly absent CD8+ T cell compartment (Figure S4C). Faint Id2 and Id3 floxed bands were observed in deletion analysis of splenic Id2f/fId3f/fCd4cre+ CD4+ T cells, suggesting that there may be some peripheral selective pressure for any cells escaping complete deletion (Figure S4D). However, comparison to a heterozygous control verified that the majority of peripheral T cells had undergone efficient deletion of both Id2 and Id3, as expected from the efficient deletion observed in the thymus (Figure S4A).
To further characterize the CD4 SP population in the Id2f/fId3f/fCd4cre+ mice and to determine which events during the DP to SP transition are Id-dependent, we examined the expression dynamics of surface receptors that are functionally important during this transition (Carpenter and Bosselut, 2010). While expression of the chemokine receptor CXCR4 was properly down-regulated in Id2f/fId3f/fCd4cre+ CD4 SP thymocytes, both the chemokine receptor CCR7 and cytokine receptor IL7Rα failed to be efficiently up-regulated in the absence of Id2 and Id3 (Figure 5A and 5B). Next, we analyzed the expression of two transcription factors, KLF2 and Tox, which are normally up-regulated upon positive selection (Kuo et al., 1997; Wilkinson et al., 2002). Both KLF2 and Tox were expressed in Id2f/fId3f/fCd4cre+ CD4 SP thymocytes, comparably to cre− controls (Figure 5C). Together, these results further indicate that the CD4 SP cells in these mice have characteristics of normal SP thymocytes, but do demonstrate some defects. CCR7 and IL7Rα up-regulation upon positive selection are shown here to be Id-dependent events and likely require Id-mediated E-protein down-regulation.
Figure 5. CCR7 and IL7Rα fail to be efficiently up-regulated inId2 f/fId3f/fCd4cre+CD4 SP thymocytes.
(A) Representative staining of thymus from indicated genotypes. Plots are pre-gated on DP (CD4+CD8+) or CD4 SP (CD4+CD8−TCRβ+) thymocytes and percentages in each quadrant are displayed. Percent means ± SD for total CCR7+ thymocytes within the CD4 SP compartment were 78 ± 8.0 for cre− (n=6) and 25 ± 12 for cre+ (n=6), with a significant difference (p<0.001). Percent means ± SD for total CXCR4− thymocytes within the CD4 SP compartment were 86 ± 4.6 for cre− (n=4) and 80 ± 12 for cre+ (n=4). (B) Representative staining of thymus with histograms pre-gated on DP and CD4 SP thymocytes as described in (A). Percentages within the IL7Rα+ gate are displayed. Percent means ± SD for IL7Rα+ thymocytes within the CD4 SP compartment were 55 ± 4.1 for cre− (n=3) and 3.7 ± 1.6 for cre+ (n=3), with a significant difference (p<0.001). (C) Quantitative RT-PCR analysis of Klf2 and Tox expression in sorted DP (CD4+CD8+) and CD4 SP (CD4+CD8−TCRβ+) thymocytes. Duplicate runs from a sample sorted from an individual animal (n=3 for cre− DP, cre+ DP, cre− CD4 SP; n=4 for cre+ CD4 SP) were averaged and normalized to the expression of Gapdh. Graphed results are means with error bars representing SEM. (D) Representative staining of thymus with histograms pre-gated on DP and CD4 SP thymocytes as described in (A). Percentages within the HSAlo gate are displayed (HSA clone M1/69 was used). Percent means ± SD for HSAlo thymocytes within the CD4 SP compartment were 53 ± 6.0 for cre− (n=4) and 86 ± 8.4 for cre+ (n=4), with a significant difference (p<0.001) Plots in (A, B, and D) are from analysis of the same individuals. See also Figure S5.
The Id2f/fId3f/fCd4cre+ CD4 SP population was next analyzed for expression of HSA and TCR usage. HSA expression was down-regulated in this compartment, consistent with an SP-like phenotype (Figure 5D). However, there was an increase in the percentage of HSAlo cells within the Id2f/fId3f/fCd4cre+ CD4 SP population. This could be due to a delay in exiting the thymus and/or expansion of this population. While the CD4 SP population in these animals did not appear to simply result from clonal expansion of a few cells proceeding to the CD4 SP stage prior to complete deletion, the Id2f/fId3f/fCd4cre+ CD4 SP cells did display more variation in usage of TCRβ and TCRα chains from animal to animal when compared to cre− controls (Figure S5A). This result may reflect expansion within a fraction of the CD4 SP thymocytes or could be due to alterations in TCR selection. Nonetheless, only one out of the four mice analyzed demonstrated a dominant Vβ and Vα TCR usage among the specific chains examined, suggesting that while expansion may be occurring in a fraction of CD4 SP thymocytes, a relatively diverse population of CD4 SP thymocytes is being generated in the absence of Id2 and Id3.
