Abstract
Tumor hypoxia triggers signaling cascades that significantly affect biologic outcomes such as resistance to radiotherapy and chemotherapy in breast cancer. Hypoxic regions in solid tumor are spatially heterogeneous. Therefore, delineating the origin and extent of hypoxia in tumors is critical. In this study, we have investigated the effect of hypoxia on different metabolic pathways, such as lipid and choline metabolism, in a human breast cancer model. Human MDA-MB-231 breast cancer cells and tumors, which were genetically engineered to express red fluorescent tdTomato protein under hypoxic conditions, were used to investigate hypoxia. Our data were obtained with a novel three-dimensional multimodal molecular imaging platform that combines magnetic resonance (MR) imaging, MR spectroscopic imaging (MRSI), and optical imaging of hypoxia and necrosis. A higher concentration of noninvasively detected total choline-containing metabolites (tCho) and lipid CH3 localized in the tdTomato-fluorescing hypoxic regions indicated that hypoxia can upregulate tCho and lipid CH3 levels in this breast tumor model. The increase in tCho under hypoxia was primarily due to elevated phosphocholine levels as shown by in vitro MR spectroscopy. Elevated lipid CH3 levels detected under hypoxia were caused by an increase in mobile MR-detectable lipid droplets, as demonstrated by Nile Red staining. Our findings demonstrate that noninvasive MRSI can help delineate hypoxic regions in solid tumors by means of detecting the metabolic outcome of tumor hypoxia, which is characterized by elevated tCho and lipid CH3.
Introduction
Transcriptional regulation of several genes by hypoxia-inducible factor 1 (HIF-1) is a well-characterized adaptive response to hypoxia [1,2]. HIF-1 is a transcriptional master regulator of hypoxia-inducible genes, many of which promote tumor progression, for example, vascular endothelial growth factor (VEGF) [3] and lysyl oxidase [4]. HIF-1 modulates the transcription of target genes by binding hypoxia response elements (HREs) containing the consensus binding site 5′-RCGTG-3′ located within the promoter region of genes. Different HIF-1-regulated gene products respond to hypoxia differently. Some of these gene products such as VEGF are largely secreted into the surrounding stromal matrix and are additionally regulated by multiple other factors in breast cancer, which reduces their specificity as immunohistochemical markers of hypoxia [3]. Other HIF-1-regulated genes such as glucose transporter 1 [5] and carbonic anhydrase IX [6] are localized to the cell of origin and undergo hypoxic regulation that is predominantly due to HIF-1. HIF-1α, a subunit of the HIF-1 dimer, is overexpressed in breast cancer, and it promotes tumor progression [1,2]. HIF-1α is modulated by the oxygen status of the cell such that it is degraded during normoxia and stabilized during hypoxia [2]. Hypoxia-inducible factor 2α (HIF-2α) is also increased in human cancers [7]. It has a very similar sequence to HIF-1α, dimerizes with HIF-1β to form HIF-2, and activates the transcription of a group of target genes that partially overlaps with but is mostly different from those regulated by HIF-1 [8]. Hypoxia is one of the hallmarks of tumors and triggers multiple signaling cascades that significantly affect biologic outcomes, including angiogenesis, selection for resistance to apoptosis, resistance to radiation and chemotherapy, and increased invasion and metastasis [9–11]. Hypoxic regions in breast tumors can have different degrees of severity and can dynamically fluctuate as they arise from inadequate blood supply and blood vessel collapse, typically forming a heterogeneous spatial distribution inside of tumors [9,12].
Although cancer cells display a remarkable tolerance to hypoxia, prolonged and severe hypoxia can eventually result in anoxia, which can lead to necrotic cell death, a phenomenon seen in many human tumors including breast cancer [13]. Tumor necrosis has been proposed as a marker of poor prognosis in a variety of solid malignant tumor types including breast cancer [14–16]. Necrosis of cancer tissue may be a later consequence of hypoxic cell formation in the progression of breast cancer. However, the links between necrotic and hypoxic cells are not yet fully investigated. Expression analysis of the HIF-1 responsive genes glucose transporter 1 and carbonic anhydrase IX in 97 breast tumors demonstrated that more than 90% of necrotic and 30% of nonnecrotic tumors expressed at least one hypoxia marker [17]. Independent assessment of epithelial and stromal hypoxia can provide new information in terms of prognosis and patient outcome because stromal hypoxia can improve patient survival [17]. Therefore, being able to identify the metabolic changes that cancer cells undergo in hypoxic and necrotic tumor regions would help to provide a better understanding and evaluation of hypoxia-triggered signaling pathways in cancer.
In the present study on breast cancer, we have visualized hypoxia by means of MDA-MB-231-HRE-tdTomato, where the red fluorescent protein tdTomato is under the control of an HRE-containing promoter [18] and thus is conditionally expressed primarily during hypoxia. The tdTomato fluorescence intensity is several-fold more intense than the enhanced green fluorescent protein fluorescence intensity [19,20]. Tumoral total choline-containing metabolites (tCho; composed of phosphocholine [PC], glycerophosphocholine [GPC], and free choline [Cho]) were studied by both in vivo 1H magnetic resonance (MR) spectroscopic imaging (MRSI) and in vitro high-resolution MR spectroscopy (MRS). In this tumor model, we also analyzed mobile lipids, which are lipid droplets containing triglycerides and cholesterol esters, with MRSI/MRS [21]. Several studies have demonstrated that these lipids are closely involved in various aspects of tumor biology such as cell proliferation, necrosis, hypoxia, and drug resistance [22–25].
The spatial distribution of both tCho and mobile lipids, as detected by MRSI in malignant tumors, is frequently heterogeneous [26]. The cause of this heterogeneity is largely unknown as molecular analyses of excised tumor specimens typically report on the entire tumor specimen or section. Therefore, obtaining MRSI information on the spatial heterogeneity, especially extending measurements on two-dimensional tissue sections to three-dimensional tumor volumes, will provide a better understanding of the spatial distribution of hypoxia-triggered molecular signaling pathways in the tumor. To understand and evaluate the complexity as well as the spatial distribution of the hypoxic response of tumors, we have recently developed a three-dimensional multimodal molecular imaging platform that combines MR imaging (MRI), MRSI, and optical imaging of hypoxia and necrosis in breast cancer [27]. Here we have thoroughly analyzed the tCho and mobile lipid distribution of eight orthotopic MDA-MB-231-HRE-tdTomato tumor xenografts using our three-dimensional multimodal molecular imaging platform and begun to unravel the spatial relationship between hypoxia, necrosis, and metabolite levels. The detected hypoxia-driven molecular pathways observed in these in vivo three-dimensional tumor measurements were confirmed by in vitro studies.
