Abstract
Background
The maternally inherited α-Proteobacteria Wolbachia pipientis is an obligate endosymbiont of nematodes and arthropods, in which they induce a variety of reproductive alterations, including Cytoplasmic Incompatibility (CI) and feminization. The genome of the feminizing wVulC Wolbachia strain harboured by the isopod Armadillidium vulgare has been sequenced and is now at the final assembly step. It contains an unusually high number of ankyrin motif-containing genes, two of which are homologous to the phage-related pk1 and pk2 genes thought to contribute to the CI phenotype in Culex pipiens. These genes encode putative bacterial effectors mediating Wolbachia-host protein-protein interactions via their ankyrin motifs.
Results
To test whether these Wolbachia homologs are potentially involved in altering terrestrial isopod reproduction, we determined the distribution and expression of both pk1 and pk2 genes in the 3 Wolbachia strains that induce CI and in 5 inducing feminization of their isopod hosts. Aside from the genes being highly conserved, we found a substantial copy number variation among strains, and that is linked to prophage diversity. Transcriptional analyses revealed expression of one pk2 allele (pk2b2) only in the feminizing Wolbachia strains of isopods.
Conclusions
These results reveal the need to investigate the functions of Wolbachia ankyrin gene products, in particular those of Pk2, and their host targets with respect to host sex manipulation.
Background
The evolutionary success of the maternally inherited α-Proteobacteria Wolbachia pipientis is partly due to its ability to manipulate host reproduction to favour vertical transmission from mother to offspring. Wolbachia are also able to switch between hosts via horizontal transfer, which contributes to the impressive diversity and range of infected hosts [1]. These obligate endosymbionts are found in most filarial nematodes and are estimated to be present in ~60% of arthropod species [2-4]. In arthropods, Wolbachia are considered to be sex-parasites because they alter compatibility between eggs and sperm, feminize or kill males, or induce parthenogenesis [2,5,6]. Since Wolbachia remain unculturable endosymbionts, comparative genomics and evolutionary approaches are particularly useful for identifying putative bacterial determinants involved in Wolbachia-host interactions.
Recent genome analyses of different Wolbachia strains revealed a surprisingly high number of ankyrin domain-containing genes (ank genes) [7-11]. Their presence is suggested to be the result of lateral gene transfer since they are mostly found in eukaryotes but in few bacterial and viral genomes [12,13]. The 33-residue ankyrin repeats (ANK) form tandem arrays that mediate specific protein-protein interactions and have diverse functions in transcription initiation, cell cycle regulation and signalling, cytoskeleton integrity, ion transport, inflammatory responses and development [12,14]. The two closely related intracellular bacteria Anaplasma phagocytophilum and Ehrlichia chaffeensis secrete ankyrin proteins (AnkA and p200, respectively) that bind to host DNA and/or proteins [15,16]. It has been demonstrated that AnkA plays an important role in facilitating intracellular infection [17] whereas p200 is thought to affect host cell gene transcription and promote the survival of the pathogen [16]. Hence it has been suggested that ank genes encode Wolbachia effectors that alter host biology [18,19].
Several studies have suggested that Wolbachia ANK proteins were implicated in the molecular basis of Cytoplasmic Incompatibility (CI) [8,9,20-23]. Sequence divergence between closely related Wolbachia strains causing distinct CI types in Culex pipiens mosquito populations has been found in two ank genes (pk1 and pk2), among a subset of phylogenetic markers [20,22-25]. Thus, these polymorphic pk1 and pk2 ank genes, located within a so-called WO prophage region of Wolbachia genome, are suggested to contribute to the CI phenotype. Consistent with this argument, expression of the pk2 gene occurred specifically in female mosquitoes [8,22,23]. Moreover, a premature stop codon was found in the pk2 gene of the Wolbachia strain (wAu) that is unable to cause CI in D. simulans[21].
