Abstract
Background
Giardia duodenalis is a common protozoan parasite of humans and animals. Genetic characterization of single loci indicates the existence of eight groups called assemblages, which differ in their host distribution. Molecular analyses challenged the idea that G. duodenalis is a strictly clonal diplomonad by providing evidence of recombination within and between assemblages. Particularly, inter-assemblage recombination events would complicate the interpretation of multi-locus genotyping data from field isolates: where is a host infected with multiple Giardia genotypes or with a single, recombined Giardia genotype.
Methods
Population genetic analyses on the single and multiple-locus level on an extensive dataset of G. duodenalis isolates from humans and animals were performed.
Results
Our analyses indicate that recombination between isolates from different assemblages are apparently very rare or absent in the natural population of Giardia duodenalis. At the multi-locus level, our statistical analyses are more congruent with clonal reproduction and can equally well be explained with the presence of multiple G. duodenalis genotypes within one field isolate.
Conclusions
We conclude that recombination between G. duodenalis assemblages is either very rare or absent. Recombination between genotypes from the same assemblage and genetic exchange between the nuclei of a single cyst needs further investigation.
Keywords: Giardia lamblia, Giardia intestinalis, Giardia duodenalis, Genetic recombination, Population genetics
Background
Giardia duodenalis (syn. G. lambliaG. intestinalis) is the etiological agent of giardiasis, a gastrointestinal infection of humans, companion animals, livestock and wildlife. Symptoms of a G. duodenalis infection range from asymptomatic to severe diarrhea as well as chronic disease [1]. G. duodenalis has a simple life cycle comprising rapidly multiplying, non-invasive trophozoites on the mucosal surface of the intestine, and the production of environmentally resistant cysts that are shed with the host faces. Infectious cysts are transmitted by the faecal-oral route either by direct contact or by ingestion of contaminated food or water [2]. G. duodenalis is considered as a species complex, whose members show little variation in their morphology, yet can be assigned to eight distinct assemblages (A to H) based on enzyme electrophoretic and genetic studies [3,4]. Assemblages A and B can infect and multiply in humans and are also found in a wide range of mammals. The remaining assemblages show more restricted host ranges: C and D are predominantly found in canids, E in livestock, F in cats, G in rodents and H in marine vertebrates (seal and gull) [5,6]. In endemic areas where humans and animals live closely together, transmission from human to animals or vice versa may occur [7-9]. Direct evidence for transmission from animals to human is lacking, because Giardia cysts are shed into the environment, making it very difficult to determine the primary source of the infection. Genetic characterization has been extensively used to assess the role of animals in the epidemiology of human infection and to develop tools for tracing sources of infection. However, the zoonotic potential of G. duodenalis remains a major and unresolved issue [1,10,11].
Many molecular epidemiological studies have been based on the analysis of a single marker, often from a limited number of isolates. Using single locus approaches, the zoonotic potential of G. duodenalis assemblage A and B appears to be high: Irrespective of the genetic marker used, sequences from human and animal field isolates frequently appeared similar, if not identical [12]. In order to increase the accuracy of genotyping of G. duodenalis isolates, multi-locus sequence typing strategies were introduced [13-18]. When genotypes from field isolates were defined using a multi-locus sequence typing scheme, only 2 from the 84 multi-locus genotypes (MLG) of assemblage A and none MLGs (n = 99) of assemblage B appear to have a zoonotic potential [12]. Surprisingly, the genotypes of Giardia field isolates repeatedly constituted a combination of loci derived from different assemblages [12]. The latter finding can be explained by two biological phenomena.
One explanation is that a Giardia field isolate is not a singular clone, but consists of a mixture of different Giardia genotypes. A Giardia field isolate is often not more than a DNA extract, obtained either directly from a stool sample or indirectly after (immune-) isolation of faecal cysts. For this situation, the uptake of genetically different Giardia cysts from the environment by a host, or subsequent infection of an already infected host, likely without overt symptoms, with a different Giardia genotype, must occur. As starting in vitro cultures from field samples prove to be very difficult due to variations in excitation and adaptation, and the ever-present bacterial and fungal contamination, it is hard to affirm the clonality of Giardia field isolates [19,20]. Alternatively, the Giardia isolates are clonal, but the mixing of loci from different assemblages have arisen by (para) sexual recombination, i.e. genetic exchange, between G. duodenalis assemblages. Although G. duodenalis shows no cytological evidence of meiotic and sexual recombination, several studies challenged the idea that G. duodenalis is a strictly clonal organism [21,22]. These studies have demonstrated: (i) the presence in the G. duodenalis genome of true homologs of genes involved in meiosis in other eukaryotes [23,24]; (ii) the exchange of genetic material in different chromosomal regions among human isolates of the parasite [25,26]; (iii) the fusion between cyst nuclei (karyogamy) and the transfer of genetic material (episomal plasmids) between them [27]. These results are pivotal for the existence of sexual recombination. Recombination may take place at three levels within G. duodenalis: (i) between the two nuclei at an individual level, (ii) between individuals of the same assemblage and (iii) between individuals of different assemblages.
