Abstract
Verrucarin A (VA) is a small molecule derived from the fungal plant pathogen Myrothecium verrucaria and was identified as a selective inhibitor of clear cell renal cell carcinoma (CCRCC) cell proliferation in a high-throughput screen of a library of naturally occurring small molecules. CCRCC arises as a result of loss-of-function mutations in the von Hippel-Lindau (VHL) gene. Here we show that VA inhibits protein translation initiation culminating in apoptosis through the extrinsic signaling pathway. Reintroduction of the VHL gene in CCRCC cells afforded resistance to VA's apoptotic effects. This resistance is mediated in part by the formation of stress granules that entrap signaling molecules that initiate the apoptotic signaling cascade. The VHL gene product was found to be a component of stress granules that develop as result of VA treatment. These findings reveal an important role for the VHL gene product in cytotoxic stress response and have important implications for the rational development of VA-related compounds in chemotherapeutic targeting of CCRCC.
Introduction
A lack of clear early warning signs, a wide range of symptoms and resistance to radiation and chemotherapy all characterize von Hippel-Lindau (VHL)-associated clear cell renal cell carcinoma (CCRCC). Underlying the disease are loss-of-function mutations in the VHL tumor suppressor gene [1]. Currently, several drug discovery approaches are being explored for targeting the disease, and recently, we reported on the use of a high-throughput screen (HTS) that identified several natural molecules that showed selective cytotoxicity to VHL-defective (VHL-/-) CCRCC cells [2]. Verrucarin A (VA) was one of the compounds identified in this HTS campaign. The compound, also known as muconomycin A, had previously been described as a protein synthesis inhibitor [3]. However, several protein synthesis inhibitors that were part of the screening library did not demonstrate differential cytotoxicity between paired VHL-/- and VHL-competent (VHL+/+) CCRCC cells. We, therefore, undertook this study to better characterize the mechanism behind the toxicity of VA in CCRCC cells and to examine the role played by the VHL gene product (pVHL) in mediating response to cytotoxic stress induced by VA.
Materials and Methods
Reagents
VA, purchased from Sigma, St Louis, MO (catalog no. V4877), was dissolved in dimethyl sulfoxide (DMSO) and stored as 20-mM aliquots at -20°C. Cycloheximide (CHX; catalog no. C4859), verrucarol (catalog no. V1628), and heparin (catalog no. H4784) were also obtained from Sigma. Ultrapure sucrose (catalog no. 15503) and Tris-HCl (catalog no. 15567) were purchased from Invitrogen (Grand Island, NY). Didemnin B and 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide were obtained from the Drug Synthesis and Chemistry Branch (National Cancer Institute, Bethesda, MD).
Cells and Cell Culture
786-O, UOK-121, RCC4, and UOK-127 cell lines, characterization, and growth and propagation conditions have been described previously [2]. Throughout their use, cell morphology, growth curve, and possible mycoplasma contamination were regularly monitored. 786-ONLS-VHL and 786-ONES-VHL cells were generated through transient transfection of 786-O cells with the previously described NLS-VHL-ef-IRES-puro and NES-VHL-ef-IRES-Puro constructs, respectively [4].
Western Blot Analysis
Cell lysates for Western blot analysis were prepared as described earlier [2]. Briefly, 30 µg of protein from lysates was separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, blocked overnight in Odyssey blocking buffer (Li-Cor, Lincoln, NE) at 4°C, and incubated with primary antibodies obtained from Cell Signaling, Danvers, MA (cleaved caspase 3, cleaved caspase 8, eIF2α, p-eIF2α, p38, p-p38, PARP, S6, p-S6, survivin, VHL) and Santa Cruz Biotechnology, Santa Cruz, CA (cFLIPS/L, JNK, pJNK). After incubating with primary antibodies, membranes were washed and incubated with anti-mouse IRDye 680 and anti-rabbit IRDye 800 secondary antibodies (Li-Cor). Blots were scanned using an Odyssey infrared imaging system, and band intensities were determined using the Odyssey band quantitation software after background subtraction using the median top-bottom method. Intensities of bands of interest were normalized to the signals from the corresponding loading control bands of β-actin, α-tubulin, or GAPDH. In addition, Ponceau S staining of membranes was also used as loading control.
Flow Cytometry Analysis
After treatment with 10 nM VA for 4 hours, cells were harvested using Accutase (catalog no. A11105; Invitrogen) and stained with Annexin V-propidium iodide (PI) (catalog no. V13245; Invitrogen), anti-CD262 (catalog no. 12-9908)-7AAD (catalog no. 559925; BD Pharmingen, Franklin Lakes, NJ), anti-CD95 (catalog no. 12-0959; eBioscience, San Diego, CA)-7AAD, anti-CD178 (catalog no. 12-9919)-7AAD, or mouse IgG1 K Isotype control (catalog no. 12-4714)-7AAD. Data were acquired on an Accuri C6 flow cytometer (BD) and exported into FCS express 4 software for analysis.
