Abstract
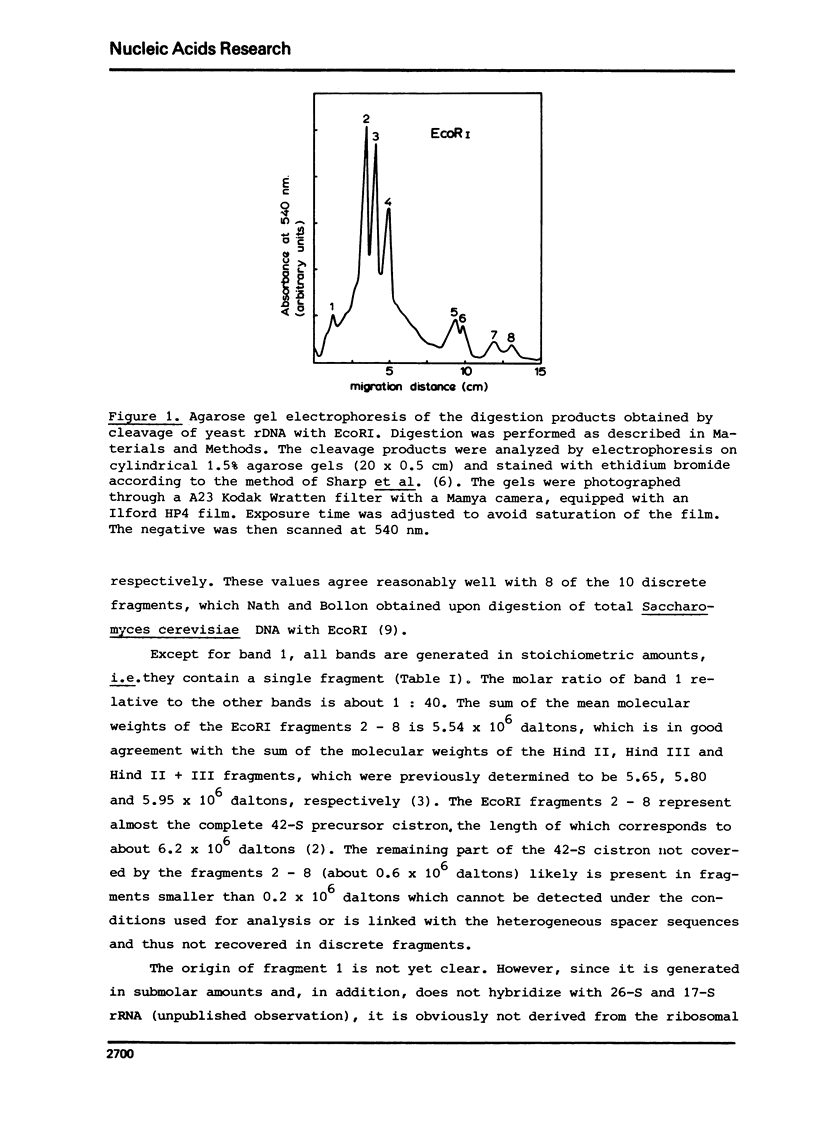
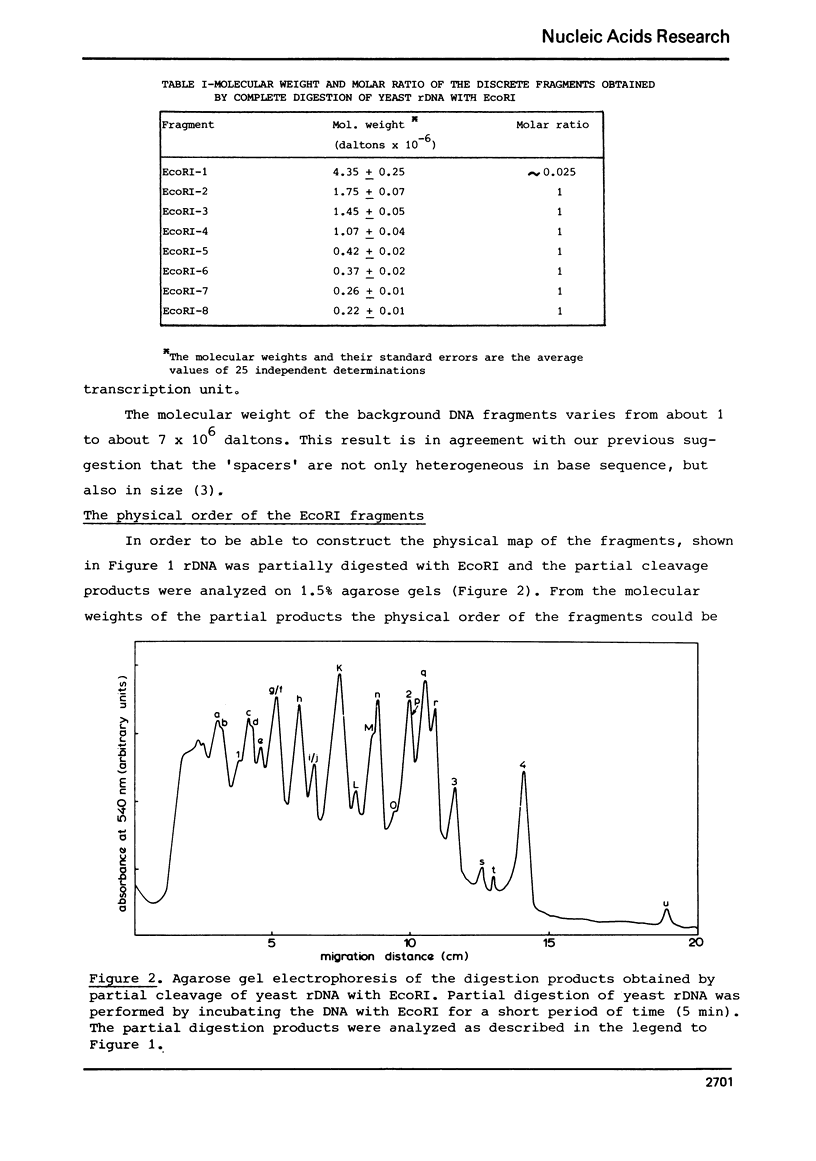
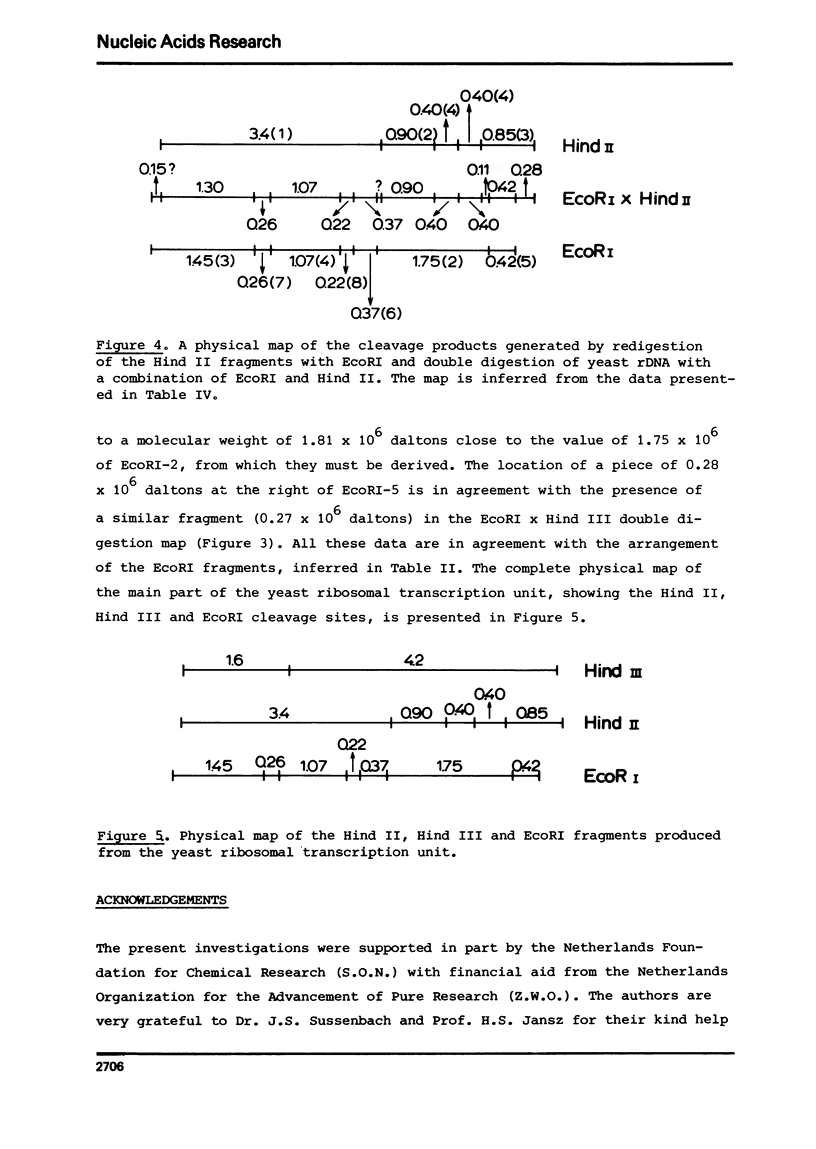
Yeast ribosomal DNA (rDNA) was digested with the restriction endonuclease EcoRI. Eight distinct fragments were obtained with a molecular weight of 4.35 (1), 1.75 (2), 1.45 (3), 1.07 (4), 0.42 (5), 0.37 (6), 0.26 (7) and 0.22 x 10(6) (8) daltons, respectively. Except for fragment 1 with a molecular weight of 4.35 x 10(6) daltons, all fragments are derived from the multiple ribosomal transcription units. The 'spacer' sequences, on the other hand, gave rise to digestion products which are very heterogeneous in size. By analysis of the partial digestion products which are very heterogeneous