In this study, we aimed to determine whether the prophage pk1 and pk2 ankyrin genes were involved in the CI phenotype described in three Wolbachia-infected species of terrestrial isopods. We also investigated whether these genes were conserved and expressed in Wolbachia strains inducing feminization, the main Wolbachia phenotype described for this group of hosts [2]. From the genome of the feminizing wVulC Wolbachia strain that infects the isopod Armadillidium vulgare (the genome completion is currently being done by our group in the frame of the European Wolbachia project: EuWol), we annotated the pk1 and pk2 alleles among all ank genes identified from the wVulC contigs. We investigated the distribution, copy number and expression patterns of both genes in seven additional Wolbachia strains that induce either CI or feminization in isopods. We identified a large copy number variation of the pk1 and pk2 genes among Wolbachia strains, which is probably coupled to prophage evolution. Surprisingly, our results also revealed that expression of one pk2 allele (pk2b2) is only detected in feminizing Wolbachia strains and never in the three CI-inducing strains of isopods.
Results
Characterization and distribution of pk1 and pk2 genes
Six copies of the pk1 gene and three copies of the pk2 gene were identified in the contig assembly of the wVulC genome (Table 1). Each of the six putative prophage regions of the assembly contains one pk1 allele and three of these prophages also harbour one pk2 allele (Table 1). Two wVulC pk1 alleles (ANK46a/b and ANK60a/b) and one pk2 allele (ANK40a/b) were each found in two identical copies. These results were confirmed by Southern blotting (Additional file 1: Figure S1) and are consistent with the sequencing of PCR products (Table 1).
Table 1.
List ofpk1andpk2alleles associated to their prophages in differentWolbachiagenomes
| Wolbachia strain | Super group | Host species | Genome | Prophage | pk1 | pk2 | Reference | ||
|---|---|---|---|---|---|---|---|---|---|
|
wCauB |
B |
Cadra cautella |
ABA78515 |
WOCauB2 |
B2gp13 |
|
- |
|
[26] |
| |
|
|
ABA78516 |
WOCauB3 |
B2gp14 |
|
- |
|
[26] |
|
wMel |
A |
Drosophila melanogaster |
NC_002978 |
WOMelB |
WD0596 |
|
WD0636 |
|
[11] |
|
wPip-BfB |
B |
Culex pipiens pipiens |
- |
- |
AM397076 |
pk1b |
AM397073 |
pk2c |
[24] |
|
wPip-EpA |
B |
Culex pipiens quinquefasciatus |
- |
- |
AM397075 |
pk1a |
AM397074 |
pk2d |
[24] |
|
wPip-Is |
B |
Culex pipiens pipiens |
- |
- |
AM397078 |
pk1d |
DQ000474 |
pk2a |
[25] |
|
wPip-KaC |
B |
Culex pipiens quinquefasciatus |
- |
- |
AM397079 |
pk1e |
DQ000472 |
pk2a |
[24] |
|
wPip-Lv |
B |
Culex pipiens pipiens |
- |
- |
AM397077 |
pk1c |
DQ000472 |
pk2a |
[25] |
|
wPip-Pel |
B |
Culex pipiens quinquefasciatus |
NC_010981 |
WOPip1 |
WPa_0256 |
ANK08 |
- |
|
[8] |
| |
|
|
|
WOPip2 |
WPa_0315 |
ANK14 |
WPa_0299 |
ANK12 |
[8] |
| |
|
|
|
WOPip3 |
- |
|
