Abstract
Direct simulations reveal key mechanistic features of early-stage protein translocation and membrane integration via the Sec-translocon channel. We present a novel computational protocol that combines non-equilibrium growth of the nascent protein with microsecond-timescale molecular dynamics trajectories. Analysis of multiple, long-timescale simulations elucidates molecular features of protein insertion into the translocon, including signal-peptide docking at the translocon lateral gate (LG), large-lengthscale conformational rearrangement of the translocon LG helices, and partial membrane integration of hydrophobic nascent-protein sequences. Furthermore, the simulations demonstrate the role of specific molecular interactions in the regulation of protein secretion, membrane integration, and integral membrane protein topology. Salt-bridge contacts between the nascent-protein N-terminus, cytosolic translocon residues, and phospholipid head groups are shown to favor conformations of the nascent protein upon early-stage insertion that are consistent with the Type II (Ncyt/Cexo) integral membrane protein topology; and extended hydrophobic contacts between the nascent protein and the membrane lipid bilayer are shown to stabilize configurations that are consistent with the Type III (Nexo/Ccyt) topology. These results provide a detailed, mechanistic basis for understanding experimentally observed correlations between integral membrane protein topology, translocon mutagenesis, and nascent-protein sequence.
Introduction
The Sec translocon is a central component of the cellular machinery for protein secretion and membrane targeting. 1 Comprised of a hetero-trimer of membrane-bound proteins (SecYEG), the translocon operates as a passive channel for co-translational and post-translational protein translocation2,3 and for the insertion of transmembrane (TM) domains of integral membrane proteins. 4,5
Structural6–10 and cross-linking 11–15 studies provide evidence for large-scale conformational changes in the translocon that facilitate its protein targeting functions, and extensive biochemical research has established the effects of translocon residue mutations and nascent protein primary structure on Sec-facilitated protein targeting. 16–24 In particular, quantitative assays have revealed that the hydrophobicity of a nascent-protein sequence determines its stop-transfer efficiency, i.e., its relative propensity to undergo membrane integration, rather than translocation, upon co-translational insertion into the Sec translocon. 20,21 And the fraction of single-spanning TM domains that adopt either Type II (cytoplasmic-N-terminal/exoplasmic-C-terminal) or Type III (exoplasmic-N-terminal/cytoplasmic-C-terminal) membrane topology is strongly influenced by the physicochemical properties of the translocon and the nascent-protein sequence. 25–32
Yet despite advances in the experimental characterization of the Sec translocon, important aspects of this cellular machinery remain inaccessible to direct interrogation. Particularly little is known about the dynamics of the translocon and nascent protein during the earliest stages of protein translocation, a critical period in the regulation of protein secretion, membrane integration, and membrane protein topology. Outstanding questions relate to nascent-protein conformations that are visited in the early stages of insertion, molecular mechanisms that connect features of the translocon and nascent-protein residues to its targeted destination and topology, and the initiation of nascent-protein secondary structure. These issues are difficult to experimentally probe because they involve transient interactions and processes, confined molecular environments, and membrane-bound complexes that create challenges for high-resolution approaches.
In this study, we introduce a protocol for directly modeling the dynamics of nascent-protein insertion into the translocon, and we leverage the specialized Anton computing system 33,34 to perform microsecond-timescale simulations of early-stage protein translocation and membrane integration. The reported simulations, although short in comparison to second-minute timescales of the biological process, are nonetheless extremely long by the standards of state-of-the-art molecular dynamics (MD) studies and provide a powerful exploratory tool for investigating the early-stage dynamics of nascent-protein insertion into the translocon. Insertion of both hydrophobic and hydrophilic nascent-protein domains is modeled, and quantitative metrics are employed to characterize nascent-protein and translocon conformational changes, the formation of salt bridges and specific interactions, and the development of large-lengthscale hydrophobic contacts. These simulations, when interpreted in combination with experimental studies and previous nanosecond-timescale MD simulation studies of the translocon, 35–40 offer new insights into the mechanistic details of Sec-facilitated protein translocation and membrane integration.
Methods
Using microsecond-timescale MD trajectories with more than 120,000 atoms, we explicitly model the insertion of nascent-protein residues into the Thermotoga maritima SecYEG channel via the SecA ATPase molecular motor (Figure 1). The structure of the SecA-SecYEG complex has been obtained via crystallography. 7 The all-atom simulation cell (Figure 1, middle panel) includes explicit solvent, counterions, and 222 palmitoyloleoylphosphatidylcholine lipid molecules. Long MD trajectories are performed using the special-purpose Anton computing system; 33,34 to meet the system-size limitations of the Anton hardware, SecA is truncated at a distance of 15 Å from the translocon, and the heavy atoms of SecA are harmonically restrained to their corresponding positions in the crystal structure. The residues of SecYEG and all other atoms in the system are unrestrained. Full details of the simulations are described in Supporting Information.
Figure 1.
