Abstract
Background
The pbr resistance operon from Cupriavidus metallidurans CH34 plasmid pMOL30 confers resistance to Pb(II) salts, and is regulated by the Pb(II) responsive regulator PbrR, which is a MerR family activator. In other metal sensing MerR family regulators, such as MerR, CueR, and ZntR the cognate regulator binds to a promoter with an unusually long spacer between the −35 and −10 sequences, and activates transcription of resistance genes as a consequence of binding the appropriate metal. Cysteine residues in these regulators are essential for metal ion coordination and activation of expression from their cognate promoter. In this study we investigated the interaction of PbrR with the promoter for the structural pbr resistance genes, PpbrA, effects on transcriptional activation of altering the DNA sequence of PpbrA, and effects on Pb(II)-induced activation of PpbrA when cysteine residues in PbrR were mutated to serine.
Results
Gel retardation and footprinting assays using purified PbrR show that it binds to, and protects from DNase I digestion, the PpbrA promoter, which has a 19 bp spacer between its −35 and −10 sites. Using β-galactosidase assays in C. metallidurans, we show that when PpbrA is changed to an 18 bp spacer, there is an increase in transcriptional activation both in the presence and absence of Pb(II) salts up to a maximum induction equivalent to that seen in the fully-induced wild-type promoter. Changes to the −10 sequence of PpbrA from TTAAAT to the consensus E. coli −10 sequence (TATAAT) increased transcriptional activation from PpbrA, whilst changing the −10 sequence to that of the Tn501 mer promoter (TAAGGT) also increased the transcriptional response, but only in the presence of Pb(II). Individual PbrR mutants C14S, C55S, C79S, C114S, C123S, C132S and C134S, and a double mutant C132S/C134S, were tested for Pb(II) response from PpbrA, using β-galactosidase assays in C. metallidurans. The PbrR C14S, C79S, C134S, and C132S/C134S mutants were defective in Pb(II)-induced activation of PpbrA.
Conclusions
These data show that the metal-dependent activation of PbrR occurs by a similar mechanism to that of MerR, but that metal ion coordination is through cysteines which differ from those seen in other MerR family regulators, and that the DNA sequence of the −10 promoter affects expression levels of the lead resistance genes.
Keywords: Metal-resistance, Metal-protein interactions, Metalloregulation, Bacterial gene expression
Background
Lead (Pb) is a widely distributed, environmentally persistent, toxic metal. Most bacteria that are tolerant or resistant to lead either precipitate Pb in an insoluble form, or actively export it [1]. Although some metal efflux ATPases, such as ZntA from Escherichia coli, and CadA from Staphylococcus aureus plasmid pI258, can export Pb(II) as well as Zn(II) and Cd(II) [2,3], the only characterized bacterial Pb(II) specific resistance system is from Cupriavidus (formerly Wautersia and Ralstonia) metallidurans CH34 [4,5] - a Gram-negative, multiply metal-resistant, β-proteobacterium originally isolated from a decantation basin at a Belgian zinc production plant (and originally identified as Alcaligenes eutrophus CH34; [6]). Over 150 genes in CH34 are involved in metal resistance, of which at least 70 are carried on the plasmids pMOL28 (171 kb) or pMOL30 (234 kb), and the remainder are carried on the 3.92 Mb chromosome or on a 2.58 Mb second chromosome [7]. Plasmid pMOL30 carries the czc (Cd(II), Zn(II), Co(II)), mer (Hg(II)), sil (Ag(I)), cop (Cu(II)) and pbr Pb(II) resistance operons [4,8].
The pbr lead resistance operon from pMOL30 was originally predicted to contain structural genes which encode PbrT, a putative Pb(II) uptake protein belonging to the ILT (Iron Lead Transporter) family [9], a P-type efflux ATPase (PbrA), a predicted inner–membrane protein (PbrB), a predicted prelipoprotein signal peptidase PbrC and a Pb(II) binding protein, PbrD. The regulator of the pbr operon was shown to be PbrR, which is a MerR family regulator [4,10] Subsequent work has shown that the pbr operon also contains an interrupted orf; pbrU upstream of pbrT[11,12] which is predicted to encode a putative inner membrane (Major Facilitator Family MFS1) permease gene, which is probably inactive, but still part of the pbr operon; and that PbrB/PbrC is a fusion protein [11,12], and encodes an inner membrane bound undecaprenyl pyrophosphate (C55-PP) phosphatase [5]. The pbr operon contains a predicted MerR-like promoter from which pbrRTU are transcribed on one DNA strand, and the pbrABCD genes are transcribed as a polycistronic message on the other [4,12]. The most recent work on the mechanism of lead resistance encoded by the pMOL30 pbr operon has proposed a model where Pb2+ induces expression of the pMOL30-encoded PbrABCD by PbrR, as well as expression of zinc and cadmium efflux ATPase homologs ZntA and CadA which are carried on the chromosome or second chromosome. Each of these three ATPases is involved in exporting Pb2+ into the periplasm where inorganic phosphates produced by PbrB are involved in precipitating Pb2+ as insoluble lead phosphate. This model finds no role for PbrT, C, and D, yet some reports suggest PbrC may be required for the maturation or activity of phosphatase in the periplasm[5]. PbrR from pMOL30 (Rmet_5946) is related to several other PbrR-like regulators that have been identified in the C. metallidurans CH34 chromosome, including pbrR2 (Rmet_2303 also known as pbr691[13,14] which is believed to regulate a cadA and a pbrC homolog on the chromosome, and pbrR3 (Rmet_3456 also known as pbr710) believed to regulate a zntA homolog on the second chromosome, both of which are believed to be involved in Pb2+ export [12]. There is evidence for only very low levels of cross-regulation of the pMOL30 PpbrA promoter by PbrR2 or PbrR3 [15].
Other metal-sensing MerR family members include those responding to cadmium (CadR; [16,17]), copper (CueR; [18-20], ActP; [21], SctR; [22]), zinc (ZntR, [23,24]; ZccR (Zn, Co, Cd), [25]) and gold (GolS, [26]). Metal-sensing MerR family regulators share many common features: they bind to and activate gene expression from promoters with unusually long spacer sequences of 19-20 bp between the −35 and −10 sequences, and contain cysteine and other amino acids that are essential in coordinating metals and activating gene expression [10,16,20,27-29].
The objectives of this study were to 1) Characterize the interaction between PbrR and the pbrA promoter, and study the effects on transcription of shortening the 19 bp spacer between the −35 and −10 sequences, and altering the −10 sequence of PpbrA; and 2) to investigate the importance of cysteine residues in PbrR activation of PpbrA in response to Pb(II) ions. To this end each of the cysteine residues in PbrR (C14, C55, C79, C114, C123, C132 and C134) were individually changed to serine residues and a double mutant (C132S, C134S) was created. The effects of these mutations on in vivo transcriptional activation in response to Pb(II) were determined in C. metallidurans using β-galactosidase assays.