WPa_0340 |
ANK16 |
[8] |
| |
|
|
|
WOPip4 |
- |
|
WPa_0413 |
ANK25 |
[8] |
| |
|
|
|
WOPip5 |
WPa_1308 |
ANK56 |
- |
|
[8] |
|
wRi |
A |
Drosophila simulans |
NC_012416 |
WORiA1 |
WRi_005620 |
ANK16 |
WRi_005440 |
ANK14 |
[9] |
| |
|
|
|
WORiA2 |
WRi_010280 |
ANK35 |
WRi_010100 |
ANK33 |
[9] |
| |
|
|
|
WORiB |
WRi_p07220 |
ANK29 |
- |
|
[9] |
| |
|
|
|
WORiB |
WRi_p07650 |
ANK31 |
- |
|
[9] |
|
wPip-Sl |
B |
Culex pipiens quinquefasciatus |
- |
- |
AM397076 |
pk1b |
DQ000471 |
pk2b |
[25] |
|
wVitA |
A |
Nasonia vitripennis |
HQ906662 |
WOVitA1 |
|
pk1-1 |
|
pk2-1 |
[27] |
| |
|
|
HQ906663 |
WOVitA2 |
|
pk1-2 |
|
- |
[27] |
|
wVulC |
B |
Armadillidium vulgare |
- |
WOVulC1 |
HM452371 |
ANK46b |
- |
|
This study |
| |
|
|
|
WOVulC2 |
HM452372 |
ANK32 |
- |
|
This study |
| |
|
|
|
WOVulC3 |
HM452373 |
ANK60b |
HM452376 |
ANK40a |
This study |
| |
|
|
|
WOVulC4 |
HM452374 |
ANK25 |
HM452376 |
ANK40b |
This study |
| |
|
|
|
WOVulC5 |
HM452371 |
ANK46a |
HM452375 |
ANK48 |
This study |
| WOVulC6 | HM452373 | ANK60a | - | This study |
Accession numbers from this study are in bold.
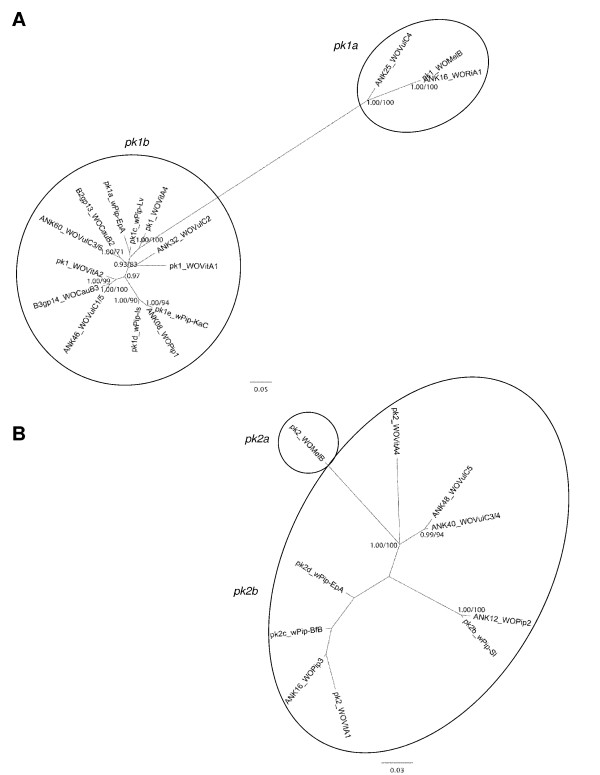
Relationships between these alleles and all the published sequences of Wolbachia pk1 and pk2 genes were assessed based on the DNA sequences encoding ANK-repeats. No evidence of recombination events has been detected in the alignments of pk1 and pk2 gene regions encoding ANK motifs (see Methods). In the pk1 sequences, the number of variable sites is 467 out of 1068, of which 408 are informative. Similarly, there are 66 informative sites in the 292 bp-long pk2 sequence alignment. The resulting genetic networks show that pk1 and pk2 sequences group in two clusters (Figure 1). Based on these relationships, we defined two different types of pk1 genes (named pk1a and pk1b) and two different types of pk2 genes (named pk2a and pk2b) (Figure 1). The wVulC genome contains a single copy of the pk1a type (ANK25) and 5 copies belonging to the pk1b type: ANK32, ANK46a/b and ANK60a/b (Figure 1A). The wVulC pk1a gene clusters together with the pk1a type gene of wMel (Figure 1A, 15.9% divergent) whereas it shares 55.2 to 65.2 % identity with the five wVulC pk1b type sequences. The region encoding ANK repeats in the wVulC ANK60a/b alleles is related to the pk1 sequence from the wCauB2 prophage (Figure 1A). wVulC ANK46a/b alleles are closely related to the pk1 gene from the wCauB3 prophage (Figure 1A). ANK32 seems somehow related to the pk1 gene from the WOVitA1 prophage (Figure 1A). The wVulC genome also harbours three genes of the pk2b type, ANK48 and ANK40a/b, further called pk2b1 and pk2b2 alleles respectively. In contrast to the pk1 gene, all three wVulC pk2 alleles form a cluster in the gene network (Figure 1B). Their closest relative is the pk2 gene harboured by the WOVitA4 prophage of the Wolbachia strain endosymbiont of Nasonia vitripennis (Figure 1B).