in size. By analysis of the partial digestion products, together with the data obtained by digestion with a combination of two restriction enzymes (EcoRI and Hind II or Hind III) and redigestion of the Hind II-and Hind III-fragments with EcoRI, the physical map of the EcoRI cleavage sites in the ribosomal transcription unit could be established.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Meyerink J. H., Retèl J., Planta R. J., Heidekamp F. Topographical analysis of yeast ribosomal DNA by cleavage with restriction endonucleases. Mol Biol Rep. 1976 Apr;2(5):393–400. doi: 10.1007/BF00366261. [DOI] [PubMed] [Google Scholar]
- Nath K., Bollon A. P. Generation of discrete yeast DNA fragments by endonuclease RI. Nature. 1975 Sep 11;257(5522):155–157. doi: 10.1038/257155a0. [DOI] [PubMed] [Google Scholar]
- Retèl J., Planta R. J. Nuclear satellite DNAs of yeast. Biochim Biophys Acta. 1972 Oct 27;281(3):299–309. doi: 10.1016/0005-2787(72)90442-x. [DOI] [PubMed] [Google Scholar]
- Retèl J., Van Keulen H. Characterization of yeast ribosomal DNA. Eur J Biochem. 1975 Oct 1;58(1):51–57. doi: 10.1111/j.1432-1033.1975.tb02347.x. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- van den Bos R. C., Planta R. J. Studies on the role of rapidly labeled 20-S RNA in the biosynthesis of ribosomal RNA in yeast. Biochim Biophys Acta. 1971 Sep 30;247(1):175–180. doi: 10.1016/0005-2787(71)90822-7. [DOI] [PubMed] [Google Scholar]