Methods
Bacterial strains, plasmids and growth media
Bacterial strains and plasmids used in this study are shown in Table 1. Escherichia coli strains were grown in LB broth [30] at 37°C. C. metallidurans strains were grown at 30°C in 869 medium, 284 Tris or 284 MOPS medium [4,6]. For β-galactosidase assays of PbrR-regulated PpbrA promoter activity, C. metallidurans strains were grown in 284 MOPS medium [4] minimising any Pb(II) precipitation during growth. C. metallidurans strains were grown in SOB medium without MgSO4[30] prior to electroporation of plasmids, and SOB medium containing MgSO4 after electroporation. Pb(II) induction was achieved by growth in PbNO3, and antibiotics were used at the following concentrations:- for E. coli: carbenicillin (Melford laboratories, UK), 200 μg/ml; chloramphenicol 25 μg/ml; kanamycin, 50 μg/ml and trimethoprim lactate 30 μg/ml (all from Sigma Chemical UK); for C. metallidurans: trimethoprim lactate 500 μg/ml.
Table 1.
Bacterial strains and plasmids
| Bacterial strain | Properties or Genotype | Reference or source |
|---|---|---|
|
E. coli |
|
|
| TG2 |
supE hsdΔ5 thiΔ (lac-proAB) F' Δ(srl-recA)306::Tn10(Tcr) lacZΔM15 |
[31] |
| BL21(DE3)pLysS |
F-ompT hsdSB (rB- mB-) gal dcm (DE3) pLysS (Cmr) |
Novagen |
|
C. metallidurans |
|
|
| CH34 |
Zn, Cd, Co, Pb, Cu, Hg, Ni and Cr resistance |
[6] |
| AE104 |
Plasmid-cured C. metallidurans strain- sensitive to toxic metals |
[6] |
| Plasmid |
Description |
Reference or source |
| pET32LIC |
Apr Overexpression plasmid for ligation-independent cloning |
Novagen |
| pET32LIC pbrR |
AprpbrR cloned into pET32LIC |
This study |
| pMa5/8 |
Apr Cms Mutagenesis vector |
[32] |
| pMc5/8 |
Aps Cmr Mutagenesis vector |
[32] |
| pMaPbrR/PpbrA |
Apr Cms Mutagenesis vector with pbrR/PpbrA cloned in to it |
This study |
| pMOL1139 |
Kmr, The pbr operon cloned into plasmid pRK415 |
B. Borremans |
| pMU2385 |
Tpr 13.3 kb low copy number lacZ reporter plasmid |
[33] |
| pMUPpbrA |
Tpr pMU2385 containing the PpbrA promoter directing lacZ transcription |
This study |
| pMUPpbrA-1 |
Tpr pMU2385 containing the PpbrA promoter with a 1 bp deletion |
This study |
| pMUPpbrAcon |
Tpr As pMUPpbrA, but −10 sequence changed to E. coli consensus |
This study |
| pMUPpbrAmer |
Tpr As pMUPpbrA, but −10 sequence changed to mer promoter |
This study |
| pMUPbrR/PpbrA |
Tpr, pMU2385 containing pbrR, PpbrA ΔpbrA directing lacZ transcription |
This study |
| pMUPbrRC14S/PpbrA |
As pMUPbrRPpbrA, but PbrR C14S |
This study |
| pMUPbrRC55S/PpbrA |
As pMUPbrRPpbrA, but PbrR C55S |
This study |
| pMUPbrRC79S/PpbrA |
As pMUPbrRPpbrA, but PbrR C79S |
This study |
| pMUPbrRC114S/PpbrA |
As pMUPbrRPpbrA, but PbrR C114S |
This study |
| pMUPbrRC132S/PpbrA |
As pMUPbrRPpbrA, but PbrR C132S |
This study |
| pMUPbrRC134S/PpbrA |
As pMUPbrRPpbrA, but PbrR C134S |
This study |
| pMUPbrRC132,134 S/PpbrA |
As pMUPbrRPpbrA, but PbrR C132S/C134S |
This study |
| pUC21 |
Apr, high copy number cloning vector; ColE1 replicon |
[34] |
| pUK21 |
Kmr, intermediate copy number cloning vector; p15A replicon |
[34] |
| pUK21pbr1 | Kmr, HindIII/SalI pbrR/PpbrA/ΔpbrA from pMOL1139 cloned into pUK21 | This study |
DNA manipulations
DNA manipulations were as described by [30]. Oligonucleotides were synthesized by Alta Bioscience, the University of Birmingham; or MWG Biotech, Germany. The DNA sequence of all mutants and cloned PCR products were confirmed by sequencing using a PE Applied Biosystems Big Dye version 2.0 sequencing kit according to the manufacturer’s protocol, followed by analysis on an ABI 3700 sequencer in the Functional Genomics Laboratory, School of Biosciences, the University of Birmingham. The primers used for sequencing were: pMUforward and pMUreverse, complementary to the sequences flanking the multiple cloning site of pMU2385, and PbrApe for pMapbrR/PpbrA clones (Table 2).
Table 2.
Oligonucleotides used for site directed mutagenesis, and overexpression
| Oligonucleotide | Sequence | Description or reference |
|---|---|---|
|
pbrR C14S |
5’ CCA CCG GGG ATG CGG TGC 3’ |
PbrR mutagenesis primer |
|
pbrR C55S |
5’ CCA GAG ACC GGG AGT GAC G 3’ |
PbrR mutagenesis primer |
|
pbrR C79S |
5’ GAC TTC ACC GGA ATC CTG G 3’ |
PbrR mutagenesis primer |
|
pbrR C114S |
5’ GGC ACC AGA AGA GGC TTC G 3’ |
PbrR mutagenesis primer |
|
pbrR C123S |
5’ GCA GAA TCC CGG ACG ATT G 3’ |
PbrR mutagenesis primer |
|
pbrR C132S |
5’ CGT ATC ACA CAC GGA GTC CGA C 3’ |
PbrR mutagenesis primer |
|
pbrR C134S |
5’ CGT ATC AGA CAC GCA GTC CGA C 3’ |
PbrR mutagenesis primer |
|
pbrR C132S/C134S |
5’ CGT ATC AGA CAC GGA GTC CGA C 3’ |
PbrR mutagenesis primer |
|
pbrApe |
5’ GCG CCA ACC GTG CTC GGT TCT GGG 3’ |
Primer extension/sequencing primer [4] |
| pbrBstEII |
5’ GCG AAT GGT CAC CAC CGG 3’ |
Primer to amplify PpbrA |
| pbrNruI |
5’ GCT TGT CGC GAA TCA GCG 3’ |
Primer to amplify PpbrA |
| pMU forward |
5’ GAT TCT CCC CAC ATC ACC AG 3’ |
Sequencing primer for pMU2385 |
| pMU reverse |
5’ TGC CAG CAT TTC ATA ACC AA 3’ |
Sequencing primer for pMU2385 |
| M13-F |
5’ CGC CAG GGT TTT CCC AGT CAC GAC 3’ |
Sequencing primer for pUK plasmids |
| M13-R |
5’ GAG CGG ATA ACA ATT TCA CAC AGG 3’ |
Sequencing primer for pUK plasmids |
| conpbr: |
5’ CTAGAGGGTTAATCGGCAAC 3’ |
PpbrA mutagenesis primer |
| merpbr: |
5’ CTAGAGGGTGTAAGGTCGGCAAC 3’ |
PpbrA mutagenesis primer |
| -1EcoPbr |
5’ GGG GAA TTC GAA GCT TGC T 3’ (3’ primer) |
PpbrA mutagenesis primer |
| -1CentreBam |
5’ GCC GAT TTA AAC CCT CTA GT 3’ (primer B) |
PpbrA mutagenesis primer |
| -1CentreEco |
5’ CGG CTA AAT TTG GGA GAT CA 3’ (primer A) |
PpbrA mutagenesis primer |
| -1BamPbr |
5’ CAG TAT ACC TAG GCA GCT GG 3’ (5’ primer) |
PpbrA mutagenesis primer |
|
pbrR ATG (LIC) |
5’ GAC GAC GAC AAG ATG AAT ATC CAG ATC GGC GAG C 3’ |
PbrR cloning and overexpression primer |
|
pbrR TAG (LIC) |
5’ GAG GAG AAG CCC GGT CTA GTC GCT TGG ATG GGC 3’ |
PbrR cloning and overexpression primer |
| T7 terminator | 5’ CGA TCA ATA ACG AGT CGC C 3’ | Sequencing primer |
Underlined bases highlight alteration from the wild-type sequence.
