Abstract
Objective
Myoclonus is characterized by sudden, brief involuntary movements and its presence is debilitating. We identified a family suffering from adult-onset, cortical myoclonus without associated seizures. We performed clinical, electrophysiological, and genetic studies to define this phenotype.
Methods
A large, four-generation family with history of myoclonus underwent careful questioning, examination, and electrophysiological testing. Thirty-five family members donated blood samples for genetic analysis, which included SNP mapping, microsatellite linkage, targeted massively parallel sequencing, and Sanger sequencing. In silico and in vitro experiments were performed to investigate functional significance of the mutation.
Results
We identified 11 members of a Canadian Mennonite family suffering from adult-onset, slowly progressive, disabling, multifocal myoclonus. Somatosensory evoked potentials indicated a cortical origin of the myoclonus. There were no associated seizures. Some severely affected individuals developed signs of progressive cerebellar ataxia of variable severity late in the course of their illness. The phenotype was inherited in an autosomal dominant fashion. We demonstrated linkage to chromosome 16q21-22.1. We then sequenced all coding sequence in the critical region, identifying only a single co-segregating, novel, nonsynonymous mutation, which resides in the gene NOL3. Furthermore, this mutation was found to alter post-translational modification of NOL3 protein in vitro.
Interpretation
We propose that Familial Cortical Myoclonus (FCM) is a novel movement disorder that may be caused by mutation in NOL3. Further investigation of the role of NOL3 in neuronal physiology may shed light on neuronal membrane hyperexcitability and pathophysiology of myoclonus and related disorders.
INTRODUCTION
Myoclonus is characterized by sudden, brief involuntary movements1. It can be severely debilitating. Myoclonus is thought to arise from spinal, subcortical, or cortical neuronal hyperexcitability, and electrophysiologic examination can readily distinguish amongst these foci of aberrant excitation1. However, beyond anatomical localization little is known about the mechanisms of hyperexcitability.
Cortical myoclonus is characterized by focal or multifocal jerks and is commonly induced by movement or somatosensory stimulation1. Cortical myoclonus has numerous etiologies including post-hypoxic myoclonus (Lance-Adams syndrome), metabolic abnormalities, as well as CJD and various neurodegenerative disorders2. Cortical myoclonus can be dominantly inherited in epileptic disorders such as Familial Adult Myoclonic Epilepsy (FAME)3 (also known as Familial Cortical Myoclonic Tremor with Epilepsy, amongst other names). No causative mutations for FAME have yet been described, although linkage has been demonstrated to chromosomes 8q23.3-q24.11, 2p11.1-q12.2 and 5p15.31-p15refs 4–6. Myoclonus can also be dominantly inherited in Myoclonus-Dystonia Syndrome (MDS, or DYT11), usually resulting from mutations in the SCGE gene on chromosome 7 (although there is also evidence of linkage to chromosome 18p11)7. Notably, in addition to myoclonus MDS patients usually suffer from dystonia and psychiatric comorbidity, and the myoclonus is subcortical, as evidenced by normal somatosensory evoked potentials (SSEPs)8–11.
Familial cortical myoclonus without epilepsy or other neurological deficits has not been described. Here we present a large family suffering from autosomal dominant, slowly progressive, multifocal, cortical myoclonus evoked by somatosensory stimuli. The myoclonus is not associated with seizures or other neurological deficits aside from mild cerebellar ataxia late in the course of the illness. The family’s phenotype is clinically and electrophysiologically distinct from other familial disorders in which myoclonus is prominent, including FAME and MDS. We used genome-wide SNP genotyping, linkage, Sanger-sequencing, and targeted massively parallel sequencing to demonstrate linkage to chromosome 16q21-22.1 and to identify a mutation in NOL3. Furthermore, we showed that the mutation alters post-translational modification of NOL3 protein in vitro. In total, these findings describe a novel Mendelian movement disorder, expand the differential diagnosis for myoclonus, illustrate the power of massively parallel genotyping and sequencing technologies, and provide a compelling candidate gene for investigating mechanisms of neuronal membrane hyperexcitability.
PATIENTS AND METHODS
Patient Phenotyping and DNA Preparation
This study was approved by the University of Western Ontario Health Sciences Research Ethics Board, and all family members provided written informed consent prior to undergoing personal interview and complete neurological examination. An individual was identified as affected with myoclonus if there were symptoms of recurrent sudden, involuntary rapid movements of the face, extremities or body, or if myoclonus was observed upon examination. Nine of 11 affected individuals, and 12 unaffected relatives (Supporting Figure S1), underwent somatosensory evoked potential (SSEP) studies, performed by median nerve stimulation at 5.1 Hz as a square wave pulse of 0.2ms duration. An SSEP was determined to be “giant” if the P25-N33 amplitude difference was greater than 8.6μV, as defined by Shibasaki1. Ten affected patients and 25 unaffected relatives donated DNA for genetic studies.