Early-stage protein insertion into the Sec translocon. (Middle Panel) The all-atom system employed in the MD simulations, including the translocon (gray surface, with the helices TM2b and TM7 in green, the pore residues in orange, and the plug moiety in violet), the truncated SecA protein (white surface), and the nascent protein undergoing insertion (yellow, blue, red). (Top Panel) Expanded view of the interface region between SecA and the translocon, with the two nascent protein insertion points (IP1 and IP2) indicated. The SecA two-helix-finger and β-sheet domains are indicated in light green and light blue, respectively, and the nascent protein residues are presented with same color scheme as in the middle panel. (Bottom Panel) Schematic illustration of the simulation protocol used to model non-equilibrium protein insertion. The translocon is shown in gray, with the LG region indicated in green, the pore residues in orange, and the plug moiety in violet. SecA is shown in white, and the nascent-protein sequence is shown using the same coloring scheme as in the middle panel. Each period of nascent-protein growth is followed by a microsecond-timescale trajectory at fixed protein length.
We introduce a non-equilibrium simulation protocol to describe the SecA-driven insertion of nascent-protein residues into the translocon channel. The novel protocol includes nanosecond-timescale growth of the nascent amino acid chain at the cytosolic mouth of the translocon followed by microsecond-timescale evolution of the system (Figure 1, bottom panel). Long MD trajectories are performed using the special-purpose Anton computing system 33,34 under conditions of constant pressure and temperature. As is emphasized in the final paragraph of the Methods section, the presented insertion protocol describes necessary features of SecA-driven nascent protein insertion, including the sequential introduction of nascent protein residues and molecular confinement at the cytosolic mouth of the translocon channel; however, the detailed mechanism of the SecA driving force remains an open question. 4,41–43
The key features of the insertion protocol are as follows. After initial equilibration of the SecA-SecYEG complex, a nascent protein composed of n + 4 amino acid residues is introduced, with the four residues at the N-terminus aligned with the axis of the translocon channel and with the center-of-mass position of the remaining n residues placed at an “insertion point” at the cytosolic mouth of the translocon channel. Each of the n residues on the C-terminal end of the nascent protein exists in either an off-state, in which non-bonding interactions between each residue and the rest of the system are excluded, or an on-state, in which all interactions are included; residues in the off-state are tethered to the insertion point via harmonic restraints. We model the SecA-driven protein insertion using a repeated two-step cycle composed of (i) growth, in which residues at the C-terminal end of the nascent protein are sequentially switched from the off-state to the on-state at a pace of one residue per five nanoseconds, and (ii) evolution, in which the system is evolved without growth using standard MD. The simulations presented in the main text include three growth/evolution cycles (Figure 1, bottom panel), with each growth period leading to the insertion of fifteen new protein residues, followed by an evolution period of 0:9 – 1:8 μs in time (Table 1). The Supporting Information provides a full description of the simulation protocol employed for the nascent protein growth period, including the details of off- to on-state switching. Also presented are additional insertion simulations in which each growth period leads to the insertion of two new protein residues and each evolution period extends 100 ns in time.
Table 1.
Summary of the insertion trajectories.a
| Trajectory | IP | Mature Domain | Evolution period (μs) | Total Length (μs) | ||
|---|---|---|---|---|---|---|
| #1 | #2 | #3 | ||||
| T1 | 1 | L30 | 0.90 | 1.54 | 1.41 | 4.05 |
| T2 | 1 | Q30 | 0.90 | 1.67 | 1.41 | 4.18 |
| T3 | 2 | L30 | 1.38 | 1.50 | 1.80 | 4.88 |
| T4 | 2 | Q30 | 1.38 | 1.50 | 1.80 | 4.88 |
In each trajectory, the first growth period spans 0.055 μs, whereas the latter two span 0.075 μs.
The insertion protocol is used to obtain four microsecond-timescale simulations of early-stage protein translocation and membrane integration via the translocon. Each of the simulated insertion trajectories (T1–T4) employs one of two different insertion points and one of two different nascent-protein sequences (Table 1). The nascent-protein sequence is composed of a hydrophobic, N-terminal signal peptide (SP) and a C-terminal mature domain sequence. In all cases, the SP sequence (MGPRL11, with residues listed from the N-terminus) matches the 15-residue synthetic SP that was employed by Spiess and coworkers for the investigation of integral membrane protein topogenesis; 32 the C-terminal mature domain for the nascent protein is comprised of either a purely hydrophilic 30-mer of glutamine (Q30) or a purely hydrophobic 30-mer of leucine (L30). The first insertion point (IP1) employed in these simulations is positioned close to the highly conserved β-sheet that connects the NBD1 and PPXD domains of SecA and that is thought to be the binding site for the nascent protein 44 (Figure 1, top panel); the second insertion point (IP2) is positioned close to the loop of the two-helix-finger domain of SecA, which has been suggested to mechanically push the nascent protein through the translocon. 7,42 Exact coordinates for the insertion points are provided in the Supporting Information.
