SUMMARY
The nuclear pore complex encloses a central channel for nucleocytoplasmic transport, which is thought to consist of three nucleoporins, Nup54, Nup58, and Nup62. However, the structure and composition of the channel are elusive. We determined the crystal structures of the interacting domains between these nucleoporins and pieced together the molecular architecture of the mammalian transport channel. Located in the channel midplane is a flexible Nup54·Nup58 ring that can undergo large rearrangements yielding diameter changes from ~20 to ~40 nm. Nup62·Nup54 triple helices project alternately up and down from either side of the midplane ring and form nucleoplasmic and cytoplasmic entries. The channel consists of as many as 224 copies of the three nucleoporins, amounting to a molar mass of 12.3 MDa and contributing 256 phenylalanine-glycine repeat regions. We propose that the occupancy of these repeat regions with transport receptors modulates ring diameter and transport activity.
INTRODUCTION
The nuclear pore complex (NPC) houses the most versatile transport conduit in eukaryotes. With an estimated molar mass of more than 100 MDa (Reichelt et al., 1990), the NPC facilitates bidirectional transport of macromolecules of various sizes between nucleus and cytoplasm.
The mammalian NPC contains multiple copies of ~30 nucleoporins (nups) (Cronshaw et al., 2002) and is anchored in the pore membrane domain of the nuclear envelope by distinct integral membrane proteins. The core of the NPC exhibits 2-fold symmetry in the plane of the nuclear envelope and 8-fold symmetry in the nucleocytoplasmic direction (Gall, 1967; Unwin and Milligan, 1982). Attached to both sides of this symmetric core are filaments that are bundled into a “nuclear basket” on the nucleoplasmic side. Schematically, the symmetric core can be viewed as a series of concentric cylinders: an outermost “pore membrane protein,” a “coat,” an “adaptor,” and an innermost “transport” cylinder (Hsia et al., 2007). The symmetric core houses the central transport channel with a reported diameter of 40–50 nm in midplane (Panté and Kann, 2002; Frenkiel-Krispin et al., 2010). Large-scale shape changes of the NPC were shown by electron microscopy, particularly for its channel during transport events (Beck et al., 2004; Kiseleva et al., 1998). The molecular basis for these large morphological changes is presently unknown.
Only three of the mammalian nups—Nup62, Nup54, and Nup58—have been localized by immunoelectron microscopy to the center of the NPC (Guan et al., 1995; Hu et al., 1996) along with Nup45, which is a splice variant of Nup58 with an identical α-helical region (Hu and Gerace, 1998). How they form the transport channel is not known.
These channel nups contain predicted α-helical regions of about 150–200 residues and natively unfolded regions of up to ~300 residues that are marked by repeating phenylalanine-glycine (FG) motifs (Figure 1A; Hu et al., 1996). FG repeat regions, present in one-third of all nups, collectively function as a permeability barrier (Rout et al., 2000). They provide binding sites for transport receptors (Radu et al., 1995; Bayliss et al., 2000; Isgro and Schulten, 2005) and reversibly compact upon binding of transport factors (Lim et al., 2007).
Figure 1. Mapping of Interacting Domains.
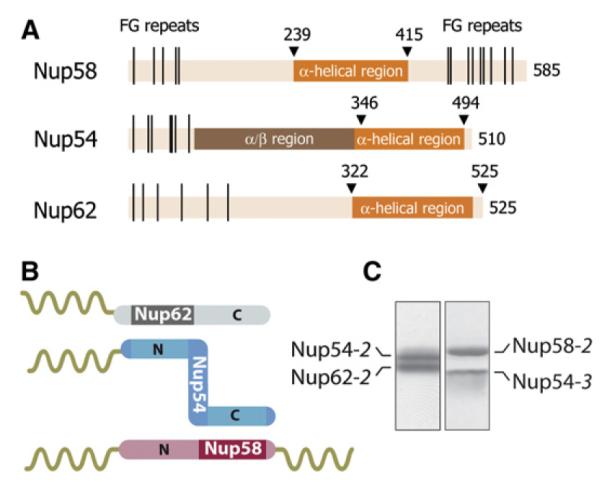
(A) Domain representation of Nup58, Nup54, and Nup62 from Rattus norvegicus indicating α-helical (dark orange), α/β (brown), and unstructured regions (light orange) with FG repeats (black lines) (Melčák et al., 2007).
(B) Schematic representation of binding sites on Nup54 (blue) for Nup62 (gray) and Nup58 (pink). The α-helical and the unstructured regions with FG repeats are visualized as rods and coils, respectively.
(C) Coexpressed and purified Nup62-2·Nup54-2 (left) and Nup54-3·Nup58-2 (right) complexes were analyzed by SDS-PAGE and stained by Coomassie blue. See also Figure S1.
Channel nups can be isolated from NPCs as a complex, but there is no agreement on their stoichiometry. In various studies, the reported ratios for Nup62:Nup54:Nup58 were 4:4:1 (Finlay et al., 1991) or 1:2:1 (Buss and Stewart, 1995; Kita et al., 1993). In the case of Nup62:Nup54:Nup58:Nup45, the ratios were 1:1:1:1 (Guan et al., 1995) or 1:2–3:3:2 (Cronshaw et al., 2002). It is conceivable that these discrepancies result from differential losses of nups due to variations in the isolation procedures.
Nup62, Nup54, and Nup58 are essential for nuclear transport (Finlay et al., 1991). Moreover, an autosomal-recessive mutation in human Nup62 (glutamine 391 to proline) (Basel-Vanagaite et al., 2006) causes a defect in brain development and leads to infantile bilateral striatal necrosis (IBSN) (Mito et al., 1986). The molecular consequences of this mutation are not known.
A major step toward understanding the molecular design of the transport channel is to determine the structure of the interactome of the channel nups. So far, crystal structures of a Nup58 α-helical region, forming a tetramer of two dimers, were reported. It was proposed that, in a circular arrangement of eight such tetramers, sliding between the dimeric interfaces would collectively vary the diameter of the channel by 3 nm (Melčák et al., 2007).
Here, we report the crystal structures of the Nup54·Nup58 and Nup62·Nup54 interacting domains and reconstruct the molecular architecture of the NPC transport channel. Its salient feature is a Nup54·Nup58 midplane ring that can undergo large-scale and reversible expansion. Nup62·Nup54 heterotrimers project vertically from the midplane ring and constitute the nucleoplasmic and cytoplasmic entries of the transport channel. We propose mechanisms for regulation of the channel diameter and formation of a “lateral gate” for transport of integral membrane proteins.
RESULTS
Previously, a complex of the α-helical regions of Nup62, Nup54, and Nup58 was assembled by coexpression in E. coli (Nup62-1, Nup54-1, and Nup58-1) (for definition of these and derived fragments used in this study, see Figure S1 available online; Melčák et al., 2007). In order to define the minimal interacting α-helical regions among the three channel nups, here, a series of deletion constructs was coexpressed, and the resulting complexes were purified. We identified two distinct regions of Nup54, one binding to Nup58 and the other to Nup62 (Figure 1B). Specifically, a C-terminal part of the α-helical region of Nup54 (Nup54-3) interacts with a C-terminal part of the α-helical region of Nup58 (Nup58-2), and an N-terminal part of the α-helical region of Nup54 (Nup54-2) interacts with an N-terminal part of the α-helical region of Nup62 (Nup62-2)(Figure 1C). The interacting regions of Nup54 are separated by an ~40 residues long linker. We conclude that the Nup62·Nup54·Nup58 complex is composed of two independently interacting modules: Nup54·Nup58 and Nup62·Nup54.