Genome-wide SNP Genotyping Arrays and Linkage Using Microsatellite Markers
DNA samples from 10 affected patients and 2 unaffected family members (marked with asterisks in Figure 2A) were genotyped for 264,422 SNPs using the GeneChip Human Mapping 250K Nsp Array (Affymetrix, Santa Clara, CA), according to manufacturer’s instructions. All samples passed quality control (call rate > 95%, correct sex determination), and genotype calling was performed with Affymetrix software (Supporting Table S1). We performed linkage analysis on 10 affected patients and 25 unaffected family members using microsatellite short tandem repeat markers for the three putative linkage regions (Supporting Table S2) and standard techniques (Supporting Information).
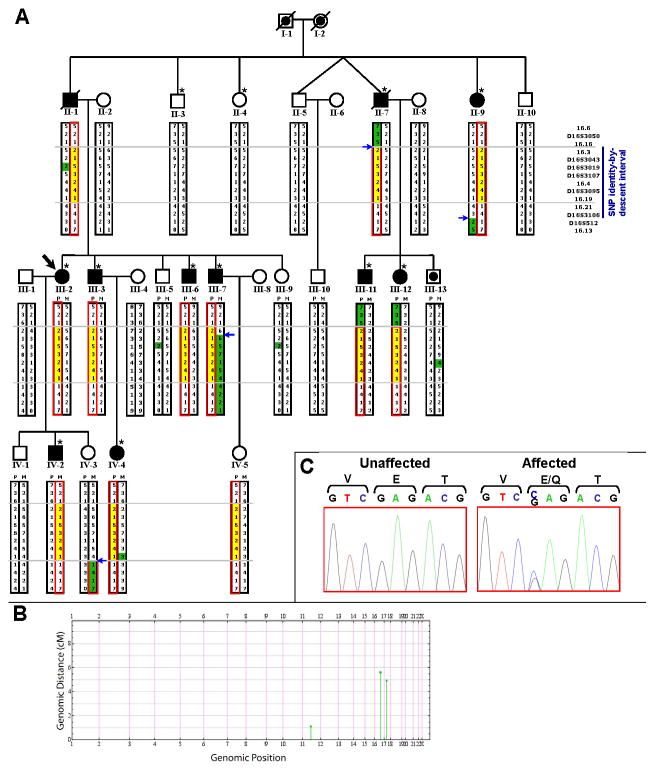
Figure 2. Pedigree with chromosome 16q21-22.1 haplotypes, and NOL3 mutation.
A) Each microsatellite allele for a given marker is denoted by a different integer value. The red box encloses the mutant haplotype. The yellow box encloses the critical region of the mutant haplotype. Blue arrows denote recombination events. Alleles are highlighted in green if they differ from the remainder of that haplotype, because of either a recombination or non-Mendelian inheritance (II-1 and progeny, III-13, IV-4; likely due to spontaneous repeat contraction/expansion). II-1 and II-7 are now deceased but were examined prior to death. IV-3 was an asymptomatic adult with normal SSEP. The dot (III-13) denotes unknown phenotype; III-13 refused SSEP. IV-5 was an asymptomatic 26-year-old at exam (refused SSEP), and is predicted to be a carrier although incomplete penetrance is possible. Forty-two relatives were omitted to preserve anonymity. Asterisks denote subjects who were genotyped for genome-wide SNP identity-by-descent mapping.
B) Genome-wide SNP mapping identified 3 candidate linkage regions (green). IBD block size (cM) is plotted against genomic position. Blocks that were not identical-by-descent have been omitted.
C) Sanger-sequencing chromatographs confirmed co-segregation of the NM_001185058.1:c.61G>C variant, predicted to cause an E21Q mutation in NOL3.
Massively Parallel Sequencing and Sanger-Sequencing of Additional Coding Sequences
We designed a capture library to target the coding sequence of every RefSeq gene within the critical region, performed library construction and hybrid-selection according to manufacturers’ protocols, and performed single-end library sequencing. Average coverage of the target region was 634x (Supporting Table S3A, Supporting Figure S2). All exons and splice junctions of all known or predicted genes that were covered less than 10x (post-duplicate removal) by massively parallel sequencing were subsequently sequenced by bidirectional Sanger-sequencing of PCR amplicons (Supporting Table S4). Full details are described at length in the Supporting Information.
In silico Homology Modelling, NOL3 Constructs, Cell Lines, Western Blots
In silico homology modelling was performed using the structure of the Nod1 CARD domain (PDB structure 2DBD) as the template. Modelling was performed with SWISS-MODEL and analyzed with UCSF Chimera software. The E21Q mutation was introduced into a pcDNA3.1 vector containing human NOL3 cDNA sequence with a C-terminal FLAG tag12. HEK293 stable cell lines generation and Western blot analyses of cell lysates were performed using standard techniques, described at length in the Supporting Information.