Here, we only address the latter situation, i.e. recombination between assemblages, as this mostly complicates the interpretation of the molecular epidemiological data. Genetic exchange between different assemblages may occur in laboratory cultures or in nature, but it remains to be determined to what extent this occurs in natural populations [28]. Genetic exchange between isolates of different assemblages was addressed using two approaches: The detection of mosaic sequences in three loci and the performance of several tests for clonal reproduction at the population level.
Methods
Data collection
A European network of public and veterinary health Institutions that focused on zoonotic protozoan parasites (the ZOOnotic Protozoa Network, ZOOPNET) was established [12]. Within this consortium, a molecular epidemiological database was built, and currently contains information on 3351 Giardia isolates, which encompass 4954 sequences from 5 different loci. Although GenBank sequences constitute approximately 45% of the database, limited epidemiological data (mainly country and source of isolation) are available for those isolates. All molecular epidemiological data were stored and analyzed in Bionumerics (Version 6.10; Applied Math, Belgium). A selection of these sequences was made using the same strategy as previously described [4] . For example, sequences that were too short to cover regions of variation within any given assemblage were used only for analysis at the level of that assemblage, but not at the level of sub-assemblage. In addition, when multiple, identical sequences from any given isolate were deposited in GenBank, only the longest available sequence was retrieved.
DNA-sequence analysis
The loci beta-giardin (BG), glutamate dehydrogenase (GDH), and triose phosphate isomerase (TPI) were sorted into their different genes, assemblages, and sub-assemblages (AI, AII and AIII) as well as alignments along the gene using previously defined references [12]. Tests for intra-assemblage recombination events were performed essentially as described [26]. The four-gamete test, MaxChi, and the four additional tests offered in RDP V3.41, RDP, GENCONV, Chimera, and SiScan [29] were performed. All tests rely on the premise that recombination will produce mosaic sequences. Because the region and length of GDH sequenced varied widely among the isolates, two GDH genealogies were produced; a GDH1, which used sequences that spanned nucleotides 186–1,159 and a GDH2, which used sequences that spanned nucleotides 258–647 where nucleotide positions correspond to the complete sequence of GDH from the WB:C6 genome [30]. Potential recombinants identified by these algorithms, were discarded if both parental sequences were from the same assemblage (potential intra-assemblage recombination).
Sequences were aligned by using MAFFT [31], distance-based analyses were conducted by using Kimura 2-parameter distance estimates, and trees were constructed by using the Neighbor-Joining algorithm, implemented in the Bionumerics program. Bootstrap proportions were calculated by the analysis of 500 replicates for neighbor-joining trees.
Probability of observing no offspring of recombinant types in the sample
Occurrence of a recombinant of B assemblage and E assemblage at TPI locus of a human isolate is an evidence of genetic exchange between the two assemblages. Under the hypothesis of selective neutrality, we calculated the probability of our observation; each recombinant type was found in only one sample at the particular locus, to statistically test the hypothesis, we made an assumption that a recombinant and a non-recombinant sequence type are selectively neutral, and calculated the probability that each recombinant type was observed exactly once in the samples of DNA sequences under the assumption of selective neutrality. The probability was calculated using the formula [32],
| (1) |
where the symbol n is the number of samples at a given locus, the symbol k is the number of distinct recombinant types and a non-recombinant type, the function s(n,k) is the stirling numbers of the first kind, and the bracket |.| indicates an absolute value.
Criteria for clonality: Identical genotypes (test d)
The extent to which the predominant genotype was overrepresented can be quantitatively evaluated by calculating the probability P of observing as many or more individuals of the particular genotype as actually observed in the sample,
| (2) |
where x = probability of the multi-locus genotype under the null hypothesis of full recombination, estimated by multiplying the observed frequency of the single-locus genotypes; n = number of individuals sampled; and m = number of individuals in the sample with the particular genotype [28].