We further investigated the MHC restriction of these Id-deficient CD4 SP thymocytes. First, mouse CD1d tetramer staining demonstrated that the majority of TCRβ+ CD4 SP thymocytes in Id2f/fId3f/fCd4cre+ animals are not invariant NK T cells (Figure S5B). Second, upon Id2f/fId3f/fCd4cre+ bone marrow transfer to congenic MHC II-deficient recipients, donor cells are capable of efficiently reconstituting the DP but neither the CD8 SP nor CD4 SP compartment (Figure S5C). A small amount of donor derived TCRβ+ CD4 SP thymocytes were observed in the MHC-II-deficient recipients, but the percentage was greatly diminished when compared to the TCRβ+ CD4 SP population generated from the same donor cells when transferred to control wild-type or Rag2-deficient recipients. Together, this data implies that the majority of TCRβ+ CD4 SP thymocytes developing in the Id2f/fId3f/fCd4cre+ mice are MHC II-restricted.
The block in CD8 SP production and permission of CD4 SP development upon removal of Id2 and Id3 further support a differential role for E-proteins in regulating CD4+ vs. CD8+ lineage development. The Id2f/fId3f/fCd4cre model suggests that, in addition to E-proteins being required for CD4+ lineage choice, disruption of the amount and/or timing of E-protein down-regulation may be sufficient to drive CD4+ lineage production over CD8+ development.
E-proteins are required prior to CD4+ lineage specification
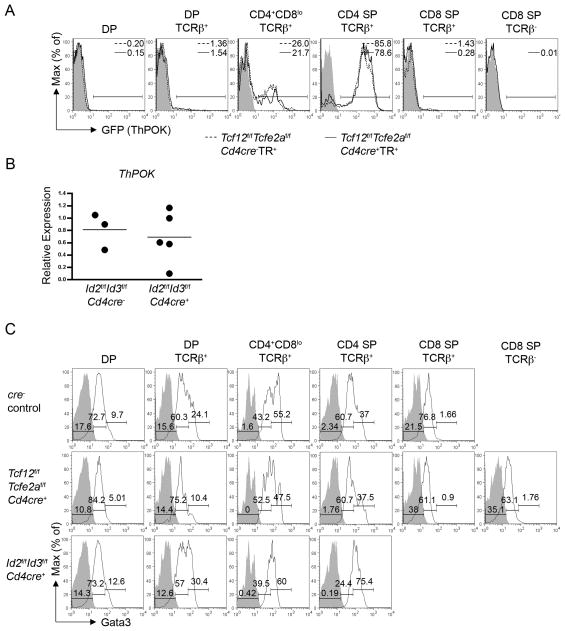
The phenotypes of the E-protein-deficient and Id-deficient mouse models both support a role for E-proteins positively regulating the generation of CD4+ T cells. We next sought to determine at which point E-proteins are required during the CD4+ lineage developmental program. For this purpose, we first analyzed expression of the CD4-specific transcriptional regulator ThPOK. ThPOK expression is initiated and maintained after TCR-mediated positive selection for the commitment of MHC II-restricted thymocytes to the CD4+lin eage (He et al., 2005; He et al., 2008; Sun et al., 2005). Therefore, ThPOK expression can be used as a marker of CD4+ lineage identity. We crossed Tcf12f/fTcfe2af/fCd4cre+mice with a ThPOK reporter strain (denoted here as TR) that expresses GFP under the control of Thpok regulatory elements (He et al., 2008). Analysis of Tcf12f/fTcfe2af/fCd4cre+TR+ mice demonstrated no significant GFP+ population within the CD8 SP TCRβ+ or CD8 SP TCRβ− gates above the background observed in cre−TR+ CD8 SP TCRβ+ controls (Figure 6A). This result suggested that there were no cells initially adopting the CD4+ lineage fate within the CD8 SP compartment. Similar to the Tcf12f/fTcfe2af/fCd4cre−TR+ control mice, GFP expression in Tcf12f/fTcfe2af/fCd4cre+TR+ mice was primarily restricted to the CD4+CD8lo and CD4 SP compartments. While there are very few cells within the CD4+CD8lo and CD4 SP compartments in E-protein-deficient mice, we did detect ThPOK expression among these cells, consistent with a previous finding that E-proteins were not required for maintenance of ThPOK expression in mature T cells (data not shown). However, the vast majority of cells, which lost E-protein activity and by-passed the requirement for positive selection, were unable to activate ThPOK expression and were unable to undergo CD4+ lineage development.
Figure 6. E-proteins function up-stream of ThPOK and Gata3 induction.