SSEP Studies in Mice
Mice were anesthetized with ketamine/xylazine and an electrode headmount was affixed to the skull. Two screws on each side of the brain served as a pair of recording electrodes straddling each hemisphere’s somatosensory cortex. A needle electrode was inserted into the gastrocnemius muscle, and the muscle was stimulated with 1msec pulses until a response was observed in the recording electrodes, defining the threshold. The stimulus intensity was then set to 125% of threshold and at least 10 sweeps at this intensity were recorded with an inter-stimulus interval of 10 seconds. Sweeps were averaged, normalized to peak amplitude, and overlaid onto a single figure (Supporting Figure S4) for comparison. Methods are described in more detail in the Supporting Information.
RESULTS
Clinical Presentation
We identified a large Canadian Mennonite family (Supporting Figure S1) with a history of myoclonus. Case Reports are presented in the Supporting Information. In total, eleven family members (7 male, 4 female) exhibited stimulus-evoked, multifocal myoclonus in the face, arms and legs (Table 1). Onset ranged from the second to the seventh decade. Inheritance was autosomal dominant. Myoclonus was triggered by action, sudden movements, and/or by inadvertent somatosensory stimuli, but not by light, noise or startle. Symptoms reportedly were aggravated by fatigue, exertion, sleep deprivation, emotion, and hunger. Five individuals observed that their symptoms were alleviated by a diet of frequent small meals taken throughout the day. Alcohol reportedly had no effect except for one individual in whom myoclonus was ameliorated and in another in whom myoclonus was exacerbated by alcohol. Most (8 of 11) individuals had sustained multiple sudden falls without loss of consciousness that were provoked either by sudden movements or by walking on uneven ground. Patients could not prevent these falls nor could they protect themselves and thus they often sustained severe injuries. Generally, they could get up immediately after and function with normal strength. (Table 1)
Table 1.
Clinical characteristics in 11 affected family members.
| ID | Age of Onset | Myoclonus
|
Falls | Speech disturbance | Other | Effective treatment | Median SSEPs | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| action | touch | motions | insomnia | fatigue, exertion | lack of food | Location | Alcohol effect | P25-N33 (μV) | ||||||
| II-1 | 50 | + | + | + | + | + | + | UE, LE | none | + | − | mild ataxia | clonazepam | 69.4 |
| II-7 | 50s | + | + | + | F,UE,LE | none | + | − | multi-infarct dementia | valproate | not done | |||
| II-9 | 60s | + | UE,LE | none | + | − | action tremor | 79.1 | ||||||
| III-2 | late teens | + | + | + | + | F,UE,LE | none | + | + | clonazepam | 20.3 | |||
| III-3 | 30s | + | + | + | + | + | UE,LE | decreased | + | − | mild ataxia | clonazepam, valproate | 39.4 | |
| III-6 | 40s | + | + | + | + | F,UE | none | − | − | 55.0 | ||||
| III-7 | 18 | + | + | + | + | + | + | UE,LE | increased | + | − | mild ataxia | valproate | 42.9 |
| III-11 | 40 | + | + | UE,LE | none | + | − | mild ataxia | refused | |||||
| III-12 | 40 | + | + | + | UE | none | − | − | 21.8 | |||||
| IV-2 | 20 | + | + | + | + | + | + | F,UE,LE | none | + | + | giant, data unavailable | ||
| IV-4 | late 20s | + | + | + | + | + | UE,LE | none | − | + | 21.2 | |||
Abbreviations are: +, presence, −, absence; F, face; UE, upper extremity; LE, lower extremity; FNT, finger nose test.