Although it is generally agreed that SecA utilizes ATP hydrolysis to drive the nascent protein across the translocon channel, 45,46 questions remain regarding the details of this process, including the oligomeric state of SecA, 47–49 the nature of SecA conformational changes that generate the driving force for nascent-protein insertion, 7,44,50,51 and the exact roles of ATP binding and hydrolysis events. 41,43,52,53 The goal of the current study is not to investigate the detailed mechanism of the SecA motor action; rather, we aim to characterize the conformational dynamics and mechanisms associated with nascent-protein insertion into the translocon. We thus model only the most fundamental roles of SecA in the insertion process: providing confinement of the nascent protein at the cytosolic mouth of the translocon channel and enforcing sequential insertion of the nascent protein into the translocon. Although the extent to which this simplification impacts any conclusions about post-translational protein translocation is difficult to assess without a better understanding of the SecA mechanism, we note that co-translational (ribosome-driven) nascent-protein insertion does not involve explicit coupling of a molecular motor to conformational changes in the translocon; 54 the insertion protocol employed here is thus at least relevant for the co-translational pathway. Furthermore, the fundamental issues that are the focus of this study, including the conformational dynamics of the nascent protein and translocon, are expected to arise in all biological pathways for Sec-facilitated protein translocation. 2,55
Results and Discussion
Translocon conformational response
The insertion simulations reveal mechanistic features of both early-stage protein translocation and membrane integration. Figure 2 presents snapshots of trajectories T1 and T2, respectively, at various times during protein insertion. The system is viewed from the perspective of the lipid bilayer, with the translocon LG helices (TM2b and TM7) in green, the pore residues in orange, and the plug moiety in red. Prior to the introduction of the mature domain residues at 0.9 μ s, the trajectories are identical, exhibiting configurations for which the two LG helices are in close proximity. At longer insertion times, it is seen in both trajectories that hydrophobic residues (blue) of the nascent protein localize at the translocon LG helices, which undergo significant separation.
Figure 2.
Structural features of the nascent protein and translocon at various times along the insertion trajectories T1 and T2. The translocon is shown in gray surface, with the two LG helices in green, the pore residues in orange and the plug moiety in violet. The nascent-protein SP and the hydrophobic mature domain of the nascent protein are colored in blue, while the hydrophilic mature domain is colored in red.
Figure 3 quantifies the LG conformational changes as a function of simulation time. The LG is characterized in terms of its width profile along the channel axis, η, which lies perpendicular to the plane of the membrane. As is illustrated in Figure 3A, the LG width profile measures the minimum horizontal distance between helices TM7 and TM8 (green) on one side of the translocon LG and helices TM2b and TM3 (yellow) on the other; detailed definitions for both the channel axis and the LG width profile in terms of the molecular coordinates are provided in the Supporting Information. Figure 3B presents the LG width profile obtained at various times during trajectories T1 and T2. The two profiles coincide for the initial stages of insertion (t = 0.5 μs), with the LG width narrowing in the region of the translocon pore residues (η≈ − 18) and widening at the cytosolic (η> − 15) and lumenal (η< − 20) openings. Significant changes in the width profiles for these two trajectories emerge at longer times. Comparison of the width profiles for trajectory T2 (red) at t = 2.0 μs and t = 3.5 μs reveals nearly uniform widening of the LG along the channel axis, including the region of the pore residues. Trajectory T1 (blue) also shows extensive widening of the translocon channel at the cytoslic opening, although it is accompanied by contraction of the LG width in the regions of the pore residues and the lumenal opening. For all insertion times, Figure 3C presents the difference between the channel width profile for trajectories T1 and T2, which emphasizes the differing extent to which the LG opens during nascent-protein insertion.
Figure 3.
Translocon LG width profiles along trajectories T1 and T2. (A) Illustration of the LG width profile, which is indicated with red arrows. The coordinate associated with the channel axis is indicated at left. (B) The LG width profiles for trajectories T1 (blue) and T2 (red) at various times. The data at time 0.5 μs is repeated in the dashed black curve. (C) The difference in the LG width profiles between trajectories T1 and T2.