Materials and Methods
Breast Cancer Cells
An estrogen-independent human breast cancer cell line, MDA-MB-231 [28,29], derived from a metastatic lesion of a breast adenocarcinoma in the mammary gland, was obtained from the American Type Culture Collection (ATCC, Rockville, MD). To generate MDA-MB-231 cells that express tdTomato under hypoxic conditions as an internal, HIF-1α-driven hypoxia sensor, MDA-MB-231 cells were stably transfected with a construct containing HREs of the human VEGF gene ligated to the complementary DNA of tdTomato, which generated MDA-MB-231-HRE-tdTomato cells as previously described [18]. MDA-MB-231-HRE-tdTomato cells were cultured in RPMI-1640 (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum (Gemini Bio-Products, Woodland, CA), 100 U/ml penicillin, 100 µg/ml streptomycin (Sigma-Aldrich), and 400 µg/ml G418 (Sigma-Aldrich). tdTomato protein expression in normoxic control and hypoxic MDA-MB-231-HRE-tdTomato cell cultures was detected by fluorescence microscopy using a 20x objective attached to a Nikon inverted microscope, equipped with a filter set for 528 to 553 nm excitation and 600 to 660 nm emission and a Nikon COOLPIX digital camera (Nikon Instruments, Inc, Melville, NY).
Breast Tumor Xenografts
All experimental animal protocols were approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine. Solid MDA-MB-231-HRE-tdTomato tumor xenografts were generated by inoculating 2 x 106 MDA-MB-231-HRE-tdTomato cells in 0.1 ml of Hank balanced salt solution (Sigma-Aldrich) orthotopically into the left second mammary pad of female athymic nude mice (catalog no. 01B74; National Cancer Institute, Frederick, MD). Tumor xenografts reached their final experimental size of approximately 8 x 8 x 8 mm3 within 8 weeks, and mice weighed between 24 and 28 g.
Three-dimensional In Vivo T1-Weighted MRI and MRSI
Tumor-bearing mice were anesthetized by intraperitoneal injection of ketamine (25 mg/kg; Phoenix Scientific, Inc, Encinitas, CA) and acepromazine (2.5 mg/kg; Aveco, Phoenix Scientific) diluted in saline as previously described [27]. All eight mice were scanned in vivo on a 9.4-T Bruker Avance Small Animal MRI Scanner (Bruker, Billerica, MA) in the Broadway Research Building Molecular Imaging Center at the Johns Hopkins University School of Medicine. The tumors of anesthetized mice were placed inside a home-built 1H solenoid coil mounted on a plastic cradle with a water-heating pad lying on the bottom to maintain the normal body temperature of the animal. A rapid-acquisition-with-relaxation-enhancement (RARE) spin-echo fast imaging sequence was used to acquire three-dimensional reference T1-weighted images. Three-dimensional RARE was performed with the following parameters: echo time (TE) of 7.2 milliseconds, repetition time of 500 milliseconds, RARE factor of 4, flip angle of 90°, field of view (FOV) of 1.0 x 1.0 x 1.0 cm, 64-phase encode steps (64 x 64 x 64 voxels), and number of averages (NA) of 4. Total acquisition time was 13 minutes. The reconstruction of MRI reference images was performed using ParaVision 5.0 software (Bruker).
Three-dimensional MRSI data were acquired with a three-dimensional spin-echo chemical shift imaging (CSI) sequence using the same coil [30]. Water-suppressed MRSI was performed using VAPOR on a 1.0 x 1.0 x 1.0-cm FOV enclosing the tumor with the same geometry as used for the corresponding three-dimensional RARE acquisition, with an in-plane resolution of 1.25 x 1.25 x 1.25-mm per pixel and the following parameters: repetition time of 1000 milliseconds, TE of 82 milliseconds, sweep width of 4000 Hz, and spectral resolution 3.9 Hz/point. Standard k-space sampling was used with a matrix size of 8 x 8 x 8 (zero filled to 64 x 64 x 64) and NA of 4 resulting in a total measurement time of 33 minutes. Corresponding three-dimensional CSI images of the unsuppressed water signal and lipids were acquired of the same volume with TE of 15 milliseconds and NA of 2, and all other parameters remaining the same as for the water-suppressed MRSI. After these MRI/MRSI studies, mice were killed, and each tumor was marked with a novel fiducial marker system [27,31] and cut into 2-mm-thick sections for hypoxia imaging as described in our previous report [27,31].
Ex Vivo Microscopic Imaging
For microscopic measurements, a given slide holding an embedded, fiducially marked [31] tumor slice was taken out of the icebox and placed under the microscope for imaging. In between microscopic measurements, the fresh tumor slices were returned to the icebox to keep the tissue at 4°C, and they were periodically moistened with saline to avoid dehydration of the tissue. Both bright-field and fluorescence microscopy images with the same FOV were obtained using a 1x objective attached to the Nikon microscope described above to visualize hypoxia. After microscopy, samples were stored in a -80°C freezer until sets of 10-µm-thick cryosections were cut across the tumor at an interval of 500 µm from top to body wall throughout all 2-mm-thick slices. The 10-µm sections were stained with hematoxylin and eosin (H&E) for identification of tumor necrosis and were imaged by the same microscope with 1x magnification.
Fusion of MRI/MRSI, Optical, and Histologic Imaging
Both water-unsuppressed and water-suppressed MR spectroscopic images were reconstructed by an in-house IDL program, which performed Fourier transformations over both spatial and spectral axes. The tCho signal at 3.2 ppm from the water-suppressed MRSI data (TE = 82 milliseconds) and the lipid CH3 signal at 0.9 ppm from the water-unsuppressed MRSI data (TE = 15 milliseconds) were used. The concentration maps of these two metabolite signals were quantified by normalizing their signals to the water signal in the corresponding water-unsuppressed MRSI data [32].
A previously developed multimodal molecular imaging platform [27] was applied, which performs fiducial marker-based three-dimensional reconstruction of optical images, which are then coregistered with three-dimensional MRI/MRSI. To visualize the tumor boundary and hypoxic regions, three-dimensional reconstruction of bright-field and fluorescence images was performed by using this platform [27] and displayed in Amira 5.2.1 (Mercury Computer Systems, Chelmsford, MA). Similarly, three-dimensional reconstruction of H&E-stained images to extract the tumor boundary and necrotic region was completed [27]. Coregistration of three-dimensional T1-weighted MR images, three-dimensional optical bright-field/fluorescence images, and three-dimensional H&E-stained images was also achieved by using this platform [27].