Myoclonus became progressively more frequent and widespread over decades and thereby interfered with activities of daily living. Older patients became wheelchair-dependent. Four showed signs of mild cerebellar limb and gait ataxia, late in the course of their illness. There were no signs of cognitive decline, spasticity or dystonia apart from one individual who had several strokes and developed multi-infarct dementia, likely independent of the myoclonus which had been present for many years. Moreover, overt seizures were not observed, nor was there a history of psychiatric illness or treatment with psychotropic medication. Myoclonus was effectively suppressed by clonazepam or valproic acid. (Table 1)
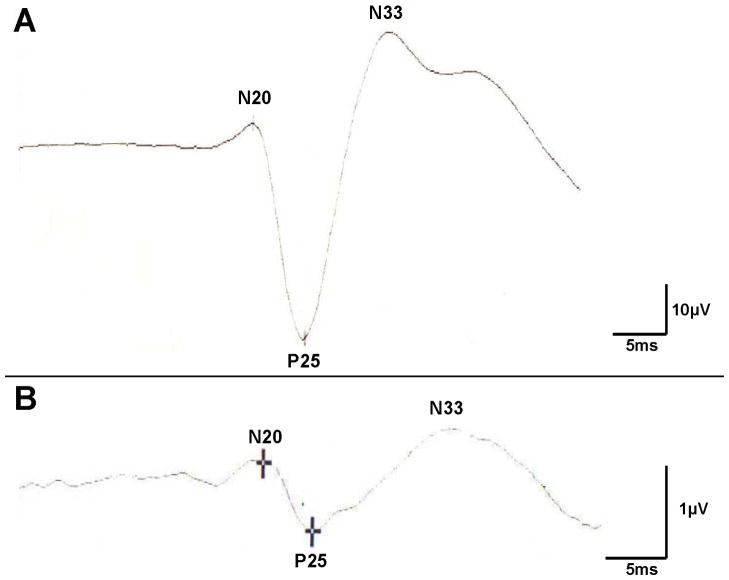
SSEP studies, using median nerve electrical depolarization as the stimulus, and recording electrodes to measure ensuing brain electrical activity, were performed. SSEPs in 9 of 9 affected patients showed giant cortical responses, the sine qua non of cortical myoclonus1,2 (Figure 1A). All 12 unaffected relatives who were tested with SSEPs showed normal cortical evoked responses of low amplitude (Figure 1B). Repeated EEG recordings in several affected family members were always normal with no epileptiform activity; thus, the infrequent falls were unlikely to be caused by seizures.
Figure 1. Somatosensory-evoked potentials in the FCM family.
Somatosensory-evoked potential (SSEP) waveforms were measured following electrical stimulation of the median nerve at the wrist in a representative affected (A, patient II-1, 69.4μV) and unaffected (B, 1.1μV) family member.
Mapping the Disease Locus
To identify the mutation underlying this autosomal dominant disorder, ten affected and two unaffected family members (Figure 2A) were genotyped by genome-wide SNP mapping. Mapping using identity-by-descent (IBD) analysis, a statistical technique for analyzing SNP data to identify chromosomal regions of shared haplotypes, was conducted essentially as described13. This approach identified three putative linkage regions (Figure 2B, Supporting Table S2), two of which were ruled out by classic microsatellite linkage analysis. In contrast, there was positive linkage to the third region, an 8 Mb interval on chromosome 16q21-22.1 which was subsequently refined by fine mapping of recombinants to a 5.57 Mb critical region (Figure 2A). The peak pairwise LOD score was 4.54.
Massively Parallel Sequencing to Identify NOL3 Mutation
The 5.57 Mb critical region contained 113 candidate genes. To identify putative disease-causing variants, we utilized next-generation sequencing technologies in performing targeted DNA capture of the critical region in one affected patient (IV-2) followed by massively parallel sequencing14 (Supporting Table S3). Mean coverage over 98.9% of targeted base-pairs (bp) was 634x (Supporting Table S3). A number of GC-rich or highly repetitive exons were absent from the arrays used for targeted DNA capture, or were present on the arrays but sequenced at less than 10x coverage (post-duplicate removal), so we individually Sanger-sequenced all of these exons (Supporting Table S4). Thus, all known or predicted coding sequences (including splice junctions) in the critical region were either bidirectionally Sanger-sequenced or sequenced at mean per-base coverage of 634x (Supporting Table S3). The sensitivity to detect heterozygous variants via massively parallel sequencing approaches 100% at per-base coverage of 20x or greater15, so our complementary sequencing approaches are very likely to have identified all coding variants in the critical region.
Seven variants met our quality criteria as putative disease-causing variants (Supporting Table S5), but only one variant co-segregated with affected status, was not present in any genome database or controls, and was predicted to cause an amino acid change. This variant was NM_001185058.1:c.61G>C in the gene nucleolar protein 3 (NOL3), predicted to cause an E21Q missense mutation. This variant is not present in any SNP database or in 1,094 genomes from multiple ethnicities studied in the 1000 Genomes Project, was not detected upon Sanger-sequencing in 252 unrelated Caucasian control subjects, and was absent from an additional 4,246 normal controls analyzed by Sequenom MassArray16. Thus, having exhaustively sequenced all coding sequences in the critical region and identified only a single novel, nonsynonymous coding mutation, the E21Q mutation, which is extremely rare (absent from more than 10,000 control chromosomes), we conclude that the NOL3 mutation is likely causative of this family’s clinical presentation.