Ribosome Isolation and Polysome Profile Analysis
For ribosome isolation, untreated cells and cells treated for 10 minutes at 37°C with the cross-linker bis-maleimidohexane (90 µM in phosphate-buffered saline [PBS]; Molecular Biosciences, Boulder, CO) were lysed after pelleting by resuspending in low-salt ribosome extraction buffer (20 mM Tris-HCl pH 7.5, 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, Halt protease inhibitor cocktail, 100 µg/ml CHX, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 200 µg/ml heparin in diethylpyrocarbonate (DEPC)-treated water). Highs-salt ribosome extraction buffer contained 500 mM KCl instead. After incubating on ice for 10 minutes, the lysate was centrifuged at 10,000g for 10 minutes at 4°C and the supernatant (cytosolic extract) was used. This cytosolic extract was centrifuged at 100,000g for 2 hours on a 15% sucrose cushion for isolation of ribosomes and ribosomal subunits. The pellet, which included polysomes, monosomes, and ribosomal subunits, was suspended in a NuPAGE (Invitrogen) sample loading buffer, whereas the supernatant was also resuspended in the same loading buffer after precipitation of proteins with 10% trichloroacetic acid.
For polysome profile analysis, equal numbers of cells seeded into two T-225 flasks and incubated overnight were used (70%–80% confluent). Cells in one flask were treated with 5 µg/ml VA, whereas cells in the second flask received DMSO treatment. Both flasks were incubated for 15 minutes at 37°C and treated with CHX at a final concentration of 100 µg/ml for 15 minutes. After incubating at 37°C, cells were trypsinized in PBS containing CHX. Trypsinized cells were rinsed with ice-cold PBS + CHX and lysed with 400 µl of ribosome extraction buffer. Lysates were then cleared by centrifugation at 10,000g for 10 minutes, and cleared supernatants were loaded onto sucrose gradients. These gradients were prepared by layering 2 ml each of 47%, 37%, 27%, 17%, and 7% sucrose in ribosome extraction buffer (without heparin and Triton X-100) and allowed to stand at 4°C for 16 hours. Loaded gradients were then subjected to ultra-centrifugation (Beckman L8-80M, 39,000 rpm, SW41Ti rotor; Beckman, Palo Alto, CA) for 165 minutes. While monitoring absorbance at 254 nm, fractions were collected from the top through displacement by injecting 60% sucrose at the bottom of the gradient at 1.0 ml/min (ISCO 640 density gradient fractionator; ISCO, Lincoln, NE).
Immunocytochemistry
Cells pretreated with either DMSO or VA for 24 hours were harvested and allowed to attach to glass chips with Y-shaped fibronectin micropatterns (catalog no. 11-012-10-12; CYTOO, Grenoble, France). After allowing them to spread, cells were fixed with 4% fresh para-formaldehyde and permeabilized with 0.1% Triton X-100 in PBS. Samples were blocked using Odyssey blocking buffer overnight at 4°C. Anti-TIA-1 (1:100, catalog no. sc-1751; Santa Cruz Biotechnology) or anti-VHL (1:100, catalog no. 2738; Cell Signaling) antibodies were incubated with samples overnight at 4°C. Finally, samples were washed and incubated for 45 minutes with Alexa Fluor 488 goat anti-rabbit (catalog no. A31628; Invitrogen) and Alexa Fluor 594 goat anti-mouse (catalog no. A31624; Invitrogen). Counterstaining was done using ProLong Gold antifade reagent with DAPI (catalog no. P36931; Invitrogen) as the mounting medium. Images were acquired using Zeiss LSM 510 microscopes at 63x magnification (Zeiss, Thornwood, NY). Colocalization studies were performed as described previously using ImageJ software [2].
Statistical Analysis
Student's t test was performed to determine the statistical significance of differences between treated and control groups. P < .05 was calculated in all experiments where applicable and was considered statistically significant. Error bars represent SEM of three independent experiments run in parallel in triplicate in all plots.