Criteria for clonality: Absence of recombinant genotypes (test e)
This criterion is the probability of observing as few different multi-locus types as actually observed, given the size of the samples and expected multi-locus type frequencies under the hypothesis of full recombination [28]. The probability was calculated by a Monte Carlo approach by drawing ten-thousand random samples from the multinomial distribution with the vector of expected frequencies in Table 1. The probability was defined to be the fraction of random samples for which the number of distinct multi-locus types are less than or equal to the number of multi-locus types actually observed.
Table 1.
Frequencies of three-locus types under the hypothesis of free recombination
|
Host |
Human (n = 369) |
Livestock (n = 45) |
||||
|---|---|---|---|---|---|---|
| Assemblage* | Num | Obs.% | Exp.% | Num | Obs.% | Exp.% |
| 000 |
140 |
38 |
6 |
11 |
34 |
2 |
| 001 |
4 |
1 |
10 |
0 |
0 |
5 |
| 010 |
1 |
0 |
10 |
0 |
0 |
5 |
| 011 |
6 |
2 |
15 |
0 |
0 |
14 |
| 100 |
0 |
0 |
9 |
0 |
0 |
5 |
| 101 |
0 |
0 |
14 |
0 |
0 |
14 |
| 110 |
1 |
0 |
14 |
0 |
0 |
14 |
| 111 |
217 |
59 |
22 |
34 |
64 |
43 |
|
Host |
Cat (n = 17) |
Dog (n = 55) |
||||
|
Assemblage* |
Num |
Obs.% |
Exp.% |
Num |
Obs.% |
Exp.% |
| 000 |
7 |
41 |
7 |
24 |
44 |
11 |
| 001 |
0 |
0 |
10 |
1 |
2 |
11 |
| 010 |
0 |
0 |
10 |
0 |
0 |
12 |
| 011 |
0 |
0 |
14 |
0 |
0 |
11 |
| 100 |
0 |
0 |
10 |
1 |
2 |
14 |
| 101 |
0 |
0 |
14 |
1 |
2 |
13 |
| 110 |
0 |
0 |
14 |
3 |
5 |
14 |
| 111 | 10 | 59 | 20 | 25 | 45 | 14 |
* Letter 0 is a wildcard that designates either A (human, livestock, and cat assemblages) or C (dog). Similarly, letter 1 designates one of B (human), E (livestock), D (dog), or F (cat). Expected (Exp.) frequencies were calculated by multiplying the frequencies of genotype 0 at each locus with the frequencies of genotype 1 at each locus in eight possible ways. For comparison, the frequency of observed (Obs.) is displayed as well.
Criteria for clonality: Linkage disequilibrium (test f)
This is a standard test for non random association between loci, under the assumptions of random mating, and non-overlapping generations [33]. In our database of G. duodenalis, two genotypes were recorded at each locus. Hence we refer to one genotype by a letter 0 and to the other genotype by a letter 1. Linkage disequilibrium D12 between locus 1 and locus 2 was calculated using the equation [33],
| (3) |
where p00 is the frequency of congruent assemblages 00, p0* is the frequency of assemblage whose locus 1 is the type 0 and locus 2 is either 0 or 1, and p*0 is the frequency of assemblage whose locus 2 is the type 0 and locus 1 is either 0 or 1.
In the case of three-locus typing, linkage disequilibrium was calculated using the equation 1 in [34],
| (4) |
where D23D13 and D12 are pair-wise linkage disequilibrium between the three loci 1, 2, and 3, p000 is the frequency of multi-locus types 000. The symbol * is a wildcard for either genotype 0 or 1. Thus, p00* is the frequency of the assemblages having genotype 0 at the first and at the second loci and genotype at the third locus is either 0 or 1. P-values for linkage disequilibrium were obtained by a Monte Carlo approach as described in the test e but using D as the test statistic.
Index of association
Index of association for G. duodenalis was calculated using the observed frequencies of two genotypes at each of the three loci (Table 1) using the equation 3 in [35]. Variance of the index of association was calculated using the equation 4 in [35].