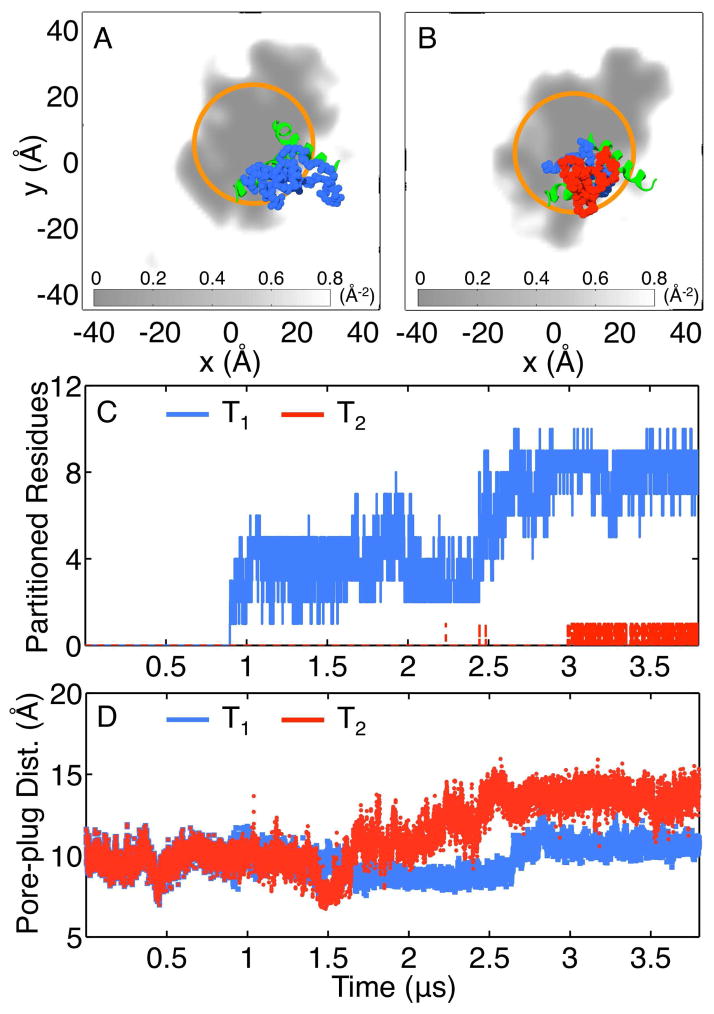
Figure 4 provides insight into the mechanistic basis for LG opening in Figure 3. Figures 4A and 4B present snapshots from insertion trajectories T1 and T2, respectively, after t = 3.5 μs of simulation time; the simulation cell is viewed along the channel axis from the cytosolic side of the membrane, and the density field of the membrane lipid tails is shown in grayscale. The density field represents the number density of heavy atoms in the hydrophobic lipid tails projected onto the x – y plane of the simulation cell; it is plotted with Gaussian smoothing on a lengthscale of 2 Å. For clarity, only the LG residues of the translocon (green) and the residues of the nascent protein (blue, red) are shown explicitly, and the set of points at a distance of 18 Å from the channel axis are indicated (orange). In both trajectories, the nascent protein is localized in the region of the LG helices, with hydrophobic residues (blue) in close contact with the hydrophobic lipid tails. The more hydrophobic nascent protein (Figure 4A) partially exits the translocon channel in favor of the membrane interior.
Figure 4.
Early stage membrane integration. (A, B) Representative configurations from trajectories T1 and T2 after t = 3.5 μs of simulation time. The nascent-protein residues (hydrophobic in blue, hydrophilic in red) and the translocon LG helices (green) are shown in atomistic detail. The density field for the hydrophobic lipid tails is projected onto the x – y plane, with gray indicating low density and white for high density. The orange circles indicate positions that are 18 Å from the center of the channel axis. (C) The time-evolution of the number of nascent-protein residues,
 , that partition into the membrane during the insertion simulations. (D) The time-evolution of the pore-plug distance in the insertion simulations.
, that partition into the membrane during the insertion simulations. (D) The time-evolution of the pore-plug distance in the insertion simulations.
To quantify the relative degrees to which trajectories T1 and T2 exhibit membrane integration, Figure 4C plots the number of nascent-protein residues,
 , that exit the translocon channel as a function of simulation time; specifically, the figure reports the number of residues for which the corresponding α-carbon lies beyond 18 Å from the channel axis (indicated in orange in Figures 4A and 4B) and falls between −30 and 0 Å along the channel axis (indicated in Figure 3A). Markedly different behaviors are seen for the two trajectories, with insertion of the more hydrophobic peptide leading to a much greater degree of membrane integration.
, that exit the translocon channel as a function of simulation time; specifically, the figure reports the number of residues for which the corresponding α-carbon lies beyond 18 Å from the channel axis (indicated in orange in Figures 4A and 4B) and falls between −30 and 0 Å along the channel axis (indicated in Figure 3A). Markedly different behaviors are seen for the two trajectories, with insertion of the more hydrophobic peptide leading to a much greater degree of membrane integration.
For trajectories T1 and T2, the differences in the channel width profiles seen in Figure 3 correlate with the differing degree of membrane integration in Figure 4. For both trajectories, the hydrophobic SP binds at the LG, leading to partial opening, as was predicted in earlier free energy calculations of the translocon conformational landscape in the presence of a hydrophobic substrate. 40 In trajectory T1, the more hydrophobic nascent protein partitions directly from the cytosolic region of the channel, without inducing any widening of the LG in the channel pore or lumenal regions (Figure 3B, blue). In trajectory T2, the hydrophilic nascent protein does not partition into the membrane interior and instead remains localized in the translocon channel; to accommodate the volume of the growing protein, the LG widens along the entire channel axis, including the region of the pore residues (Figure 3B, red).
Finally, Figure 4D plots the structural response of the translocon plug moiety during the insertion trajectories, with the distance between the pore and plug residues plotted as a function of simulation time. This distance measures the minimal separation between the α-carbon atoms of the six residues of the translocon pore (residues 82, 86, 187, 191, 274, 396) and the α-carbon atoms for the residues in the plug moiety (residues 65–74). For trajectory T1, the results show little change in the pore-plug distance, despite the significant degree of membrane integration observed.
