Abstract
Hepatitis C virus (HCV) infection is closely tied to the lipid metabolism of liver cells. Here, we identify the triglyceride-synthesizing enzyme DGAT1 as an important host factor for HCV infection; DGAT1 interacts with the viral nucleocapsid core and is required for the trafficking of core to lipid droplets. Inhibition of DGAT1 activity or RNAi-mediated knockdown severely impairs infectious virion production, implicating DGAT1 as a new target for antiviral therapy.
Over 160 million individuals are infected with HCV, and in ~80% of cases, infection persists and can lead to severe liver disease. No vaccine is available, and current treatments are not effective against the strains most prevalent in the US and Europe. After the virus enters liver cells, a viral polyprotein is translated at the endoplasmic reticulum (ER) from a single positive-stranded RNA genome and processed into structural and nonstructural proteins1. Nonstructural proteins autonomously replicate viral RNA, which together with structural proteins, the nucleocapsid core and E1 and E2 envelope proteins, is packaged into progeny virions. Both RNA replication and infectious particle assembly are thought to occur at ER membranes. However, recent reports showed a critical involvement of lipid droplets (LDs) in infectious virion production2–4, and viral budding requires the lipoprotein secretion pathway5–7.
LDs form when triglycerides and cholesterol esters accumulate between the bilayer of the ER membrane and are surrounded by a phospholipid monolayer and associated proteins8. DGAT1 and DGAT2 enzymes catalyze the final step in triglyceride biosynthesis and are critical in LD biogenesis9. Both enzymes are ER-resident, and upon uptake of fatty acids, DGAT2, but not DGAT1, localizes to LDs9. Both enzymes have similar activities in vitro; however, DGAT1 has broader substrate specificity9, and only DGAT2 is essential in vivo in vivo10,11.
To determine if DGAT enzymes influence the HCV life cycle, shRNAs directed against DGAT1 or DGAT2 were introduced into HCV-permissive hepatoma cells12–14. Transduced cultures were inoculated with low concentrations of infectious HCV expressing eGFP as a marker for infection (eGFP-Jc1), and viral spread was monitored by flow cytometry (Fig. 1a). Spreading infection was suppressed by 65–90% with two DGAT1 shRNAs (day 9 post infection). Expression of shRNA-resistant DGAT1 rescued the suppressive effect of the DGAT1 knockdown on viral infection (Supplementary Fig. S1). No change was seen with DGAT2 shRNA pointing to a unique role of DGAT1 in HCV infection. Knockdown of DGAT expression was verified by real-time RT-PCR and, for DGAT1, by western blotting (Fig. 1b).
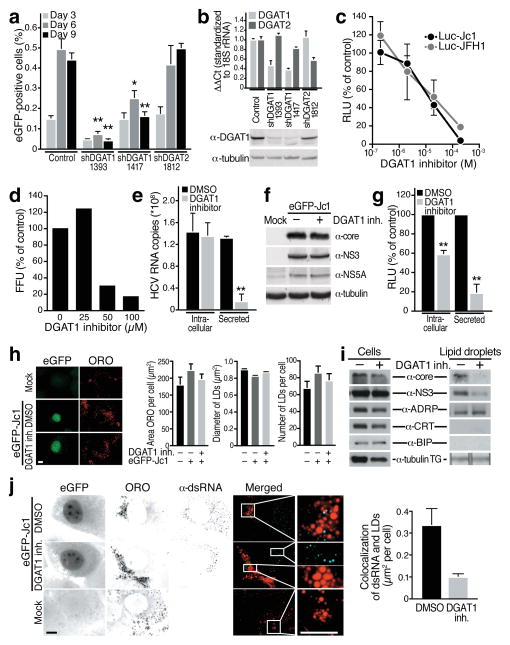
Fig. 1. DGAT1 activity is necessary for HCV particle assembly at LDs.