Structure of the Nup62·Nup54 Interacting Domain
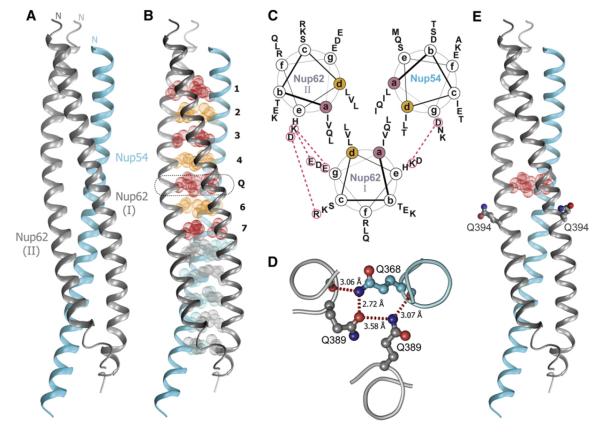
The Nup62-2·Nup54-2 complex crystallized in the space group P212121. The structure was solved by single-wavelength anomalous dispersion (SAD). The final model was refined to 2.8Å resolution, with an Rwork of 23.5% and an Rfree of 27.7% (Table S1). The asymmetric unit of the crystal is composed of one Nup54-2 and two Nup62-2 molecules (Figure 2A). The model contains residues 346–402 of Nup54 and residues 364–419 (molecule I) and residues 364–417 (molecule II) of two Nup62-2 molecules. No electron density was observed for residues 403–407 of Nup54-2 and for terminal residues of Nup62-2. One α helix of Nup54-2 and one α helix each of two Nup62-2 molecules form a parallel triple helical bundle (Figure 2A). The Nup62-2·Nup54-2 complex is primarily held together by numerous hydrophobic van der Waals contacts, burying ~6,240Å2 of surface area. The overall dimensions of the complex are approximately 93Å x 23Å x 23Å.
Figure 2. Structure of Nup62-2·Nup54-2.
(A) Ribbon representation of the asymmetric unit consisting of one Nup54-2 α helix (cyan) and two Nup62-2 α helices (gray). The N termini are labeled.
(B) Buried residues of the heterotrimer are shown as van der Waals spheres. Residues of the coiled-coil interface at a and d position of heptad repeat are colored in red and orange, respectively. Layers of the coiled-coil region are labeled from 1 to 7. Layer 5 is labeled as “Q” and is highlighted by a dashed box.
(C) Helical wheel representation of the coiled-coil region of the Nup62-2·Nup54-2 complex, viewed from the N terminus. The side chains of residues at a and d positions are segregated into nonequivalent layers, forming a predominantly hydrophobic core. Charged residues on the surface of the coiled coil that are involved in ionic interactions between adjacent α helices (red dashed lines) are marked as red circles.
(D) Cross-section of the Q layer showing hydrogen bond network. Side chains of the Gln residues and two main chain oxygen atoms of the Q layer are shown in stick-and-ball representation; backbone is shown as a main chain Ca trace colored as in (A). Hydrogen bonds are shown as red dashed lines, and bond lengths are indicated.
(E) Ribbon representation of the Nup62-2·Nup54-2 structure colored as in (A), showing the position of Q394. Residues Q394 of two Nup62 molecules are shown in stick-and-ball representation, and the residues of the Q layer are shown as red van der Waals spheres.
See also Figures S2 and S3 and Tables S1, S2, and S3.
The Coiled-Coil Interface of the Nup62-2·Nup54-2 Complex
The triple helix of the Nup62-2·Nup54-2 complex exhibits two distinct interfaces: its N-terminal half displays a “knob-into-hole” packing characteristic of coiled-coil proteins (Crick, 1953), whereas the packing of residues in its C-terminal half is similar to the more general class of α-helical bundle proteins (Figure 2B). The coiled-coil region consists of seven layers perpendicular to the long axis of the triple helix (Figure 2B). The core of layers 1–4 and of layer 7 consists exclusively of nonpolar residues (Figures 2C and S2). Interestingly, the core of layer 5 is comprised exclusively of polar glutamine residues (termed Q layer), which form a network of hydrogen bonds (Figure 2D). The core of layer 6 contains two nonpolar residues (Leu392 of two Nup62) and the polar residue Thr371 of Nup54 (Figures 2C and S2).
The outer surface of the Nup62-2·Nup54-2 triple helix includes a network of salt bridges that interconnect α helices and contribute to the specificity of Nup62-2·Nup54-2 interactions. Many of these residues as well as core residues of the heterotrimer are evolutionarily conserved in eukaryotes (Figure S2).
In a Human Disease, a Genetic Mutation Destabilizes the Nup62-2·Nup54-2 Complex
IBSN disease is caused by Gln391 to Pro substitution in human Nup62 (Basel-Vanagaite et al., 2006). Our work here revealed that the corresponding residue in rat Nup62, Gln394 (Figure S2A), is a surface residue of the coiled-coil region and is localized in the vicinity of the Q layer (Figure 2E).
To analyze the effect of this mutation on the stability of the Nup62-2·Nup54-2 complex, we mutated Gln394 to Pro. Pull-down experiments of coexpressed His6-tagged Nup54-2 either with wild-type (WT) Nup62-2 or with the mutant Nup62-2/Q394P showed a greatly diminished interaction of Nup54-2 with Nup62-2/Q394P (Figure S3A), suggesting reduced binding affinity. The molar mass of WT and mutant Nup62-2/Q394P·Nup54-2 (termed Q394P) complexes was determined by size exclusion chromatography coupled to multiangle light scattering (Table S2). At 4°C, freshly purified WT and Q394P complexes exhibited molar masses (Figure S3B and Table S2A) that correspond to a heterotrimer (Figure 2A). After a 15 min incubation of both complexes at 25°C prior to being analyzed at 4°C, the molar mass of the WT complex remained the same. However, the molar mass of the Q394P complex shifted from 23.5 ± 0.1 kDa to 7.7 ± 0.3 kDa, demonstrating the dissociation of the complex into Nup62-2/Q394P and Nup54-2 monomers (Figure S3B and Table S2A).
The structural stability of WT and Q394P complexes was also probed by circular dichroism (CD) spectroscopy. CD wavelength spectra of α-helical proteins show characteristic minima at 222 and 208 nm, and the ratio of molar ellipticities at 222 and 208 nm is expected to be close to 1 for coiled coils (Lau et al., 1984). Based on these two criteria, CD spectra indicated that the WT complex forms a stable α-helical coiled coil at the temperatures tested, whereas the Q394P complex exhibits a marked loss in α-helical structure at physiological temperatures (Figure S3C) and is similar to the individual proteins (Table S3). The thermal unfolding profile of the WT complex, recorded by CD at 222 nm, revealed a melting temperature (Tm) of 36.8°C (Figure S3D). The Tm of the mutant complex is much lower (21.5°C) and similar to that of individual proteins (Figure S3D and Table S3). This confirms that the WT complex is more stable than the mutant Q394P complex.
Polar residues in the core of coiled coils often have destabilizing effects (Lumb and Kim, 1995). Therefore, we probed the role of the polar Q layer in the observed reduced stability of the Q394P complex. We replaced the three glutamines in the Q layer in both the WT and the mutant Q394P complex with nonpolar residues by generating a Q389L mutant of Nup62 and a Q368I mutant of Nup54 (Figure S3E). We refer to these two new complexes as “LLI-Q” and “LLI-P,” respectively. Similarly to the WT complex, both LLI-Q and LLI-P form stable heterotrimers (Table S2B), and their CD wavelength spectra are in agreement with typical coiled-coil secondary structure (Table S3). However, CD thermal unfolding profiles revealed that both LLI-Q and LLI-P have significantly higher Tm (60.8°C and 47.3°C, respectively) than the WT (36.8°C) (Figure S3D). A range of thermal stability was noted, from LLI-Q as the most stable complex and the “IBSN” mutant, Q394P, as the least stable (thermal stability of LLI-Q > LLI-P > WT > Q394P based on Tm: 60.8°C > 47.3°C> 36.8°C > 21.5°C, respectively).