Results
Frequency of inter-assemblage recombination events within three loci
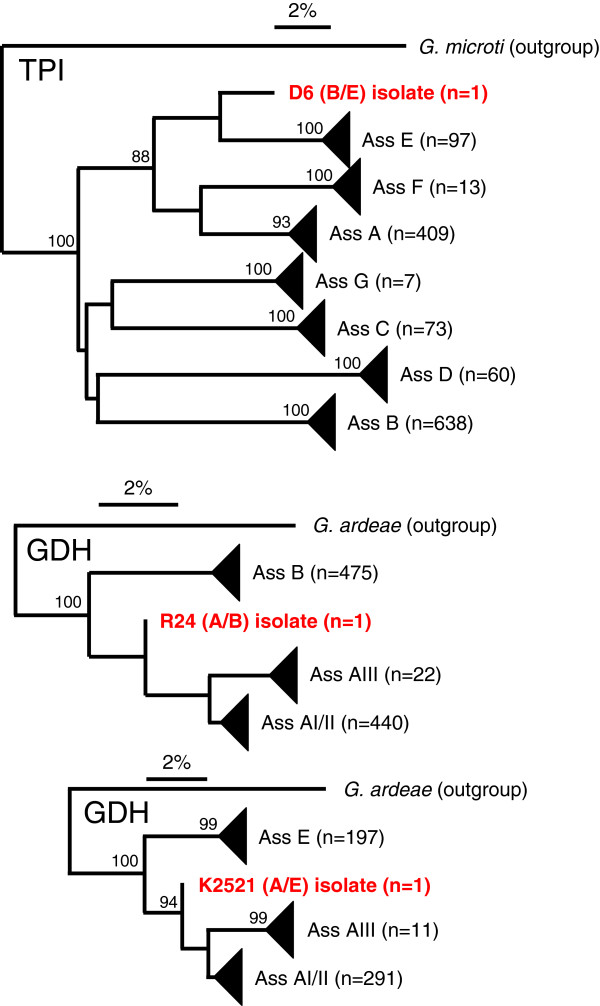
In a recent study, recombination between homologous loci from different assemblages was identified in at least three G. duodenalis isolates [26]. In this study, a limited number of sequences from two loci (a selection from GenBank entries until 2008) were analyzed. We performed the same approaches [26] to analyze the 4582 sequences from three loci stored in our database. Here, only potential recombinant events where parental sequences could be identified were taken into account. In the 1633 sequences from the BG locus, no inter-assemblage recombination events were detected. From the 1325 TPI sequences, only one isolate (D6) displayed a recombination event between assemblages B and E. In the GDH sequences (n = 1624) of two isolates, K2521 and R24, potential inter-assemblage recombination events occurred (Table 2, Additional file 1: Table S1). Both recombination events were identified previously. One 551-bp GDH sequence (GQ337967) in GenBank displayed a recombination event, where the last ~95 bp did not align with any other G.duodenalis, but were similar to H. sapiens RIO kinase 2 (AK225348) [36]. No inter-assemblage recombination was identified in the GDH sequences of the K4016 and SweCat17 isolates. These two were identified previously as potential recombinants [26], but turned out to be identical to other members of sub-assemblage AIII [13].
Table 2.
Isolates with potential inter-assemblage recombination within loci
| Source (Isolate ID) | Locus | Mixed | Reference | Nested-PCR | Description |
|---|---|---|---|---|---|
| Cattle (K2521) |
GDH |
A/E |
DQ182604 [37] |
Yes |
18SRDNA sequence of this rectal faecal sample was identified as Ass E. Other study samples contained Ass AI, AIII or Ass E. |
| Surface water (R24) |
GDH |
A/B |
EU350516 [38] |
Yes |
Environmental water sample,which contained very few cysts. |
| Human (D6) |
TPI |
B/E |
EU272164 [39] |
Yes |
Faecal sample derived from rural community in Egypt, where people commonly live in close contact with their livestock. |
| Cattle (K4016) |
GDH |
Is Ass AIII |
DQ182607 [37] |
Yes |
AIII is excluded as recombinant from AI and AII as they latter two could not be identified as parental strains. |
| Cat (Swecat171) | GDH | Is Ass AIII | EU769223 [14] | Yes |
Lack of inter-assemblage recombination will eventually result in the independent and divergent evolution of alleles from different assemblages [42]. Phylogenetic analyses of the available sequences of GDH and TPI generally display divergent and independent evolution (not shown, but see [4,12]). The sequences of the three isolates which underwent potential recombination events (Table 2) turned out to be exceptional, as they do not cluster significantly with any of the other sequences from the same locus (Figure 1). We conjectured that a successful recombinant is (at least) equally fit as its parents, resulting in offspring. To statistically test this hypothesis, we made an assumption that a recombinant and a non-recombinant type are selectively neutral in producing offspring, and calculated the probability that each recombinant type in the sample was observed exactly once in the sample. For GDH locus, we counted two recombinant types each occurring exactly once in the sample and counted the rest of 1622 non-recombinant types. The probability of this observation under the assumption of selective neutrality was estimated to be 0.5%. For TPI locus, we counted one recombinant type occurring exactly once in the sample and counted the rest of 1324 non-recombinant types. The probability of this observation under the assumption of selective neutrality was 6% (see Methods for calculation of the probability).