(a) Infection of shRNA-expressing Huh7.5 cells with low concentrations of eGFP-Jc1 viral stock. Spreading viral infection was measured by flow cytometry of eGFP (mean ± s.e.m.; n = 4; *p < 0.05, **p < 0.01). (b) Real-time RT-PCR analysis of DGAT1 or DGAT2 mRNAs in knockdown cells (mean ± s.e.m.; n = 4) or western blot analysis of DGAT1 protein expression. Available antibodies against DGAT2 do not reliably detect endogenous human DGAT2 in our hands. (c) Dose-dependent decrease of infectious titres in Huh7.5 cells transfected with Luciferase-Jc1 or Luciferase-JFH1 RNA and incubated with increasing concentrations of the DGAT1 inhibitor or DMSO for 48 h. Naïve Huh7.5 cells were infected with cell supernatants of treated cells and lysed 48 h post-infection to analyze luciferase activity (expressed as percent relative to DMSO control; mean ± s.e.m.: n = 3). (d) Dose-dependent decrease of infectious virus titers released from freshly isolated primary human hepatocytes infected with HCV-Jc1 viral stock and treated with increasing amounts of the DGAT1 inhibitor or DMSO for 3 days. Shown are infectivity titers (in FFU) expressed as percent of DMSO control. A single experiment is shown. (e) Real-time RT-PCR analysis of HCV RNA isolated from cells or from supernatants of Huh7.5 cells electroporated with eGFP-Jc1 RNA and treated with DGAT1 inhibitor (20 μM) or DMSO. Results are expressed as HCV RNA copy numbers per 1 μg total cellular RNA normalized to 18S rRNA (Intracellular) or per 1 ml culture supernatant (Secreted) at day 4 after transfection (mean ± s.d.; n = 6; **p < 0.01). (f) Western blot analysis of cell extracts described in e) lysed at day 4 after transfection. (g) Infection of naïve Huh7.5 cells with either intracellular or secreted viral particles isolated from Huh7.5 transfected with Luciferase-Jc1 RNA and treated with the DGAT1 inhibitor (20 μM) or DMSO. Shown are luciferase values expressed as percent of DMSO control (mean ± s.d.; n = 3; **p < 0.01). (h) Representative images and quantification of epifluorescence of Huh7 Lunet cells electroporated with eGFP-Jc1 RNA and treated with DMSO or DGAT1 inhibitor (20 μM) after oil-red-O (ORO) staining. LD area: mean of 1000 cells ± s.e.m.; LD diameter: mean of > 1600 LDs ± s.e.m.; LD number: mean of > 50 cells ± s.e.m. (i) Western blot analysis of cell extracts or isolated LD fractions from cells described in h). TG: extracted triglycerides analyzed by thin layer chromatography. (j) Indirect immunofluorescence of double-stranded RNA (α-dsRNA) at LDs (ORO) in cells described in h) (scale bar = 10 μm) and quantification of overlap of the signals for double-stranded RNAs and ORO per cell (mean of 30 cells ± s.e.m.).
Similar results were obtained in infected hepatoma cells treated with a DGAT1-specific small molecule inhibitor (further described in Supplementary Fig. S2). Increasing concentrations of DGAT1 inhibitor caused a gradual decline in infectious viral particle production for infectious HCV-Jc1 and HCV-JFH1 expressing firefly luciferase as infection marker (Fig. 1c; IC50 Luc-Jc1 = 7.3 μM; Luc-JFH1 = 7.4 μM). Experiments with the DGAT1 inhibitor were also performed in freshly isolated primary human hepatocytes15. Cells were inoculated with infectious HCV-Jc1, and focus-forming units were measured after transfer of infectious supernatant from primary cells to permissive hepatoma cells. Primary hepatocytes were effectively infected and produced viral titers similar to hepatoma cells (Supplementary Fig. S3a). Production of infectious viral particles was suppressed by DGAT1 inhibitor treatment, albeit at higher concentrations than in hepatoma cells (Fig. 1d). These data show that DGAT1 activity is necessary for efficient propagation of HCV in transformed and primary human liver cells.