We conclude, first, that the buried Q layer dramatically reduces the thermal stability of the Nup62·Nup54 complex; second, that the Q394P substitution in Nup62 itself has a significant destabilizing effect; and third, that the destabilizing effects of the Q layer and the Q394P mutation are additive. Hence, in human Nup62, the Q391P mutation near the Q layer would result in destabilization of the interaction between Nup62 and Nup54 in IBSN disease.
We noticed that other human genetic diseases also involve mutations to Pro near a layer of buried polar residues in coiled-coil proteins such as lamin A, desmin, and myosin heavy chain 7 (Figure S3F). Therefore, we suggest that these mutations may have similar destabilizing effects.
Structure of the Nup54·Nup58 Interacting Domain
The Nup54-3·Nup58-2 complex crystallized in the space group P4122, and the structure was solved by the SAD phasing method. The final model was refined to 2.5Å resolution with an Rwork of 25.0% and an Rfree of 27.2% (Table S1) and contains residues 456–494 of Nup54 and residues 327–412 of Nup58. No electron density was observed for residues 445–455 of Nup54 and for residues 413–415 of Nup58.
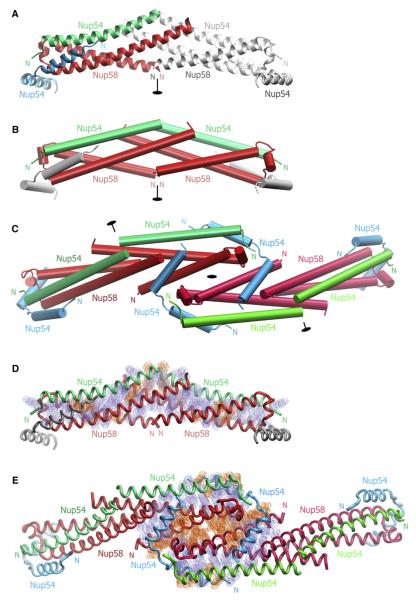
The asymmetric unit of the crystal is composed of one Nup58-2 and two Nup54-3 molecules (Figure 3A). Nup58-2 folds into a hairpin containing a shorter N-terminal helix (N helix; residues 328–357) and a longer C-terminal helix (C helix; residues 368–410) connected by a loop with one α-helical turn (Figure S4A). Notably, Nup54-3 appears in two distinct conformations: a single helix and a helix-loop-helix, termed “straight” and “bent” Nup54-3, respectively. Whereas the straight conformer consists of a 34 residues long α helix (residues 460– 493), the bent conformer is composed of two α helices, each 13 residues long, connected by a central loop (N helix, residues 460–472; C helix, residues 477–489; Figure S4B). The straight Nup54-3 α helix is aligned in parallel with the C helix of the Nup58-2 hairpin, altogether forming a Nup54-3·Nup58-2 three-helix bundle. One end of the three-helix bundle, which contains the hairpin turn of Nup58-2, is designated the “head,” and the other is designated the “tail.” In the asymmetric unit, the N helix of the bent Nup54-3 conformer intercalates and “caps” the head region of the three-helix bundle (Figure 3A).
Figure 3. Structure of Nup54-3·Nup58-2.
(A) The asymmetric unit consists of one Nup58-2 (red) protomer and two Nup54-3 protomers of different conformation (straight Nup54-3, green; bent Nup54-3, cyan). A symmetry-related unit is shown in gray and 2-fold axis of symmetry in black.
(B) Cartoon representation of the crystallographic hexamer, in which a heterotetramer consisting of two straight Nup54-3 conformers (green) and two Nup58-2 hairpins (red) forms a compact substructure. Nup54-3 caps are shown in gray.
(C) Structure of the crystallographic dodecamer. Three 2-fold axes of symmetry are indicated in black. Due to 2-fold symmetry, the dodecamer defines all interhexameric contacts.
(D) Residues that mediate hydrophobic (blue) and electrostatic (orange) interactions in the Nup54-3·Nup58-2 heterotetramer are shown in van der Waals sphere representation. The backbones of the heterotetramer (Nup58-2, red; straight Nup54-3, green) and bent Nup54-3 caps (gray) are indicated.
(E) Nature of the interactions between two Nup58-2·Nup54-3 heterotetramers and the two Nup54-3 clamps within the dodecamer. Interacting residues are indicated as in (D).
Two three-helix bundles are intertwined with each other in a tail-to-tail orientation and are capped by two bent Nup54-3 protomers to form a hexamer that is a crystallographic oligomerization module (Figures 3A and 3B). Two such modules form a crystallographic dodecamer (Figure 3C), and multiple modules build a higher-order structure in the crystal lattice (see below, Figure 6B).
Within the hexamer, a tetramer consisting of two Nup58-2 hairpins and two straight Nup54-3 molecules forms a compact substructure (Figure 3B). This Nup54-3·Nup58-2 heterotetramer is primarily held together by hydrophobic van der Waals contacts and several hydrogen bond interactions (Figure 3D). In the heterotetramer, the buried surface area of the interacting Nup54-3 and Nup58-2 protomers is 6,913Å2, and that of the interface between two Nup54-3·Nup58-2 three-helix bundles is 2,710Å2. The distribution of B factors suggests that the interface between two three-helix bundles is the most rigid region of the whole structure (see below, Figure 5F).
In the dodecamer, two crystallographic oligomerization modules are associated by lateral head-to-head interaction (Figure 3C). This interaction occurs via antiparallel association of Nup58-2 α helices from two adjacent heterotetramers and is strengthened by two bent Nup54-3 molecules acting as clamps. Clamping involves both α helices of the bent Nup54-3: the N helix interacts with the head region of the heterotetramer, whereas the C helix of bent Nup54-3 interacts with two neighboring Nup58-2, engaging mainly the N helix of one Nup58-2 and the C helix of the other Nup58-2 (Figure 3C).
In the crystallographic dodecamer, the contacts between Nup58-2 molecules of adjacent heterotetramers are predominantly mediated via polar residues (Figure 3E) and bury a surface area of 1,312 A2. In contrast, clamping by bent Nup54-3 involves primarily nonpolar residues, and two such Nup54-3 molecules bury a surface area of 8,076Å2. Many interacting residues are evolutionarily conserved (Figure S4), suggesting that the interfaces are physiologically relevant.
We conclude that the distinct conformers of Nup54-3 play distinct structural roles; straight Nup54-3 molecules and Nup58-2 hairpins form compact heterotetramers, which are clamped together into higher-order structures by bent Nup54-3 protomers.
Mutational Analyses of Nup54·Nup58 Interaction
In the knob-into-hole pattern of packing, a knob is usually a nonpolar side chain of one α helix, which fits into a hole composed of four nonpolar side chains on a neighboring α helix (Crick, 1953). In the interface between two Nup54-3·Nup58-2 three-helix bundles, we identified two symmetry-related layers of three knob residues each (Figure S5A). Two knobs are located on the C helices of adjacent Nup58-2 molecules (Tyr406 and Val396) and a third one on Nup54-3 (Leu488; Figure S5B).
Polar knobs compromise the stability of coiled-coil interactions (Lumb and Kim, 1995). One of the knobs, Tyr406, on the C helix of Nup58-2, fits into a hole generated by Val396, Leu399, Ile392, and the aliphatic moiety of Asn395 on the C helix of the adjacent Nup58-2 (Figure S5C). The polar hydroxyl group of Tyr406 is exposed to solvent and binds to an upstream Gln402 via a water-bridged hydrogen bond. As the polar hydroxyl moiety is likely to destabilize the knob-into-hole contact between the α helices, we mutated residue Tyr406 of Nup58-2 to either nonpolar Phe or charged Asp. We coexpressed these mutants together with Nup54-4 (a variant of Nup54-3, shortened by eight N-terminal residues that were found to be disordered in the crystal structure; Figure S1). Affinity pull-down experiments showed that the amount of Nup54-4 bound to the His6-tagged Nup58-2 was higher for the Y406F mutant and lower for the Y406D mutant compared to the WT (Figure S5D). This indicates that the Y406F mutation stabilized the interaction, whereas the Y406D mutation weakened it, suggesting that Tyr406 plays a critical balancing role in the stability of the Nup54·Nup58 complex. Moreover, deletion of the last ten residues of Nup58-2 (Tyr406 to Leu415) further destabilizes binding to Nup54-4 (Figure S5D). Together, these data suggest that multiple contacts between two Nup54-3·Nup58-2 three-helix bundles within the heterotetramer are critical for the stability of the Nup54·Nup58 interaction.