Figure 1.
Phylogenetic analysis based on single gene sequences of TPI and GDH, as obtained using neighbour joining. Bootstrap values > 85 are indicated. Terminal branches were collapsed for clarity. The three recombinant sequences (D6, R24 and K2521) form singular branches between the existing assemblage structures.
Frequency of inter-assemblage recombination events between loci of two assemblages
Absence of recombination within the three loci is insufficient to rule out inter-assemblage recombination, as these loci are relatively conserved regions and have an average size of only ~ 500 base pairs. The probability of detecting inter-assemblage recombination becomes significantly higher when multiple, independent loci on different chromosomes are used. Potential recombination events were assessed between pairs of assemblages, which occur most frequently together, i.e. which share similar host categories. Recombination between assemblages A and B was studied in G. duodenalis isolates from humans, between assemblages C and D in dog isolates, between A and F in cat isolates and between A and E in isolates from livestock. The presence of a particular multi-locus type in great excess is often the most robust and significant evidence of clonal reproduction, i.e. lack of recombination [28]. Here, a multi-locus type of a G. duodenalis isolate was based on the concatenated assemblage-typing of the three different loci BG, GDH and TPI. Only two multi-locus types are found in great excess in humans, namely AAA (38%) and BBB (59%). Of the six possible combinations of A and B in three-locus genotypes, only 3% are found (Table 1). The same was observed in the three other host categories. Two multi-locus types are found in great excess: AAA and EEE in livestock, AAA and FFF in cats, and CCC and DDD in dogs (Table 1). The contrast to the observed frequencies of three-locus genotypes is clear when the expected frequencies of three-locus types under the hypothesis of free recombination is calculated (Table 1), supporting the assertion that sexual reproduction between assemblages in G. duodenalis did not leave its trace on the population structure.
Three formal tests for clonality were performed and all three yielded highly significant results in the four G. duodenalis populations isolated from human, livestock, cat, and dog (Table 3). These results support that clonal reproduction had indeed shaped the observed population structures, independent of the origins from which the genetic materials of G. duodenalis were isolated. A fourth statistical test, Index of association (IA), was originally developed to index the extent of clonality within bacterial populations [35]. Small estimates for the variance indicate that the index values were significantly different from zero, and hence that recombination has been rare or absent.
Table 3.
Tests for clonality applied to the three-locus typing datasets
| Criterion | Human | Livestock | Cat | Dog |
|---|---|---|---|---|
| Identical genotypes widespread (d) |
<10-9 |
10-6 |
10-4 |
10-9 |
| Absence of recombinant genotypes (e) |
<10-4 |
<10-4 |
<10-4 |
<10-4 |
| Linkage disequilibrium (f) |
0.38* |
0.24* |
0.38* |
0.42* |
| Index of association (IA) | 1.8 ± 0.00 | 2.0 ± 0.02 | 2.0 ± 0.04 | 1.5 ± 0.02 |
* P value < 10-4. Italic letters in parentheses corresponds to the same tests in [43]. IA-values which are significantly different from zero indicates that recombination has been rare or absent.
Discussion
Occurrence of one recombination event between assemblage B and assemblage E at the TPI locus of a human isolate is an indication that genetic exchange between the two assemblages has occurred. However, detection of a single recombinant is important but not sufficient information to assert the nature of recombination between Giardia duodenalis assemblages. In fact, in a considerable number of isolates present in the database, this recombinant was found only once. The test under the assumption of selective neutrality between a recombinant and a non-recombinant type supported that a Giardia recombinant might contribute genetically little to the population structure of Giardia duodenalis. It could be argued that a zoonotic transmission event between a human individual and domestic livestock is unlikely to produce a novel viable G. duodenalis assemblage by a recombination of existing assemblages at TPI locus. Molecular typing of the Giardia isolates at GDH locus in addition indicated that a recombinant at the GDH locus contributes genetically very little to the population structure of Giardia duodenalis. The observation of a recombination event between a GDH DNA-sequence (GQ337967) and a DNA sequence probably derived from its host (sheep) is highly anomalous. A PCR-artifact or sequencing error seems more likely.