(A) Tcf12f/fTcfe2af/fCd4cre+ mice were crossed onto a ThPOK reporter (TR) background. Staining of indicated genotypes for reporter induced GFP expression. Controls (shaded) are from wild-type (B6) mice. Histograms are pre-gated on specified thymic populations, and the percent GFP+ is indicated and was determined using the gate shown. Percent means ± SD for the GFP+ compartment were 0.86 ± 0.53 for cre−TR+ CD8 SP TCRβ+ (n=3); 0.19 ± 0.072 for cre+TR+ CD8 SP TCRβ+ (n=4); 0.027 ± 0.021 for cre+TR+ CD8 SP TCRβ− (n=4). CD8 SP TCRβ+ wild-type cells were used for the background control staining shown in both the CD8 SP TCRβ+ and CD8 SP TCRβ− plots. Total thymocyte cell number means ± SD (x108) for 1–2 month old mice were 1.4 ± 0.67 for cre−TR+ (n=3) and 0.88 ± 0.42 for cre+TR+ (n=3). (B) Quantitative RT-PCR analysis of ThPOK expression in sorted CD4 SP TCRβ+ thymocytes from Id2f/fId3f/fCd4cre− and Id2f/fId3f/fCd4cre+ mice (gating strategy shown in Figure S6A). Samples were normalized to the expression of Gapdh. Each dot represents the average of triplicate runs for a sample sorted from an individual animal, with n=3 for cre− and n=5 for cre+. The line displays the mean value for each genotype. Serving as a negative control, ThPOK product was undetectable in cre− and cre+ DP samples (n=3) by this method. (C) Intracellular staining for Gata3 expression in Tcf12f/fTcfe2af/fCd4cre+, Id2f/fId3f/fCd4cre+, and Id2f/fId3f/fCd4cre− control mice. Plots are pre-gated on specified thymic populations and display isotype control staining (shaded) and anti-Gata3 staining (line). Id2f/fId3f/fCd4cre− isotype controls were used for all subsets shown except for the CD8 SP TCRβ− plot, which displays the Tcf12f/fTcfe2af/fCd4cre+ isotype control. Histograms are subdivided into three increasing amounts of Gata3 expression. Percentages for anti-Gata3 staining in each of these gates are displayed. Data are representative of three independent experiments. See also Figure S6.
Analysis of Id2f/fId3f/fCd4cre+ mice demonstrated that phenotypically, the development of CD4 SP TCRβ+ thymocytes was intact (Figure 4). However, to ensure that these cells were transcriptionally initiating the CD4+ lineage fate, expression of ThPOK within Id2f/fId3f/fCd4cre+ CD4 SP cells was examined as well. As expected, ThPOK expression was detected in CD4 SP thymocytes from Id2f/fId3f/fCd4cre+ mice and was within the range of ThPOK expression from Id2f/fId3f/fCd4cre− controls for 4 of the 5 animals examined (Figure 6B and S6A). This data suggests that activation of ThPOK expression can occur independently of TCR-mediated Id up-regulation.
The transcription factor Gata3 has been shown to regulate CD4+ lineage specification and does so in part by allowing activation of ThPOK expression (Wang et al., 2008). In addition, Gata3 expression is up-regulated during the DP to SP transition primarily in MHC II-restricted cells entering the CD4+ lineage (Hendriks et al., 1999; Hernandez-Hoyos et al., 2003). Therefore, we next investigated if E-proteins are required up-stream or down-stream of Gata3 up-regulation by analyzing Gata3 expression in our E-protein-deficient and Id-deficient mouse models. Intracellular staining of Gata3 in Tcf12f/fTcfe2af/fCd4cre+ mice displayed a decrease in the percentage of positively selected thymocytes expressing higher amounts of Gata3, indicating a defect in specification of CD4+ lineage thymocytes (Figure 6C and S6B and S6C). In addition, cells expressing high amounts of Gata3 were not detected in the CD8 SP compartment. These results indicate that Gata3 up-regulation fails to be induced in the absence of HEB and E2A.
Complimentary to the E-protein deficient analysis, Gata3 up-regulation was efficiently induced in Id2f/fId3f/fCd4cre+ mice for the development of CD4 SP thymocytes. After initiation, more cells expressed higher amounts of Gata3 (Figure 6C). Overlay of the recently selected populations suggested that the amount of Gata3 in Id2f/fId3f/fCd4cre+ CD4+CD8lo cells had already started to resolve, as it did in the CD4 SP stage in the control cells (Figure S6C). However, this intermediate amount of expression persisted through the CD4 SP stage in the Id-deficient animals. Together, these results further support that E-proteins play a positive role in regulating Gata3 expression. Cells receiving a TCR-mediated positive selection signal in the absence of Id2 and Id3 can initiate Gata3 up-regulation, consistent with a role for HEB and E2A up-stream of initiation of CD4+ lineage specification.