To identify which step in the HCV life cycle requires DGAT1, we quantified intracellular and secreted HCV RNA in infected hepatoma cells. Treatment with the DGAT1 inhibitor reduced viral RNA released into the culture supernatant by ~80%, while intracellular viral RNA concentrations remained unchanged (Fig. 1e, Supplementary Fig. S3c). No change in intracellular viral protein expression was observed (Fig. 1f). Thus, DGAT1 activity is not required for viral RNA replication and translation, but likely controls HCV infection at the level of virion assembly or release.
To differentiate between these possibilities, viral particles were isolated from infected cells and used to inoculate naïve cells. Intracellular infectivity was decreased in DGAT1 inhibitor-treated cultures, indicating that viral assembly is the step in the viral life cycle mainly regulated by DGAT1, although additional effects on viral release cannot be excluded (Fig. 1g).
Because LDs are critically involved in HCV assembly, we speculated that loss of DGAT1 activity reduced cellular LD levels and thus caused the reduction in infectious particle production. Unexpectedly, no change in overall neutral lipid content, LD size, or LD numbers was observed in DGAT1 inhibitor-treated hepatoma cells (Fig. 1h). Indeed, levels of triglycerides, cholesterol esters, phospholipids, and secreted lipoproteins remained unchanged in cells treated with the DGAT1 inhibitor or shRNAs against DGAT1 or DGAT2, while knockdown of both DGAT enzymes together efficiently reduced neutral lipid content and LD numbers (Supplementary Fig. S4). These results show that the two DGAT enzymes have redundant functions in lipid synthesis in hepatoma cells and that a more complex mechanism than the mere reduction of LDs is responsible for the inhibitory effect on HCV infection.
We focused our efforts on the viral core protein because it localizes to LDs, and this localization plays a critical role in HCV particle assembly2,4,16. Core represents the first 191 amino acids of the viral polyprotein and anchors the polyprotein at the ER membrane with its C-terminal signal peptide17. After two sequential C-terminal cleavages, processed core remains attached to the cytoplasmic leaflet of the ER membrane and from there gains access to the surface of LDs18.
To determine if DGAT1 regulates core association with LDs, LD fractions were isolated from infected cells. Core was present in the LD fraction isolated from control-treated, but not DGAT1 inhibitor-treated cells, demonstrating that core association with LDs depends on DGAT1 (Fig. 1i). Similar results were obtained with the viral NS3 protein, which together with core localizes to LDs during active HCV particle production (Fig. 1i),2,19. LD fractions contained similar amounts of the LD-resident adipose differentiation-related protein (ADRP), while ER-resident proteins calreticulin and GRP78/BiP were absent.
Immunofluorescence microscopy confirmed that the core protein was retained at the ER in hepatoma cells treated with the DGAT1 inhibitor or expressing shRNAs against DGAT1, but not DGAT2 (Supplementary Fig. S5a,c). In contrast, overexpression of DGAT1 in HeLa cells, which harbor very few LDs, was sufficient to localize core from the ER to LDs, a process reversible by the addition of the DGAT1 inhibitor (Supplementary Fig. S6).
Since the localization of core to LDs is critical to recruit viral RNA replication complexes to the close proximity of LDs2, we stained HCV-infected cells with antibodies detecting viral double-stranded (ds) RNA20. While in vehicle-treated cells, a subset of LDs was decorated with signals for dsRNAs, very little overlap between LD- and dsRNA-generated signals was seen in DGAT1 inhibitor-treated cells (Fig. 1j). These results support the model that packaging of viral genomes into progeny virions requires DGAT1.