Results
VA Inhibits CCRCC Cell Proliferation and Induces Apoptosis
While screening for inhibitors of VHL-defective cell proliferation [2], VA, a trichothecene isolated from yeast, was found to selectively inhibit the proliferation of pVHL-defective (VHL-/-) CCRCC cells 786-O, RCC4, UOK-121 and UOK-127 with half-maximal inhibitory concentrations (IC50) between 10 and 100 nM. VA was also found to significantly impact clonogenicity of CCRCC cells (Figure 1A). Its IC50 value in pVHL-competent (VHL+/+) 786-OVHL, RCC4VHL, UOK-121VHL, and UOK-127VHL cells was found to be between 10 and 100 µM. We then proceeded to determine whether this level of activity was specific to CCRCC cells by testing VA against a panel of VHL+/+ non-CCRCC renal cancer cell lines. Evaluation of its impact on cell proliferation in these cell lines also gave IC50 values between 3 and 30 µM, confirming that it possessed a selective effect in VHL-/- CCRCC cells (Table W1).
Figure 1.
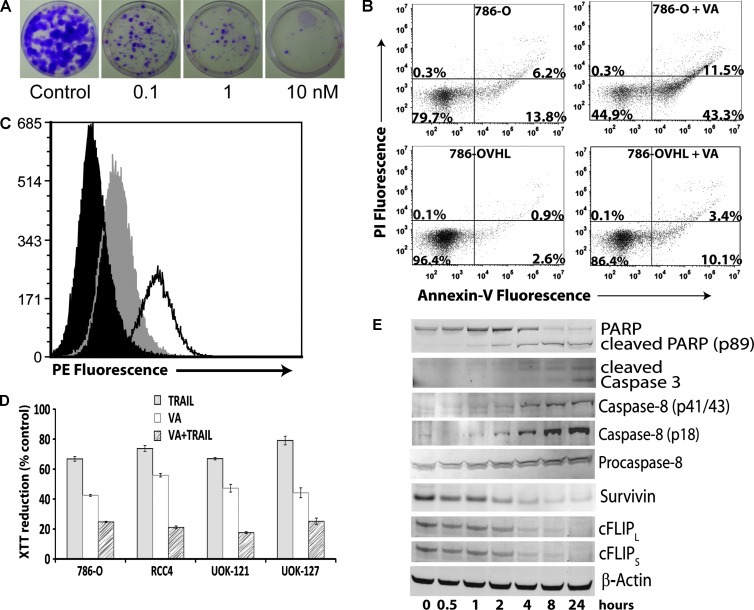
Inhibition of proliferation of CCRCC cells by VA and induction of apoptosis. (A) Clonogenic survival assay of VA-treated 786-O cells. 786-O cells were treated with increasing concentrations of VA for 24 hours. After harvesting, cells were seeded at 100 cells/dish in 60-mm dishes in complete medium and allowed to grow for 14 days. Colonies were fixed and stained with crystal violet. (B) Flow cytometric analysis of PI and Annexin V double-stained 786-O and 786-OVHL cells. Cells were treated for 24 hours with 10 nM VA or DMSO. Percentages of cells in each quadrant shown are from one representative experiment. (C) 786-O cells were analyzed by flow cytometry for cell surface expression of DR5. Cells were stained with phycoerythrin (PE)-conjugated isotype control (black) or DR5-PE after control (gray) or VA (unshaded, 10 nM) treatment. (D) CCRCC cells were treated with control, VA (10 nM), TRAIL (20 ng/ml), or VA + TRAIL for 24 hours. Cell viability was assessed using 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide reduction. (E) Time course of Western blot analysis of apoptosis signaling proteins in 786-O cells treated with 10 nM VA. A representative β-actin blot is shown as a loading control.
Direct observation of VA-treated CCRCC cells under a microscope revealed cell shrinkage and membrane blebbing, suggesting apoptotic cell death. Analysis of Annexin V and PI double-stained 786-O and 786-OVHL cells by flow cytometry after treatment with VA confirmed this observation (Figure 1B). To better characterize the mechanism by which VA curtailed CCRCC cell proliferation, its effect on multiple apoptosis pathway components was examined.
No appreciable changes in the level of the Bcl-2 family of proteins were seen on VA treatment (data not shown), suggesting that apoptosis may be initiated through the extrinsic signaling pathway. Therefore, we next looked at cell surface changes of death receptor pathway components including Fas (CD95), Fas ligand (CD178), and TRAIL death receptors 1 (DR4) and 2 (DR5). Using flow cytometry analysis, we detected a significant increase only in the cell surface expression levels of DR5 (Figure 1C).
CCRCC cells are known to be TRAIL resistant [5]. As a result, we sought further confirmation for increased cell surface expression of DR5 by carrying out a TRAIL sensitization experiment. It was found that pretreatment of CCRCC cells with VA increased their sensitivity to TRAIL (Figure 1D). Also, because resistance to TRAIL-induced apoptosis in CCRCC has been linked to the high level of expression of the antiapoptotic molecule survivin [6], we examined whether VA treatment affected the expression level of this protein. Western blot analysis data revealed that VA treatment downregulated survivin levels in CCRCC cells (Figure 1E). Overexpression of survivin in CCRCC cells, on the other hand, increased their resistance to VA (Figure W1).