Quantitative Analysis of MRI/MRSI, Optical, and Histologic Imaging
Once three-dimensional reconstruction and co-registration was completed, two different quantification methods, a volume percentage comparison and the average metabolite concentration for each region, were applied to the obtained co-registered three-dimensional data sets to measure the co-localization of the detected metabolites with the normoxic, hypoxic, and necrotic regions in the tumor. Volume percentage was defined as the number of hypoxic voxels that overlap with a given metabolite normalized to total hypoxic voxels. The volume percentage of metabolite overlap with normoxic and necrotic voxels was calculated analogously. This quantification method depends on the threshold of signal intensity, which was set to capture tumor regions of the highest metabolite intensities. To select this threshold, the histogram of a given metabolite image was calculated, and the threshold was automatically set to include the highest 5% of the area under the histogram. The average metabolite concentrations in normoxic, hypoxic, and necrotic regions were calculated by averaging all voxel values of a given metabolite within each of these three regions. In both of these two quantification methods, the hypoxic region was determined by setting a threshold of the highest 5% of the area under the histogram in the fluorescence images. The tumor area was determined by edge detection of the tumor boundaries in three-dimensional MRI RARE images. The necrotic region was also determined by edge detection of the necrosis boundaries in the H&E-stained images.
Dual-Phase Extraction of Cells
Approximately 108 MDA-MB-231-HRE-tdTomato cells were exposed for 24 hours to standard normoxic conditions in a regular incubator (control) or to hypoxic conditions in a hypoxia cell culture chamber containing PO2 less than 1% (Biospherix, Lacona, NY). As a third condition, hypoxia was mimicked by treating cells for 24 hours with 200 µM CoCl2. Cells under control or experimental conditions were incubated with regular cell culture medium containing all supplements as described above. Treated cancer cells were washed, centrifuged, and separated into lipid and water-soluble extracts using a dual-phase extraction method based on methanol/chloroform/water (1:1:1, vol/vol/vol) as previously described [29,33,34].
MRS Data Acquisition and Processing
The lyophilized water-soluble cell extracts were dissolved in D2O containing the concentration and chemical shift reference 3-(trimethylsilyl) propionic-2, 2, 3, 3-d4 acid (TSP; Sigma-Aldrich). Lipid cell extracts were dissolved in 0.6 ml of CDCl3/CD3OD (2:1, vol/vol) containing tetramethylsilane (TMS) as an internal concentration and chemical shift standard (CDCl3 and CD3OD were premixed with TMS by the manufacturer; Cambridge Isotope Laboratories, Inc, Andover, MA). Fully relaxed 1H MR spectra of the water-soluble and lipid extracts were acquired on a Bruker Avance 500 nuclear magnetic resonance spectrometer operating at 11.7 T using a 5-mm HX inverse probe as previously described [29,33]. The Cho peak at 3.209 ppm, the PC peak at 3.227 ppm, the GPC peak at 3.236 ppm, and the TSP peak at 0 ppm in the 1H MR spectra of the water-soluble extracts, and the lipid CH3 peak at 0.9 ppm and the TMS peak at 0 ppm in the 1H MR spectra of the lipid extracts were fitted and integrated by the software MestRec 4.9 (Mestrelab Research, Escondido, CA) and normalized to the cell number, volume, and the TSP or TMS concentration standard as previously described [29,33].
Nile Red Staining, Confocal Fluorescence Microscopy, and Quantitative Image Analysis
MDA-MB-231-HRE-tdTomato cells were exposed to normoxic, hypoxic, and CoCl2 conditions for 24 hours as described above. Cells were washed with PBS and incubated with a 1:1000 dilution in PBS of a 1-mg/ml stock solution of Nile Red (9-diethylamino-5H-benzo[a]phenoxazine-5-one; Sigma-Aldrich) in acetone for 10 minutes at room temperature, which stains lipid droplets [35–37]. Cell nuclei were counterstained with Hoechst H-33342 (Molecular Probes, Eugene, OR). Cells were washed and mounted using Faramount aqueous mounting medium (DakoCytomation, Carpinteria, CA). Fluorescence microscopy was done with a confocal Zeiss LSM 710 META confocal laser scanning microscope (Carl Zeiss, Inc, Thornwood, NY) using a Plan-Apochromat 63/1.4 oil immersion lens (Zeiss) in the Johns Hopkins University School of Medicine Microscope Facility. Nile Red and H-33342 were excited at 488 and 405 nm, respectively, and fluorescence emission was detected with photomultipliers in the range of 550 to 620 nm and 420 to 480 nm, respectively. Confocal z-stacks of sections were imaged. Twenty-one randomly selected FOVs of 1-µm thickness containing the maximal number of lipid droplets were imaged for quantification. Numbers and diameters of lipid droplets per cell were quantified using our in-house Lyso software as previously described [38]. This Lyso software detected the boundaries of the nucleus and the lipid droplets in the microscopic confocal fluorescence images and calculated the center point and effective diameter of each detected lipid droplet based on the detected boundaries.
Statistical Analysis of Experiments
An unpaired one-tailed t test (α = 0.05) was conducted to detect significant differences between experimental groups using Excel 2007 (Microsoft Corp, Redmond, WA). P < .05 was considered to be significant in the in vitro dual-phase extraction and Nile Red staining experiments. P < .1 was considered to be significant in the MRSI and optical imaging experiments.
Results
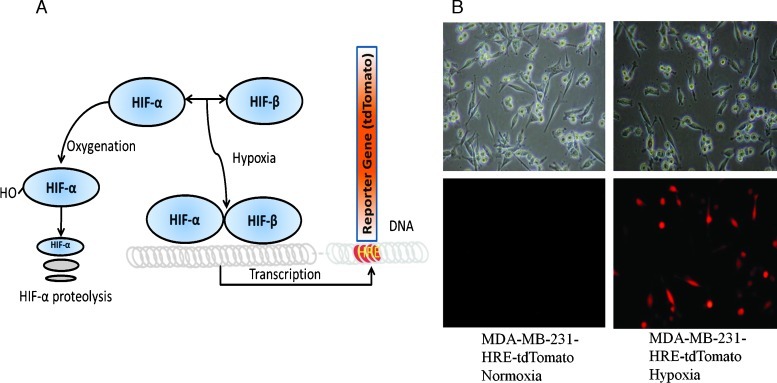
After exposure of cells to 24 hours of hypoxia (0.3%–0.5% O2), human MDA-MB-231-HRE-tdTomato cells stably transfected with the HRE-tdTomato construct (Figure 1A) exhibited bright tdTomato fluorescence as shown in Figure 1B. Exposure of MDA-MB-231-HRE-tdTomato cells to normoxic control conditions resulted in undetectable tdTomato fluorescence signal (Figure 1B). These findings prove that tdTomato fluorescent protein is only expressed under hypoxic condition in MDA-MB-231-HRE-tdTomato cells.
Figure 1.