The insertion trajectories presented in Figures 3 and 4 exhibit important mechanistic features of early stage membrane integration (trajectory T1) and protein translocation (trajectory T2). The corresponding analysis of trajectories T3 and T4 reveals similar mechanistic features (Figures S1–S3). Although it is important to avoid over-interpreting the small number of illustrative MD trajectories presented here, these long-timescale simulations nonetheless reveal details of the conformational changes that are central to regulation of stop-transfer efficiency in Sec-facilitated protein translocation. In particular, we note that the results in Figures 3 and 4 are consistent with the observation that cross-linking of the LG helices inhibits the protein translocation pathway, 14 since trajectory T2 exhibits significant opening of the LG. Figure 4D is also consistent with experimental evidence that the conformation of the plug moiety is not significantly altered upon membrane integration, 15 as well as the observation that deletion of the plug moiety has little effect on stop-transfer efficiency.21 In addition to finding little movement of the plug upon membrane integration, Ref. 15 reports more significant displacement of the plug during protein translocation; we note that Figure 4D also shows a greater degree of displacement of the plug moiety for trajectory T2 than for trajectory T1, although our simulations probe stages of the protein translocation that are too early to exhibit the full degree of plug displacement.
Nascent-protein hydrophobic contacts
In addition to its role in the regulation of stop-transfer efficiency, the Sec translocon influences the orientation, or topology, of integral membrane proteins. One such effect is that increasing SP hydrophobicity leads to a diminished fraction of proteins that undergo integration in the Type II orientation, 29–32 suggesting that hydrophobic contacts involving the nascent protein play a role in regulating integral membrane protein topogenesis. 56,57 Here, we explore this effect by characterizing the degree to which the insertion simulations exhibit hydrophobic contacts that stabilize nascent-protein configurations that are consistent with the early stages of either Type II or Type III membrane integration.
Figure 5 illustrates the nascent-protein conformational dynamics that accompany early-stage membrane integration. Figures 5A–D present snapshots of the trajectories T1 and T3 after 3.5 μs of simulation, with parts A and B showing the configuration of the SP relative to the translocon LG and parts C and D characterizing the solvation environment of the SP. The corresponding results for trajectories T2 and T4 lead to similar conclusions and are presented in Figure S4.
Figure 5.
The SP adopts configurations that differ with respect to orientation, secondary structure, and solvation environment along the simulated insertion trajectories T1 and T3. (A, B) The conformation of the nascent protein in trajectories T1 (part A) and in trajectory T3 (part B). The nascent protein is presented in blue, with the N-terminal residue highlighted in yellow. The translocon is shown as a gray surface. (C, D) Formation of the hydrophobic interface between the SP (blue) and the lipid bilayer. Water within 8 Å of the SP is shown as a light blue surface. (E) The hydrophobic contact area between the SP and the surrounding lipid molecules, plotted as a function of time in trajectories T1 (blue) and T3 (red).
Figures 5A and 5B illustrate strikingly different configurations for the nascent protein following early-stage insertion into the translocon. In both cases, the SP intercalates between the two LG helices. However, in part A, the SP adopts a partially helical conformation with the N-terminus buried inside the translocon channel. This orientation of the nascent protein enables the hydrophobic residues of the SP to extend across the LG and to make contact with the membrane hydrophobic lipid tails. In part B, the SP remains disordered, with the charged N-terminus exposed to the lipid phosphate head groups; the remainder of the SP occupies the interior of the translocon, and the LG helices widen to a lesser degree than in part A. The nascent protein in part B adopts a looped configuration that has been anticipated for early-stage Type II membrane integration; 58 in contrast, the buried N-terminal configuration for the nascent protein in part A is more consistent with the early stages of Type III membrane integration. 56
Figures 5C and 5D present the nascent-protein solvation environment for these two configurations, including the density of water molecules (light blue) within 8 Å of the SP (dark blue). In part C, the absence of solvent density at the interface between the SP and the lipid tails is clear; the LG helices separate to make room for this hydrophobic contact, and water molecules at the cytosolic mouth of the translocon evacuate the space between the hydrophobic residues of the SP and the interior of the lipid membrane. Part D reveals a different solvation environment for the SP, with water molecules solvating the LG region due to the presence of the charged SP N-terminus and the hydrophilic lipid heads.
Figure 5E quantifies the magnitude and time-dependence of the hydrophobic contact between the SP and the membrane interior. For both trajectories, the contact surface area between the SP residues and the lipid molecules are plotted as a function of simulation time; details of the surface area calculation are provided in the Supporting Information. For trajectory T1, in which the N-terminus of the SP is buried in the channel interior, the hydrophobic contact area increases markedly with simulation time; sharp increases in the curve correspond to the periods of peptide growth in the insertion simulation protocol. In contrast, the loop configuration adopted by the SP in trajectory T3 leads to consistently small hydrophobic contact area at all times.
These results suggest that nascent-protein configurations that are on-pathway for Type III integration exhibit significant hydrophobic contact between the SP and the membrane interior, whereas configurations that are consistent with early-stage Type II integration exhibit aqueous solvation of the LG region. It follows that increased hydrophobicity of the SP residues will preferentially stabilize configurations of the kind shown in Figures 5A and 5C, enhancing the degree to which the nascent proteins undergo Type III integration. Similarly, mutation of positively charged residues on the N-terminus of the nascent-protein SP will destabilize configurations of the kind shown in Figures 5B and 5D, decreasing the degree to which nascent proteins undergo Type II integration. Both of these trends have been experimentally observed. 29–32 The simulation results presented here suggest a simple mechanistic basis for understanding the sensitivity of integral membrane protein topology to hydrophobic residues in the nascent-protein SP sequence.