Because the cellular-FLICE inhibitory protein (cFLIP) acts as an immediate inhibitor of death receptor-mediated proapoptotic signaling, we also looked at changes in its level in response to VA treatment. VA was found to decrease the levels of both the long and short splice variants of cFLIP (Figure 1E). Probing changes in the levels of other components of the death-inducing signaling complex through Western blot analysis revealed that, although VA treatment did not affect caspase 9 levels, it induced caspase 8 cleavage (Figure 1E). Thus, based on the above findings, it seems that VA treatment triggers death receptor-induced cell death through changes in the level of components of the death-inducing signaling complex in CCRCC cells.
VA Inhibits Translation in CCRCC Cells
Previous studies carried out with VA in yeast have shown that it inhibits protein synthesis in vitro [7]. Therefore, we asked whether its proapoptotic effects in CCRCC cells were related to its effect on protein synthesis. First, its impact on overall protein levels in cells was assessed using a fluorescent-labeling experiment for incorporation of l-azidohomoalanine, a methionine analog, in nascent protein synthesis. This showed that VA caused a decrease in total protein levels in cells in a dose dependent manner. Interestingly, it caused a smaller decrease in total protein levels in VHL+/+ cells compared to VHL-/- cells at all concentrations tested (Figure W2A).
To determine whether the differential impact on proliferation between VHL-/- and VHL+/+ CCRCC cells was specific to VHL-/- CCRCC cells or whether it is a property shared by protein synthesis inhibitors in general, we also tested inhibitors of translation initiation (hippuristanol, verrucarol) and translational elongation (anisomycin, puromycin, didemnin B, and CHX). Although anisomycin and didemnin B showed selectivity toward inhibition of cell proliferation in VHL-/- cells, their activity was significantly weaker than that observed with VA, suggesting that VA had a specific and selective activity in VHL-/- CCRCC cells (Figure W2B).
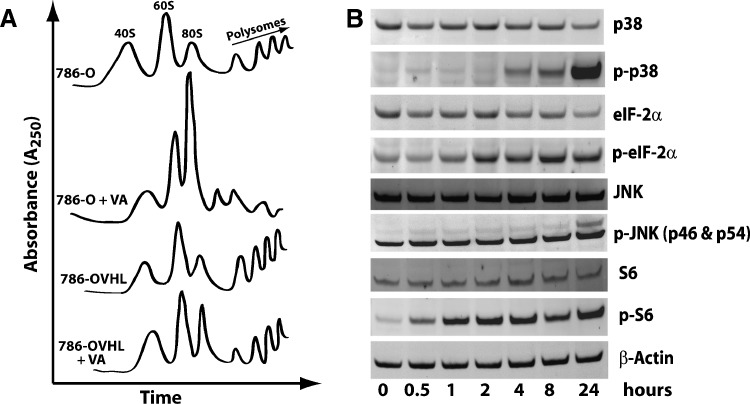
We also examined the specific effects of VA on the protein synthesis machinery using ultracentrifugation and sedimentation of ribosomal extracts over a sucrose gradient. Compared with DMSO-treated control cells, VA-treated 786-O cells displayed elevated 80S peaks with significant reduction in polysomal peaks (Figure 2A). Although VA also caused an increase in 80S peaks in 786-OVHL cells, this increase was not as dramatic as its effect in 786-O cells. It also had a less pronounced effect on polysomes in 786-OVHL cells. Further evidence on its effect on the ribosome came from Western blot analysis data (Figure 2B), showing induction of phospho-ribosomal protein S6, phospho-eIF2α, phospho-p38, and phospho-JNK [8]. However, pretreatment with JNK or p38 inhibitors did not reverse or attenuate apoptotic effects of VA, suggesting that, although it induces cellular stress, signaling through p38 or SAPK/JNK was not required for its effects.
Figure 2.
Effect of VA on protein synthesis and stress induction. (A) Representative sucrose density gradient sedimentation polysome profiles of ribosomes extracted from control and VA-treated 786-O and 786-OVHL cells. (B) Time course of Western blot analysis of stress signaling pathway proteins in 786-O cells treated with 10 nM VA. A representative β-actin blot is shown as a loading control.