Figure 1.
Networks ofpk1andpk2genes. Networks of (A) pk1 and (B) pk2 sequences encoding ANK motif clusters. Identical sequences were merged and are represented under one unique label comprising information on the gene and prophage when available: (A) ANK16_WORiA1 corresponds also to ANK35_WORiA2, ANK08_WOPip1 = ANK14_WOPip2 = ANK56_WOPip5 = pk1b_wPip-BfB/Sl (B) pk2_WOMelB = ANK14_WORiA1 = ANK33_WORiA2, ANK12_WOPip2 = ANK25_WOPip4 = pk2_wPip-Is/Lv/KaC (see Table 1). Node support: ML bootstrap/MrBayes posterior probability, values <70 or 0.70 are not shown.
Using the same primer set as for wVulC ( Additional file 1: Table S1), the taxonomic distribution of pk1 and pk2 genes was extended by PCR to seven Wolbachia strains that induce either CI or feminization in isopods. All these strains of isopods are known to belong to the B-supergroup of Wolbachia whatever the phylogenetic marker used [2]. They do not form separate monophyletic clades according to the phenotype they induce in their hosts based on the wsp gene ( Additional file 1: Figure S2). We also investigated the copy number variation by Southern blot analyses of EcoRI or BamHI digested DNA using pk1apk1b and pk2b1 probes which, according to sequence identities, preferentially hybridized on pk1apk1b and pk2b types, respectively (Table 2 & Additional file 1: Figure S1). In congruence with amplification and sequencing data, the pk1a and pk1b probes revealed two to six copies of the pk1 gene in the studied strains (Table 2). By direct sequencing of the PCR products, we found that the pk1a gene of Wolbachia strains of C. convexusP. pruinosusA. vulgare (wVulM) and A. nasatum harboured 1, 1, 2 and 3 EcoRI sites, respectively, explaining the discrepancy between the number of bands observed by Southern blots, and the number of different sequences obtained (Table 2 & Additional file 1: Figure S1). Similarly, two pk1b alleles of the Wolbachia strain of A. nasatum contained one BamHI restriction site. Each of the two more intense Southern Blot signals ( Additional file 1: Figure S1) revealed the presence of two identical copies wVulC pk1b alleles, as confirmed by the analysis of contigs. Furthermore, Southern blots using a pk2b1 probe in combination with sequencing data revealed three copies of the pk2 gene in all strains tested except one (Table 2 & Additional file 1: Figure S1). In the Wolbachia strain of P. pruinosus, sequences of PCR products revealed two identical pk2 alleles, each containing one BamHI restriction site explaining the five signals obtained by Southern blotting (Table 2 & Additional file 1: Figure S1). Moreover, no signal was obtained from digested and undigested DNA of Wolbachia-free ovaries of isopod (non-infected population from Nice, France), which confirmed the Wolbachia origin of the pk1 and pk2 genes.
Table 2.