The absence of ThPOK and Gata3 expression in the CD8+ T cell compartment of E-protein deficient mice suggests that CD4+ lineage development was blocked prior to induction of these factors and demonstrates that the phenotypic CD8+ compartment did not contain any transcriptionally-defined CD4+ lineage cells. To further analyze the transcriptional profile of these cells, we examined expression of the CD8+ lineage transcriptional regulator Runx3 (Collins et al., 2009). Runx3 expression was assessed by amplifying transcripts from the distal Runx3 promoter, specific to the CD8+ lineage (Egawa et al., 2007). CD8 SP thymocytes from Tcf12f/fTcfe2af/fCd4cre+ mice did express Runx3, and amounts were comparable to that of cre− control CD8 SP cells (Figure S6D). In addition, a faint Runx3 product was detected in the DP compartment in one of the two Tcf12f/fTcfe2af/fCd4cre+ animals analyzed, above that of the cre− and B6 controls. This finding is consistent with the pre-mature CD8 SP-like development occurring in Tcf12f/fTcfe2af/fCd4cre+ DP thymocytes (Jones and Zhuang, 2007).
E-proteins promote CD4+ lineage development beyond regulation of CD4 expression
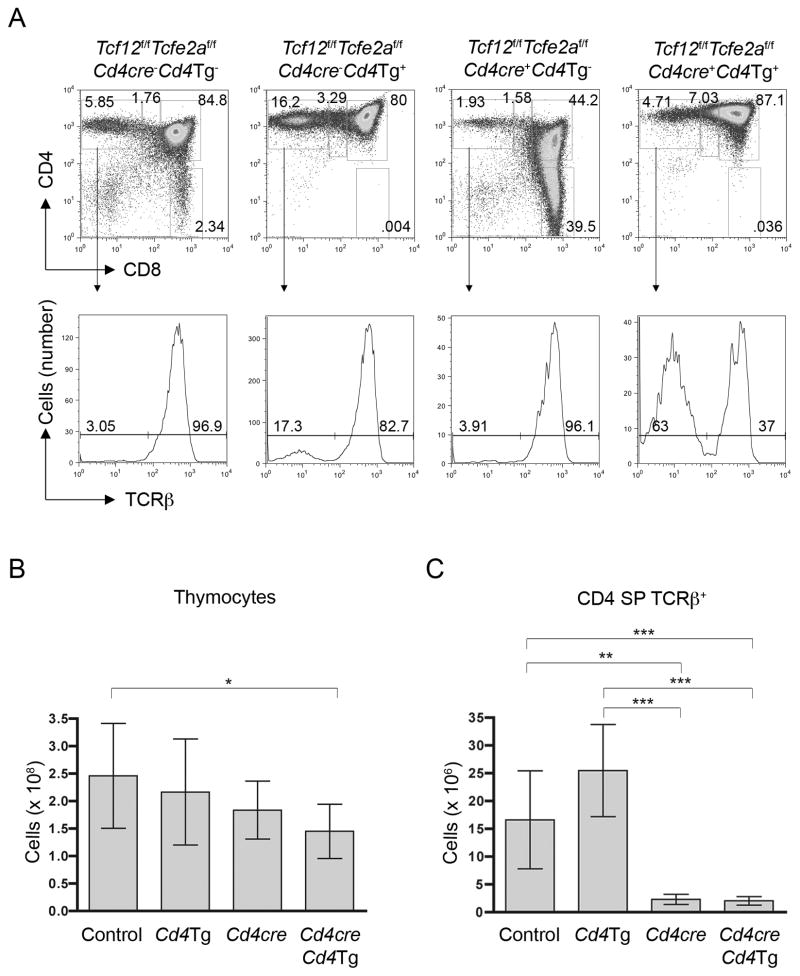
E-proteins have previously been shown to regulate expression of the TCR co-receptor CD4 (Sawada and Littman, 1993), and maintenance of CD4 expression is important for signaling CD4+ lineage differentiation (Sarafova et al., 2005). Therefore, we investigated if a defect in CD4 expression upon removal of HEB and E2A was responsible for the block in CD4+ T cell development. For this purpose, we tested if the addition of a Cd4 transgene (Van Laethem et al., 2007) could rescue CD4 development in the Tcf12f/fTcfe2af/fCd4cre+ mice.
If a Cd4 transgene were able to rescue CD4+ lineage development in the absence of HEB and E2A, we would expect to see an increase in the CD4 SP population, potentially at the expense of CD8+ lineage development. Even though Tcf12f/fTcfe2af/fCd4cre+Cd4Tg+ mice did show a slight increase in the percentage of CD4+CD8− thymocytes compared to Tcf12f/fTcfe2af/fCd4cre+ mice, CD4 SP TCRβ+ cells made up only a fraction of the cells in that gate (Figure 7A). In mice that carry the Cd4 transgene, which is expressed throughout T cell development, the CD4 SP gate also contained double negative thymocytes, and the DP gate also contained CD8 SP cells. While the total number of thymocytes was slightly reduced in Tcf12f/fTcfe2af/fCd4cre+Cd4Tg+ mice compared to controls, the number of CD4 SP TCRβ+ cells in these animals was significantly reduced from that of Tcf12f/fTcfe2af/fCd4cre−Cd4Tg− and Tcf12f/fTcfe2af/fCd4cre−Cd4Tg+ mice (Figure 7B and 7C). Additionally, there was no increase in the CD4 SP TCRβ+ cell number above that of Tcf12f/fTcfe2af/fCd4cre+ mice, clearly indicating that the Cd4 transgene could not rescue CD4+ lineage development.