Identifying additional NOL3 alleles that are associated with FCM or FCM-like phenotypes would support the conclusion that NOL3 mutation causes FCM. We identified 7 subjects from 5 kindreds suffering from cortical myoclonus that may be familial, but extensive Sanger-sequencing failed to yield any NOL3 variants (data not shown). However, the clinical phenotypes of the 7 subjects were not identical to the FCM family, and none of the 5 kindreds exhibited parent-to-child transmission characteristic of dominant inheritance. We anticipate that additional families with FCM will come to light upon publication of this article.
Functional Effects of NOL3 Mutation
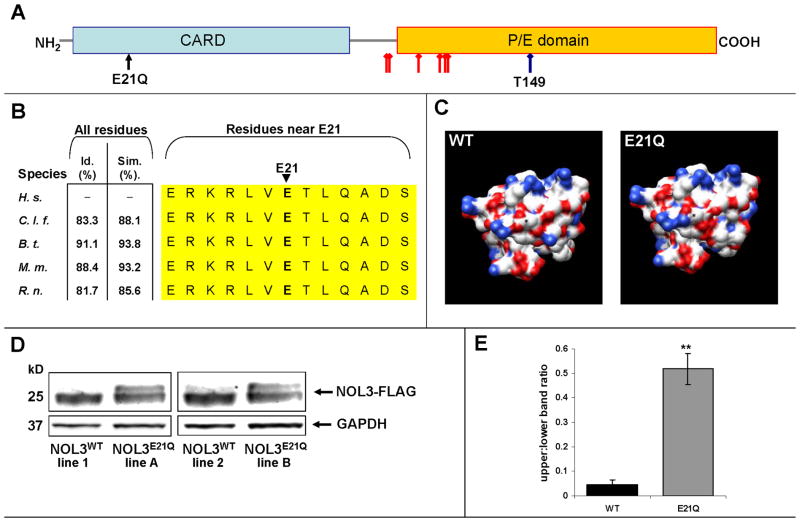
NOL3 encodes a 208 amino acid protein that is expressed in heart, skeletal muscle, and brain12,17–19. The mutated residue, E21, resides in the N-terminal Caspase Activation Recruitment Domain (CARD), a motif that mediates protein-protein binding via electrostatic interactions20,21 (Figure 3A). Because the 3-dimensional protein structure is not known, we performed in silico homology modeling, which predicted that the E21Q mutation (which changes an acidic glutamate to a neutral glutamine residue) alters the electrostatic surface potential of the NOL3 CARD (Figure 3C). In NOL3 homologs, which can only be identified in higher vertebrates, the CARD sequence is extremely conserved and the E21 residue is 100% conserved (Figure 3B).
Figure 3. E21Q mutation in NOL3 alters post-translational modification of NOL3 protein.
A) NOL3 protein contains a C-terminal proline(P)/glutamate(E) domain, and an N-terminal Caspase Activation Recruitment Domain (CARD). The CARD mediates protein-protein binding via electrostatic interactions. The E21Q mutation converts an acidic residue (E) to a neutral residue (Q). Known phospho-residues are denoted by red diamond-head arrows; T149 is a well-characterized phospho-residue (blue diamond-head arrow).
B) NOL3 is conserved in all known homologues, and the CARD residues near E21 are 100% conserved (yellow). Id. denotes aa identity; Sim., aa similarity; H. s., Homo sapiens; C. l. f., Canis lupus familiaris (canine); B.t., Bos taurus (cattle); M. m., Mus musculus (mouse); R. n., Rattus norvegicus (rat).
C) In silico homology modelling of WT and E21Q NOL3 protein structure predicts that E21Q alters the electrostatic surface potential of NOL3. Asterisk denotes aa 21; red is negative charge, blue is positive charge.
D) NOL3E21Q-FLAG exhibits altered post-translational modification in HEK293 cells. Shown is representative experiment from four independently-generated stable cell lines probed with anti-FLAG antibody. GAPDH is loading control.
E) Quantification of upper:lower ratio of FLAG bands in NOL3WT-FLAG vs. NOL3E21Q-FLAG stable cell lines. Shown is mean of three biological replicates; error bars denote standard deviation. The paired two-tailed t-test P-value is 0.0078.
To verify the functional consequences of this mutation, we generated HEK293 cell lines with stably-incorporated cDNA transgenes encoding FLAG-tagged wild-type (WT) or mutant (E21Q) NOL3. Upon harvesting protein and immunoblotting, we observed two distinct bands in NOL3E21Q cells but only a very faint upper band in NOL3WT cells (Figure 3D). By densitometry, the higher molecular weight species is ~10-fold enriched in NOL3E21Q cells (Figure 3E). This was observed for multiple independent stable cell lines, for transiently transfected HEK293 cells, when probing with either anti-FLAG (Figure 3D) or anti-NOL3 antibody, and for the COS7 cell line. In total, these results indicate that the E21Q mutation alters post-translational modification of NOL3 protein in cells.