Copy number of pk1 and pk2 alleles according to their ‘type’ in different Wolbachia strains
| Wolbachia strain | Host |
pk1 |
alleles |
pk2 |
alleles |
|---|---|---|---|---|---|
| a | b | a | b | ||
|
wAse |
Oniscus asellus |
1 |
3 |
0 |
3 |
|
wCauB |
Cadra cautella |
0 |
2 |
0 |
0 |
|
wConV |
Cylisticus convexus |
1 |
3 |
0 |
3 |
|
wDil |
Porcellio dilatatus dilatatus |
1 |
4 |
0 |
3 |
|
wMel |
Drosophila melanogaster |
1 |
0 |
1 |
0 |
|
wNas |
Armadillidium nasatum |
1 |
5 |
0 |
3 |
|
wPet |
Porcellio dilatatus petiti |
1 |
1 |
0 |
3 |
|
wPip-Pel |
Culex pipiens |
0 |
3 |
0 |
3 |
|
wPrull |
Porcellionides pruinosus |
1 |
5 |
0 |
3 |
|
wRi |
Drosophila simulans |
3(2) |
1 |
2 |
0 |
|
wVitA |
Nasonia vitripennis |
3 |
0 |
2 |
0 |
|
wVulC |
Armadillidium vulgare |
1 |
5 |
0 |
3 |
| wVulM | Armadillidium vulgare | 1 | 5 | 0 | 3 |
The number of pseudogenes is indicated in brackets. ND: non determined.
Molecular evolution of pk1 and pk2genes
The GC content of wVulC pk1 alleles (mean ± SE, 33.9 ± 0.3%) is similar to that of the whole genome assembly (34.5%) whereas the GC content of wVulC pk2 alleles (ANK40a/b: 36.8%, ANK48: 36.3%) is significantly greater. Similar results were obtained considering pk1 and pk2 genes of all Wolbachia genomes (pk1: 34.0 ± 0.1%; pk2: 37.2 ± 0.2%; genomes: 34.8 ± 0.3%) (paired t-test, t = 13.79, df = 15, p = 6.3e-10) ( Additional file 1: Table S2). Interestingly, the GC content of pk1 and pk2 sequences is significantly different from the whole prophage sequences, which comprise an intermediate GC content of 35.8 ± 0.2% (paired t-tests; prophage vs. pk1, t = 12.60, df = 11, p = 7.0e-8; prophage vs. pk2, t = 3.85, df = 8, p = 4.9e-3) ( Additional file 1: Table S2). ANK motif-encoding sequence analysis indicated no recombination and Ka/Ks (the ratio of the rate of non-synonymous substitutions (Ka) to the rate of synonymous substitutions (Ks)) of all positions was 0.211 ± 0.009 for Pk1 and 0.245 ± 0.020 for Pk2. Purifying selection is thus acting on these domain-encoding sequences and no sites are under positive selection.
All translated pk1 full-length sequences are predicted to harbour two transmembrane domains in their C-terminal region but a variable number of ANK motifs ranging from 8 to 10 ( Additional file 1: Figure S3). In wVulC, ANK46a/b and ANK60a/b sequences (pk1b type) are shorter in their N-terminal region than the other Pk1 translated sequences (42 and 62 amino acids, respectively). One indel at position 117 of the DNA sequence of wVulC ANK46a/b is responsible for a frame shift, which splits the gene into two ORFs homologous to the full-length pk1 of other strains. ANK60a/b sequences are shortened by a transposase gene insertion in the 5′ region. In contrast, pk2 translated sequences are more conserved (84.5 to 100% identity) among Wolbachia strains than pk1. All Pk2 amino acid sequences harbour 3 ANK motifs except in the wAu strain (host: D. simulans) in which a premature stop codon disrupts the third motif ( Additional file 1: Figure S3).