PbrR overexpression and purification
The pbrR gene was amplified from pMOL1139 using VentR® DNA polymerase (New England Biolabs) and the primers: pbrRATG (LIC) and pbrRTAG (LIC) (Table 2). The pbrR PCR product was annealed with plasmid pET32LIC (Novagen), according to manufacturers’ recommendations. DNA sequencing using the primer T7 reverse (Table 2) was used to confirm the nucleotide sequence of the cloned fragment. The thioredoxin-PbrR fusion protein was overexpressed in E. coli BL21 (DE3) pLysS, purified and stored under reducing conditions as described in [23]. The thioredoxin- S tag was cleaved from the fusion protein using enterokinase, according to the manufacturer’s protocol (Novagen) and removed using S-tag affinity agarose. PbrR purity was estimated by PAGE analysis. The concentration of the purified protein was determined by Bradford assay [35].
Gel retardation and DNAse I protection assays of PpbrA with PbrR
Gel retardation experiments were as described in [36], with initial experiments to determine PbrR DNA binding using a 1144 bp HindIII/SalI fragment from pMOL1139 containing pbrR, PpbrA and a truncated pbrA (positions 409 and 1553 on the pbr operon) [4] cloned into pUK21 [34] to make plasmid pUKpbr1. pUKpbr1 was digested with NruI/BstEII and end labelled with [γ32P]-dATP for gel retardations. Further gel retardation and footprinting experiments used a 296 bp PpbrA PCR product, amplified from pMOL1139 using the primers pbrBstEII and pbrNruI (Table 2) and labelled using [γ32P]-dATP. DNAse I protection assays of PpbrA with PbrR were as described by [37], using the 296 bp PpbrA promoter PCR product detailed above. The DNA sequence of the region was obtained from the 296 bp PpbrA PCR product using the pbrApe primer (Table 2) [4] and run alongside the DNAase I footprint (Figure 1B).
Figure 1 .
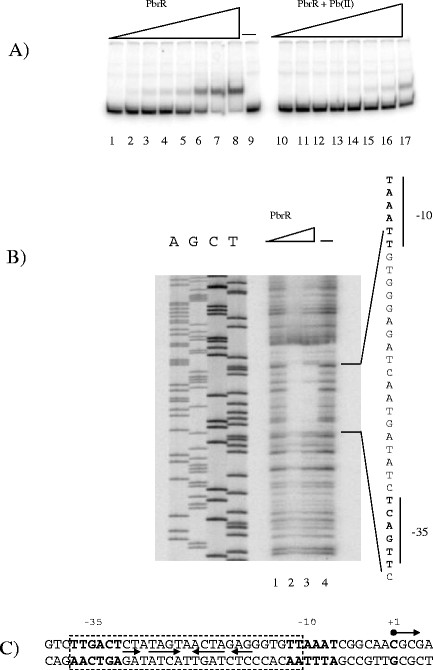
(a) Gel retardation of PpbrAwith PbrR. Each reaction contained the same amount of 32P-end-labelled 296 bp PpbrA PCR product (60 fmol). Lanes 1, 9 and 10 contained no PbrR. PbrR concentrations in lanes 2–8 and 11–17 increase 2-fold from 0.3 to 19.2 pmol. Lanes 10–17 contained 10 μM Pb(II). (b) DNase I protection assay of PbrR bound to the 296 bp PCR product containing the PbrA promoter. Lanes AGCT, DNA sequence of the 296 bp PCR product pbrA promoter, using the pbrApe primer. Lanes 1 and 4, no added pbrR, lane 2 and 3 increasing amounts of added PbrR. (c) Diagram of the PpbrA promoter. The transcript start site is marked in bold and indicated with an arrow [4]. The region of the promoter protected by PbrR from DNAase I digestion is marked with a box. The predicted −35 and −10 sequences are marked in bold, and the dyad symmetrical sequence is marked with arrows.
Cloning of pbrR-PpbrA-ΔpbrA and mutagenesis of the PbrR cysteines
All cloning and mutagenesis work was done in E. coli K-12 TG2. The 1144 bp pbrR-PpbrA-ΔpbrA DNA fragment described above was cloned into pMa5/8 [32] from pUK21pbr1 using the flanking EcoRI and BamHI sites to make pMaPbrR/PpbrA. Gapped duplex mutagenesis of each of the cysteine residues in pbrR was as previously described [32] using the primers pbrRC14S, pbrRC55S, pbrRC79S, pbrRC114S, pbrRC123S, pbrRC132S, pbrRC134S, or pbrRC132S, C134S (Table 2), and mutants verified by DNA sequencing as described [15]. The wild type and mutant pbrR genes on the 1144 bp pbrR-PpbrA-ΔpbrA DNA fragment were individually sub-cloned as EcoRI - BamHI fragments into pMU2385 [33] as described previously [15]. The resulting constructs contained a self-regulating transcriptional unit, with PbrR controlling the transcription of pbrR through PpbrR and regulating transcription of lacZ in pMU2385 on the other DNA strand through PpbrA. These constructs were the basis of the studies of the regulation of PpbrA by PbrR in C. metallidurans AE104.