To study the molecular mechanism how DGAT1 regulates core trafficking to LDs, we performed coimmunoprecipitation experiments. Core associated with Flag-Dgat1, but not Flag-Dgat1, after coexpression in 293T cells; the interaction was not interrupted by the DGAT1 inhibitor (Fig. 2a). In addition, core efficiently coimmunoprecipitated with catalytically inactive Dgat1 (H426A), excluding that the interaction requires the enzymatic activity of DGAT1 (Supplementary Fig. S7). Core also coimmunoprecipitated with endogenous DGAT1 after lentiviral expression in hepatoma cells (Fig. 2b) and in two permissive hepatoma cell lines infected with HCV-Jc1 (Fig. 2c). However, despite the protein-protein interaction, no marked colocalization of both proteins was observed in immunofluorescence microscopy. Endogenous DGAT1 showed a reticular localization while the core protein mainly localized to punctate structures corresponding to LDs (Fig. 2d). After core expression, DGAT1 appeared more concentrated around LDs carrying core, but no significant overlap was observed. Notably, treatment with DGAT1 inhibitor or knockdown of DGAT2 did not affect the subcellular localization of DGAT1 (Supplementary Fig. S5b,d).
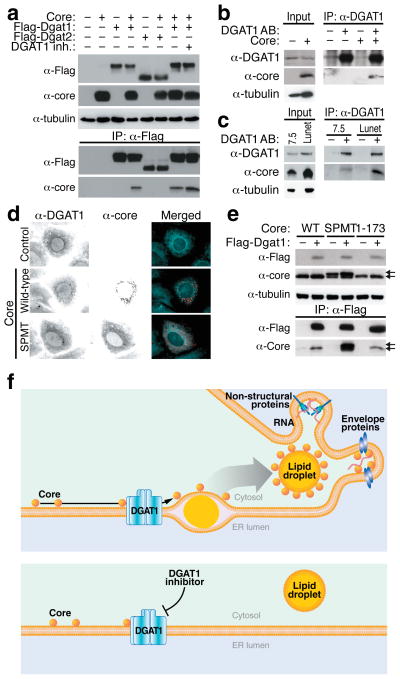
Fig. 2. Specific interaction of the HCV core protein with DGAT1.
(a) Coimmunoprecipitation assays in 293T cells transfected with expression vectors for the HCV core protein and Flag-DGAT1 or Flag-DGAT2 proteins and treated with DGAT1 inhibitor (20 μM) or DMSO. After immunoprecipitation with α-Flag agarose, the core protein was detected by western blotting with α-core antibodies. (b) Coimmunoprecipitation of the HCV core protein with endogenous DGAT1 in Huh7 hepatoma cells transduced with a core-expressing lentiviral vector. (c) Coimmunoprecipitation of the HCV core protein with endogenous DGAT1 in Huh7.5 and Huh7 Lunet cells electroporated with eGFP-Jc1 RNA. (d) Indirect immunofluorescence of core and endogenous DGAT1 in Huh7 cells transfected with wild-type or mutant (SPMT) core expression vectors (scale bar = 10 μm). (e) Coimmunoprecipitation assays in 293T cells transfected with expression vectors for wild-type (WT), mutant (SPMT) or truncated (1–173) core protein and the Flag-DGAT1-expressing plasmid. Arrows mark unprocessed (upper) and processed (lower) core protein. (f) Model of HCV core recruitment to DGAT1-generated LDs. See text for details.
We speculated that core only transiently interacts with DGAT1 when produced at the ER before migrating to LDs and examined a core mutant (SPMT) that cannot access LDs18. The SPMT mutant colocalized with endogenous DGAT1 in support of the model that both proteins interact at the ER (Fig. 2d). In agreement with this model, Flag-Dgat1 interacted stronger with the ER-retained SPMT mutant than with wild-type core in coimmunoprecipitation experiments (Fig. 2e). A truncated core mutant lacking the C-terminal signal peptide (core 1–173) coimmunoprecipitated with Dgat1, showing that this region does not mediate the interaction.