(A) Diagram depicting the MDA-MB-231-HRE-tdTomato construct, which is activated by HIF-1α stabilization under hypoxic conditions, whereas HIF-1α is continuously being degraded under normoxic conditions. HIF-1 binds HREs in the HRE-tdTomato construct resulting in tdTomato expression in hypoxic regions, which can be detected by fluorescence microscopy and imaging. (B) Brightfield (top) and tdTomato fluorescence (bottom) microscopic images (20x lens) of the same FOVs from normoxic (left) and hypoxic (right) live MDA-MB-231-HRE-tdTomato cells.
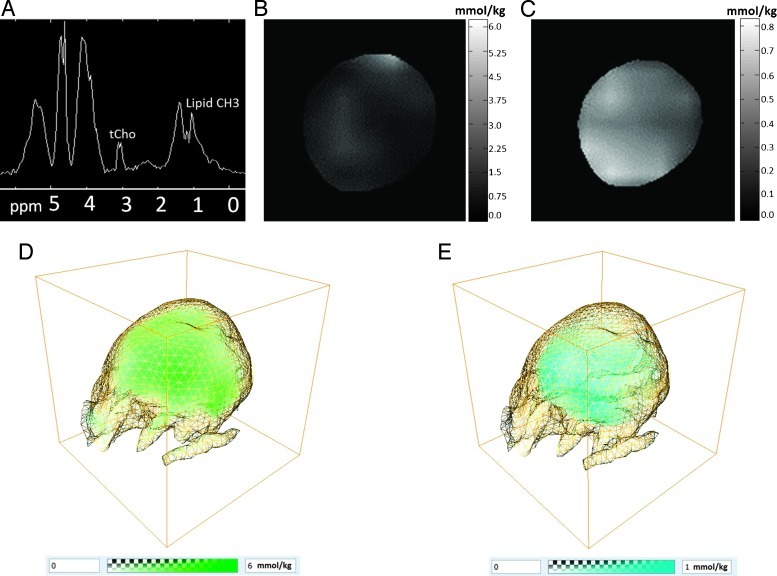
To evaluate the relationship between hypoxia and the 1H MRS-detectable tCho and lipid CH3 signals, eight MDA-MB-231-HRE-tdTomato breast tumor xenografts were studied by in vivo MRSI in three dimensions to detect tCho and lipid CH3, followed by ex vivo optical imaging to detect hypoxia and ex vivo H&E histology imaging to detect necrosis. A representative CSI MRSI spectrum from an MDA-MB-231-HRE-tdTomato tumor with tCho at 3.2 ppm and lipid CH3 at 0.9 ppm is shown in Figure 2A. Representative two-dimensional tCho and lipid CH3 concentration maps were reconstructed, and these are displayed in Figure 2, B and C, respectively. Representative three-dimensional tCho and lipid CH3 concentration volumes were reconstructed, and these are displayed in Figure 2, D and E, respectively. The spatial distributions of both tCho and lipid CH3 were heterogeneous in this breast tumor model.
Figure 2.
(A) Example of a water-suppressed 1H CSI MRSI spectrum from a representative MDA-MB-231-HRE-tdTomato tumor (approximately 360 mm3) obtained with a spatial resolution of 0.9 x 0.9 x 0.8 mm showing tCho at 3.2 ppm and lipid CH3 at 0.9 ppm. (B) Example of a two-dimensional tCho concentration map obtained from the signal at 3.2 ppm in the water-suppressed MRSI data (TE = 82 milliseconds). (C) Example of a two-dimensional lipid CH3 concentration map obtained from the signal at 0.9 ppm in the water-unsuppressed MRSI data (TE = 15 milliseconds). Example of (D) a three-dimensional tCho concentration volume (green) and (E) a three-dimensional lipid CH3 concentration volume (cyan) with the tumor boundary shown as a yellow grid.
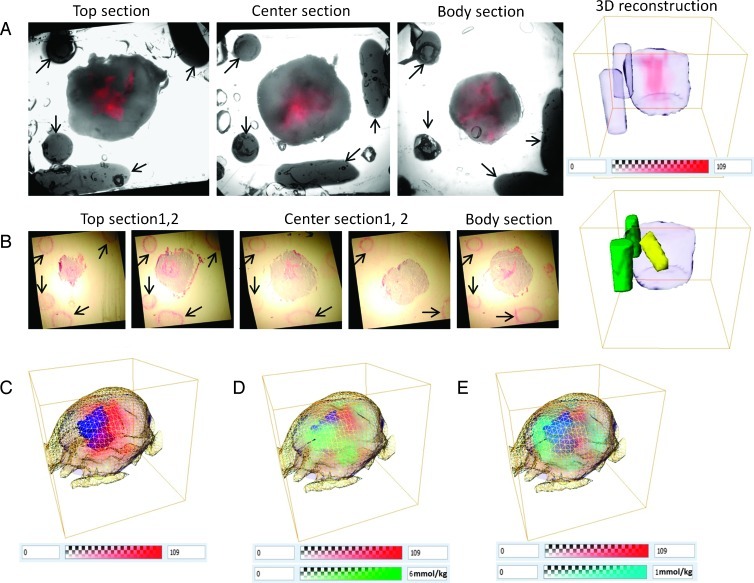
After completion of the MRI/MRSI measurements, mice were killed and tumors removed and sectioned into 2-mm-thick fresh slices for microscopy. Figure 3A shows an example of bright-field and fluorescence images of the top, center, and body sections from a representative MDA-MB-231-HRE-tdTomato tumor and the corresponding three-dimensional reconstruction of the tumor boundary, fiducial markers, and the tdTomato fluorescence in hypoxic regions. All MDA-MB-231-HRE-tdTomato tumors displayed a heterogeneous spatial distribution of hypoxic regions within the tumor. Figure 3B shows an example of the H&E-stained images from the same tumor as shown in Figure 3A, and the corresponding three-dimensional reconstruction of necrotic regions (yellow) and fiducial markers (green). Figure 3C displays an overlay of the hypoxic (red) and necrotic regions (blue) with the MRI-derived tumor boundary (yellow grid) and the bright-field-derived tumor boundary (purple). The necrotic region in this tumor is adjacent to the hypoxic region in the center of the tumor, indicating that an extreme lack of oxygen leads to cell death. Figure 3D shows an overlay of tCho (green), hypoxic regions (red), and necrotic regions (blue) including the tumor boundary obtained after application of our coregistration and three-dimensional reconstruction platform. Figure 3E displays an overlay of lipid CH3 (cyan), hypoxic regions (red), and necrotic regions (blue) including the tumor boundary.
Figure 3.