The lack of observation of selectively neutral or fitter recombinant types was not due to a limited capacity of detecting a recombination event that could be associated with a single locus typing method. The analysis of the three-locus sequence typing data by applying a battery of population genetic tests supported that the contribution of recombination events between the three loci to the population structure of G. duodenalis is also limited. These population genetic tests have been applied to a number of microorganisms, to support a clonal population structure of Leishmania as well as Trypanosoma, and to reject a clonal population structure of Candida albicans[28]. Judging by the weight of the evidence provided by the population genetic analyses of the molecular typing datasets of G. duodenalis, both a single and multi-locus based typing method, we opt for the hypothesis that the population structure of G. duodenalis assemblages is clonal. This conclusion is by no means in conflict with the existence of recombination within the two nuclei of a cyst at the genomic and the cellular level [21,22], nor does our analysis exclude the possibility of intra-assemblage recombination.
Our molecular epidemiological database consists of 1312 human and animal isolates with two or more loci. Fifteen percent (n = 197) of these isolates constitute a combination of loci derived from different assemblages. Based on the analyses of this study, we conclude that these isolates are a mixture of different Giardia genotypes from different assemblages. This is probably an underestimation, as mixed infections are not always detected. Unequal loads or stability of the microorganisms of interest in the original sample, preferential immuno-purification or preferential binding of primer pairs to one of microorganisms will result in the detection of only one of them. PCR assays based on the use of assemblage-specific primers have been developed to show the frequency of mixed infections [8,44]. It also is possible that another fraction of the field-isolates in this database consists of a mixture of different Giardia genotypes from the same assemblage. Besides allelic sequence heterogeneity, the latter may be an additional explanation for the presence of ambiguous nucleotides (“double peaks”) present in the sequences of many isolates. Unfortunately, it is still not possible to distinguish between a clonal isolate and an isolate consisting of a mixture of the same assemblage. A PCR-based approach using a generic set of primers appears to be unreliable for the detection of mixed infections.
Conclusion
Molecular epidemiology is probably the best method available to study transmission dynamics of G. duodenalis in and between human and animal populations. The relatively poor genetic resolution of the available single loci used for these studies could not be solved by combining several of these loci to a multi-locus genotyping scheme. Multilocus-sequence typing directly on field isolates is hampered by the occurrence of mixed infections in both animals and humans. Our data imply that genetic recombination between genotypes of different assemblages is either very rare or absent in the G. duodenalis population. Thus, multilocus sequence types from field isolates, which consist of more than one assemblage, should be interpreted as a mixed infection. Improved in vitro culturing methods and novel molecular typing methods, such as single cyst PCR, are required to investigate the directionality and frequency of animal to human transmission and to determine the frequency of intra-assemblage recombination events [45].
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
TM, ML, SC, and HS performed data collection and sequence analysis. KT and AS performed population genetic and mathematical analysis. HS performed phylogenetic analysis. KT, SC, EL and HS were involved in the study design and interpretation of the results. KT and HS drafted the manuscript and wrote the final version. All authors read and approved the final manuscript.
Supplementary Material
Table S1. Inter-assemblage recombination events within three loci.
Contributor Information
Katsuhisa Takumi, Email: katsuhisa.takumi@rivm.nl.
Arno Swart, Email: arno.swart@rivm.nl.
Theo Mank, Email: t.mank@streeklabhaarlem.nl.
Erica Lasek-Nesselquist, Email: elasekness@gmail.com.
Marianne Lebbad, Email: marianne.lebbad@smi.se.
Simone M Cacciò, Email: simone.caccio@iss.it.
Hein Sprong, Email: hein.sprong@rivm.nl.