Absence of Neuronal Hyperexcitability in NOL3 knockout mice
Since NOL3 mutation causes a human disorder of neuronal hyperexcitability, we investigated the phenotype of a previously-generated Nol3 knockout (Nol3−/−) mouse line22. On inspection, Nol3−/− mice did not exhibit any overt motor phenotype including myoclonus. To investigate cortical excitability, we measured SSEPs in aged (5 months old) Nol3−/− mice and litter-mate WT (Nol3+/+) controls. SSEPs were recorded with cortical surface electrodes in response to stimulation with brief current pulses delivered via a needle electrode to the hindlimb. Mean SSEP time to peak (WT: 59.4±0.4 msec [n=4]; KO 58.3 ± 0.1 msec [n=3]; p=0.67 t-test) and 10–90% rise time (WT: 41.8±0.1 msec [n=4]; KO: 44.2 ± 0.3 msec [n=3]; p=0.21 t-test) were statistically equivalent between genotypes (Supporting Figure S4). These data indicate normal conduction velocity of signals from the periphery to the cortex, and normal population responses of the somatosensory cortex to peripheral stimulation.
DISCUSSION
We have described a large family suffering from autosomal dominant, cortical myoclonus. Cortical myoclonus is dominantly inherited in epileptic disorders such as FAME3. However, the clinical presentation of FAME is distinctly different from that observed in our family where myoclonus was slowly progressive and became disabling late in the course of the illness, and overt seizures were not observed. Moreover, linkage to all known FAME loci4–6 was ruled out. The presentation in our family is not consistent with a progressive myoclonic epilepsy syndrome because there were neither seizures nor progressive dementia. The phenotype would be more consistent with the syndrome of progressive myoclonic ataxia defined by infrequent seizures and little or no cognitive dysfunction23, although ataxia was mild and occurred late in the illness, and we could not detect even infrequent seizures. Another possible diagnosis is hereditary hyperekplexia, an autosomal dominant disorder characterized by an excessive startle reaction. However, hereditary hyperekplexia is clinically distinct and neurophysiologically characterized by hyperexcitability of brain stem origin: giant SSEP cortical responses are not observed24.
The presentation in our family is also distinct from Myoclonus Dystonia Syndrome (MDS), which is, to our knowledge, the sole hereditary essential myoclonus syndrome hitherto described. MDS is characterized by juvenile-onset essential myoclonus, dystonia, and psychiatric comorbidity8,9, whereas in this family myoclonus was adult-onset and not accompanied by dystonia or psychiatric comorbidity. Furthermore, in MDS the symptoms are clearly ameliorated by alcohol, which was not the case in this family. Additionally, in MDS the myoclonus is of subcortical origin and is not stimulus-sensitive, and giant SSEPs are never observed10,11. Finally, in this family we excluded linkage to known MDS chromosomal regions7,8, and we sequenced but found no mutations in the SCGE gene8,9.
Hence, by exclusion we conclude that this family represents the first report of a novel clinical syndrome characterized by autosomal dominant, adult-onset, cortical myoclonus without associated seizures. We propose for this disorder the term Familial Cortical Myoclonus (FCM).
We used SNP mapping, linkage, and targeted massively parallel sequencing to identify a mutation in NOL3. Although to date we have been unable to identify additional FCM kindreds with mutations in NOL3, we have presented substantial evidence that NOL3 is the disease gene underlying FCM. First, we sequenced all known or predicted splice junctions and coding sequence within the critical region at 634x average coverage and detected only a single novel variant, the E21Q mutation in NOL3, which co-segregated with the phenotype. Second, the E21Q mutation was absent in over 10,000 control chromosomes; given the deluge of sequencing data recently deposited in public databases, the absence of the E21Q mutation in these databases constitutes additional evidence that it is extremely rare and therefore likely causative of this family’s phenotype. Finally, we found bioinformatic and in vitro evidence suggesting that the E21Q mutation has functional effects on NOL3 protein.
Nevertheless, we do concede the possibility that the E21Q variant in NOL3 is a private variant that is linked to the actual disease mutation but is not itself causative. Some plausible explanations to explain this scenario are the following: (1) the causative mutation resides in coding sequence within the critical region that is currently not known or predicted; (2) the causative mutation resides in non-coding sequence within the critical region, e.g. a UTR or promoter mutation; (3) the causative mutation was not detected due to systematic sequencing error. However remote these possibilities, we are pursuing two parallel lines of investigation in order to definitively demonstrate that NOL3 is the causative gene underlying FCM. The first is characterization of a knock-in mouse model harboring an E21Q missense mutation. The second is identification of independent NOL3 alleles that cause FCM, or FCM-like phenotypes such as FAME, SCGE mutation-negative MDS, and even idiopathic generalized epilepsy. We invite collaborations to study patients suffering from either familial or sporadic undiagnosed cortical myoclonus or a related phenotype.