Figure 7. Addition of a Cd4 transgene fails to rescue the CD4+ T cell defect in E-protein deficient mice.
Tcf12f/fTcfe2af/fCd4cre+ mice were crossed onto a Cd4 transgenic background. (A)Representative staining of thymus from indicated genotypes. Histograms are pre-gated on the CD4 SP gate, as shown. Percentages in each gated population are displayed. (B) Cell number in the thymus of 1–2 month old mice. (C) Number of CD4 SP TCRβ+ thymocytes in 1–2 month old mice. (B and C) Control n=9, cd4Tg n=8, Cd4cre n=6, Cd4creCd4Tg n=10. For cell counts, control and Cd4cre animals were either +/+ or +/− for endogenous Cd4, and Cd4Tg and Cd4creCd4Tg animals were +/+, +/−, or −/− for endogenous Cd4. We detected no effect on cell numbers with these variations in the endogenous Cd4 background. ***p<0.001, **p=0.0018, and *p=0.0091. Graphed results are means with error bars representing SD. See also Figure S7.
Analysis of peripheral T cell populations also demonstrated this failure of the Cd4 transgene to rescue CD4+ T cells in the absence of HEB and E2A (Figure S7). In addition, the large populations of CD8 TCRβ+ and TCRβ − cells detected in Tcf12f/fTcfe2af/fCd4cre+ mice continued to be produced in Tcf12f/fTcfe2af/fCd4cre+Cd4Tg+ mice. Therefore, the requirement for E-proteins that allows for CD4+ lineage differentiation is distinct from E-proteins’ role in regulating CD4 expression.
Discussion
Here, we have demonstrated a specific requirement for E-proteins in the generation of CD4+ lineage T cells. Previous analysis had shown that E-proteins play a pivotal role in regulating DP survival and positive selection, but it is clear from the present study that E-protein function extends to the regulation of CD4+ vs. CD8+ lineage choice as well. Not only are E-proteins required for the initiation of CD4+lin eage development, but they are also required to prevent entry of MHC II-restricted thymocytes into the CD8+ lineage. Therefore, in addition to enforcing the requirement for a TCR at the positive selection step, HEB and E2A also enforce matching of the TCR with the appropriate co-receptor.
The conditional removal of both Id2 and Id3 at the DP stage revealed a phenotype supporting these findings, which had not previously been seen with either Id2- or Id3- deficiency alone (Guo et al., 2011; Pan et al., 1999; Rivera et al., 2000; Yokota et al., 1999). Removal of Id proteins is anticipated to disrupt the down-regulation of E-proteins in response to a TCR-mediated positive selection event. The finding that CD4+ development, albeit limited, can proceed, whereas CD8 development is blocked, suggests that persistent E-protein activity during positive selection permits CD4+lin eage development while preventing CD8+ lineage development.
The identification of a phenotype dependent on the deletion of both Id2 and Id3 is reminiscent of that seen with the analysis of the Tcf12 and Tcfe2a double conditional model (Jones and Zhuang, 2007; Wojciechowski et al., 2007). In both cases, functions are revealed that were previously masked by redundant factors. Just as HEB and E2A serve redundant function to regulate the DP to SP transition, Id2 and Id3 likely serve redundant function, at least in part, to regulate E-protein down-regulation during this developmental window. Even though the kinetics of Id2 up-regulation are likely slower (Schmitz et al., 2003) and Id2 expression was not altered during αβ T cell development in the absence of Id3 (data not shown), Id2 is sufficient in the absence of Id3 to allow for CD8+ lineage development. The fact that CD4+ T cells continue to develop in the absence of both Id2 and Id3, but CD8+ T cells do not, suggests that CD4+ lineage differentiation and CD8+ lineage differentiation may have differential requirements for the complete down-regulation of E-proteins. While this is a topic of interest that we are continuing to pursue, the findings here from the Id2f/fId3f/fCd4cre+ model strongly correlate with a role for E-proteins in driving CD4+ development and blocking CD8+ development. We anticipate that the overall dosage of E-proteins and Id proteins is critical to properly regulate development of the CD4+ and CD8+ lineages.
The Id2f/fId3f/fCd4cre model also provides a means to investigate which events during the DP to SP transition are Id-dependent vs. Id-independent. In addition, comparison of the results from this work with the previous data from the Tcf12f/fTcfe2af/fCd4cre model (Jones and Zhuang, 2007) allows us to begin identifying which targets are E-protein dependent. For example, the up-regulation of both CCR7 and IL7Rα as cells enter the SP stage were shown here to be Id-dependent events. Because the up-regulation in expression of these receptors was blocked in the absence of Id proteins and induced upon removal of E-proteins, CCR7 and IL7Rα are likely repressed by E-protein activity and dependent on E-protein down-regulation for their induction. We anticipate that this combinatorial approach of using both Id-deficient and E-protein-deficient mouse models will help to identify which E-protein targets may be involved in coordinating lineage determination.