pVHL Binds the Ribosome
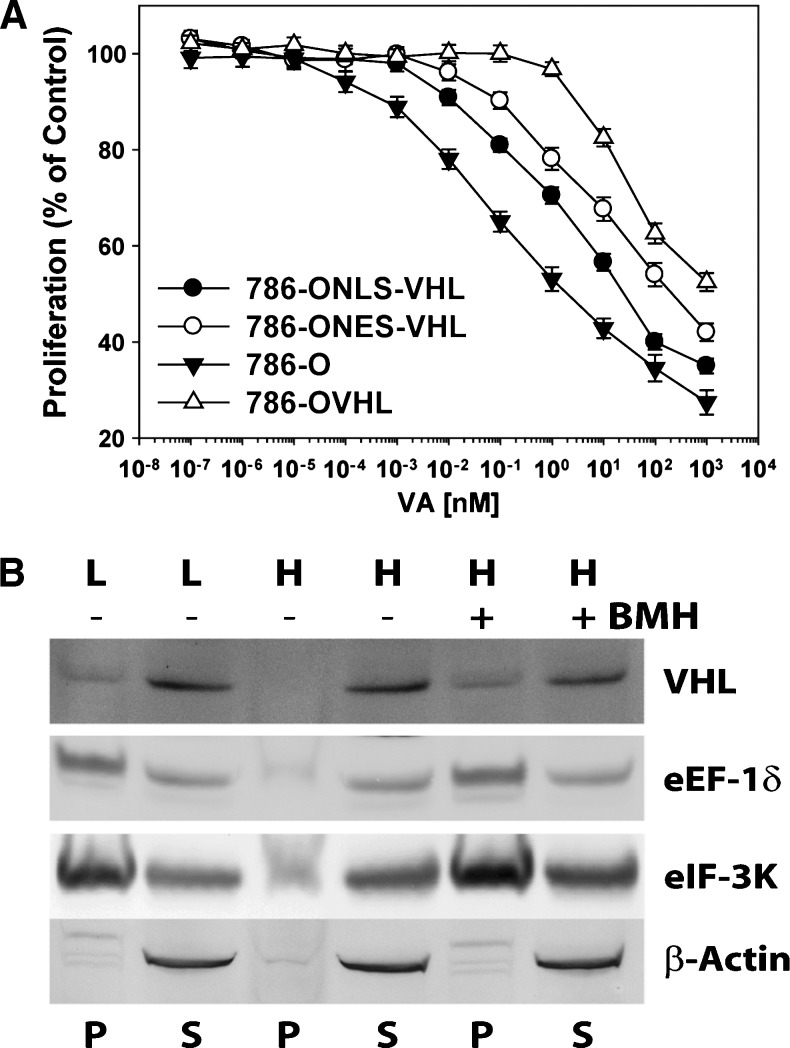
pVHL is known to shuttle between the cytosol and the nucleus in cells [9]. Therefore, we next asked whether its nuclear or cytosolic effects were responsible for resistance to VA. To answer this question, 786-ONLS-VHL and 786-ONES-VHL cells were generated through transfection with VHL constructs where the VHL gene was tagged with either a nuclear localization signal (NLS-VHL) or a nuclear export signal (NES-VHL), respectively. Evaluation of VA in these cell lines showed that, compared with 786-O empty vector-transfected cells, both cells lines were more resistant to VA, suggesting that both nuclear and cytosolic effects of pVHL play a role in resistance. Interestingly, however, 786-ONES-VHL cells seemed more resistant to VA than 786-ONLS-VHL cells (Figure 3A).
Figure 3.
pVHL's cytosolic role in VA-treated 786-OVHL cells. (A) The effects of nuclear and cytosolic pVHL on resistance to VA's effects on cell proliferation. (B) Western blot analysis of protein components in pellets (P) and supernatant (S) obtained by centrifugation of ribosomes extracted with low- (L) and high- (H) salt buffers from control- (-) and VA- (+) treated cells. The protein cross-linking reagent bis-maleimidohexane (BMH) was used to confirm pVHL binding of ribosomes extracted with high salt.
As a result, we asked whether resistance to VA in VHL+/+ cells was due, in part, to pVHL's direct role on the protein synthesis machinery. First, we examined whether pVHL binds the ribosome and found that it sedimented along with proteins of the ribosomal fraction of CCRCC cells prepared with a low-salt-containing ribosomal extraction buffer that maintains total ribosomal integrity (Figure 3B). In contrast, when ribosomal integrity was disrupted with high-salt-containing ribosomal extraction buffer, pVHL was absent from the protein pellet. Pretreatment of cellular proteins with a protein cross-linking reagent before extraction of ribosomes with a high-salt-containing buffer, on the other hand, led to the detection of pVHL in the protein pellet from ribosomes fractionated over a sucrose cushion. Taken together, the above data suggest that pVHL binds the ribosome.