The molecular mechanism by which NOL3 mutation may cause FCM remains an open question. One possibility is that the E21Q mutation alters NOL3 phosphorylation, because NOL3 has 7 known phospho-residues25 (Figure 2A), one of which, T149, has been extensively characterized26 (Figure 2A). T149 is dephosphorylated by the phosphatase calcineurin27,28, and it is interesting to speculate whether the diverse neurotoxicities of calcineurin inhibitors, which include seizures, may be related to alterations in phospho-NOL3. Since the mutation occurs in a protein-protein interaction motif and is predicted to alter the electrostatic surface potential (Figure 2C), one plausible hypothesis is that the E21Q mutation alters binding of NOL3 to its kinase and/or phosphatase, thereby increasing the level of phospho-NOL3 in cells. However, at this time we cannot rule out other NOL3 post-translational modifications.
The absence of an excitability phenotype in Nol3−/− mice, in concert with the observation that one neurologically normal human control subject harbors a heterozygous deletion spanning NOL3ref 29, strongly argues against a haploinsufficiency or dominant negative mechanism causing this autosomal dominant disorder. Instead, it seems more plausible that NOL3 mutation causes FCM by a gain-of-function or neomorphic mechanism, although at this time the exact mechanistic link between NOL3 and neuronal hyperexcitability remains speculative.
In conclusion, we identified a family suffering from a novel movement disorder for which we propose the term Familial Cortical Myoclonus (FCM). FCM is characterized by autosomal dominant, adult-onset, slowly progressive, multifocal, cortical myoclonus. We utilized unbiased, genome-wide approaches to identify a NOL3 mutation that likely causes FCM. We anticipate that this work will enhance diagnosis and genetic counseling in patients with myoclonus. Furthermore, as has proved the case for many other Mendelian diseases, further investigation to understand the role of NOL3 in FCM pathophysiology may provide insight into mechanisms of membrane hyperexcitability relevant to other forms of myoclonus and epilepsy.
Supplementary Material
Acknowledgments
We are indebted to the patients and their family for their participation. We thank G. Nunez and P. Kuffa for pcDNA3 constructs; K. Ishikawa for BEAN/tk2/FLJ10116 primer sequences; and R. Nussbaum, S.-Y. Howng and W.C. Hallows for helpful discussions. This work was supported, in part, by National Institutes of Health (NIH) grants NS044379 and U54 RR19481 (L.J.P.); NIH grants T32GM07618 and F31NS077533 (J.F.R.); and the Sandler Fund for Neurogenetics (Y.H.F. and L.J.P.). R.P.L. and L.J.P. are Investigators of the Howard Hughes Medical Institute.
References
- 1.Shibasaki H, Hallett M. Electrophysiological studies of myoclonus. Muscle Nerve. 2005;31:157–174. doi: 10.1002/mus.20234. [DOI] [PubMed] [Google Scholar]
- 2.Caviness J, Brown P. Myoclonus: current concepts and recent advances. Lancet Neurol. 2004;3:598–607. doi: 10.1016/S1474-4422(04)00880-4. [DOI] [PubMed] [Google Scholar]
- 3.Uyama E, Fu YH, Ptácek L. Familial adult myoclonic epilepsy (FAME) Adv Neurol. 2005;95:281–288. [PubMed] [Google Scholar]
- 4.Plaster NM, Uyama E, Uchino M, et al. Genetic localization of the familial adult myoclonic epilepsy (FAME) gene to chromosome 8q24. Neurology. 1999;53:1180–1183. doi: 10.1212/wnl.53.6.1180. [DOI] [PubMed] [Google Scholar]
- 5.Striano P, Zara F, Striano S. Autosomal dominant cortical tremor, myoclonus and epilepsy: many syndromes, one phenotype. Acta Neurol Scand. 2005;111:211–217. doi: 10.1111/j.1600-0404.2005.00385.x. [DOI] [PubMed] [Google Scholar]
- 6.Depienne C, Magnin E, Bouteiller D, et al. Familial cortical myoclonic tremor with epilepsy: the third locus (FCMTE3) maps to 5p. Neurology. 2010;74:2000–2003. doi: 10.1212/WNL.0b013e3181e396a8. [DOI] [PubMed] [Google Scholar]
- 7.Grimes DA, Han F, Lang AE, et al. A novel locus for inherited myoclonus-dystonia on 18p11. Neurology. 2002;59:1183–1186. doi: 10.1212/wnl.59.8.1183. [DOI] [PubMed] [Google Scholar]
- 8.Roze E, Apartis E, Clot F, et al. Myoclonus-dystonia: clinical and electrophysiologic pattern related to SGCE mutations. Neurology. 2008;70:1010–1016. doi: 10.1212/01.wnl.0000297516.98574.c0. [DOI] [PubMed] [Google Scholar]