References
- Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam RD. Biology of Giardia lamblia. Clin Microbiol Rev. 2001;14:447–475. doi: 10.1128/CMR.14.3.447-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monis PT, Andrews RH, Mayrhofer G, Ey PL. Genetic diversity within the morphological species Giardia intestinalis and its relationship to host origin. Infect Genet Evol. 2003;3:29–38. doi: 10.1016/S1567-1348(02)00149-1. [DOI] [PubMed] [Google Scholar]
- Wielinga CM, Thompson RC. Comparative evaluation of Giardia duodenalis sequence data. Parasitology. 2007;134:1795–1821. doi: 10.1017/S0031182007003071. [DOI] [PubMed] [Google Scholar]
- Caccio SM, Ryan U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E, Welch DM, Sogin ML. The identification of a new Giardia duodenalis assemblage in marine vertebrates and a preliminary analysis of G. duodenalis population biology in marine systems. Int J Parasitol. 2010;40:1063–1074. doi: 10.1016/j.ijpara.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins RM, Meloni BP, Groth DM, Wetherall JD, Reynoldson JA, Thompson RC. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J Parasitol. 1997;83:44–51. doi: 10.2307/3284315. [DOI] [PubMed] [Google Scholar]
- Geurden T, Levecke B, Caccio SM, Visser A, De Groote G, Casaert S, Vercruysse J, Claerebout E. Multilocus genotyping of Cryptosporidium and Giardia in non-outbreak related cases of diarrhoea in human patients in Belgium. Parasitology. 2009;136:1161–1168. doi: 10.1017/S0031182009990436. [DOI] [PubMed] [Google Scholar]
- Traub RJ, Monis PT, Robertson I, Irwin P, Mencke N, Thompson RC. Epidemiological and molecular evidence supports the zoonotic transmission of Giardia among humans and dogs living in the same community. Parasitology. 2004;128:253–262. doi: 10.1017/S0031182003004505. [DOI] [PubMed] [Google Scholar]
- Isaac-Renton JL, Lewis LF, Ong CS, Nulsen MF. A second community outbreak of waterborne giardiasis in Canada and serological investigation of patients. Trans R Soc Trop Med Hyg. 1994;88:395–399. doi: 10.1016/0035-9203(94)90397-2. [DOI] [PubMed] [Google Scholar]
- Isaac-Renton JL, Cordeiro C, Sarafis K, Shahriari H. Characterization of Giardia duodenalis isolates from a waterborne outbreak. J Infect Dis. 1993;167:431–440. doi: 10.1093/infdis/167.2.431. [DOI] [PubMed] [Google Scholar]
- Sprong H, Caccio SM, van der Giessen JW. Identification of zoonotic genotypes of Giardia duodenalis. PLoS Negl Trop Dis. 2009;3:e558. doi: 10.1371/journal.pntd.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccio SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008;38:1523–1531. doi: 10.1016/j.ijpara.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Lebbad M, Mattsson JG, Christensson B, Ljungstrom B, Backhans A, Andersson JO, Svard SG. From mouse to moose: multilocus genotyping of Giardia isolates from various animal species. Vet Parasitol. 2010;168:231–239. doi: 10.1016/j.vetpar.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Martinez-Diaz RA, Sansano-Maestre J, Martinez-Herrero Mdel C, Ponce-Gordo F, Gomez-Munoz MT. Occurrence and genetic characterization of Giardia duodenalis from captive nonhuman primates by multi-locus sequence analysis. Parasitol Res. 2011;109:539–544. doi: 10.1007/s00436-011-2281-z. [DOI] [PubMed] [Google Scholar]
- Lebbad M, Petersson I, Karlsson L, Botero-Kleiven S, Andersson JO, Svenungsson B, Svard SG. Multilocus genotyping of human Giardia isolates suggests limited zoonotic transmission and association between assemblage B and flatulence in children. PLoS Negl Trop Dis. 2011;5:e1262. doi: 10.1371/journal.pntd.0001262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Sprong H, Pozio E, Caccio SM. Genotyping Giardia duodenalis Isolates from Dogs: Lessons from a Multilocus Sequence Typing Study. Vector Borne Zoonotic Dis. 2012;12:206–213. doi: 10.1089/vbz.2011.0751. [DOI] [PubMed] [Google Scholar]
- Solarczyk P, Majewska AC. Prevalence and multilocus genotyping of Giardia from animals at the zoo of Poznan, Poland. Wiad Parazytol. 2011;57:169–173. [PubMed] [Google Scholar]