Collectively, our observations identify a hitherto unrecognized function of DGAT1 as a host factor for HCV infection. They demonstrate that infectious HCV particle production does not randomly occur in the vicinity of LDs but requires DGAT1-mediated LD formation. DGAT1 binds the HCV core protein and localizes core to DGAT1-generated LDs, thereby recruiting viral RNA replication complexes to the proper site of viral assembly (Fig. 2f). In cells treated with the DGAT1 inhibitor, core still interacts with DGAT1 but cannot access LDs as DGAT1-generated LDs no longer form. As a consequence, production of infectious viral particles is impaired despite the presence of LDs generated by DGAT2.
Why HCV selectively targets DGAT1, and not DGAT2, remains unknown at this point. However, the finding that DGAT2-generated LDs form normally in DGAT1 inhibitor-treated cells points to a unique opportunity for therapeutic intervention. DGAT1 inhibitors, currently in early clinical trials for obesity-associated diseases, may serve as novel antiviral therapeutics that selectively suppress HCV’s interaction with LDs without compromising the overall formation of lipid droplets in liver cells.
Supplementary Material
Acknowledgments
We thank Eric Yen for Sf9 cell lysates, Ralf Bartenschlager, Charles M. Rice, Francis V. Chisari, John McLauchlan, and Takaji Wakita for cell lines, infectious HCV constructs and helpful discussions, Matt Spindler for the pSicoRMS, and members of the Ott and Farese laboratories for support. This work was supported by funds from the Gladstone Institutes, the Hellman Family Foundation (M.O.) and the NIH (R03 AI069090 (M.O.); R01 DK056084 (R.F.); P30 DK026743 (UCSF Liver Center). We gratefully acknowledge support through fellowships from the Human Frontiers Science Program (E.H.), the INRS (Ce.H. and A.C.), and a Training Grant from NIDDK (Ch.H.).
Footnotes
Author contribution
E.H. designed, performed and analyzed most of the experiments and wrote the manuscript. Ch.H. and R.V.F. provided the DGAT1 inhibitor. Ch.H. also performed DGAT activity assays, analyzed data and edited the manuscript. Ce.H. and A.C. performed the experiments in human primary hepatocytes. K.K. constructed the DGAT1 mutants and performed some of the immunoprecipitation experiments. A.R.R., R.V.F., and M.O. supervised this work, analyzed data and edited (A.R.R. and R.V.F.) or wrote (M.O.) the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Moradpour D, Penin F, Rice CM. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 2.Miyanari Y, et al. Nat Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 3.Roingeard P, Hourioux C, Blanchard E, Prensier G. Histochem Cell Biol. 2008;130:561–566. doi: 10.1007/s00418-008-0447-2. [DOI] [PubMed] [Google Scholar]
- 4.Boulant S, Targett-Adams P, McLauchlan J. J Gen Virol. 2007;88:2204–2213. doi: 10.1099/vir.0.82898-0. [DOI] [PubMed] [Google Scholar]
- 5.Huang H, et al. Proc Natl Acad Sci USA. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang KS, Jiang J, Cai Z, Luo G. J Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gastaminza P, et al. J Virol. 2008;82:2120–2129. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farese RV, Jr, Walther TC. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith SJ, et al. Nat Genet. 2000;25:87–90. doi: 10.1038/75651. [DOI] [PubMed] [Google Scholar]
- 11.Stone SJ, et al. J Biol Chem. 2004;279:11767–11776. doi: 10.1074/jbc.M311000200. [DOI] [PubMed] [Google Scholar]
- 12.Wakita T, et al. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindenbach BD, et al. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 14.Zhong J, et al. Proc Natl Acad Sci USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Podevin P, et al. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.06.058. [DOI] [Google Scholar]
- 16.Barba G, et al. Proc Natl Acad Sci USA. 1997;94:1200–1205. doi: 10.1073/pnas.94.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selby MJ, et al. J Gen Virol. 1993;74(Pt 6):1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- 18.McLauchlan J, Lemberg MK, Hope G, Martoglio B. EMBO J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y, Yates J, Liang Y, Lemon SM, Yi M. J Virol. 2008;82:7624–7639. doi: 10.1128/JVI.00724-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Targett-Adams P, Boulant S, McLauchlan J. J Virol. 2008;82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.