- 9.Nardocci N, Zorzi G, Barzaghi C, et al. Myoclonus-dystonia syndrome: clinical presentation, disease course, and genetic features in 11 families. Mov Disord. 2008;23:28–34. doi: 10.1002/mds.21715. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Cunic D, Paradiso G, et al. Electrophysiological features of myoclonus-dystonia. Mov Disord. 2008;23:2055–2061. doi: 10.1002/mds.22273. [DOI] [PubMed] [Google Scholar]
- 11.Marelli C, Canafoglia L, Zibordi F, et al. A neurophysiological study of myoclonus in patients with DYT11 myoclonus-dystonia syndrome. Mov Disord. 2008;23:2041–2048. doi: 10.1002/mds.22256. [DOI] [PubMed] [Google Scholar]
- 12.Koseki T, Inohara N, Chen S, Núñez G. ARC, an inhibitor of apoptosis expressed in skeletal muscle and heart that interacts selectively with caspases. Proc Natl Acad Sci USA. 1998;95:5156–5160. doi: 10.1073/pnas.95.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shojaee S, Sina F, Banihosseini SS, et al. Genome-wide linkage analysis of a Parkinsonian-pyramidal syndrome pedigree by 500K SNP arrays. Am J Hum Genet. 2008;82:1375–1384. doi: 10.1016/j.ajhg.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi M, Scholl U, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;60:2.12.1–2.12.18. doi: 10.1002/0471142905.hg0212s60. [DOI] [PubMed] [Google Scholar]
- 17.Engidawork E, Gulesserian T, Yoo BC, et al. Alteration of caspases and apoptosis-related proteins in brains of patients with Alzheimer’s disease. Biochem Biophys Res Commun. 2001;281:84–93. doi: 10.1006/bbrc.2001.4306. [DOI] [PubMed] [Google Scholar]
- 18.Gulesserian T, Engidawork E, Yoo BC, et al. Alteration of caspases and other apoptosis regulatory proteins in Down syndrome. J Neural Trans. 2001;(Suppl):163–179. doi: 10.1007/978-3-7091-6262-0_13. [DOI] [PubMed] [Google Scholar]
- 19.Shelke RR, Leeuwenburgh C. Lifelong caloric restriction increases expression of apoptosis repressor with a caspase recruitment domain (ARC) in the brain. FASEB J. 2003;17:494–496. doi: 10.1096/fj.02-0803fje. [DOI] [PubMed] [Google Scholar]
- 20.Chou JJ, Matsuo H, Duan H, Wagner G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell. 1998;94:171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- 21.Zhou P, Chou J, Olea RS, et al. Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: a structural basis for specific adaptor/caspase interaction. Proc Natl Acad Sci USA. 1999;96:11265–11270. doi: 10.1073/pnas.96.20.11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donath S, Li P, Willenbockel C, et al. Apoptosis repressor with caspase recruitment domain is required for cardioprotection in response to biomechanical and ischemic stress. Circulation. 2006;113:1203–1212. doi: 10.1161/CIRCULATIONAHA.105.576785. [DOI] [PubMed] [Google Scholar]
- 23.Marseille Consensus Group. Classification of progressive myoclonic epilepsies and related disorders. Ann Neurol. 1990;28:113–116. doi: 10.1002/ana.410280129. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto J, Fuhr P, Nigro M, Hallett M. Physiological abnormalities in hereditary hyperekplexia. Ann Neurol. 1992;32:41–50. doi: 10.1002/ana.410320108. [DOI] [PubMed] [Google Scholar]
- 25.Olsen JV, Vermeulen M, Santamaria A, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 26.Li PF, Li J, Müller EC, et al. Phosphorylation by protein kinase CK2: a signaling switch for the caspase-inhibiting protein ARC. Mol Cell. 2002;10:247–258. doi: 10.1016/s1097-2765(02)00600-7. [DOI] [PubMed] [Google Scholar]
- 27.Tan WQ, Wang JX, Lin ZQ, et al. Novel cardiac apoptotic pathway: the dephosphorylation of apoptosis repressor with caspase recruitment domain by calcineurin. Circulation. 2008;118:2268–2276. doi: 10.1161/CIRCULATIONAHA.107.750869. [DOI] [PubMed] [Google Scholar]
- 28.Lu X, Moore PG, Liu H, Schaefer S. Phosphorylation of ARC is a critical element in the antiapoptotic effect of anesthetic preconditioning. Anesth Analg. 2011;112:525–531. doi: 10.1213/ANE.0b013e318205689b. [DOI] [PubMed] [Google Scholar]
- 29.Park H, Kim JI, Ju YS, et al. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat Genet. 2010;42:400–405. doi: 10.1038/ng.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.