Cloning and mutagenesis of PpbrA
A 266 bp SphI - NruI fragment containing the PpbrA promoter (positions 1062 and 1328 of the pbr operon) was cloned from pMOL1139, into the HindIII site of pUK21, by rendering the vector and insert blunt-ended using T4 DNA polymerase. The cloned PpbrA DNA fragment was sub-cloned as an EcoRI - BamHI fragment into pMa5/8 for site directed mutagenesis. The −10 sequence of PpbrA was mutated as described above using the primers conpbr and merpbr (Table 2) to change the PpbrA −10 sequence from TTAAAT (wild type) to TATAAT (consensus) or TAAGGT (mer-like). The mutant PpbrA promoters were cloned into pMU2385 using EcoRI and BamHI, creating plasmids pMUPpbrA, pMUPpbrA(con) and pMUPpbrA(mer) in which the pbrA promoter regulates expression of the lacZ gene. After DNA sequencing, the activity of these mutant promoters was assayed in C. metallidurans CH34.
Construction of the PpbrA −1 mutant
Mutagenic PCR [38] of the 1144 bp pbrR-PpbrA-ΔpbrA DNA fragment from pMapbrR/PpbrA was used to construct the −1 promoter mutant of PpbrA, using the primers -1CentreEco and -1CenterBam to introduce the −1 deletion, and primers -1EcoPbr and -1BamPbr as flanking primers (Table 2). The PCR product containing the -1PpbrA promoter was digested with EcoRI and BamHI and subcloned into the multiple cloning site of pMU2385. The DNA sequence of the pbrR-PpbrA-ΔpbrA DNA fragment containing the −1 deletion in PpbrA was confirmed, and this plasmid provided the mutant promoter for the assay in C. metallidurans AE104.
β-galactosidase assays in C. Metallidurans
pMU2385 plasmid constructs were electroporated into C. metallidurans, and cultures containing pMU2385 derivatives were assayed for ß-galactosidase activity as described in [39] with modifications described by [15].
Results
PbrR binds to the pbrA promoter and pb(II) decreases the binding affinity of PbrR to PpbrA in vitro
PbrR was overexpressed as a thioredoxin-his Tag-S tag-fusion protein using the pET32-LIC expression system, purified and released after enterokinase digestion as untagged, full length PbrR, as described in Materials and Methods. The PbrR preparation was estimated as being >95% pure PbrR by Coomassie Blue staining of standard SDS-PAGE gels (data not shown). We had originally identified a candidate PpbrA promoter based on sequence similarity to other MerR family promters, and on run-off transcription studies of the pbr operon [4] and studied PbrR interactions with this region of the pbr operon. Initial PbrR gel retardation assays on 32P-end-labelled DNA from pUK21pbr1, which contained pbrR/PpbrA/ΔpbrA, had been digested with BstEII and NruI showed retardation only of the 282 bp BstEII/NruI DNA fragment containing the previously identified PpbrA promoter region, and no other fragments from the plasmid (data not shown). Addition of PbrR to the end-labelled 296 bp PpbrA PCR product retarded this fragment, and addition of Pb(II) to PbrR and PpbrA increased the amount of PbrR required to retard the PpbrA DNA fragment (Figure 1A) indicating that PbrR-Pb(II) had a lower affinity in vitro with PpbrA than did apo-PbrR did, as is the case with MerR and Hg(II) (reviewed in [10]).
PbrR protects the pbrA promoter from DNAse I digestion in vitro
The 296 bp PpbrA PCR product described above was also used to determine the PbrR binding site on the promoter by DNase I protection assay. Figure 1B shows the autoradiograph of the PbrR DNase I footprint on PpbrA. The region protected by PbrR on PpbrA includes the −35 and −10 sequences as well as the 19 bp spacer containing an imperfect dyad symmetrical sequence between them, and is consistent with DNAse I protection results for MerR, CueR and ZntR [18,20,23,24,40].
The transcription start site [4], the predicted −35 and −10 sites, and the region of the PpbrA promoter protected by PbrR are shown in Figure 1C. The PpbrA promoter has a −35 sequence (TTGACT) that is identical to those for PmerT from Tn501 and PzntA from E. coli K-12 (Figure 2) and shares 5/6 identity with the consensus E. coli −35 sequence. The predicted PpbrA −10 sequence (TTAAAT) has a 4/6 identity to the consensus E. coli −10 sequence (TATAAT) and the spacing between the −35 and −10 sequences is 19 bp, as is the case with other MerR family regulatory regions except ZntR (20 bp; [23]).
Figure 2 .

Alignment of selected promoters for structural genes regulated by MerR family metal responsive regulators: PbrR[4]; MerR[10], ZntR[23], CueR[20]. The −35 and −10 sequences are marked in BOLD. Arrows show dyad symmetrical DNA sequences within the promoters.
Promoter DNA mutations alter PpbrA activity in C. Metallidurans
The importance to promoter functionality of the number of nucleotides between the −35 and −10 sequences of the PpbrA promoter, and the effects of altering the DNA sequence of the PbrR binding site or −10 sequence of PpbrA were investigated using pMUPbrR/PpbrA −1 in C. metallidurans AE104. The PpbrA −1 mutant (Figure 3A), in which the spacer between the −35 and −10 sequences was shortened in such a way that the −35 and −10 sequences were not altered, and the dyad symmetrical sequences in the spacer between the −35 and −10 were retained, showed increased promoter activity in the absence of Pb(II) (Figure 3A) compared to the wild type promoter, but no induction beyond the maximum level seen for the wt promoter with 100 μM Pb(II). These results are similar to those seen for the MerR activated promoter PmerT −1 from Tn501[41], which is constitutively transcriptionally active in both the presence and absence of Hg(II). Changes to the pbrA promoter −10 sequence, so that it more closely resembled the consensus sequence for an E. coli promoter [42], caused up-regulation of PpbrA activity both in the absence and presence of Pb(II). Changes made in PpbrA so that it resembled the Tn501 merT promoter −10 sequence resulted in promoter activity remaining repressed in the absence of Pb(II), but strongly induced in its presence to expression levels 5-fold higher than the wild-type pbrA promoter (Figure 3B). These differences in promoter sequence are likely to alter RNA polymerase binding to the promoter, which could in turn affect the structure of the PbrR-RNA polymerase-DNA ternary complex.
Figure 3 .