(A) Example of bright-field and fluorescence images of top, center, and body sections from a representative MDA-MB-231-HRE-tdTomato tumor (approximately 360 mm3) and the corresponding three-dimensional reconstruction of tumor boundary, fiducial markers, and tdTomato-fluorescing hypoxic regions. Arrows point out the fiducial markers. (B) H&E-stained images of the same tumor as shown in A and corresponding three-dimensional reconstruction of necrotic regions and fiducial markers. Arrows point out the fiducial markers. (C) Overlay of hypoxic (red) and necrotic (blue) regions with MRI tumor boundary (yellow grid) and bright-field tumor boundary (purple). (D) Overlay of tCho (green), hypoxic (red), and necrotic (blue) regions with tumor boundary. (E) Overlay of lipid CH3 (cyan), hypoxic (red), and necrotic (blue) regions with tumor boundary.
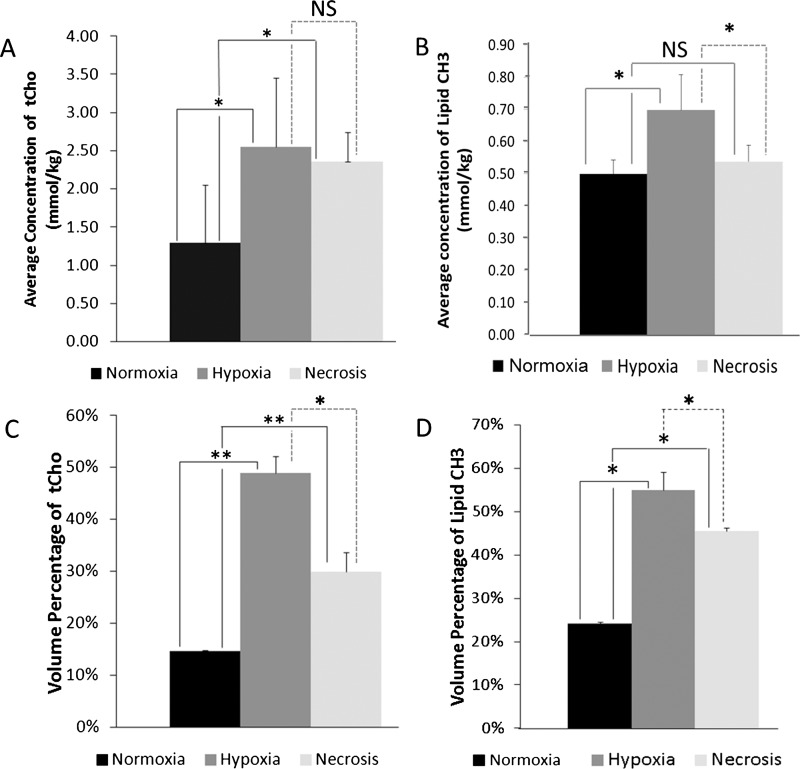
The quantification of overlaps of hypoxic or necrotic regions with tCho or lipid CH3 was compared to the overlaps of normoxic regions with tCho or lipid CH3 in eight MDA-MB-231-HRE-tdTomato breast tumors, all of which contained hypoxic regions and four of which contained necrotic regions. Figure 4A illustrates the average concentration of tCho in normoxic, hypoxic, and necrotic regions. The average concentration of tCho in the normoxic regions was significantly (P = .01, n = 8 for comparison between hypoxia and normoxia; P = .04, n = 4 for comparison between hypoxia and necrosis) lower than that in the hypoxic and necrotic regions. The average concentration of lipid CH3 was significantly (P = .08, n = 8 for comparison between hypoxia and normoxia; P = .07, n = 4 for comparison between hypoxia and necrosis) higher in hypoxic regions than that in normoxic and necrotic regions (Figure 4B). Figure 4C presents the volume percentage of tCho in hypoxic, normoxic, and necrotic regions. The volume percentage of tCho in the hypoxic and necrotic regions was significantly (P = .0006, n = 8 for comparison between hypoxia and normoxia; P = .006, n = 4 for comparison between necrosis and normoxia) higher than that in the normoxic regions, whereas the volume percentage of tCho in hypoxic regions was significantly higher than that in necrotic regions (P = .09, n = 4). Similarly, the volume percentage of lipid CH3 was higher in hypoxic and necrotic regions compared with that in normoxic regions (P = .01, n = 8 for comparison between hypoxia and normoxia; P = .02, n = 4 for comparison between necrosis and normoxia), whereas the volume percentage of lipid CH3 was higher in hypoxic than in necrotic regions (P = .05, n = 4; Figure 4D). In summary, our results showed that the highest-volume percentages and the highest concentrations of tCho and lipid CH3 were detected in hypoxic regions of breast tumor xenografts.
Figure 4.
Average concentration of (A) tCho and (B) lipid CH3 in normoxic (black), hypoxic (gray), and necrotic regions (light gray) in millimoles per kilogram. Volume percentage of (C) tCho and (D) lipid CH3 in normoxic (black), hypoxic (gray), and necrotic regions (light gray). Values are mean ± SE. *P < .1, **P < .01. NS indicates not significant.
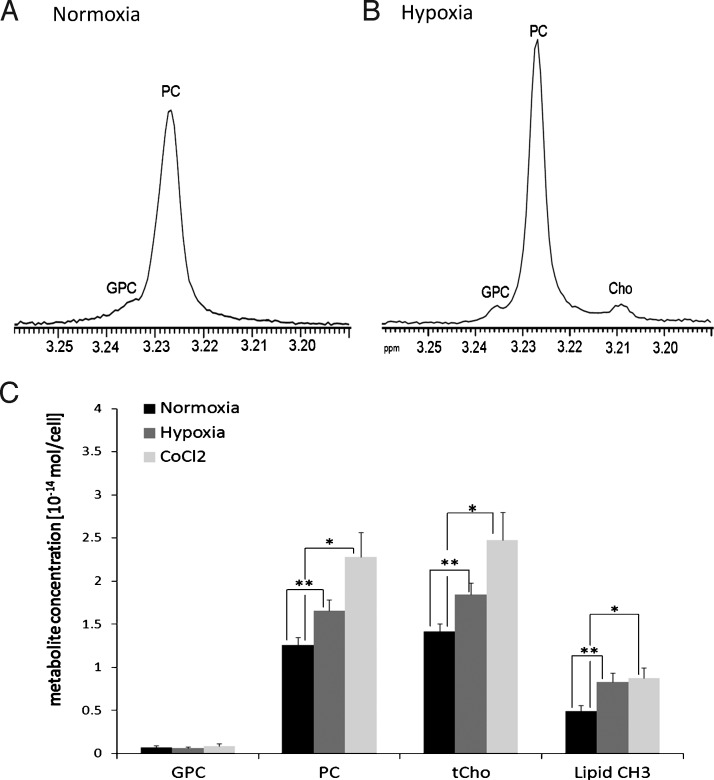
To further examine if hypoxia can increase both tCho and lipid CH3 levels, we measured cellular PC, GPC, and tCho levels in hypoxic versus normoxic MDA-MB-231-HRE-tdTomato cells in culture by using high-resolution 1H MRS. Figure 5, A and B, presents representative 1H MRS spectra after hypoxic and normoxic exposure, respectively. Figure 5C shows the quantification analysis of these high-resolution 1H MRS measurements, indicating that PC (P = .008, n = 6) as well astCho(P = .008, n = 6) significantly increased after hypoxic exposure compared with normoxic controls. Lipid CH3 also increased after hypoxia compared to normoxia (P = .008, n = 6). Similarly, CoCl2-treated cells, in which hypoxia was mimicked by CoCl2, showed significantly higher PC (P = .04, n = 6), tCho (P = .05, n = 6), and lipid CH3 (P = .01, n = 6) levels in these cell culture experiments (Figure 5C).