Nascent-protein salt-bridge formation
Finally, we investigate the mechanism by which nascent-protein salt-bridge formation influences the topology of integral membrane protein TM domains. The mutation of negatively-charged residues at the cytosolic mouth of the translocon alters observed fractions of Type II and Type III integral membrane proteins, suggesting that electrostatic interactions involving the nascent protein play a role in conferring integral membrane protein topology. 26 Furthermore, favorable interactions involving the translocon are thought to facilitate the translocation of Arg-containing peptide sequences 59,60 and to reconcile large discrepancies between the experimentally observed stop-transfer efficiency of Arg-containing peptides and computed water/membrane transfer free energies. 20–22,61–63 We explore these effects by characterizing the interactions of the translocon with positively charged residues in the nascent protein during insertion.
Figure 6A presents representative configurations from the insertion trajectories, viewed along the channel axis from the cytosolic side of the membrane. These snapshots reveal salt-bridge contacts that are formed between the Arg residue of the nascent-protein SP (blue) and either negatively charged residues on the translocon (E330, E110, and D404; yellow) or the phosphate head groups of the lipid bilayer (red). The configurations in panels E330, E110, and D404 are obtained from trajectory T2 after 0.5, 1.4 and 2.6 μs of simulation time, respectively, whereas panel corresponds to trajectory T3 after 2.6 μs. Figure 6B presents the time-dependence of salt-bridge contacts in the simulations. The contacts are defined to include configurations for which the protonated nitrogen atom of either the Arg residue or the N-terminus of the SP is within 4 Å of the anionic oxygen atom of the corresponding translocon residue or phosphate group. The corresponding time-series plots for trajectories T1 and T4 are provided in Figure S5. The structural alignment used to determine the eukaryotic homologues of residues E330, E110 and D404 is presented in Figure S6.
Figure 6.
Formation of salt bridges involving the N-terminus of the nascent protein. (A) Representative configurations associated with salt bridges that are observed in the insertion trajectories. The SP is shown in blue, with its Arg residue shown in the space-filling representation. The translocon is shown in white ribbon, with the two LG helices in green. The negatively charged residues on the translocon are shown in yellow, and the lipid head-groups are shown in orange and red. (B) The time-evolution of the salt bridges formed during trajectories T2 (red) and T3 (blue).
It is clear from Figure 6B that salt-bridge contacts form almost immediately upon nascent-protein insertion trajectories and persist over microsecond timescales. In the first microsecond of trajectory T2, which corresponds to the initial translation of the hydrophobic SP sequence, the nascent protein forms transient contact with the lipid head groups, as well as residues E330 and E110 of the translocon. At longer times, following translation of mature-domain residues, trajectory T2 exhibits extended salt-bridge contact with residue D404. In contrast, trajectory T3 immediately forms salt-bridge contact with the phosphate head groups of the membrane lipid molecules that persist throughout the length of the simulation.
The observed salt-bridge contacts involving the nascent protein are consistent with experimental observations that negatively-charged translocon residues play a role in establishing the orientation of integral membrane protein TM domains. In particular, we see the N-terminus of the nascent protein interacting at the earliest stages of insertion with the homologue of residue E110; mutation of this residue was experimentally found to decrease membrane integration for TM domains in the Type III orientation and increase membrane integration of TM domains in the Type II orientation. 26 Furthermore, long-lived salt-bridge contacts between the nascent protein and residue D404 are observed in trajectory T2; mutation of the homologues of residues D404 and E330 is experimentally found to increase membrane integration of TM domains in the Type III configuration and decrease integration of TM domains in the Type II orientation. 26 The position of residue D404 and E330 at the cytoslic mouth of the translocon (Figure S6) suggests that these residues favor configurations that are consistent with Type II membrane integration; the results for trajectory T3 in Figure 6B, as well as in Figures 5B and 5D, suggest that a similar effect may arise from interactions of the N-terminus with the phosphate head groups of the membrane bilayer. 56,64
Conclusions
We have introduced a simulation protocol for modeling the non-equilibrium dynamics of nascent-protein insertion into the Sec translocon. The approach is employed in combination with microsecond-timescale MD trajectories to investigate early-stage Sec-facilitated protein translocation and membrane integration. Analysis of multiple, long-timescale simulations reveals important molecular features of protein insertion into the translocon, including SP docking at the translocon LG, large-lengthscale conformational rearrangement of the translocon LG helices, and partial membrane integration of hydrophobic nascent-protein sequences.