Comparative analysis of pk1 and pk2 mRNA expression in CI and feminizing Wolbachia strains
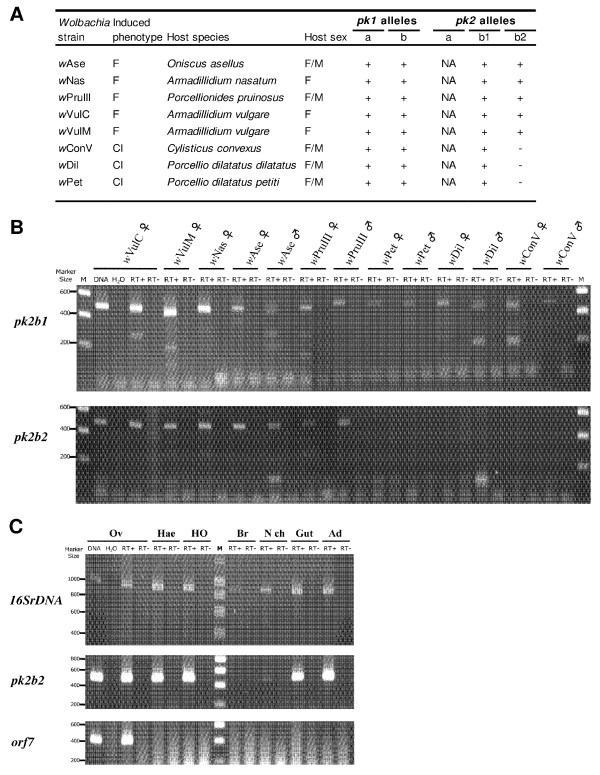
RT-PCR using allele-specific primers was performed to examine the expression patterns of pk1 and pk2 mRNA in adult gonads of isopods harbouring CI-inducing or feminizing Wolbachia strains (Figure 2). Evidence of expression was observed for all copies of pk1 and pk2 genes except for one allele of the pk2b type (Figure 2A). Although the pk2 alleles were amplified by PCR using DNA of all the three isopod CI-inducing Wolbachia strains (positive controls only shown for wVulC), RT-PCR did not yield any detectable products in these strains using the pk2b2 allele-specific primers (Figure 2B). In contrast, the pk2b2 allele was clearly expressed in all the feminizing Wolbachia strains (Figure 2B). In hosts where both males and females are infected by CI-inducing or feminizing strains, no clear sex-specific differences were observed in pk1 and pk2 expression (Figure 2A). We further examined the expression of pk2b2 and another prophage gene, orf7 which encodes the phage capsid, in several tissues of A. vulgare females harbouring the feminizing wVulC strain (Figure 2C). While orf7 was expressed only in ovaries, the host tissue where the density of Wolbachia is higher, transcription of pk2b2 was revealed in all tissues tested (except the brain) (Figure 2C).
Figure 2.
Transcriptional analyses ofpk1andpk2alleles. (A) Transcriptional results of the pk1 and pk2 alleles obtained from gonads of eight isopod species harbouring either feminizing (F) or CI-inducing (CI) Wolbachia strains. Plus or minus signals indicate expression, or not, of the copy(ies). Distinction is made between the two different pk2 alleles named pk2b1 and pk2b2 within the pk2b type. F: female; M: male. NA: no pk2a type alleles were amplified in these strains. (B) Transcriptional results of pk2b1 and pk2b2 alleles are shown from ovaries or testes (when infected) of eight isopod species. Primers used are shown in ( Additional file 1: Table S1). The DNA template control (only wVulC presented) shows the intensity and specificity of the band detected with each pair of primers. RT + and RT- indicate, respectively, the presence or the absence of reverse transcriptase in the reactions. M: DNA size markers. (C) Transcriptional results of the 16S rDNA, pk2b2 and orf7 genes in seven different tissues of A. vulgare harbouring the wVulC Wolbachia strain. Ov: ovaries; Hae: haemocytes; HO: hematopoietic organ; Br: brain; N ch: nerve chain; gut; Ad: adipose tissue.