Figure 5.
Expanded regions of representative high-resolution 1H MR spectra of MDA-MB-231-HRE-tdTomato breast cancer cell extracts after 24 hours of (A) normoxia or (B) hypoxia. Both samples are from approximately 3 x 107 cells. (C) Average concentration of GPC, PC, PC/GPC, tCho, and lipid CH3 quantified from 1H MR spectra obtained from MDA-MB-231-HRE-tdTomato cells exposed to normoxic control conditions, hypoxia, or CoCl2. Values are mean ± SE. *P < .05, **P < .01.
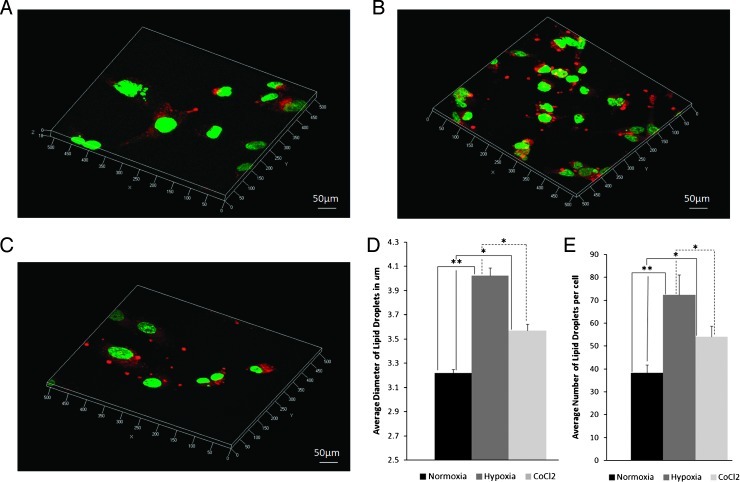
Figure 6, A to C, shows three-dimensional confocal microscopic z-stack images of Nile Red-stained MDA-MB-231-HRE-tdTomato cells after 24 hours of normoxic, hypoxic, and CoCl2 exposure. Both average diameter and average number of lipid droplets in 21 randomly selected FOVs were calculated by our in-house software. Figure 6D demonstrates that the average diameters of lipid droplets were increased in hypoxic and CoCl2 exposed cells compared with normoxic control cells (P = .025, n = 21 for comparison between CoCl2 and normoxia; P = .0002, n = 21 for comparison between hypoxia and normoxia). Figure 6E shows that the average number of lipid droplets was increased in hypoxic- and CoCl2-exposed cells compared with normoxic control cells (P = .019, n = 21 for comparison between CoCl2 and normoxia; P = .0008, n = 21 for comparison between hypoxia and normoxia).
Figure 6.
Three-dimensional confocal microscopic z-stack images of Nile Red-stained MDA-MB-231-HRE-tdTomato breast cancer cells after 24 hours of (A) normoxia, (B) hypoxia, and (C) CoCl2 exposure. (D) Average diameter and (E) average number of lipid droplets after 24 hours of normoxia, hypoxia, or CoCl2 exposure. Quantitative analysis was performed from 21 randomly selected FOVs. Values are mean ± SE. *P < .05, **P < .01.
Discussion
Volume percentages and concentrations of tCho and lipid CH3 significantly increased in hypoxic breast tumor regions compared with normoxic regions, as determined by our three-dimensional multi-modal molecular imaging platform and quantification. The increase in tCho was due to a hypoxia-induced increase in PC as shown in our cell culture experiments. These findings are in good agreement with our previous study, which showed that choline kinase α (Chk-α) expression is regulated by means of HIF-1 binding to the Chk-α promoter in prostate cancer cells [26]. This study suggested that the putative Chk-α promoter region contains at least six HREs and that HIF-1 binding of these HREs can increase Chk-α expression within hypoxic environments, consequently increasing cellular PC and tCho levels within these environments [26]. However, it is not clear if all of the HREs in the putative Chk-α promoter region are involved in upregulating Chk-α expression within hypoxic environments because HRE-7 may be involved in transcriptional down-regulation of Chk-α as indicated by mutation studies [39]. Necrotic tumor regions also contained elevated tCho levels in our study, which was most likely due to persisting elevated tCho levels from previous severe hypoxia in these regions.
Our finding of increased lipid droplets leading to an increased lipid CH3 signal in hypoxic and necrotic breast tumor regions is in good agreement with a study in rodent brain tumors, which attributed the presence of mobile lipids in viable cells primarily to the existence of hypoxic regions adjacent to necrotic areas [22]. In this study, lipid droplets were located in necrotic tumor regions, and severely hypoxic cells accumulated small lipid droplets in C6 rat brain gliomas [22]. We showed that the highest lipid CH3 levels were found in hypoxic regions of MDA-MB-231-HRE-tdTomato tumors, whereas necrotic regions contained less lipid CH3, making lipid CH3 a good marker of hypoxic regions. In addition, we detected an increase in the volume percentage of lipid CH3 in necrotic regions compared to control, which probably stems from a gradual transition of severely hypoxic regions to necrotic tissue. This necrotic tissue then still contains some of the metabolites that were produced, while the cells underwent a period of severe hypoxia before undergoing necrotic cell death. The increased volume percentage of lipid CH3 in necrotic regions of our breast tumor model is in good agreement with comparable data obtained in human brain tumors [25].