The molar mass of the stabilized Nup54-4·Nup58-2/Y406F complex (purified by affinity, ion exchange, and size exclusion chromatography) was determined (31.2 ± 0.2 kDa; Figure S5E) and matches closely the calculated molar mass of the heterotetramer (32.2 kDa; Figure S5F). These data suggest that the bent Nup54-4 caps are not stably associated with the Nup54-4·Nup58-2 heterotetramer and were depleted during the multiple purification processes. Corresponding experiments with the purified WT Nup54-4·Nup58-2 complex required much higher concentrations to achieve a similar molar mass as the mutant complex (Table S2C), consistent with a lower stability of the WT complex.
Detection of Higher-Order Oligomers in Solution
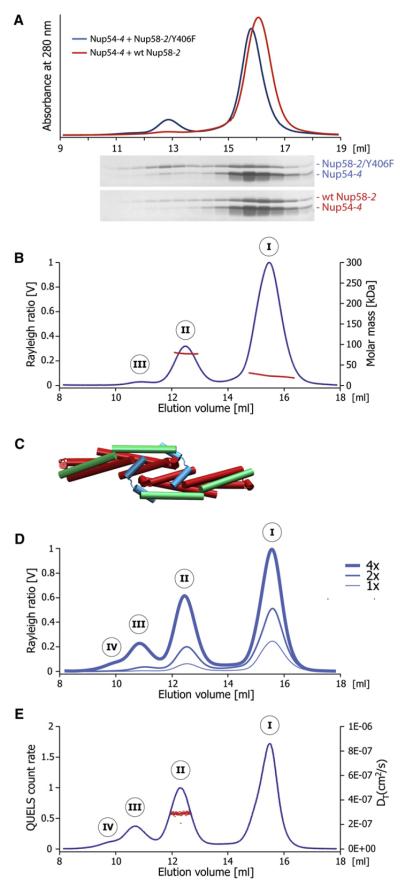
To investigate whether higher-order oligomers can be assembled in vitro, we purified individually expressed WT Nup58-2, Nup58-2/Y406F, and Nup54-4. We then mixed Nup54-4 with either WT Nup58-2 or Nup58-2/Y406F in 2:1 molar ratio and analyzed the mixture by size exclusion chromatography. An additional peak with higher molar mass was detected in the elution profile. This peak was enhanced for the mixture with Nup58-2/Y406F compared to the WT Nup58-2 (Figure 4A). Therefore, for further analyses of Nup54-4·Nup58-2 higher-order oligomers, we used the Y406F mutant of Nup58-2, as it forms a more stable complex with Nup54-4.
Figure 4. Analysis of Higher-Order Assemblies in Solution.
(A) Size exclusion chromatography (top) of WT Nup58-2 (red) or Nup58-2/ Y406F (blue), each mixed with Nup54-4, in 1:2 molar ratio (2 mg/ml each); SDS-PAGE analyses of fractions are shown in the bottom panels.
(B) Nup58-2/Y406F and Nup54-4 were mixed (2 mg/ml each) and analyzed by size exclusion chromatography coupled to multiangle light scattering, yielding three peaks (I, II, and III). The Rayleigh ratio (blue) and the molar mass (red) versus the elution volume for one experiment are shown.
(C) Cartoon representation of the decamer.
(D) Concentration-dependent higher-order Nup54·Nup58 oligomerization. Nup58-2/Y406F and Nup54-4 were mixed at increasing concentrations (1x corresponds to 2 mg/ml each) and analyzed as in (B).
(E) Nup58-2/Y406F and Nup54-4 were mixed (at 8 mg/ml each) and analyzed by size exclusion chromatography coupled to dynamic light scattering. The dynamic light scattering (QUELS) count rate (blue) and translational diffusion coefficient DT (red) versus the elution volume for one experiment are shown.
See also Figures S5 and S6 and Table S2.
To determine the molar mass of Nup54-4·Nup58-2/Y406F higher-order oligomers, we coupled the same size exclusion chromatography experiment to multiangle light scattering and detected three peaks, designated I, II, and III. The molar mass distribution within peak I extends from 18 to 31 kDa (Figure 4B), suggesting a mixture of homo- and hetero-oligomers of Nup58-2 and Nup54-4 (see Tables S2C and S2D and Figure S5; for solution studies of Nup58 homo-oligomers, see Melčák et al., 2007). The molar mass of peak II is 77.9 ± 1.6 kDa (Figure 4B). This matches closely to a decamer (Figure 4C) consisting of two Nup54-4·Nup58-2/Y406F tetramers clamped together by two bent Nup54-4 protomers (calculated molar mass, 75.2 kDa).
As expected, with increasing protein concentration (2- and 4-fold), larger amounts of higher-order oligomeric forms were detected. At high concentration, peak III becomes more prominent, and a shoulder (likely a peak IV) appears (Figure 4D). The appearance of these additional peaks suggests that further oligomerization proceeds by consecutive association of Nup54-4·Nup58-2 heterotetramers via Nup54-4 clamps.
The critical role of the Nup54-4 clamps for the formation of higher-order assemblies was confirmed by mutational analysis of the loop region. When the conserved Gly476 (Figure S4B), which contributes to loop formation in bent Nup54-4 conformer, was mutated to Ala, the yield of higher-order oligomers was decreased (Figure S6).
Using size exclusion chromatography coupled to dynamic light scattering, we also measured the diffusion coefficient (DT(avg) = 2.9 ± 0.01 × 10−7 cm2/s; Figure 4E), from which the hydrodynamic radius of the oligomers in peak II was obtained (RH = 4.6 nm). The experimental data match closely those calculated from the atomic coordinates of the decamer (calculated DT = 3.1 × 10−7 cm2/s) and confirm the elongated shape of the decamer (the RH of a 78 kDa globular protein would be 3.8 nm). We conclude that the higher-order oligomers that are observed in the crystal structure also exist in solution.
Structural Comparison between Nup58 Homo-Oligomers and Nup54·Nup58 Hetero-Oligomers
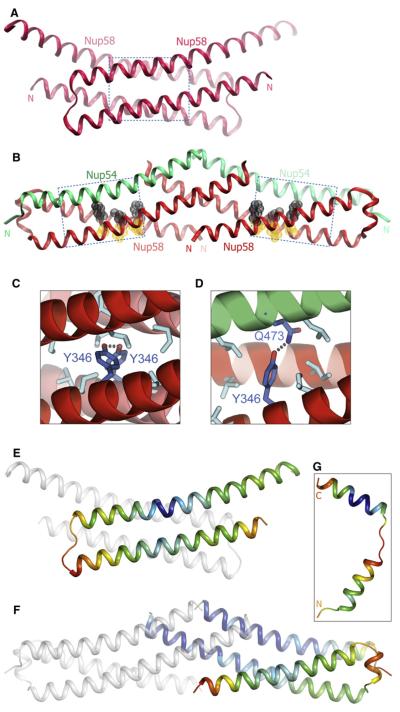
Comparison between the Nup58-2 homo-oligomers (Melčák et al., 2007) and the Nup54-3·Nup58-2 hetero-oligomers revealed substantial differences in the structure of Nup58. Each of the 82Å long hairpin dimers (Figure 5A) in the Nup58-2 homotetramer is laterally displaced by Nup54-3 to form a Nup54-3·Nup58-2 heterotetramer that is 105Å long (Figure 5B). This extensive lateral displacement converts a Nup58-2 four-helix bundle without coiled-coil interactions (Figure 5A) into a structure in which each Nup58-2 molecule folds into an intramolecular coiled coil (Figure 5B). Nup58 residues at the a position (Phe339, Tyr346, Leu385) and at the d position (Gln342, Thr381, Gln388) of heptad repeats exhibit pairwise complementary knob-into-hole interactions (Figure 5B). Moreover, the loop of Nup58-2 is expanded (Figure S4A), and the N and C helices are tilted by 150 degrees, thereby exposing numerous residues for interaction with Nup54-3.