All-atom simulations reveal the role of specific molecular interactions in the regulation of protein secretion, membrane integration, and integral membrane protein topology. In particular, it is shown that hydrophobic nascent-protein domains stabilize open configurations of the translocon LG and facilitate partitioning of the nascent protein into the membrane lipid bilayer. Furthermore, we find that particular salt-bridge contacts between the nascent-protein N-terminus, cytosolic translocon residues, and phospholipid head groups favor conformations of the nascent protein that are consistent with the Type II topology, whereas increased SP hydrophobicity stabilizes nascent-protein configurations that are consistent with the Type III topology.
This work reports new insights obtained from detailed, microsecond-timescale MD simulations, and it provides a mechanistic basis for understanding experimentally observed correlations between integral membrane protein topology, translocon mutagenesis, and nascent-protein primary sequence. However, we also emphasize the limitations of all-atom MD trajectories for studying slower (i.e., second- to minute-timescale) features of Sec-facilitated protein translocation that give rise to experimentally observed kinetic effects, 32,65 and more generally, the biological timescales of protein biogenesis and transport. Regardless of important recent advances in the computation of MD trajectories, the accessible timescales for atomistic simulations will remain many orders of magnitude shorter than biologically relevant timescales for the foreseeable future. Ongoing efforts to understand protein biogenesis and transport must also involve the development of new methods and strategies for coarse-grained theoretical descriptions of the protein translocation machinery.
Supplementary Material
Acknowledgments
This research was supported in part by the U.S. Office of Naval Research (USONR) under Grant No. N00014-10-1-0884 and the Department of Energy under Grant No. DE-SC0006598. Additionally, T.F.M. acknowledges an Alfred P. Sloan Foundation fellowship. We acknowledge use of the Anton supercomputer system that is hosted by the National Resource for Biomedical Supercomputing (NRBSC) at the Pittsburgh Supercomputing Center (PSC), with funding from the National Institute of General Medical Sciences under grant RC2GM093307, as well as computing resources from the National Science Foundation under Grant No. CHE-1040558.
Abbreviations
- TM
transmembrane
- MD
molecular dynamics
- LG
lateral gate
- SP
signal peptide
Footnotes
Supporting Information. Detailed simulation protocols and supplementary trajectory analysis are provided.
References
- 1.Wickner W, Schekman R. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport TA. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 3.Driessen AJM, Nouwen N. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 4.Duong F, Wickner W. EMBO J. 1998;17:696–705. doi: 10.1093/emboj/17.3.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heinrich SU, Mothes W, Brunner J, Rapoport TA. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- 6.Berg Bvd, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 7.Zimmer J, Nam Y, Rapoport TA. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egea PF, Stroud RM. Proc Natl Acad Sci USA. 2011;107:17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, Beckmann R. Nat Struct Mol Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam PC, Maillard AP, Chan KK, Duong F. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junne T, Schwede T, Goder V, Spiess M. Mol Biol Cell. 2006;17:4063–4068. doi: 10.1091/mbc.E06-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li W, Schulman S, Boyd D, Erlandson K, Beckwith J, Rapoport TA. Mol Cell. 2007;26:511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 14.du Plessis DJ, Berrelkamp G, Nouwen N, Driessen AJ. J Biol Chem. 2009;284:15805–15814. doi: 10.1074/jbc.M901855200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lycklama ANJA, Wu ZC, Driessen AJ. J Biol Chem. 2011;286:43881–43890. doi: 10.1074/jbc.M111.297507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emr SD, Hanley-Way S, Silhavy TJ. Cell. 1981;23:79–88. doi: 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- 17.Bieker KL, Silhavy TJ. Cell. 1990;61:833–842. doi: 10.1016/0092-8674(90)90193-i. [DOI] [PubMed] [Google Scholar]
- 18.Kim SJ, Mitra D, Salerno JR, Hegde RS. Dev Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- 19.Smith MA, Clemons WM, DeMars CJ, Flower AM. J Bacteriol. 2005;187:6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 21.Junne T, Kocik L, Spiess M. Mol Biol Cell. 2010;21:1662–1670. doi: 10.1091/mbc.E10-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ojemalm K, Higuchi T, Jiang Y, Langel U, Nilsson I, White SH, Suga H, von Heijne G. Proc Natl Acad Sci USA. 2011;108:E359–364. doi: 10.1073/pnas.1100120108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trueman SF, Mandon EC, Gilmore R. Mol Biol Cell. 2011;22:2983–2993. doi: 10.1091/mbc.E11-01-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devaraneni PK, Conti B, Matsumura Y, Yang Z, Johnson AE, Skach WR. Cell. 2011;146:134–147. doi: 10.1016/j.cell.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Heijne G. Nature. 1989;341:456–458. doi: 10.1038/341456a0. [DOI] [PubMed] [Google Scholar]