Hypoxic regions of breast tumors have long proven to be resilient and unresponsive to both radiotherapy and chemotherapy [40]. Such resistance to treatment contributes to the incidence of cancer recurrence [40]. Our results indicate that three-dimensional MRSI detection of tCho and lipid CH3 and its heterogeneity might prove useful as biomarkers not only of breast tumor aggressiveness and progression but also of tumor hypoxia, which is consistent with our previous two-dimensional MRSI report in a prostate cancer model [26]. Although we observed that elevated tCho and lipid CH3 overlapped with a large portion of hypoxic regions, regions of high tCho and lipid CH3 without fluorescence were also occasionally observed. The latter indicates that factors other than hypoxia may have also contributed to the spatial heterogeneity of the tumoral tCho and lipid CH3 levels. Effects of such other factors and conditions of the tumor microenvironment, such as acidic extracellular pH and nutrient deprivation, stromal cell contributions, and contributions from extracellular matrix components, should be further investigated in the future. In addition, the relatively long half-life of the tdTomato fluorescent protein in our hypoxia model may have limitations in representing the dynamic fluctuating nature of tumor hypoxia.
Visualizing the spatial distribution of MR-visible tCho and lipids in three dimensions could also be useful for monitoring treatment response. For example, tCho and lipids were mapped in a rat brain containing a herpes simplex virus thymidine kinase-positive BT4C glioma undergoing apoptosis-inducing ganciclovir treatment [41]. Spatially distinct changes of both signals were observed on day 8 of treatment with an increased mobile lipid concentration and decreased tCho levels [41].
Because of the broader line widths that result from magnetic field inhomogeneities in in vivo MRSI, it is currently not possible to resolve Cho, PC, and GPC, even at the highest available magnetic field strengths. As a result, in this study, only tCho and lipid CH3 were observed. To study more biomolecules, such as lipids, proteins, and peptides, which showed an altered expression or content under hypoxic conditions or necrosis, it may be helpful in future studies to use an approach that combines ex vivo mass spectrometry (MS) with in vivo MS imaging [42,43]. Additional sections that were cut adjacent to the H&E-stained sections can be used to perform MS imaging, and our multimodal molecular imaging platform, in the future, can be adapted for studying altered biomolecules in hypoxic and necrotic regions in breast cancers.
Acknowledgments
The authors thank Vaddapuram P. Chacko for expert technical support with the MRI studies on the Bruker 9.4 T Small Animal Scanner. The authors also thank Menglin Cheng for technical laboratory support.
Abbreviations
- Chk
choline kinase
- Cho
free choline
- CSI
chemical shift imaging
- FOV
field of view
- GPC
glycerophosphocholine
- H&E
hematoxylin and eosin
- HIF
hypoxia-inducible factor
- HRE
hypoxia response element
- MR
magnetic resonance
- MRS
magnetic resonance spectroscopy
- MRI
magnetic resonance imaging
- MRSI
magnetic resonance spectroscopic imaging
- MS
mass spectrometry
- NA
number of averages
- PC
phosphocholine
- RARE
rapid acquisition with relaxation enhancement
- tCho
total choline-containing compounds
- TE
echo time
- TMS
tetramethylsilane
- TSP
3-(trimethylsilyl) propionic-2, 2, 3, 3-d4 acid
- VEGF
vascular endothelial growth factor
Footnotes
This work was supported by the National Institutes of Health (grant R01 CA134695).
References
- 1.Semenza GL. Expression of hypoxia-inducible factor 1: mechanisms and consequences. Biochem Pharmacol. 2000;59:47–53. doi: 10.1016/s0006-2952(99)00292-0. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JTA, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- 5.Smith TAD. Facilitative glucose transporter expression in human cancer tissue. Br J Biomed Sci. 1999;56:285–292. [PubMed] [Google Scholar]
- 6.Ivanov S, Liao SY, Ivanova A, Danilkovitch-Miagkova A, Tarasova N, Weirich G, Merrill MJ, Proescholdt MA, Oldfield EH, Lee J, et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158:905–919. doi: 10.1016/S0002-9440(10)64038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–421. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau KW, Tian YM, Raval RR, Ratcliffe PJ, Pugh CW. Target gene selectivity of hypoxia-inducible factor-α in renal cancer cells is conveyed by post-DNA-binding mechanisms. Br J Cancer. 2007;96:1284–1292. doi: 10.1038/sj.bjc.6603675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hockel M, Vaupel P. Tumor hypoxia: definitions and current clinical, biologic, and molecular aspects. J Natl Cancer Inst. 2001;93:266–276. doi: 10.1093/jnci/93.4.266. [DOI] [PubMed] [Google Scholar]
- 10.Tatum JL, Kelloff GJ, Gillies RJ, Arbeit JM, Brown JM, Chao KS, Chapman JD, Eckelman WC, Fyles AW, Giaccia AJ, et al. Hypoxia: importance in tumor biology, noninvasive measurement by imaging, and value of its measurement in the management of cancer therapy. Int J Radiat Biol. 2006;82:699–757. doi: 10.1080/09553000601002324. [DOI] [PubMed] [Google Scholar]
- 11.Williams KJ, Cowen RL, Stratford IJ. Hypoxia and oxidative stress. Tumour hypoxia—therapeutic considerations. Breast Cancer Res. 2001;3:328–331. doi: 10.1186/bcr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iakovlev VV, Pintilie M, Morrison A, Fyles AW, Hill RP, Hedley DW. Effect of distributional heterogeneity on the analysis of tumor hypoxia based on carbonic anhydrase IX. Lab Invest. 2007;87:1206–1217. doi: 10.1038/labinvest.3700680. [DOI] [PubMed] [Google Scholar]
- 13.Bussolati G, Bongiovanni M, Cassoni P, Sapino A. Assessment of necrosis and hypoxia in ductal carcinoma in situ of the breast: basis for a new classification. Virchows Arch. 2000;437:360–364. doi: 10.1007/s004280000267. [DOI] [PubMed] [Google Scholar]
- 14.Kessler R, Gasser B, Massard G, Roeslin N, Meyer P, Wihlm JM, Morand G. Blood vessel invasion is a major prognostic factor in resected non-small cell lung cancer. Ann Thorac Surg. 1996;62:1489–1493. doi: 10.1016/0003-4975(96)00540-1. [DOI] [PubMed] [Google Scholar]
- 15.Shahab I, Fraire AE, Greenberg SD, Johnson EH, Langston C, Roggli VL. Morphometric quantitation of tumor necrosis in stage-1 non-small-cell carcinoma of lung—prognostic implications. Mod Pathol. 1992;5:521–524. [PubMed] [Google Scholar]