VA Induces Stress Granule Formation in VHL+/+ CCRCC Cells
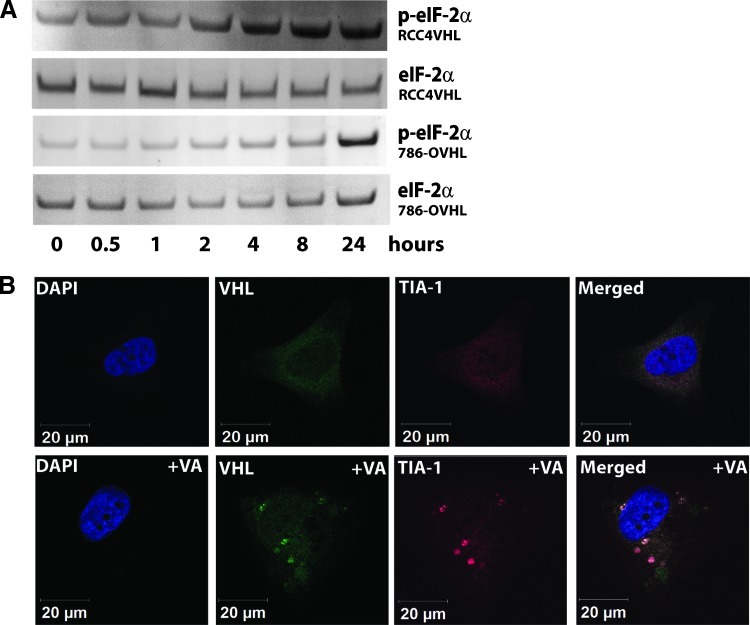
VHL+/+ cells were monitored under a microscope for any changes in their appearance upon VA treatment. The images obtained showed the appearance of punctate cytosolic structures, absent in VA-treated VHL-/- cells, reminiscent of stress granules (SGs) as early as 30 minutes after treatment with 1 µM VA. In Western blots, we found indirect evidence that VA causes SG formation in VHL+/+ cells where up-regulation of phosphorylated eIF2α (Figure 4A), often a trigger for stress-induced formation of SG [10], was observed.
Figure 4.
Induction of stress by VA in 786-OVHL cells leads to stress granule formation. (A) Western blot analysis of time course lysates from VA-treated 786-OVHL cells probed with anti-eIF2α and anti-p-eIF2α antibodies. (B) Subcellular localization of pVHL (green) and the stress granule marker TIA-1 (red) in 786-OVHL cells. Cells were treated with either VA or DMSO control before staining. Images shown are from a representative cell obtained after allowing cells to adhere to Y-shaped fibronectin micropatterns that allow normalization of cell shape, position, polarity, and internal organelle positioning. Colocalization analysis was undertaken as described in the Materials and Methods section.
We also sought support for SG formation through immunofluorescence using TIA-1 as a marker for SGs. VA-treated VHL+/+ cells probed with an antibody to TIA-1 showed the expected positive staining in these structures. Next, because SGs are absent in VA-treated 786-O cells and because pVHL was found to bind ribosomes, we asked whether pVHL was a component of SGs in VA-treated VHL+/+ cells. We carried out immunofluorescence colocalization experiments on VA-treated cells attached to Y-shaped fibronectin micropatterns on glass chips. This allowed for better quantification through normalization of cell shape and internal cell organization. The results showed that, in 786-OVHL cells stained with both anti-pVHL and antiTIA-1 antibodies, 73% ± 8.2% (median ± median absolute deviation) of SG compartments showed VHL signals while 19% ± 7.3% of total VHL signals overlapped the TIA-1 staining (Figure 4B). These findings suggest that VA treatment induces SG formation in VHL+/+ CCRCC cells and that pVHL is a component of, and might play a role in, SG formation.
Discussion
The number of cases where CCRCC continues to be refractory to conventional treatments still remains high. As a result, there is an urgent need to identify new chemotherapeutic agents. Selective inhibitors of VHL-defective cell proliferation thus represent an attractive class of compounds as they exploit vulnerabilities introduced in CCRCC cells as a result of the loss of functional pVHL. Furthermore, such compounds will not only serve as tools to better characterize the pathobiology behind CCRCC but can also be potential starting points for the development of small molecule-based therapeutic agents. We recently reported carrying out an HTS campaign to identify such natural small molecules [2]. One of the hit compounds to show potent and selective inhibition was VA, a compound derived from the yeast M. verrucaria. Through work primarily done in yeast, the compound has been shown to be an inhibitor of protein synthesis and has been used as such in many studies with mammalian cells [7]. Despite the range of cell lines where this effect has been documented, the compound shows selectivity in inhibiting the proliferation of renal carcinoma cells, which lack a functional product of the VHL gene. In this study, we provide some novel insights into selective death receptor-mediated apoptosis in CCRCC cells and the role of pVHL in mediating response to cytotoxic stress by an inhibitor of protein translation initiation—VA.