Figure 5. Conformational Changes of Nup58 Induced by Binding to Nup54.
(A) Structure of the Nup58-2 dimer (Melčák et al., 2007).
(B) In the Nup58-2·Nup54-3 heterotetramer, each Nup58-2 molecule (red) folds into a coiled coil. a position residues (orange) and d position residues (gray) are shown as van der Waals spheres. Nup54-3 molecules are shown in green. Boxed regions in (A) and (B) are expanded in (C) and (D), respectively.
(C) Tyr346 residues (blue) of two Nup58-2 molecules (red and pink α helices) form a hydrogen bond (dashed line) within a nonpolar interface of the Nup58 homodimer, indicated by gray residues.
(D) In the Nup54-3·Nup58-2 complex, an alternative polar partner for Tyr346 (blue) is provided by residue Gln473 (blue) of Nup54 (green α helix). Nonpolar interface is indicated by gray residues.
(E) Structure of Nup58-2 dimer (oriented as in A), in which one protomer is colored in a gradient according to the main-chain B factors. Blue and red represent the lowest (14Å2) and highest (74Å2) B factor values, respectively.
(F) Nup58-2·Nup54-3 heterotetramer (oriented as in B), in which one Nup58-2 and one Nup54-3 protomer are colored according to the main chain B factors ranging from 32Å (blue) to 115Å2 (red). (G) Bent Nup54-3 colored according to the main-chain B factors as in (F).
See also Figure S7.
Pairing of polar residues in a hydrophobic environment is considered to be a discriminating factor in coiled-coil partner selection (Akey et al., 2001). In the Nup58-2 homo-oligomer, the polar Tyr346 residues of the dimer interfaces are hydrogen bonded to each other in a nonpolar environment (Figure 5C), whereas in the Nup54-3·Nup58-2 complex, Tyr346 of Nup58-2 is hydrogen bonded to Gln473 of Nup54-3 (Figure 5D). Hence, this swap in polar partners in the nonpolar interfaces may provide the basis for the specificity of the Nup54·Nup58 interaction. These residues are conserved in all eukaryotes (Figure S4), underlining their significance for the Nup54·Nup58 interaction.
As a result of the transition, the loop region of Nup58-2 is expanded, from residues 364–367 in the homo-oligomer to residues 358–367 in the hetero-oligomer (Figure S4A). In both structures, the residues of the loop as well as the residues that undergo helix-to-loop transition in Nup58-2 have much higher B factors compared to the rest of the molecule, indicating high conformational flexibility of these regions (Figures 5E and 5F). Similarly, in the bent Nup54-3, the loop region and the adjacent residues of the N helix have higher B factors (Figure 5G). It is therefore conceivable that this loop region is also flexible and expandable (see also Discussion).
The Interaction between Nup54 and Nup58 Is Dynamic
Nup54-4 and Nup58-2 each forms homo-oligomers in solution (Figures S7A and S7B). Whereas Nup54-4 forms a homo-tetramer (Table S2D), Nup58-2 exists in dimer-tetramer equilibrium (Melčák et al., 2007). The lower stability of the Nup54-4·Nup58-2 WT complex compared to the Y406F mutant (Table S2C) suggests that Nup54-4 and Nup58-2 associate transiently and readily undergo disassembly and reassembly to form homo- or hetero-oligomers. Nup58-2 and Nup54-4 were identified as minimal interacting regions between Nup58 and Nup54 within the triple Nup62·Nup54·Nup58 complex. We therefore tested whether the complete α-helical regions of Nup54 and Nup58 in the triple complex would interfere with transitions between homo- and hetero-oligomers. A purified Nup62-1·Nup58-1·Nup54-1 complex was incubated with either Nup54-4 or Nup58-2 and then analyzed by size exclusion chromatography (Figure S7). Indeed, Nup58-2 competed with and replaced the longer Nup58-1 in the triple complex (Figure S7D). In contrast, Nup54-4 pulled out Nup58-1 from the complex without incorporating itself (Figure S7E). This is consistent with our mapping data showing that Nup54-4 represents only a binding site for Nup58-2 and therefore does not replace Nup54-1 that contains also a binding site for Nup62-1 (see Figure 1B). Furthermore, these exchange data indicate a dynamic equilibrium between homo-and hetero-oligomers of Nup54-4 and Nup58-2.
DISCUSSION
Prior to our studies, structure and composition of the NPC central channel were unknown. By mapping the interactome of the three channel nucleoporins—Nup62, Nup58, and Nup54— we identified two distinct complexes: Nup54·Nup58 and Nup54·Nup62 (Figure 1B). We determined the crystal structures of these complexes and carried out structure-informed studies that allowed us to deduce the molecular design of the transport channel. The centerpiece of this channel is a highly flexible mid-plane ring with alternating projections to the nucleoplasmic and cytoplasmic sides. We formulate proposals of how changes in channel diameter accommodate varying cellular demands for transport activity.
Structure of Nup62·Nup54 and Nup54·Nup58
The Nup62·Nup54 complex forms a parallel triple helix with a coiled-coil region (Figure 2). This region contains a naturally destabilizing element, a buried layer of glutamine residues. The Nup62·Nup54 complex is further destabilized by a genetic mutation in Nup62 that is the cause of IBSN disease (Figure S3).
The crystal structure of the Nup54·Nup58 complex revealed that Nup54 exists in two different conformations, termed as straight and bent Nup54 (Figure 3). Two straight Nup54 conformers and two Nup58 molecules form a compact Nup54·Nup58 heterotetramer. Higher-order oligomerization proceeds by consecutive association of heterotetramers via bent Nup54 protomers that act as clamps (Figures 6A and 6B).
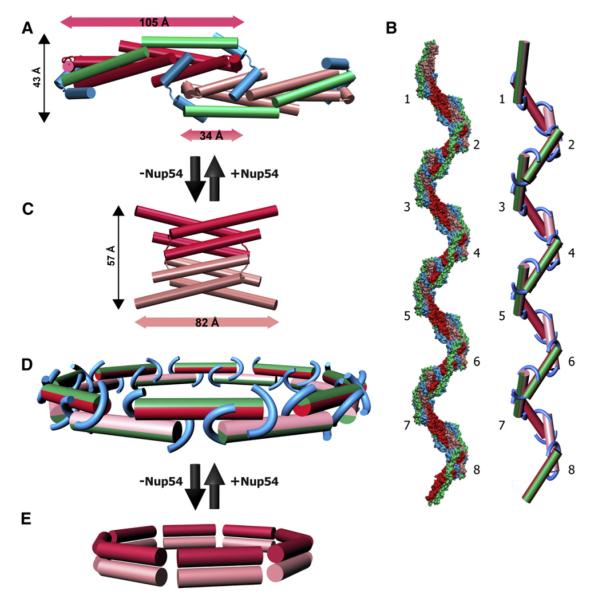
Figure 6. Alternative Interactions of Nup58 and Nup54 Can Give Rise to Large Diameter Changes within the NPC.
(A) Cartoon representation of a Nup54·Nup58 dodecamer, which corresponds to 1/8 segment of the midplane ring. Within the dodecamer, two Nup54·Nup58 heterotetramers (Nup58 molecules of consecutive tetramers in red or pink; straight Nup54 conformers, green) are clamped together by bent Nup54 (cyan). The dimensions of the heterotetramer(s) and their overlap are indicated (measured between main-chain Ca atoms).