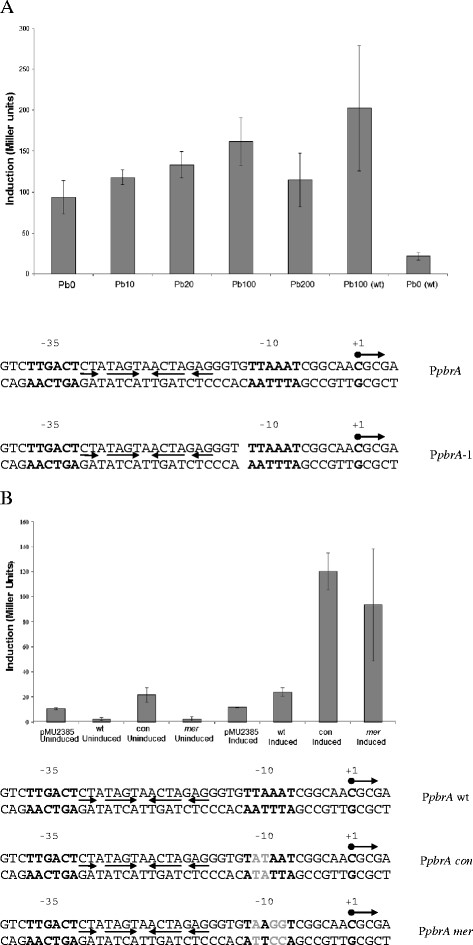
(A) β-galactosidase assay measurement of the activation of PpbrA, containing a 1 nt deletion in the 19 bp promoter spacer, to increasing levels of Pb(II) inC. metalliduransAE104 carrying pMUPbrRpbrA-1. Micromolar Pb(II) concentrations are indicated by the suffix to Pb on the abscissa. Pb0 contains no added Pb(II), Pb200 contains 200 μM Pb(II) . The sequence of wild-type PpbrA and the −1 mutant PpbrA are shown below the graph. The −35 and −10 sequences are marked in BOLD. Arrows show dyad symmetrical DNA sequences within the promoters. (B) β-galactosidase assay measurement of the activation of −10 sequence mutant PpbrA clones in pMU2385 in response to no added Pb(II) or 100 μM Pb(II). WT denotes wild-type −10 sequence (TTAAAT), CON denotes the E. coli consensus promoter −10 sequence (TATAAT) and MER the Tn501 PmerT promoter −10 sequence (TAAGGT). The sequences of the wild-type (PpbrA wt), consensus (PpbrA con), and PmerT-like promoters (PpbrA mer) are shown below the graph. The −35 and −10 sequences are marked in BOLD. Arrows show dyad symmetrical DNA sequences within the promoters, and altered bases are marked in Gray.
Cysteines 14, 79 and 134 in PbrR are essential for pb(II) responsive transcription from PpbrA in C. Metallidurans AE104
pMUPbrR/PpbrA derivatives carrying PbrR cysteine mutants (C14S, C55S, C79S, C114S, C123S, C132S, C134S, and C132S/C134S) (Table 1) were assayed for Pb(II) –dependent induction of the pbrA promoter in C. metallidurans AE104, which did not carry pMOL28 or pMOL30. These were grown in a sublethal concentration of Pb(II) (20 μM) which was sufficient to activate expression from PpbrA, without affecting growth of the Pb(II) sensitive AE104 strain. β-galactosidase assays of wild type and cysteine mutant PbrR responses to Pb(II) in C. metallidurans AE104 (Figure 4) showed cysteines C14, C79, and C134 were essential for Pb(II) induced transcriptional activation of PpbrA by PbrR. The double mutant C132S, C134S also lost Pb(II) induced activation of transcription from PpbrA, consistent with the result for the single C134S mutant.
Figure 4 .
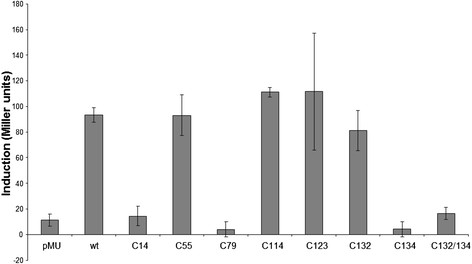
β-galactosidase assays inC. metalliduransAE104 of PpbrAactivation in response to 20 μM Pb(II) on wild-type PbrR and its cysteine mutants in pMUPbrR/PpbrA.
Discussion
PbrR is a member of the MerR family of regulators which sense metals and other environmental stimuli, and activate gene expression in response to these signals. The archetype of the family, MerR, regulates both its own expression and expression of the mercuric ion resistance genes in the polycistronic mer operon from a divergent promoter: Pmer. MerR activates expression of the structural genes at the PmerT operator/promoter (o/p) site, which has an unusually long spacer of 19 bp between the −35 and −10 sequences of the promoter (compared to the consensus E. coli σ70 promoter spacing of 16-18 bp [10]). The MerR dimer binds to a dyad-symmetrical DNA sequence within the spacer, and when three essential cysteine residues (C89, C117 and C126) in the MerR dimer coordinate to a mercuric ion in a trigonal coordination [28,29] bridging between each MerR homodimer, this change in MerR homodimer interaction is transmitted to the promoter, causing an allosteric underwinding of ~33O of the DNA at the o/p site, which realigns the −35 and −10 sequences of the promoter so that σ70 RNA polymerase can contact the promoter sequences forming the transcription open complex [43,44].
PbrR from C. metallidurans CH34 plasmid pMOL30 binds to and protects from DNAase I digestion the predicted PpbrA operator/promoter (Figure 1) (4). PpbrA has striking similarities to other metal ion-responsive MerR family promoters (Figure 2). Assays of PpbrA mutants where the spacing between the −10 and −35 sites are shortened to 18 bp, whilst the internal dyad symmetry is maintained, showed that PbrR-induced expression from PpbrA is upregulated even in the absence of Pb(II) (Figure 3). These data are all consistent with the model of activation for the MerR promoter [41,43,44]. Change of the DNA sequence of the −10 element of PpbrA to either the consensus E. coli promoter −10 sequence or the Tn501 PmerT promoter −10 sequence also caused up-regulation of promoter activity, although the PpbrA/Tn501 PmerT-like promoter still retained Pb(II) repression and induction, rather than a constitutive up-regulation seen in the −10 consensus promoter mutant. These data emphasize the importance of individual nucleotides within the promoter in affecting promoter strength, and indicate that PpbrA is suboptimal for maximum induction of the structural pbr genes. It is possible that this may represent a mechanism for fine-tuning of expression of the pbr structural genes.
In other metal ion-sensing MerR family regulators, cysteine residues are essential for metal coordination and functionality. In vivo assays of the activity of cysteine to serine mutant PbrR proteins in C. metallidurans AE104 (which lacks pMOL30) have shown that C14, C79 and C134 are essential for PbrR Pb(II) sensing and activation of PpbrA (Figure 4). PbrR C14 lies in the turn of the predicted helix-turn-helix DNA binding domain of PbrR (Figure 5) and a change of amino acid at this point could disrupt the binding of PbrR to PpbrA. Mutants in the second helix of this region of MerR have lost both activation and repression activity [45,46]. The loss of Pb(II) response in the PbrR C79S mutant is consistent with the prediction from a structure-based sequence alignment that this residue is essential for discriminating between +1 and +2 charge ions, with a cysteine being found at this position in regulators that respond to +2 ions [27]. Mutagenesis studies have all identified a cysteine residue at this position as being essential for in vivo metal-dependant activation of expression in MerR, ZntR, and ZccR.