We found that VA induces selective inhibition of the proliferation of pVHL-defective CCRCC cells, whereas reintroduction of pVHL engenders resistance to VA in CCRCC cells. Our preliminary results with VA in VHL-/- cells for hypoxia-inducible factor 2-driven luciferase expression, in chetomin-pretreated VHL-/- cells and in pretreatment with a hypoxia mimic in VHL+/+ cells, revealed that VA's effect was not hypoxia-inducible factor related, the most well-characterized target of pVHL activity. However, we found that the compound caused apoptotic and not necrotic or autophagic cell death. Lack of any significant alterations in the levels of Bcl-2, Bax, Bad, Bak, Bid, and Bim or of any detectable cytochrome c release into the cytosol indicated that apoptosis in CCRCC cells was mitochondria independent. Data from flow cytometry and Western blot analysis of components of the extrinsic apoptosis signaling pathway proteins confirmed the up-regulation of the TRAIL death receptor DR5 on the cell surface and the down-regulation of cFLIP. Previous reports show that cFLIP down-regulation can induce ligand-independent but DR5-, FADD-, and caspase-dependent apoptosis [11]. Our findings are consistent with this observation because no appreciable changes in FADD levels were detected in VA-treated cells while the CCRCC cells tested, notorious for being resistant to TRAIL-mediated apoptosis [12], became more sensitive to the effects of TRAIL on pretreatment with VA.
Also consistent with previous findings that showed VA inhibits protein synthesis, we found that the compound inhibited translation at or immediately after initiation in CCRCC cells as evidenced by a dramatic increase in monomeric 80S ribosomal content and a decrease in polysomal content. This seems to suggest that VA has a dramatic impact on cap-dependent translation initiation. However, evaluation of classical markers of ribosomal and ER stress such as degradation of 18S or 28S rRNA, up-regulation of GRP78, CHOP/GADD153 or HSP70, and Ca2+ release into the cytosol from the ER lumen were all absent in treated CCRCC cells. On the other hand, we found that VA induced up-regulation of phospho-eIF2α, a marker for “ribotoxic stress” response during treatment with the trichothecene family of compounds, of which VA is a member [13]. Further studies will be needed to elucidate the mechanism linking this nonclassic ribotoxic stress to apoptosis signaling through the extrinsic pathway.
In VA-treated VHL+/+ CCRCC cells, the ribosomal profile obtained after ultracentrifugation over sucrose gradients of ribosomal extracts revealed a slight increase in 80S ribosomal content. This suggests that cap-dependent translation initiation may also be impacted as with VHL-/- cells, albeit to a lesser extent. On the other hand, VA seemed to have little impact on polysomal content in these cells, indicating that it does not significantly affect either polysomal or cap-independent translation.
When examining any potential direct roles of pVHL on components of the translational machinery, we found that pVHL binds ribosomes. Investigations into whether its nuclear or cytosolic functions contributed to resistance to VA showed that both its nuclear and cytosolic activities contributed to resistance with cytosolic pVHL, engendering an increased resistance compared to nuclear pVHL. Findings from immunofluorescence and colocalization studies suggested that VA induced SG formation and that, in the cytosol, pVHL was found in SGs in VHL+/+ cells. SGs represent an aggregation of stalled translation initiation complexes [14] and various components of stress granules, including phospho-eIF2α, are known to mediate apoptotic signaling. Studies have shown that sequestration of these signaling molecules in SGs can lead to abrogation of stress-induced apoptosis signaling [15]. Furthermore, it has been shown that the limiting step in messenger RNA translation is the competition for limiting amounts of translation initiation factors [16]. With depletion of translation initiation complexes through SG formation, pVHL'srolein stabilizing a number of prosurvival messenger RNAs that are otherwise “weak” competitors for available translation initiation factors may be significant in preventing apoptosis [17].
Taken together, our results suggest that selective targeting of VHL-/- cells might be possible in CCRCC through the use of specific inhibitors of translation initiation. It seems that VHL+/+ cells are better able to handle the stress induced by such compounds. The evidence provided in this study suggests the presence of vulnerability in CCRCC cells that may be exploited for therapeutics development.
Supplementary Material
Acknowledgments
The authors thank Ben Roberts of the University of South Australia for providing VHL constructs with nuclear export and nuclear localization signals.
Abbreviations
- cFLIP
cellular-FLICE inhibitory protein
- CHX
cycloheximide
- CCRCC
clear cell renal cell carcinoma
- HTS
high-throughput screen
- pVHL
VHL gene protein product
- SG
stress granule
- VA
verrucarin A or muconomycin A
- VHL
von Hippel-Lindau
- VHL-/-
VHL-defective
- VHL+/+
VHL-competent
Footnotes
This project has been funded in part with federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was also supported in part by the Intramural Research Program of the National Institutes of Health, Frederick National Lab, Center for Cancer Research.