When considering the possible mechanisms by which E-proteins regulate lineage choice in positively-selected cells, one hypothesis could be that the extent of E-protein down-regulation functions as a sensor for TCR signal strength. Current models suggest that stronger or prolonged TCR signals are important for driving CD4+ lineage development (He et al., 2010). It has been proposed that signal strength positively correlates with Id3 activation, and Id3-deficient animals have been shown to have a more severe positive selection defect in mice expressing a MHC II-restricted TCR transgene over mice expressing a MHC I-restricted transgene (Alonzo et al., 2010; Bain et al., 2001; Rivera et al., 2000). However, this model cannot explain the results from our Id2 and Id3 double conditional deletion system. We have shown that the removal of Id2 and Id3 resulted in an opposite outcome, as CD8 SP development was blocked and CD4 SP development persisted. These results suggest that, in addition to sensing TCR signals, E-proteins also serve additional functions to regulate lineage choice during the DP to SP developmental window. Therefore, we propose that the timing and degree of E-protein down-regulation during the DP to SP transition is critical for the proper regulation of CD4+ vs. CD8+ lineage development. Complete removal of HEB and E2A at the DP stage in our Tcf12an d Tcfe2a double conditional deletion system prevented E-proteins from carrying out these lineage defining functions.
The requirement for E-proteins in the development of CD4+ lineage T cells could be direct or indirect. Prior to this study, one could have speculated that the reduction in CD4+T cells in the absence of E-proteins was due to a loss of CD4 expression. However, we have shown here that providing a Cd4 transgene is not sufficient to rescue CD4+ lineage development in the absence of HEB and E2A. Still, E-proteins may be directly involved in driving expression of other critical CD4+ lineage factors. For example, E-proteins may be directly required to induce Gata3 expression, as previous studies have already suggested E-protein regulation of Gata3 (Gregoire and Romeo, 1999; Schwartz et al., 2006). Even though these studies have primarily described E-proteins as repressors of Gata3 expression, the outcome of E-protein-mediated regulation of target genes can be cell-context specific. While it is possible that E-proteins may also directly activate ThPOK, there would likely be an additional requirement for activation of Gata3 or another unknown up-stream regulator because rescuing ThPOK expression in the absence of Gata3 is not sufficient to restore CD4+ lineage development (Wang et al., 2008).
Alternatively, E-proteins may promote CD4+ lineage development by repressing CD8+ lineage factors. For example, if E-proteins were responsible for repression of Runx3, activation of Runx3 by removal of HEB and E2A could contribute to inhibition of CD4+ lineage development, but would likely be only one of several events mediating the E-protein-deficient phenotype (Grueter et al., 2005; Kohu et al., 2005). In addition to regulating transcription factors, E-proteins may be regulating other factors that, for example, influence interpretation of extra-cellular signals or thymocyte migration. Cytokine signaling has been suggested to play an important role in CD8+ lineage development (Park et al., 2010), and E-proteins could function to repress CD8+ lineage development by regulating the receipt of cytokine signals or the migration of cells to cytokine-expressing environments. This is of particular interest given the defects in IL7Rα and CCR7 expression in the Id2f/fId3f/fCd4cre+ model.
E-proteins likely function to regulate CD4+ and CD8+ lineage development through activation and/or repression of multiple targets. Here, our data place E-proteins up-stream of Gata3 up-regulation, which is one of the earliest known CD4+ lineage specification events. In addition, the data show that the requirement for E-proteins to initiate CD4+ lineage development extends beyond regulation of the E-protein target, CD4. Future investigation of the potential targets described above, and others, will not only further define the role of E-proteins, but will also serve to provide mechanistic insight for how a developing thymocyte chooses CD4+ vs. CD8+ lineage fate.
Experimental Procedures
Mice
Bim-deficient, OT-II transgenic, and Id3 RFP reporter mice are from The Jackson Laboratory. MHC I and II-deficient mice are from Taconic. Tcf12 conditional, Tcfe2a conditional, Cd4cre transgenic, Id3 conditional, Cd4 transgenic, and ThPOK reporter (F2F3 line) mice have been described previously (Guo et al., 2011; He et al., 2008; Pan et al., 2002; Van Laethem et al., 2007; Wojciechowski et al., 2007; Wolfer et al., 2001). Generation of the Id2 reporter line Id2hCD5 is described below. Id2 conditional mice were generated in the laboratory of Antonio Iavarone and Anna Lasorella, and will be further described in another publication. All research with mice was performed in accordance with relevant guidelines, and protocols were approved by the Duke University Animal Care and Use Committee.