(B) Surface representation of a higher-order oligomer, derived from the X-ray structure (left) and its schematic representation (right), in which individual Nup54·Nup58 heterotetramers are depicted as single cylinders and the bent Nup54 clamps are depicted as curved tubes, colored according to (A). Eight segments are shown.
(C) Cartoon representation of the Nup58 homotetramer (dimers in pink and red) (Melčák et al., 2007).
(D) Schematic representation of a Nup54·Nup58 heterooligomer ring. Nup54·Nup58 heterotetramers are depicted as single cylinders (the green part represents two Nup54 protomers, and the red or pink part represents two Nup58 hairpins with alternating color in consecutive heterotetramers). Nup54 clamps are depicted as curved tubes (cyan).
(E) Circumferential arrangement of eight Nup58 homotetramers (dimers schematically shown as single cylinders in pink or red).
Conversion between Nup54·Nup58 Hetero- and Homo-Oligomers
The Nup58 homotetramer consists of two dimers that associate by their N-terminal helices (Figure 6C). In various conformers of the homotetramer, the electrostatic dimer-dimer interfaces were displaced by up to 11Å (Melčák et al., 2007).
The association of Nup58 with Nup54 entails large conformational changes and lateral displacements of the Nup58 molecules. In the homo-oligomer, each Nup58 dimer is 82Å long (Figure 6C), whereas in the Nup54-bound form, the length increases to 105Å (Figure 6A). The two dimers in the Nup58 homotetramer can be displaced by 11Å, whereas the corresponding two Nup54-bound Nup58 dimers are displaced by 71Å in the Nup54·Nup58 (do)decamer (Figure 6A). The transition between homo- and hetero-oligomers is reversible (Figure S7) and likely occurs via multiple intermediates. It may be facilitated by the dynamic nature of the sliding interface in the Nup58 homotetramer (Melčák et al., 2007). As a result, Nup54·Nup58 hetero-oligomers are likely converted into homotetramers of Nup54 and Nup58. Notably, both Nup54 and Nup58 interacting domains form homotetramers in solution (Table S2D; Melčák et al., 2007).
Flexibility of Nup54·Nup58 Higher-Order Oligomers Allows Ring Formation
In the Nup54·Nup58 higher-order crystal structure, contacts between heterotetramers are predominantly polar, whereas nonpolar residues primarily stabilize the Nup54 clamps. The loop regions of Nup54 clamps are likely to be flexible because they consist of a conserved glycine and hydrophilic residues (Figures S4B and S6) and due to the high B factor. Therefore, it is conceivable that the loop regions act as hinges and can accommodate displacements of the polar interface between two heterotetramers. Based on these collective structural features, we propose that the polar interface between the heterotetramers can be displaced in such a way that the crystallographically constrained spiral (Figure 6B) is rearranged into a ring structure that is indicated in Figure 6D and Movie S1.
Nup54·Nup58 Midplane Ring
Schematically, in this ring model, eight Nup54·Nup58 heterotetramers (satisfying the 8-fold symmetry of the NPC; Gall, 1967) are positioned in a circle. Two such circular structures are stacked upon each other (satisfying 2-fold symmetry for channel nups; Guan et al., 1995; Hu et al., 1996) in a staggered fashion (Figure 6D). These two segmented circular structures are clamped together by bent Nup54 molecules into a ring (Figure 6D), with an estimated height of 4 nm (Figure 6A). The diameter of the Nup54·Nup58 ring was estimated from the structure (see Experimental Procedures). Overlapping association of 105Å long Nup54·Nup58 heterotetramers (Figure 6A) yields a ring (Figure 6D) with a diameter of ~36 nm.
The Nup54·Nup58 ring mirrors the 2-fold symmetry that is found in the NPC along the plane of the nuclear envelope, and therefore we suggest that it is located in the midplane of the transport channel of the NPC.
Expandability of Nup54·Nup58 Ring
Because of the malleable nature of their connections, clamped Nup54·Nup58 heterotetramers may be laterally displaced, and therefore the midplane ring would be expandable (Movie S2).
This lateral displacement is suggested from several structural features: (1) The interfaces between heterotetramers are polar in nature and have the propensity to form alternative hydrogen bond networks as observed in Melčák et al. (2007). (2) The loop region of Nup54 clamps is likely to be flexible. (3) A loop expansion of Nup54 is suggested from the propensity of channel nups, such as Nup58, to undergo helix-to-loop transitions.
A lateral displacement of each of the 16 heterotetramers reducing their overlap by half would lead to an increase of the ring diameter from ~36 to ~45 nm. These diameter ranges are in line with the maximum size of a transport receptor-substrate complex of 39 nm that has been reported to pass through the NPC (Panté and Kann, 2002).
Large-Scale Fluctuation of Channel Diameter by Alternative Interactions
Nup54·Nup58 interacting domains exist in a dynamic equilibrium between free and bound forms (Figure S7). Therefore, we suggest that transitions between Nup54·Nup58 hetero-oligomers and homo-oligomers also occur in vivo. These transitions would be restricted to their interacting domains.
It remains to be investigated whether and to what extent the circularly arranged Nup58 homo-oligomers are laterally associated with each other. A close lateral association of Nup58 homo-tetramers would yield a ring with a diameter of ~21 nm (Figure 6E). Leaving aside a potential contribution of Nup54 homotetramers, this would reflect the channel diameter in the homo-oligomeric state. Consequently, a reversible transition between Nup54·Nup58 hetero- and homo-oligomers would change the diameter of the transport channel at midplane from ~36 to ~21 nm. Homo- and hetero-oligomers of Nup54·Nup58 would coexist in a dynamic equilibrium, resulting in a fluctuation of the channel diameter. Because the Nup54·Nup58 ring is potentially expandable (to ~45 nm), the diameter of the midplane ring may vary by more than 20 nm. Concomitantly, the diameter alteration is accompanied by a heightening of the ring to 6 nm in the Nup58 homo-oligomer and lowering to 4 nm in the Nup54·Nup58 hetero-oligomer.
Nucleoplasmic and Cytoplasmic Entries of the Transport Channel
In Nup54·Nup58 midplane ring, the N termini of the α-helical region of Nup54 project in alternating fashion above and below the plane of the ring and are connected to upstream binding sites for Nup62 via a ~40 residues long “linker.” These projections provided an important clue for alternately placing the Nup62·Nup54 complex to the nucleoplasmic and cytoplasmic side of the midplane ring (Figure 7).
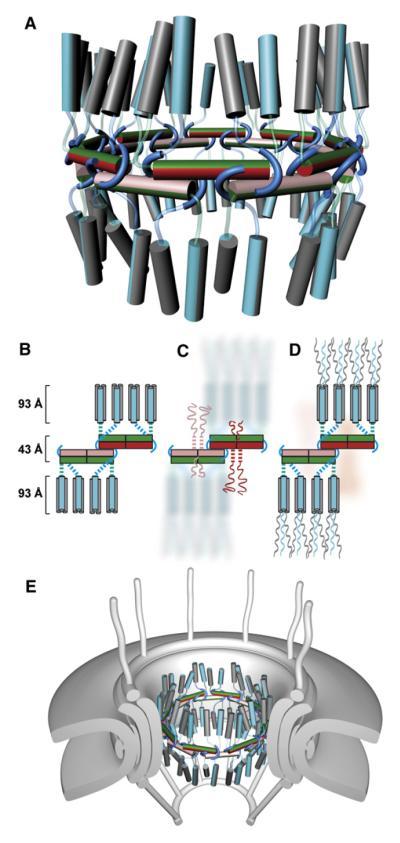
Figure 7. Architecture of the Mammalian Transport Channel Formed by the Nup62·Nup54·Nup58 Complex.