This article refers to supplementary materials, which are designated by Table W1 and Figures W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Gnarra JR, Glenn GM, Latif F, Anglard P, Lerman MI, Zbar B, Linehan WM. Molecular-genetic studies of sporadic and familial renal-cell carcinoma. Urol Clin North Am. 1993;20:207–216. [PubMed] [Google Scholar]
- 2.Woldemichael GM, Turbyville TJ, Linehan WM, McMahon JB. Carminomycin I is an apoptosis inducer that targets the Golgi complex in clear cell renal carcinoma cells. Cancer Res. 2011;71:134–142. doi: 10.1158/0008-5472.CAN-10-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leatherman DL, Middlebrook JL. Effect of emetine on T-2 toxin-induced inhibition of protein synthesis in mammalian cells. J Pharmacol Exp Ther. 1993;266:741–748. [PubMed] [Google Scholar]
- 4.Lewis MD, Roberts BJ. Role of nuclear and cytoplasmic localization in the tumour-suppressor activity of the von Hippel-Lindau protein. Oncogene. 2003;22:3992–3997. doi: 10.1038/sj.onc.1206683. [DOI] [PubMed] [Google Scholar]
- 5.VanOosten RL, Earel JK, Griffith TS. Enhancement of Ad5-TRAIL cytotoxicity against renal cell carcinoma with histone deacetylase inhibitors. Cancer Gene Ther. 2006;13:628–632. doi: 10.1038/sj.cgt.7700939. [DOI] [PubMed] [Google Scholar]
- 6.Griffith TS, Fialkov JM, Scott DL, Azuhata T, Williams RD, Wall NR, Altieri DC, Sandler AD. Induction and regulation of tumor necrosis factor-related apoptosis-inducing ligand/Apo-2 ligand-mediated apoptosis in renal cell carcinoma. Cancer Res. 2002;62:3093–3099. [PubMed] [Google Scholar]
- 7.Hernandez F, Cannon M. Inhibition of protein synthesis in Saccharomyces cerevisiae by the 12,13-epoxytrichothecenes trichodermol, diacetoxyscirpenol and verrucarin A. Reversibility of the effects. J Antibiot (Tokyo) 1982;35:875–881. doi: 10.7164/antibiotics.35.875. [DOI] [PubMed] [Google Scholar]
- 8.Yang GH, Jarvis BB, Chung YJ, Pestka JJ. Apoptosis induction by the satratoxins and other trichothecene mycotoxins: relationship to ERK, p38 MARK, and SAP/JNK activation. Toxicol Appl Pharmacol. 2000;164:149–160. doi: 10.1006/taap.1999.8888. [DOI] [PubMed] [Google Scholar]
- 9.Lee S, Chen DYT, Humphrey JS, Gnarra JR, Linehan WM, Klausner RD. Nuclear cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc Natl Acad Sci USA. 1996;93:1770–1775. doi: 10.1073/pnas.93.5.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day TW, Huang S, Safa AR. c-FLIP knockdown induces ligand-independent DR5-, FADD-, caspase-8-, and caspase-9-dependent apoptosis in breast cancer cells. Biochem Pharmacol. 2008;76:1694–1704. doi: 10.1016/j.bcp.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 13.Zhou HR, Jia QS, Pestka JJ. Ribotoxic stress response to the trichothecene deoxynivalenol in the macrophage involves the Src family kinase Hck. Toxicol Sci. 2005;85:916–926. doi: 10.1093/toxsci/kfi146. [DOI] [PubMed] [Google Scholar]
- 14.Kimball SR, Horetsky RL, Ron D, Jefferson LS, Harding HP. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am J Physiol Cell Physiol. 2003;284:C273–C284. doi: 10.1152/ajpcell.00314.2002. [DOI] [PubMed] [Google Scholar]
- 15.Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324–1332. doi: 10.1038/ncb1791. [DOI] [PubMed] [Google Scholar]
- 16.Hershey JWB, Merrick WC. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. pp. 33–88. [Google Scholar]
- 17.Danilin S, Sourbier C, Thomas L, Rothhut S, Lindner V, Helwig J-J, Jacqmin D, Lang H, Massfelder T. von Hippel-Lindau tumor suppressor gene-dependent mRNA stabilization of the survival factor parathyroid hormone-related protein in human renal cell carcinoma by the RNA-binding protein HuR. Carcinogenesis. 2009;30:387–396. doi: 10.1093/carcin/bgn275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.