Figure 5 .
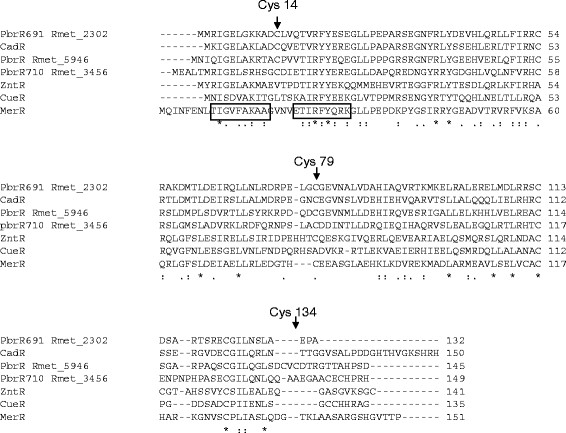
ClustalW[47,48]alignment of metal sensing MerR regulators. PbrR (Rmet_5946), PbrR691 (Rmet_2302) and PbrR710 (Rmet_3456) are from the genome of C. metallidurans CH34. CadR is from Pseudomonas stutzeri A1501. ZntR, and CueR are from the E. coli K-12 genome, and MerR is from Tn501. The helices of the Helix-Turn-Helix DNA binding domain are boxed. Essential cysteine residues (Cys14, Cys79, and Cys134 –PbrR numbering) required for activation of PpbrA by PbrR are marked. Key to symbols: * = residues in that column are identical in all sequences in the alignment. The symbol : = conserved substitutions have been observed, and the symbol . = semi-conserved substitutions are observed.
C134 in PbrR (Rmet_5496) is also essential for Pb(II) response and is part of a CVC (CXC) motif which is often found in PbrR regulators associated with orthologs of PbrABC, but not in the PbrR homologues PbrR2 (PbrR691 Rmet_2302) and PbrR3 (PbrR710 Rmet_3456), or CadR (Figure 5). A CVC motif is also found in the CadC repressor: alterations of either cysteine in this motif in CadC reduced or abolished sensing of Pb(II), Cd(II) and Zn(II) [49] and both cysteines are required for metal coordination [50,51]. Although C79 and C134 of the PbrR homodimer are essential for Pb(II) induction of PpbrA, the C132S mutant shows only a slightly reduced, not abolished, response to Pb(II). Pb(II) has been shown to have a preference for binding to cysteine residues in a tri-coordinate Pb(II)-thiol conformation [52], and Chen and coworkers have reported that the PbrR-related PbrR691 (PbrR2, Rmet_2302) regulator from the C. metallidurans genomic island 1 coordinates Pb(II) via 3 (possibly 4) cysteine coordination [14]. Pb(II) has been shown to coordinate in biological systems via a distorted trigonal planar geometry involving S and N coordination in a biomimetic N2S (alkylthiolate) compound [53], and the Pb(II), Cd(II) and Zn(II) response of the S. aureus pI258 cadmium resistance repressor CadC is dependent on three cysteine residues [49,54]. DNA footprinting suggests that like MerR, PbrR functions as a homodimer. It is possible that Pb(II) may coordinate to cysteine and histidine (or other N- side chain amino acid) residues or O-containing side chain amino-acid residues in the PbrR homodimer and C79 could provide the ligand for metal bridging between the homodimers, and in current models is thought to be necessary to trigger DNA underwinding at the regulated promoter [27]. There are histidine, glutamine, lysine and arginine residues in PbrR close to the metal-binding domain (Figure 5). In ZntR, each homodimer coordinates two zinc atoms per metal binding domain (MBD), one via C114 and C124 of the MBD, and C79 from the other monomer, whilst the other zinc atom is coordinated to C115 and H119 of the MBD, and C79 from the other monomer and both zinc atoms also coordinate to oxygen from a bridging phosphate [27,54]. Structural studies are required to understand further how Pb(II) coordinates to PbrR.
We cannot exclude the possibility that the PbrR C79S and C134S mutants we have made may have altered DNA-binding features, which may account for loss of Pb(II) response. However, mutants in the MBD of other MerR family regulators do not, but mutants in the helix-turn helix domain of these regulators do [45,46].
Conclusion
The metal-responsive MerR family transcription activators can be classified into groups which sense Hg, or Cu/Ag/Au, or Zn/Cd/Pb, and several other phylogenetically-related but uncharacterized regulator clusters [55]. PbrR (Rmet_5946) and the related PbrR691 (R_met 2302) are unusual amongst the phylogenetic cluster of related Zn(II)/Cd(II)/Pb(II)-sensing MerR family regulators that have been tested for metal specificity, because they exclusively respond to Pb(II) in plasmid based assays in C. metallidurans (PbrR: [15,56]) or using FRET (PbrR691, [13]) without any transcriptional response to Zn or Cd, whereas related MerR family regulators that have been tested respond to a greater or lesser extent to Zn(II), Cd(II) and Pb(II) [10,23,57], as do SmtB/ArsR family repressors [47,54]. However, transcriptomics experiments indicate that the pbr structural genes are also induced in the presence of other metals, arguing that expression of the pbr operon and other metal resistance operons in C. metallidurans is influenced by other factors [7,12].
Our experiments show that the mechanism of transcriptional activation by PbrR appears to be essentially identical to that of MerR family regulators that have been characterized. PbrR contains three cysteine residues that are necessary for Pb(II)-induced transcription from the pbrA promoter. C14 is in the helix-turn-helix DNA binding domain, and may be essential for the regulator/DNA interaction. C79 is essential in all divalent metal ion responsive MerR regulators tested so far, whilst C134 is not found in other characterized MerR regulators. Our data show that PbrR transcription is activated by Pb(II) using different amino acids to other divalent metal ion-activated MerR regulators, but further work is required to determine whether Pb(II) coordinates other residues in PbrR.
Abbreviations
Tp, Trimethoprim; Ap, Ampicillin; Km, Kanamycin.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JLH and DJJ carried out the experimental studies. JLH drafted the manuscript. NLB conceived and coordinated the study. All authors read and approved the manuscript.
Contributor Information
Jon L Hobman, Email: jon.hobman@nottingham.ac.uk.
Daniel J Julian, Email: Daniel.Julian@kedst.ac.uk.
Nigel L Brown, Email: Nigel.Brown@ed.ac.uk.
Acknowledgements
We gratefully acknowledge the contribution of Niels van der Lelie and Brigitte Borremans to the start of this project and to Max Mergeay for advice and training to DJJ. We thank Chris Kershaw for critical reading of the manuscript. This work was supported by the Biotechnology and Biological Sciences Research Council (research grant B10333 and a studentship to DJJ). The Birmingham Functional Genomics laboratory was supported by a Joint Infrastructure Fund grant JIF13209 and bioinformatics facilities were provided through MRC Infrastructure Award G.4600017.