(A) Cartoon representation of the transport channel with color coding and representation of the midplane ring as in Figure 6D. Nup62·Nup54 heterotrimers are shown as vertical cylinders colored in gray (two Nup62 molecules) and cyan (one Nup54). The linker regions of the Nup54 molecules are represented as cyan and green transparent tubes. See also Movie S3.
(B) Schematic representation of 1/8 segment of the transport channel. Nup54·Nup58 heterotetramers are shown as boxes in same color coding as in (A). Nup54 clamps are shown as cyan hooks. The Nup62·Nup54 heterotrimer is represented by three cylinders (Nup62 and Nup54 in gray and cyan, respectively). Dashed lines represent linker regions.
(C) Schematic arrangements of FG repeat regions of Nup58 in the transport channel. Both N-terminal (longer ribbons) and C-terminal (shorter ribbons) FG regions are colored pink or red. Dashed pink and red lines indicate N-terminal α-helical regions of Nup58 that connect Nup58 to N-terminal FG regions. Nup62 and part of Nup54 molecules are depicted in background.
(D) The position of FG repeat regions of Nup54 and Nup62. The N-terminal FG regions (ribbons) of Nup54 (cyan) and Nup62 (gray) project to cytoplasmic and nucleoplasmic sides of the midplane ring. FG regions of Nup58 are shown in the background (pink).
(E) The transport channel (as shown in A) embedded in the NPC. The core of the NPC is schematically shown as concentric cylinders and is anchored in the nuclear envelope. Attached to the core are a cytoplasmic ring with filaments and a nucleoplasmic ring with the “nuclear basket.”
The placement of Nup62·Nup54 triple helices (Figure 2A) in the transport channel is shown in Figures 7A and 7B (see also Movie S3). An association between Nup62·Nup54 heterotrimers is unlikely, as higher-order oligomerization of Nup62·Nup54 was not observed in our studies. The geometric angles as well as the distance of individual Nup62·Nup54 heterotrimers with respect to the midplane ring are likely to be variable (Figure 7A). These parameters will probably be influenced by the structure of the Nup54 linker region and by anchorage of Nup62 to other nups, such as Nup93 (Grandi et al., 1997; Miller et al., 2000). An interaction between a C-terminal region of Nup62 and Nup93 has been described for yeast homologs (Grandi et al., 1995). These Nup62 C-terminal regions project from the Nup62 helices of the heterotrimer and interact with Nup93, thereby anchoring Nup58·Nup54·Nup62 to the adaptor cylinder of the NPC (Figure 7E).
Composition of the Transport Channel
From our model, we deduce a structure-based stoichiometry of Nup62:Nup54:Nup58 to 4:2:1. Accordingly, the transport channel of the mammalian NPC consists of 32 molecules of Nup58, 64 molecules of Nup54, and 128 molecules of Nup62 (Figure 7A and Movie S3). This estimate is based on 8-fold rotational symmetry of the NPC as well as full occupancy of the two distinct Nup54 interacting sites with Nup58 and Nup62. Based on the primary structure of the participating proteins, the transport channel of the NPC yields a mass of 12.3 MDa.
The three channel nups contribute a total of 256 FG repeat regions to the NPC. Nup54 and Nup62 contain FG repeats only at their N termini, whereas Nup58 features FG regions at both its N and C terminus (Figure 1A; Hu et al., 1996). These FG regions are likely to radially project toward the center of the channel. Whereas the FG regions of Nup54 and Nup62 are located in the periphery of the transport channel, those of Nup58 are likely to be confined close to the midplane (Figures 7C and 7D).
As the predicted α-helical region of the alternatively spliced Nup45 is identical to that of Nup58, it is likely that these two nups can substitute for each other (Hu and Gerace, 1998; Melčák et al., 2007). The ratios of Nup58 and Nup45 in different cell types are variable (Hu and Gerace, 1998), indicating heterogeneity of channel composition.
The channel nups are engaged in alternative interactions, as we show for Nup54·Nup58 homo- and hetero-oligomers. Also, it has been reported for yeast homologs that Nup62 can bind directly either to Nup54 or Nup88 (Bailer et al., 2001). We suggest that the destabilizing role of the glutamine layer in the coiled-coil interface of the Nup62·Nup54 complex might play a role in lowering the energy barrier for an alternate interaction of Nup62 with Nup88.
Ultimately, alternative pairings could affect the neighboring network of nup interactions. This may lead to the differential losses in cell fractionation studies and may explain the observed inconsistencies in the abundance and stoichiometries of nups representing large populations of NPCs. Moreover, it should be noted that, for yeast NPC, substantially different models of the central pore have been proposed (Alber et al., 2007; Schrader et al., 2008).
Lateral Gating for Transport of Integral Membrane Proteins
Signal sequence- and transport receptor-mediated passage of integral membrane proteins from the outer to the inner nuclear membrane (King et al., 2006) may proceed by a mechanism in which segments of the polypeptide chain vertically slice through the NPC scaffold (Blobel, 2010; Meinema et al., 2011) while bound to their mobile transport receptors in the central channel. At present, the molecular mechanism of how to achieve such transient openings is unknown.
In the framework of an expandable Nup54·Nup58 midplane ring of the channel (Figure 7E), we propose that the lateral displacement of Nup58·Nup54 heterotetramers might proceed further and result in reversible local “derailment” and “unclamping” of stacked-upon heterotetramers, thereby providing a lateral gate. Lateral gating of the transport channel is likely accompanied by vertical parting of the NPC scaffold in order to permit passage of transport receptor-membrane protein complexes across the NPC and the adjacent pore membrane.
Regulation of Channel Diameter by Interactions between FG Repeats and Transport Receptors
Channel diameter fluctuations may play an important role to accommodate varying cellular demands for transport activity. We propose that the FG repeats of channel nups act as sensors between the rate of transport and homo- to hetero-oligomer transitions in the midplane ring. It is conceivable, for example, that in periods of high demands for transport activity, a more frequent engagement of transport receptors with FG repeats would cause widening of the channel by shifting the equilibrium toward Nup54·Nup58 hetero-oligomerization. This would result in a highly active state of the transport channel. Conversely, during periods of low demands for transport, a shift to homo-oligomerization would narrow the channel diameter and thus yield a state with a low transport activity. Reducing the channel diameter would, in turn, increase the packing density of FG repeat regions and strengthen the permeability barrier, thereby reducing leakage of molecules not intended for transport.
In summary, the analyses of the α-helical regions of the three channel nucleoporins and their arrangement in higher-order structures yielded a model for the structure for the transport channel of the mammalian NPC. Our data go a long way toward explaining the large-scale changes that have been reported by electron microscopic approaches for the morphology of the NPC, particularly for its central transport channel (Beck et al., 2004; Kiseleva et al., 1998). The existence of distinct conformers, the promiscuity of interactions, and the reversible transitions from homo- to hetero-oligomers appear to be built-in features of the channel nucleoporins. The interplay between rearrangements of α-helical regions and the engagement of FG repeat domains with numerous mobile transport receptors may enable rapid adjustments of channel activity to cellular demands for bidirectional nucleocytoplasmic transport.
EXPERIMENTAL PROCEDURES
A detailed description of the methods employed in this study is provided online in the Extended Experimental Procedures.
Protein Expression and Purification
Various expression constructs of Nup54, Nup58, and Nup62 from Rattus norvegicus were generated and expressed as described in Melčák et al. (2007) and are summarized in Figure S1. Protein complexes were produced by coexpression of the proteins in E. coli using a polycistronic expression vector. Protein complexes, single proteins, and derived mutants were purified (for crystallization and solution studies) using His6 affinity chromatography, followed by thrombin cleavage, anion exchange chromatography, and size exclusion chromatography, as described in Melčák et al. (2007).