Discussion
In this study, we found that a large copy number variation of pk1 and pk2 genes exists among Wolbachia strains, which is probably coupled to prophage dynamics and evolution. Copy number divergence in the ankyrin pk1 and pk2 is consistent with the results of previous Southern blotting analyses using the minor capsid orf7 phage gene [28]. Four different orf7 paralogs had already been identified in the wVulC strain through cloning and sequencing of heterogeneous PCR products [28]. Since multiple infections of Wolbachia in a single individual have never been observed in isopods, we can conclude that the phage WO is likely to be present in several copies in each Wolbachia strain. Our observations of Wolbachia strains of isopods suggest that dynamics of the prophage pk1 and pk2 genes is similar to that observed in the wRi and wPip-Pel genomes [8,9]. While the pk1 and pk2 ank genes are each found as a single copy within the WO-B prophage of the wMel genome [11], sequencing of the CI-inducing wPip-Pel, wRi and wVitA Wolbachia prophages revealed a duplication of these genes together with the WO-B-like prophages [8,9,27]. For instance, in the wPip-Pel genome, the three pk1 and the three pk2 genes are spread among the five different prophages which are closely related to the WO-B wMel prophage [8]. Hence, the divergence in the pk1 and pk2 gene copy number between Wolbachia strains may be explained by mechanisms related to bacterial genome organization and modulation of gene copy number [26,29-32]. As an example, two pseudogenes (wRi_ANK29 and ANK31) out the four copies of the pk1 gene in wRi, are spread in the WORiB prophage (previously annotated WO-C prophage [9], see Table 1) and may have originally been a single pk1 gene further disrupted by an insertion sequence ISWpi7. On the other hand, the high GC content of pk2 supports the occurrence of recent lateral transfers of prophage fragments containing the pk2 gene but not necessarily pk1 in the Wolbachia genomes. However, we cannot exclude the hypothesis that linkage disequilibrium occurs between pk1 and pk2 genes that are separated by at least 6.7 kilobases, representing less than 0.04% of the whole genome size. These results also highlight the genomic plasticity of the prophage region among Wolbachia strains as part of the global plasticity observed in the Wolbachia genomes [33]. Maintenance of such “mobile elements” in Wolbachia strains of arthropods may be due to the absence of, or a reduced efficiency of selection on the prophages. Nevertheless, the purifying selection acting on these pk1 and pk2 genes suggest that maintenance of sequences confers an adaptive advantage.
Besides identifying mosaic prophages, our results also reveal the differential expression of one pk2 ankyrin according to the Wolbachia phenotype they induce (CI vs. feminization). One allele (pk2b2) is only expressed in the feminizing strains and never in the three CI-inducing strains of isopods. In contrast to the observations for wPip [22,23], expression pattern of pk2b2 suggests that this allele is not involved in CI in isopods. In two recent studies, it has been shown that expression of pk1 and pk2 genes from wMel was not correlated with the CI phenotype in D. melanogaster[34,35]. Our transcriptional result rather leads to the hypothesis that this pk2b2 allele is involved in the feminization of isopod hosts. This hypothesis is strengthened by the observation that the pk2b2 allele is expressed in all A. vulgare tissues (except in the brain) whereas another prophage gene (orf7) is only expressed in ovaries. Furthermore, no differential expression of pk1 and pk2 genes was identified between sexes in isopods when either CI-inducing or feminizing Wolbachia infects both males and females. This result differs from those of Sinkins and colleagues who showed that in some CI-inducing wPip variants, the three pk2 genes (the two identical wPip_ANK12 and wPip_ANK25, and wPip_ANK16) are highly expressed in females but never in males [22,23].
Our data do not enable us to explain why pk2b2 is only expressed in feminizing strains of Wolbachia whereas its homologs, also found in CI-inducing strains, are associated with CI phenotype in mosquitoes [22,23]. First, one can suggest that this allele has been inactivated or importantly down regulated in the CI-inducing strains of isopods. Change in regulatory element repertoire and divergence in patterns of expression may occur after small-scale duplication of the genome [36]. A corollary to a change in location, paralogous and homologous pk2 copies within and among Wolbachia strains would have followed different evolutionary trajectories leading to such a phenotypic diversity. Second, genomic imprinting, process by which genes are expressed from only one parental allele due to epigenetic mechanism, can be considered as a molecular mechanism underlying the diversity of phenotypes. Recently, early changes in gene imprinting and aberrant expression of specific genes have been shown to be coupled to parthenogenesis in mice embryos [37]. Third, one can suggest that genes in the pk2 family could have diverse functions. In this way, post-transcriptional modifications and dosage of Wolbachia products, as well as genetic control by the host, cannot be dismissed. As previously suggested [38], differences in Wolbachia-induced feminization as well as the presence of the bacteria in O. asellus males, may simply result from differences in bacterial dosage or in host targets. The basic molecular mechanisms that mediate Wolbachia feminization are also still unknown although it is unlikely that this effect is driven by only one gene. In A. vulgareWolbachia effectors may target the proteinaceous androgenic hormone or its receptor, or another major sex determinant, thereby inhibiting the androgenic gland differentiation and preventing the androgenic hormone from reaching the target tissues such as gonads and tegumental epithelium [2,39,40]. This hypothesis suggests a late action of feminizing Wolbachia on host target(s) during its development, as opposed to the very early action of other Wolbachia strains that induce parthenogenesis, CI or male killing [5,41].