References
- Mire CE, Tourjee JA, O'Brien WF, Ramanujachary KV, Hecht GB. Lead precipitation by Vibrio harveyi: evidence for novel quorum-sensing interactions. Appl Environ Microbiol. 2004;70:855–864. doi: 10.1128/AEM.70.2.855-864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing C, Sun Y, Mitra B, Rosen BP. Pb(II)-translocating P-type ATPases. J Biol Chem. 1998;49:32614–32617. doi: 10.1074/jbc.273.49.32614. [DOI] [PubMed] [Google Scholar]
- Sharma R, Rensing C, Rosen BP, Mitra B. The ATP hydrolytic activity of purified ZntA, a Pb(II)/Cd(II)/Zn(II)-translocating ATPase from Escherichia coli. J Biol Chem. 2000;275:3873–3878. doi: 10.1074/jbc.275.6.3873. [DOI] [PubMed] [Google Scholar]
- Borremans B, Hobman JL, Provoost A, Corbisier P, Brown NL, van der Lelie D. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol. 2001;183:5651–5658. doi: 10.1128/JB.183.19.5651-5658.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynninen A, Touzé T, Pitkänen L, Mengin-Lecreulx D, Virta M. An efflux transporter PbrA and a phosphatase PbrB cooperate in a lead-resistance mechanism in bacteria. Mol Microbiol. 2009;74:384–394. doi: 10.1111/j.1365-2958.2009.06868.x. [DOI] [PubMed] [Google Scholar]
- Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monchy S, Benotmane MA, Wattiez R, van Aelst S, Auquier V, Borremans B, Mergeay M, Taghavi S, van der Lelie D, Vallaeys T. Transcriptomic and proteomic analyses of the pMOL30-encoded copper resistance in Cupriavidus metallidurans strain CH34. Microbiology. 2006;152:1765–1776. doi: 10.1099/mic.0.28593-0. [DOI] [PubMed] [Google Scholar]
- Mergeay M, Monchy S, Vallaeys T, Auquier V, Benotmane A, Bertin P, Taghavi S, Dunn J, van der Lelie D, Wattiez R. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol Rev. 2003;27:385–410. doi: 10.1016/S0168-6445(03)00045-7. [DOI] [PubMed] [Google Scholar]
- Debut AJ, Dumay QC, Barabote RD, Saier MH. The iron/lead supertransporter family of Fe3+/Pb2+ uptake systems. J Mol Microbiol Biotechnol. 2006;11:1–9. doi: 10.1159/000092814. [DOI] [PubMed] [Google Scholar]
- Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- Monchy S, Benotmane MA, Janssen P, Vallaeys T, Taghavi S, van der Lelie D, Mergeay M. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximum viable response to heavy metals. J Bacteriol. 2007;189:7417–7425. doi: 10.1128/JB.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi S, Lesaulnier C, Monchy S, Wattier R, Mergeay M, van der Lelie D. Lead (II) resistance in Cupriavidus metallidurans CH34: interplay between plasmid and chromosomally-located functions. Anthonie van Leeuwenhoek. 2009;96:171–182. doi: 10.1007/s10482-008-9289-0. [DOI] [PubMed] [Google Scholar]
- Chen P, Greenberg B, Taghavi S, Romano C, van der Lelie D, He C. An exceptionally selective lead(II)-regulatory protein from Ralstonia metallidurans: Development of a fluorescent lead(II) probe. Angew Chem Int Ed. 2005;44:2715–2719. doi: 10.1002/anie.200462443. [DOI] [PubMed] [Google Scholar]
- Chen PR, Wasinger EC, Zhao J, van der Lelie D, Chen LX, He C. Spectroscopic insights into lead (II) coordination by the selective lead(II)-binding protein PbrR691. J Am Chem Soc. 2007;129:12350–12351. doi: 10.1021/ja0733890. [DOI] [PubMed] [Google Scholar]
- Julian DJ, Kershaw CJ, Brown NL, Hobman JL. Transcriptional activation of MerR family promoters in Cupriavidus metallidurans CH34. Anthonie van Leeuwenhoek. 2009;96:149–159. doi: 10.1007/s10482-008-9293-4. [DOI] [PubMed] [Google Scholar]
- Brocklehurst KR, Megit SJ, Morby AP. Characterisation of CadR from Pseudomonas aeruginosa: a Cd(II)-responsive MerR homologue. Biochem Biophys Res Commun. 2003;308:234–239. doi: 10.1016/s0006-291x(03)01366-4. [DOI] [PubMed] [Google Scholar]
- Lee S-W, Glickman E, Cooksey D. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium transporting ATPase and a MerR family response regulator. Appl Environ Microbiol. 2001;67:697–702. doi: 10.1128/AEM.67.4.1437-1444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outten FW, Outten CE, Hale J, O'Halloran TV. Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, CueR. J Biol Chem. 2000;275:31024–31029. doi: 10.1074/jbc.M006508200. [DOI] [PubMed] [Google Scholar]
- Petersen C, Moller LB. Control of copper homeostasis in Escherichia coli by a P-type ATPase, CopA, and a MerR-like transcriptional activator, CopR. Gene. 2000;261:289–298. doi: 10.1016/s0378-1119(00)00509-6. [DOI] [PubMed] [Google Scholar]
- Stoyanov JV, Hobman JL, Brown NL. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol. 2001;39:502–511. doi: 10.1046/j.1365-2958.2001.02264.x. [DOI] [PubMed] [Google Scholar]
- Reeve WG, Tiwari RP, Kale NB, Dilworth MJ, Glenn AR. ActP controls copper homeostasis in Rhizobium leguminosarum bv. viciae and Sinorhizobium melliloti preventing low-pH induced copper toxicity. Mol Microbiol. 2002;43:981–991. doi: 10.1046/j.1365-2958.2002.02791.x. [DOI] [PubMed] [Google Scholar]
- Kim JS, Kim MH, Joe MH, Song SS, Lee IS, Choi SY. The SctR of Salmonella enterica serovar Typhimurium encoding a homologue of the MerR protein is involved in the copper-responsive regulation of cuiD. FEMS Microbiol Lett. 2002;210:99–103. doi: 10.1111/j.1574-6968.2002.tb11166.x. [DOI] [PubMed] [Google Scholar]
- Brocklehurst KR, Hobman JL, Lawley B, Blank L, Marshall SJ, Brown NL, Morby AP. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol. 1999;31:893–902. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- Outten CE, Outten FW, O'Halloran TV. DNA distortion mechanism for transcriptional activation by ZntR, a Zn(II)-responsive MerR homologue in Escherichia coli. J Biol Chem. 1999;274:37517–37524. doi: 10.1074/jbc.274.53.37517. [DOI] [PubMed] [Google Scholar]
- Kidd SP, Brown NL. ZccR- a MerR-like regulator from Bordetella pertussis, which responds to zinc, cadmium and cobalt. Biochem Biophys Res Comm. 2003;302:697–702. doi: 10.1016/s0006-291x(03)00249-3. [DOI] [PubMed] [Google Scholar]
- Checa SK, Espariz M, Perez Audero ME, Botta PE, Spinelli SV, Soncini FC. Bacterial sensing of and resistance to gold salts. Mol Microbiol. 2007;63:1307–1318. doi: 10.1111/j.1365-2958.2007.05590.x. [DOI] [PubMed] [Google Scholar]