Protein Crystallization, X-Ray Data Collection, and Structure Determination
Protein complexes were crystallized using the hanging-drop method. X-ray diffraction data of crystals were collected at 100 K at the NE-CAT 24-IDE beamline, APS, Chicago and at beamline 8.2.1, ALS, Berkeley. Phases were obtained from selenomethionine SAD diffraction data sets. Data collection and refinement statistics are summarized in Table S1.
Light Scattering Analysis
Purified proteins were separated by size exclusion chromatography while the column was connected to multiangle and dynamic light scattering detectors. Weight-averaged molar masses were determined by multiangle light scattering and translational diffusion coefficients (DT) by dynamic light scattering.
Circular Dichroism Spectroscopy Analysis
Secondary structure of proteins was analyzed by measuring far UV wavelength CD scans. CD data are reported as molar ellipticities [Θ]. Thermal unfolding profiles of proteins were measured by recording temperature-dependent molar ellipticity changes at 222 nm. The profiles were analyzed using a two-state denaturation model (α helix to random coil), and the apparent melting temperature (Tm) was determined from the peak of differential melting curves d[Θ222](T)/dT.
Estimation of the Diameter of the Nup54·Nup58 Ring
The diameter of the midplane ring was estimated from the structure dimensions with the approximation that the heterotetramer overlap length is not changed significantly during the spiral-to-ring transition. In the Nup54·Nup58 ring, overlapped heterotetramers form the vertices of a 16-sided regular polygon. The side length of the polygon is the length of a heterotetramer (105Å), reduced by 1/2 overlap on both ends of the tetramer (in total, 34Å; Figure 6A) and therefore amounts to 71Å. The smallest diameter of the 16-sided polygon, which is the distance between opposite vertices, is calculated as: diameter = side length/sin(11.25). With a side length of 71Å, the diameter is 36.4 nm.
Supplementary Material
ACKNOWLEDGMENTS
We thank Janisha Biyanwila Kukreja for outstanding technical assistance; Elias Coutavas for help in writing the manuscript; David King (HHMI, UC Berkeley) for mass spectrometry analysis; K. Rajashankar, Frank Murphy (APS, Chicago), and Corie Ralston (ALS, Berkeley) for excellent beamline support; Scott Banta, John Hunt, Chi Wang (Columbia University, NY), and The Rockefeller University spectroscopy and structural biology resource centers for access to instrumentation; Junseock Koh for CD-related and Sigrid Kübler (Wyatt) for helpful scientific discussions; Joe Alexander for artwork; and Elias Coutavas, Junseock Koh, Vivien Nagy, and Dragana Rogulja for critical reading of the manuscript. G.B. is an Investigator of the Howard Hughes Medical Institute.
S.R.S., R.C., G.B., and I.M. designed experiments, analyzed data, and wrote the paper. S.R.S. and I.M. performed research on the Nup54·Nup58 complex, and R.C. and I.M. performed research on the Nup62·Nup54 complex.
Footnotes
ACCESSION NUMBERS Coordinates and structure factors have been deposited in the Protein Data Bank with accession codes 3T97 (Nup62-2·Nup54-2) and 3T98 (Nup54-3·Nup58-2).
SUPPLEMENTAL INFORMATION Supplemental information includes Extended Experimental Procedures, seven figures, three tables, and three movies and can be found with this article online at doi:10.1016/j.cell.2011.09.034.
REFERENCES
- Akey DL, Malashkevich VN, Kim PS. Buried polar residues in coiled-coil interfaces. Biochemistry. 2001;40:6352–6360. doi: 10.1021/bi002829w. [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Bailer SM, Balduf C, Hurt E. The Nsp1p carboxy-terminal domain is organized into functionally distinct coiled-coil regions required for assembly of nucleoporin subcomplexes and nucleocytoplasmic transport. Mol. Cell. Biol. 2001;21:7944–7955. doi: 10.1128/MCB.21.23.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Muncher L, Straussberg R, Pasmanik-Chor M, Yahav M, Rainshtein L, Walsh CA, Magal N, Taub E, Drasinover V, et al. Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Ann. Neurol. 2006;60:214–222. doi: 10.1002/ana.20902. [DOI] [PubMed] [Google Scholar]
- Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Blobel G. Three-dimensional organization of chromatids by nuclear envelope-associated structures. Cold Spring Harb. Symp. Quant. Biol. 2010;75:545–554. doi: 10.1101/sqb.2010.75.004. [DOI] [PubMed] [Google Scholar]
- Buss F, Stewart M. Macromolecular interactions in the nucleoporin p62 complex of rat nuclear pores: binding of nucleoporin p54 to the rod domain of p62. J. Cell Biol. 1995;128:251–261. doi: 10.1083/jcb.128.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FHC. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay DR, Meier E, Bradley P, Horecka J, Forbes DJ. A complex of nuclear pore proteins required for pore function. J. Cell Biol. 1991;114:169–183. doi: 10.1083/jcb.114.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J. Mol. Biol. 2010;395:578–586. doi: 10.1016/j.jmb.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Gall JG. Octagonal nuclear pores. J. Cell Biol. 1967;32:391–399. doi: 10.1083/jcb.32.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pané N, Shevchenko A, Mann M, Forbes D, Hurt E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell. 1997;8:2017–2038. doi: 10.1091/mbc.8.10.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan T, Müller S, Klier G, Panté N, Blevitt JM, Haner M, Paschal B, Aebi U, Gerace L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol. Biol. Cell. 1995;6:1591–1603. doi: 10.1091/mbc.6.11.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Gerace L. cDNA cloning and analysis of the expression of nucleoporin p45. Gene. 1998;221:245–253. doi: 10.1016/s0378-1119(98)00467-3. [DOI] [PubMed] [Google Scholar]
- Hu T, Guan T, Gerace L. Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J. Cell Biol. 1996;134:589–601. doi: 10.1083/jcb.134.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isgro TA, Schulten K. Binding dynamics of isolated nucleoporin repeat regions to importin-beta. Structure. 2005;13:1869–1879. doi: 10.1016/j.str.2005.09.007. [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg MW, Allen TD, Akey CW. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J. Cell Sci. 1998;111:223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- Kita K, Omata S, Horigome T. Purification and characterization of a nuclear pore glycoprotein complex containing p62. J. Biochem. 1993;113:377–382. doi: 10.1093/oxfordjournals.jbchem.a124054. [DOI] [PubMed] [Google Scholar]
- Lau SY, Taneja AK, Hodges RS. Synthesis of a model protein of defined secondary and quaternary structure. Effect of chain length on the stabilization and formation of two-stranded α-helical coiled-coils. J. Biol. Chem. 1984;259:13253–13261. [PubMed] [Google Scholar]
- Lim RY, Fahrenkrog B, Köser J, Schwarz-Herion K, Deng J, Aebi U. Nanomechanical basis of selective gating by the nuclear pore complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- Lumb KJ, Kim PS. A buried polar interaction imparts structural uniqueness in a designed heterodimeric coiled coil. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, van den Bogaart G, Lusk CP, Poolman B, Veenhoff LM. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 2011;333:90–93. doi: 10.1126/science.1205741. [DOI] [PubMed] [Google Scholar]
- Melčák I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- Miller BR, Powers M, Park M, Fischer W, Forbes DJ. Identification of a new vertebrate nucleoporin, Nup188, with the use of a novel organelle trap assay. Mol. Biol. Cell. 2000;11:3381–3396. doi: 10.1091/mbc.11.10.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito T, Tanaka T, Becker LE, Takashima S, Tanaka J. Infantile bilateral striatal necrosis. Clinicopathological classification. Arch. Neurol. 1986;43:677–680. doi: 10.1001/archneur.1986.00520070035014. [DOI] [PubMed] [Google Scholar]
- Panté N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol. Biol. Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL, Jr., Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol. Cell. 2008;29:46–55. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Unwin PNT, Milligan RA. A large particle associated with the perimeter of the nuclear pore complex. J. Cell Biol. 1982;93:63–75. doi: 10.1083/jcb.93.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.