- 16.Swinson DEB, Jones JL, Richardson D, Cox G, Edwards JG, O'Byrne KJ. Tumour necrosis is an independent prognostic marker in non-small cell lung cancer: correlation with biological variables. Lung Cancer. 2002;37:235–240. doi: 10.1016/s0169-5002(02)00172-1. [DOI] [PubMed] [Google Scholar]
- 17.Tomes L, Emberley E, Niu YL, Troup S, Pastorek J, Strange K, Harris A, Watson PH. Necrosis and hypoxia in invasive breast carcinoma. Breast Cancer Res Treat. 2003;81:61–69. doi: 10.1023/A:1025476722493. [DOI] [PubMed] [Google Scholar]
- 18.Krishnamachary B, Penet MF, Nimmagadda S, Mironchik Y, Raman V, Solaiyappan M, Semenza GL, Pomper M, Bhujwalla ZM. Hypoxia regulates CD44 and its variant isoforms through HIF-1α in triple negative breast cancer. PLoS One. doi: 10.1371/journal.pone.0044078. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 21.Righi V, Mucci A, Schenetti L, Bacci A, Agati R, Leonardi M, Schiavina R, Martorana G, Liguori G, Calabrese C, et al. Identification of mobile lipids in human cancer tissues by ex vivo diffusion edited HR-MAS MRS. Oncol Rep. 2009;22:1493–1496. doi: 10.3892/or_00000592. [DOI] [PubMed] [Google Scholar]
- 22.Zoula S, Rijken PF, Peters JP, Farion R, Van der Sanden BP, Van der Kogel AJ, Decorps M, Remy C. Pimonidazole binding in C6 rat brain glioma: relation with lipid droplet detection. Br J Cancer. 2003;88:1439–1444. doi: 10.1038/sj.bjc.6600837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoula S, Herigault G, Ziegler A, Farion R, Decorps M, Remy C. Correlation between the occurrence of 1H-MRS lipid signal, necrosis and lipid droplets during C6 rat glioma development. NMR Biomed. 2003;16:199–212. doi: 10.1002/nbm.831. [DOI] [PubMed] [Google Scholar]
- 24.Delikatny EJ, Chawla S, Leung DJ, Poptani H. MR-visible lipids and the tumor microenvironment. NMR Biomed. 2011;24:592–611. doi: 10.1002/nbm.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Opstad KS, Bell BA, Griffiths JR, Howe FA. An investigation of human brain tumour lipids by high-resolution magic angle spinning 1H MRS and histological analysis. NMR Biomed. 2008;21:677–685. doi: 10.1002/nbm.1239. [DOI] [PubMed] [Google Scholar]
- 26.Glunde K, Shah T, Winnard PT, Jr, Raman V, Takagi T, Vesuna F, Artemov D, Bhujwalla ZM. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-1α signaling in a human prostate cancer model. Cancer Res. 2008;68:172–180. doi: 10.1158/0008-5472.CAN-07-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang L, Greenwood RG, Amstalden E, Chughtai K, Raman V, Winnard PT, Jr, Heeren RMA, Artemov D, Glunde K. Multimodal multi-scale molecular imaging platform for combined magnetic resonance, fluorescence, and histology imaging. NMR Biomed. (in press) [Google Scholar]
- 28.Cailleau R, Young R, Olive M, Reeves WJ., Jr Breast tumor cell lines from pleural effusions. J Natl Cancer Inst. 1974;53:661–674. doi: 10.1093/jnci/53.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glunde K, Jie C, Bhujwalla ZM. Molecular causes of the aberrant choline phospholipid metabolism in breast cancer. Cancer Res. 2004;64:4270–4276. doi: 10.1158/0008-5472.CAN-03-3829. [DOI] [PubMed] [Google Scholar]
- 30.Brown TR, Kincaid BM, Ugurbil K. NMR chemical shift imaging in three dimensions. Proc Natl Acad Sci USA. 1982;79:3523–3526. doi: 10.1073/pnas.79.11.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chughtai K, Jiang L, Greenwood TR, Klinkert I, Amstalden van Hove ER, Heeren RM, Glunde K. Fiducial markers for combined 3-dimensional mass spectrometric and optical tissue imaging. Anal Chem. 2012;84:1817–1823. doi: 10.1021/ac203373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bolan PJ, Meisamy S, Baker EH, Lin J, Emory T, Nelson M, Everson LI, Yee D, Garwood M. In vivo quantification of choline compounds in the breast with 1H MR spectroscopy. Magn Reson Med. 2003;50:1134–1143. doi: 10.1002/mrm.10654. [DOI] [PubMed] [Google Scholar]
- 33.Glunde K, Raman V, Mori N, Bhujwalla ZM. RNA interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer Res. 2005;65:11034–11043. doi: 10.1158/0008-5472.CAN-05-1807. [DOI] [PubMed] [Google Scholar]
- 34.Tyagi RK, Azrad A, Degani H, Salomon Y. Simultaneous extraction of cellular lipids and water-soluble metabolites: evaluation by NMR spectroscopy. Magn Reson Med. 1996;35:194–200. doi: 10.1002/mrm.1910350210. [DOI] [PubMed] [Google Scholar]
- 35.Fukumoto S, Fujimoto T. Deformation of lipid droplets in fixed samples. Histochem Cell Biol. 2002;118:423–428. doi: 10.1007/s00418-002-0462-7. [DOI] [PubMed] [Google Scholar]
- 36.Greenspan P, Fowler SD. Spectrofluorometric studies of the lipid probe, Nile Red. J Lipid Res. 1985;26:781–789. [PubMed] [Google Scholar]
- 37.Llewellyn BD. Nuclear staining with alum hematoxylin. Biotech Histochem. 2009;84:159–177. doi: 10.1080/10520290903052899. [DOI] [PubMed] [Google Scholar]
- 38.Glunde K, Guggino SE, Solaiyappan M, Pathak AP, Ichikawa Y, Bhujwalla ZM. Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia. 2003;5:533–545. doi: 10.1016/s1476-5586(03)80037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bansal A, Harris RA, Degrado TR. Choline phosphorylation and regulation of transcription of choline kinase α in hypoxia. J Lipid Res. 2012;53:149–157. doi: 10.1194/jlr.M021030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rockwell S, Dobrucki IT, Kim EY, Marrison ST, Vu VT. Hypoxia and radiation therapy: past history, ongoing research, and future promise. Curr Mol Med. 2009;9:442–458. doi: 10.2174/156652409788167087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liimatainen TJ, Erkkila AT, Valonen P, Vidgren H, Lakso M, Wong G, Grohn OH, Yla-Herttuala S, Hakumaki JM. 1H MR spectroscopic imaging of phospholipase-mediated membrane lipid release in apoptotic rat glioma in vivo. Magn Reson Med. 2008;59:1232–1238. doi: 10.1002/mrm.21607. [DOI] [PubMed] [Google Scholar]
- 42.van Hove ERA, Blackwell TR, Klinkert I, Eijkel GB, Heeren RMA, Glunde K. Multimodal mass spectrometric imaging of small molecules reveals distinct spatio-molecular signatures in differentially metastatic breast tumor models. Cancer Res. 2010;70:9012–9021. doi: 10.1158/0008-5472.CAN-10-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chughtai K, Heeren RMA. Mass spectrometric imaging for biomedical tissue analysis. Chem Rev. 2010;110:3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]