- Isaac-Renton JL, Shahriari H, Bowie WR. Comparison of an in vitro method and an in vivo method of Giardia excystation. Appl Environ Microbiol. 1992;58:1530–1533. doi: 10.1128/aem.58.5.1530-1533.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benere E, Geurden T, Robertson L, Van Assche T, Cos P, Maes L. Infectivity of Giardia duodenalis Assemblages A and E for the gerbil and axenisation of duodenal trophozoites. Parasitol Int. 2010;59:634–637. doi: 10.1016/j.parint.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Birky CW. Giardia sex? Yes, but how and how much? Trends Parasitol. 2010;26:70–74. doi: 10.1016/j.pt.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Caccio SM, Sprong H. Giardia duodenalis: genetic recombination and its implications for taxonomy and molecular epidemiology. Exp Parasitol. 2010;124:107–112. doi: 10.1016/j.exppara.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Ramesh MA, Malik SB, Logsdon JM. A phylogenomic inventory of meiotic genes; evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Melo SP, Gomez V, Castellanos IC, Alvarado ME, Hernandez PC, Gallego A, Wasserman M. Transcription of meiotic-like-pathway genes in Giardia intestinalis. Mem Inst Oswaldo Cruz. 2008;103:347–350. doi: 10.1590/S0074-02762008000400006. [DOI] [PubMed] [Google Scholar]
- Cooper MA, Adam RD, Worobey M, Sterling CR. Population genetics provides evidence for recombination in Giardia. Curr Biol. 2007;17:1984–1988. doi: 10.1016/j.cub.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E, Welch DM, Thompson RC, Steuart RF, Sogin ML. Genetic exchange within and between assemblages of Giardia duodenalis. J Eukaryot Microbiol. 2009;56:504–518. doi: 10.1111/j.1550-7408.2009.00443.x. [DOI] [PubMed] [Google Scholar]
- Poxleitner MK, Carpenter ML, Mancuso JJ, Wang CJ, Dawson SC, Cande WZ. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science. 2008;319:1530–1533. doi: 10.1126/science.1153752. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Breniere SF, Darde ML, Ayala FJ. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci U S A. 1991;88:5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, Best AA, Cande WZ, Chen F, Cipriano MJ. et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens WJ. The sampling theory of selectively neutral alleles. Theor Popul Biol. 1972;3:87–112. doi: 10.1016/0040-5809(72)90035-4. [DOI] [PubMed] [Google Scholar]
- Ewens W. Mathematical population genetics. Springer, Second edn; 2004. [Google Scholar]
- Long JC, Williams RC, Urbanek M. An E-M algorithm and testing strategy for multiple-locus haplotypes. Am J Hum Genet. 1995;56:799–810. [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson LJ, Gjerde BK, Furuseth Hansen E. The zoonotic potential of Giardia and Cryptosporidium in Norwegian sheep: a longitudinal investigation of 6 flocks of lambs. Vet Parasitol. 2010;171:140–145. doi: 10.1016/j.vetpar.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Langkjaer RB, Vigre H, Enemark HL, Maddox-Hyttel C. Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology. 2007;134:339–350. doi: 10.1017/S0031182006001533. [DOI] [PubMed] [Google Scholar]
- Plutzer J, Karanis P, Domokos K, Torokne A, Marialigeti K. Detection and characterisation of Giardia and Cryptosporidium in Hungarian raw, surface and sewage water samples by IFT, PCR and sequence analysis of the SSUrRNA and GDH genes. Int J Hyg Environ Health. 2008;211:524–533. doi: 10.1016/j.ijheh.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Foronda P, Bargues MD, Abreu-Acosta N, Periago MV, Valero MA, Valladares B, Mas-Coma S. Identification of genotypes of Giardia intestinalis of human isolates in Egypt. Parasitol Res. 2008;103:1177–1181. doi: 10.1007/s00436-008-1113-2. [DOI] [PubMed] [Google Scholar]
- Read CM, Monis PT, Thompson RC. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol. 2004;4:125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, Das P, Lal AA, Xiao L. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin R. Evolution of sex: The costs and benefits of sex: new insights from old asexual lineages. Nat Rev Genet. 2002;3:311–317. doi: 10.1038/nrg749. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M, Kjellberg F, Ayala FJ. A clonal theory of parasitic protozoa: the population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci U S A. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurden T, Geldhof P, Levecke B, Martens C, Berkvens D, Casaert S, Vercruysse J, Claerebout E. Mixed Giardia duodenalis assemblage A and E infections in calves. Int J Parasitol. 2008;38:259–264. doi: 10.1016/j.ijpara.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Ankarklev J, Svard SG, Lebbad M. Allelic sequence heterozygosity in single Giardia parasites. BMC Microbiol. 2012;12:65. doi: 10.1186/1471-2180-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Inter-assemblage recombination events within three loci.