- Changela A, Chen K, Xue Y, Holschen J, Outten CE, O'Halloran TV, Mondragon A. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science. 2003;301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- Helmann JD, Ballard BT, Walsh CT. The MerR Metalloregulatory Protein Binds Mercuric Ion as a Tricoordinate, Metal-Bridged Dimer. Science. 1990;248:946–948. doi: 10.1126/science.2305262. [DOI] [PubMed] [Google Scholar]
- Shewchuk LM, Verdine GL, Nash H, Walsh CT. Mutagenesis of the cysteines in the metalloregulatory protein MerR indicates that a metal-bridged dimer activates transcription. Biochemistry. 1989;28:6140–6145. doi: 10.1021/bi00441a002. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York; 1989. [Google Scholar]
- Gibson T. Studies on the Eppstein-Barr virus genome. University of Cambridge, Cambridge, U.K; 1984. [Google Scholar]
- Stanssens P, Opsomer C, McKeown YM, Kramer W, Zabeau M, Fritz HJ. Efficient oligonucleotide-directed construction of mutations in expression vectors by the gapped duplex DNA method using alternating selectable markers. Nucleic Acid Res. 1989;17:4441–4454. doi: 10.1093/nar/17.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praszkier J, Wilson IW, Pittard AJ. Mutations affecting translational coupling between the rep genes of an IncB miniplasmid. J Bacteriol. 1992;174:2376–2383. doi: 10.1128/jb.174.7.2376-2383.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene. 1991;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- Bradford MM. Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Ansari AZ, Wright JG, Brown NL, O'Halloran TV. Construction and characterization of a mercury-independent MerR activator (MerRAC): transcriptional activation in the absence of Hg(II) is accompanied by DNA distortion. EMBO J. 1993;12:413–421. doi: 10.1002/j.1460-2075.1993.tb05673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savery N, Belyaeva T, Busby S. In: Essential Techniques: Gene Transcription. Docherty K, editor. John Wiley and sons, Chichester; 1996. Protein-DNA interactions; pp. 1–33. [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in Molecular Genetics. Cold spring Harbor Laboratory Press, Cold Spring Harbor, New York; 1972. [Google Scholar]
- O’Halloran TV, Frantz B, Shin MK, Ralston DM, Wright JG. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989;56:119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Brown NL. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acid Res. 1990;18:5157–5162. doi: 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Reynolds RP. Analysis of Escherichia coli promoter sequences. Nucleic Acid Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari AZ, Bradner JE, O'Halloran TV. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995;374:371–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- Ansari AZ, Chael ML, O'Halloran TV. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature. 1995;355:87–89. doi: 10.1038/355087a0. [DOI] [PubMed] [Google Scholar]
- Ross W, Park S-J, Summers AO. Genetic analysis of transcriptional activation and repression in the Tn21 mer operon. J Bacteriol. 1989;171:4009–4018. doi: 10.1128/jb.171.7.4009-4018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewchuk LM, Helmann JD, Ross W, Park S-J, Summers AO, Walsh CT. Transcriptional switching by the MerR protein: activation and repression mutants implicate distinct DNA and mercury (II) binding domains. Biochemistry. 1989;28:2340–2344. doi: 10.1021/bi00431a053. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Hobman JL, Wilkie J, Brown NL. A design for life: prokaryotic metal-binding MerR family regulators. Biometals. 2005;18:429–436. doi: 10.1007/s10534-005-3717-7. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wong MD, Rosen BP. Role of cysteinyl residues in sensing Pb(II), Cd(II), and Zn(II) by the plasmid pI258 CadC repressor. J Biol Chem. 2001;276:14955–14960. doi: 10.1074/jbc.M010595200. [DOI] [PubMed] [Google Scholar]
- Apuy JL, Busenlehner LS, Russell DH, Giedroc DP. Ratiometric pulsed alkylation mass spectrometry as a probe of thiolate reactivity in different metalloderivatives of Staphylococcus aureus pI258 CadC. Biochemistry. 2004;43:3824–3834. doi: 10.1021/bi035668f. [DOI] [PubMed] [Google Scholar]
- Busenlehner LS, Weng T-C, Penner-Hahn JE, Giedroc DP. Elucidation of primary (α3N) and vestigial (α5) heavy metal-binding sites in Staphylococcus aureus pI258 CadC: evolutionary implications for metal ion selectivity of ArsR/SmtB metal sensor proteins. J Mol Biol. 2002;319:685–701. doi: 10.1016/S0022-2836(02)00299-1. [DOI] [PubMed] [Google Scholar]
- Magyar JS, Weng T-C, Stern CM, Dye DF, Rous BW, Payne JC, Bridgewater BM, Mijovilovich A, Parkin G, Zaleski JM, Penner-Hahn JE, Godwin HA. Reexamination of lead(II) coordination preferences in sulphur-rich sites: implications for a critical mechanism of lead poisoning. J Am Chem Soc. 2005;127:9495–9505. doi: 10.1021/ja0424530. [DOI] [PubMed] [Google Scholar]
- Anderson RJ, diTargiani RC, Hancock RD, Stern CL, Goldberg DP, Godwin HA. Characterization of the first N2S(alkylthiolate) lead compound: A model for three-coordinate lead in biological systems. Inorg Chem. 2006;45:6574–6576. doi: 10.1021/ic060497z. [DOI] [PubMed] [Google Scholar]
- Busenlehner LS, Pennella MA, Giedroc DP. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol Rev. 2003;27:131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- Permina EA, Kazakov AE, Kalinina OV, Gelfand MS. Comparative genomics of regulation of heavy metal resistance in eubacteria. BMC Microbiol. 2006;6:49. doi: 10.1186/1471-2180-6-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbisier P, van der Lelie D, Borremans B, Provoost A, de Lorenzo V, Brown NL, Lloyd JR, Hobman JL, Csoregi E, Johansson G, Mattiasson B. Whole cell and protein-based biosensors for the detection of bioavailable heavy metals in environmental samples. Anal Chim Acta. 1999;387:235–244. [Google Scholar]
- Khan S, Brocklehurst KR, Jones GW, Morby AP. The functional analysis of directed amino-acid alterations in ZntR from Escherichia coli. Biochem Biophys Res Commun. 2002;299:438–445. doi: 10.1016/s0006-291x(02)02660-8. [DOI] [PubMed] [Google Scholar]