- 26.Goder V, Junne T, Spiess M. Mol Biol Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junne T, Schwede T, Goder V, Spiess M. J Biol Chem. 2007;282:33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- 28.Seppala S, Slusky JS, Lloris-Garcera P, Rapp M, von Heijne G. Science. 2010;328:1698–1700. doi: 10.1126/science.1188950. [DOI] [PubMed] [Google Scholar]
- 29.Hikita C, Mizushima S. J Biol Chem. 1992;267:4882–4888. [PubMed] [Google Scholar]
- 30.Wahlberg JM, Spiess M. J Cell Biol. 1997;137:555–562. doi: 10.1083/jcb.137.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harley CA, Holt JA, Turner R, Tipper DJ. J Biol Chem. 1998;273:24963–24971. doi: 10.1074/jbc.273.38.24963. [DOI] [PubMed] [Google Scholar]
- 32.Goder V, Spiess M. EMBO J. 2003;22:3645–3653. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw DE, et al. SIGARCH Comput Archit News. 2007;35:1–12. [Google Scholar]
- 34.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, Bank JA, Jumper JM, Salmon JK, Shan Y, Wriggers W. Science. 2010;330:341–346. doi: 10.1126/science.1187409. [DOI] [PubMed] [Google Scholar]
- 35.Gumbart J, Schulten K. Biophys J. 2006;90:2356–2367. doi: 10.1529/biophysj.105.075291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian P, Andricioaei I. Biophys J. 2006;90:2718–2730. doi: 10.1529/biophysj.105.073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haider S, Hall BA, Sansom MS. Biochemistry. 2006;45:13018–13024. doi: 10.1021/bi061013d. [DOI] [PubMed] [Google Scholar]
- 38.Gumbart J, Schulten K. Biochemistry. 2007;46:11147–11157. doi: 10.1021/bi700835d. [DOI] [PubMed] [Google Scholar]
- 39.Bondar AN, del Val C, Freites JA, Tobias DJ, White SH. Structure. 2010;18:847–857. doi: 10.1016/j.str.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang B, Miller TF. Proc Natl Acad Sci USA. 2010;107:5399–5404. doi: 10.1073/pnas.0914752107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Wolk JP, de Wit JG, Driessen AJ. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erlandson KJ, Miller SB, Nam Y, Osborne AR, Zimmer J, Rapoport TA. Nature. 2008;455:984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang FC, Bageshwar UK, Musser SM. Mol Biol Cell. 2009;20:4256–4266. doi: 10.1091/mbc.E09-01-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimmer J, Rapoport TA. J Mol Biol. 2009;394:606–612. doi: 10.1016/j.jmb.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kusters I, Driessen AJ. Cell Mol Life Sci. 2011;68:2053–2066. doi: 10.1007/s00018-011-0681-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lycklama ANJA, Driessen AJ. Philos Trans R Soc Lond B Biol Sci. 2012;367:1016–1028. doi: 10.1098/rstb.2011.0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Or E, Boyd D, Gon S, Beckwith J, Rapoport T. J Biol Chem. 2005;280:9097–9105. doi: 10.1074/jbc.M413947200. [DOI] [PubMed] [Google Scholar]
- 48.Jilaveanu LB, Oliver D. J Bacteriol. 2006;188:335–338. doi: 10.1128/JB.188.1.335-338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kusters I, van den Bogaart G, Kedrov A, Krasnikov V, Fulyani F, Poolman B, Driessen AJ. Structure. 2011;19:430–439. doi: 10.1016/j.str.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 50.Economou A, Wickner W. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 51.Osborne AR, Clemons JWM, Rapoport TA. Proc Natl Acad Sci USA. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erlandson KJ, Or E, Osborne AR, Rapoport TA. J Biol Chem. 2008;283:15709–15715. doi: 10.1074/jbc.M710356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang FC, Bageshwar UK, Musser SM. J Biol Chem. 2012;287:12703–12714. doi: 10.1074/jbc.M111.240903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Connolly T, Gilmore R. J Cell Biol. 1986;103:2253–2261. doi: 10.1083/jcb.103.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simon SM, Peskin CS, Oster GF. Proc Natl Acad Sci USA. 1992;89:3770–3774. doi: 10.1073/pnas.89.9.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rapoport TA, Goder V, Heinrich SU, Matlack KE. Trends Cell Biol. 2004;14:568–575. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Higy M, Gander S, Spiess M. Biochemistry. 2005;44:2039–2047. doi: 10.1021/bi047976z. [DOI] [PubMed] [Google Scholar]
- 58.Shaw AS, Rottier PJ, Rose JK. Proc Natl Acad Sci USA. 1988;85:7592–7596. doi: 10.1073/pnas.85.20.7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dorairaj S, Allen TW. Proc Natl Acad Sci USA. 2007;104:4943–4948. doi: 10.1073/pnas.0610470104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johansson AC, Lindahl E. Proc Natl Acad Sci USA. 2009;106:15684–15689. doi: 10.1073/pnas.0905394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rychkova A, Vicatos S, Warshel A. Proc Natl Acad Sci USA. 2010;107:17598–17603. doi: 10.1073/pnas.1012207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gumbart J, Chipot C, Schulten K. Proc Natl Acad Sci USA. 2011;108:3596–3601. doi: 10.1073/pnas.1012758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schow EV, Freites JA, Cheng P, Bernsel A, von Heijne G, White SH, Tobias DJ. J Membr Biol. 2010 doi: 10.1007/s00232-010-9330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dowhan W, Bogdanov M. Annu Rev Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hessa T, Monne M, von Heijne G. EMBO Rep. 2003;4:178–183. doi: 10.1038/sj.embor.embor728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.