Generation of the Id2hCD5 allele
The Id2hCD5 allele was generated by creating a targeting construct that inserted an IRES hCD5 cassette intothe 3′ UTR of Id2’s exon 3. The targeting construct also contained a floxed, positive selection marker, PGKNeo cassette, and a negative selection marker, PGKDT cassette. Mouse ES cells were derived from a 129/sv strain obtained from Phillippe Soriano’s lab in 1995 and then maintained in our own lab. Id2hCD5 mice were maintained on a C57BL6 and 129/sv mixed background. The floxed PGKNeo cassette was deleted by crossing the Id2hCD5 mice with Prm-cre transgenic mice (Jackson Laboratory). All analysis here was done using mice obtained after PGKNeo deletion. Genotyping details can be found in the Supplemental Data section.
Cell staining and flow cytometry
Staining for hCD5 was done using anti-human CD5 biotin (UCHT2, BD Pharmingen, BD Biosciences), followed by SA-PECy7 staining. For intracellular staining, the Foxp3 Staining Buffer Set (eBioscience) was used for fixation and permeabilization. The following antibodies were used for analysis of intracellular Gata3 expression: mouse anti-Gata3 (L50-823, BD Pharmingen, BD Biosciences) and mouse IgG1 κ isotype control (MOPC-21, BD Phosflow, BD Biosciences). The mouse CD1d PBS57-loaded and unloaded control tetramers were from the NIH tetramer facility. Flow cytometry analysis was done using a FACSCalibur (BD Biosciences) or FACSCanto II (BD Biosciences) and FlowJo software (Tree Star). Flow cytometry plots were pre-gated on 7-aminoactinomycin D (7AAD, Molecular Probes) negative (live) lymphocytes, and doublets were eliminated when possible by using a forward scatter area vs. forward scatter height gate. FACSVantage SE with DiVa option (BD Biosciences) was used for cell sorting. All flow cytometry plots displayed together were analyzed simultaneously unless noted otherwise.
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
RNA extraction, DNase I treatment, and reverse transcription have been described previously (Lazorchak et al., 2006). Quantitative real-time PCR analysis for Klf2, Tox, and ThPOK expression was performed with a Roche LightCycler and the Fast-Start DNA master SYBR green kit I (Roche) or QuantiFast SYBR Green PCR Kit (Qiagen). The following primers were used: ThPOK primers for (5′-GTTCACAGATGCCCCAAGAG-3′) and rev1 (5′-GAAGGCCTTGTGGCACAGGTG-3), KLF2 primers (Jones and Zhuang, 2007), Tox primers (Wilkinson et al., 2002), and GAPDH primers (Lazorchak et al., 2006). Molecular weight markers used were 1 Kb and 1 Kb Plus (Invitrogen).
Statistical analysis
Statistical analysis was performed using GraphPad Prism software and statistical significance was assessed by the two-tailed Student’s t-test.
Supplementary Material
Highlights.
Deletion of HEB and E2A blocks CD4+ lineage development
CD4+-destined cells instead enter the CD8+ lineage in the absence of HEB and E2A
Deletion of Id2 and Id3 blocks CD8+ T cell development
E-proteins are required prior to CD4+ lineage specification
Acknowledgments
We thank Dr. Qi-Jing Li and Yen-Yu Lin for critical reading of the manuscript; Dr. You-Wen He for providing OT-II transgenic mice and constructs used to generate the Id2hCD5 allele; Drs. Remy Bosselut and Yumei Xiong for technical advice on Gata3 intracellular staining; Dr. Alfred Singer for providing Cd4 transgenic mice; Dr. Qi-Jing Li for providing Rag2−/− congenic mice; Dr. Pumin Zhang for assistance with Id3RFP genotyping; Drs. Yisong Wan and Yunqi Wang for providing Gata3f/+Cd4Cre samples; Meifang Dai for assistance with genotyping and i.v. injection; Ian Belle for assistance with Id2f/fId3f/fCd4cre mice; Drs. Motonari Kondo and Michael Krangel for advice and reagents; the Duke University transgenic facility for assistance with generating gene-targeted mice; the Duke Comprehensive Cancer Center Flow Cytometry facility for cell sorting; and the NIH tetramer facility for mouse CD1d tetramers. This work was supported by funding from NIH (R01GM059638) to Y.Z.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, Sant’Angelo DB. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. Journal of immunology (Baltimore, Md : 1950) 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunology and cell biology. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, Pellegrini M, Cory S, Adams JM, Strasser A. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 2002;415:922–926. doi: 10.1038/415922a. [DOI] [PubMed] [Google Scholar]
- Brugnera E, Bhandoola A, Cibotti R, Yu Q, Guinter TI, Yamashita Y, Sharrow SO, Singer A. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 2000;13:59–71. doi: 10.1016/s1074-7613(00)00008-x. [DOI] [PubMed] [Google Scholar]
- Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol. 2010;11:666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nature reviews Immunology. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. The Journal of experimental medicine. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J Exp Med. 2001;194:733–745. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire JM, Romeo PH. T-cell expression of the human GATA-3 gene is regulated by a non-lineage-specific silencer. The Journal of biological chemistry. 1999;274:6567–6578. doi: 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R, et al. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4−/CD8+ T cells. Journal of immunology (Baltimore, Md : 1950) 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- Guo Z, Li H, Han M, Xu T, Wu X, Zhuang Y. Modeling Sjogren’s syndrome with Id3 conditional knockout mice. Immunol Lett. 2011;135:34–42. doi: 10.1016/j.imlet.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- He X, Park K, Kappes DJ. The role of ThPOK in control of CD4/CD8 lineage commitment. Annual review of immunology. 2010;28:295–320. doi: 10.1146/annurev.immunol.25.022106.141715. [DOI] [PubMed] [Google Scholar]
- He X, Park K, Wang H, He X, Zhang Y, Hua X, Li Y, Kappes DJ. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Hendriks RW, Nawijn MC, Engel JD, van Doorninck H, Grosveld F, Karis A. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. European journal of immunology. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- Huang YH, Li D, Winoto A, Robey EA. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4936–4941. doi: 10.1073/pnas.0401133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohu K, Sato T, Ohno SI, Hayashi K, Uchino R, Abe N, Nakazato M, Yoshida N, Kikuchi T, Iwakura Y, et al. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage. Journal of immunology (Baltimore, Md : 1950) 2005;174:2627–2636. doi: 10.4049/jimmunol.174.5.2627. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science (New York, N Y ) 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- Lazorchak AS, Wojciechowski J, Dai M, Zhuang Y. E2A promotes the survival of precursor and mature B lymphocytes. J Immunol. 2006;177:2495–2504. doi: 10.4049/jimmunol.177.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AP, Aronow BJ, Davidson DR, Davies JA, Gaido KW, Grimmond S, Lessard JL, Little MH, Potter SS, Wilder EL, Zhang P. GUDMAP: the genitourinary developmental molecular anatomy project. J Am Soc Nephrol. 2008;19:667–671. doi: 10.1681/ASN.2007101078. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nature immunology. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Hanrahan J, Li J, Hale LP, Zhuang Y. An analysis of T cell intrinsic roles of E2A by conditional gene disruption in the thymus. J Immunol. 2002;168:3923–3932. doi: 10.4049/jimmunol.168.8.3923. [DOI] [PubMed] [Google Scholar]
- Pan L, Sato S, Frederick JP, Sun XH, Zhuang Y. Impaired immune responses and B-cell proliferation in mice lacking the Id3 gene. Molecular and cellular biology. 1999;19:5969–5980. doi: 10.1128/mcb.19.9.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, Kimura MY, Cui Y, Lucas PJ, Gress RE, et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nature immunology. 2010;11:257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]
- Sarafova SD, Erman B, Yu Q, Van Laethem F, Guinter T, Sharrow SO, Feigenbaum L, Wildt KF, Ellmeier W, Singer A. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity. 2005;23:75–87. doi: 10.1016/j.immuni.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Sawada S, Littman DR. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz I, Clayton LK, Reinherz EL. Gene expression analysis of thymocyte selection in vivo. Int Immunol. 2003;15:1237–1248. doi: 10.1093/intimm/dxg125. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- Van Laethem F, Sarafova SD, Park JH, Tai X, Pobezinsky L, Guinter TI, Adoro S, Adams A, Sharrow SO, Feigenbaum L, Singer A. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. [DOI] [PubMed] [Google Scholar]
- von Boehmer H, Kisielow P. Negative selection of the T-cell repertoire: where and when does it occur? Immunol Rev. 2006;209:284–289. doi: 10.1111/j.0105-2896.2006.00346.x. [DOI] [PubMed] [Google Scholar]
- Wang D, Claus CL, Vaccarelli G, Braunstein M, Schmitt TM, Zuniga-Pflucker JC, Rothenberg EV, Anderson MK. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol. 2006;177:109–119. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nature immunology. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom I, Forssell J, Penha-Goncalves MN, Bergqvist I, Holmberg D. A role for E2-2 at the DN3 stage of early thymopoiesis. Mol Immunol. 2008;45:3302–3311. doi: 10.1016/j.molimm.2008.02.012. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Chen JYF, Han P, Rufner KM, Goularte OD, Kaye J. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nature immunology. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB Are Required to Block Thymocyte Proliferation Prior to Pre-TCR Expression. J Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A, Bakker T, Wilson A, Nicolas M, Ioannidis V, Littman DR, Lee PP, Wilson CB, Held W, MacDonald HR, Radtke F. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat Immunol. 2001;2:235–241. doi: 10.1038/85294. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.