Conclusions
Our results highlight a large copy number variation of both pk1 and pk2 genes among strains, likely linked to prophage diversity, and also the specific expression of one pk2 allele only in the feminizing Wolbachia strains of isopods. This correlation supports the hypothesis that phenotype-related effectors or specific strain determinants in Wolbachia are likely to be encoded by prophage genes, ankyrin-repeat encoding genes, and predicted genes of unknown function [42]. Our results thus reveal the need to search for host molecules targeted by Wolbachia ankyrins and their functions with respect to host sex manipulation by Wolbachia.
Methods
Wolbachia-infected isopod species
All isopods used in this study were collected in France and reared in the laboratory. To date, CI Wolbachia have been described in three species: Cylisticus convexus (wConV, Villedaigne) [41], Porcellio dilatatus petiti (wPet, Saint-Honorat) [43] and P. d. dilatatus (wDil, Sainte-Marguerite) (Grève, unpublished results). Wolbachia strains inducing feminization have been described in A. vulgare (wVulC, Celles sur Belle and wVulM, Mery sur Cher) [44,45], A. nasatum (wNas, Poitiers) [46], Oniscus asellus (wAse, Quinçay) [38], Porcellionides pruinosus (wPruIII, Nevers) [47]. An uninfected lineage of A. vulgare (originating from Nice, France) was used as negative control for PCR and Southern blotting experiments.
Total DNA was extracted from male and female gonads of all isopod species as described previously [48]. Infection status of each individual was confirmed by a PCR-assay based on the bacterial 16S rDNA gene using Wolbachia-specific primers ( Additional file 1: Table S1) [49].
Distribution of pk1 and pk2 genes
The genome of the feminizing wVulC Wolbachia strain is at the final assembly step (whole-genome shotgun-sequencing project: European Wolbachia EuWol (contract QLK3-CT2000-01079, coordinated by K. Bourtzis, University of Ioannina, Greece). This includes phage contigs of which sequences are homologous to the Wolbachia WO prophage. Annotation of the pk1 and pk2 genes was performed by protein and DNA homology searches with BLASTP and BLASTN programs [50] using the wPip-Pel pk1 and pk2 alleles as queries (see Table 1). Ankyrin and other functional motif predictions were performed by the SMART web server [51] on protein sequences.
Specific primers were designed to amplify full-length or 200–500 bp fragments of the wVulC pk1 and pk2 alleles using a standard PCR protocol as previously described ( Additional file 1: Table S1) [52]. The purified PCR products were directly sequenced on both strands on an ABI PRISM 3100 Genetic Analyzer using Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) according to the manufacturer’s instructions.
pk1 or pk2 copy number variation among Wolbachia strains was assessed by Southern blotting. About 15 μg of total DNA were digested at 37° overnight with EcoRI or BamHI enzymes that did not cut any of the wVulC pk1 and pk2 alleles. Digested DNA as well as undigested DNA from non-infected ovaries used as controls (data not shown) was electrophoresed on 0.8% agarose gels and blotted to nylon membranes. Probes were obtained by PCR amplification of the wVulC full-length pk1 (pk1a and pk1b types) and pk2 (pk2b type) ank genes ( Additional file 1: Table S1), labelled using [α-32P]-dCTP by the random primer method and hybridized overnight to membranes. The final wash was performed at 52° in 0.1X SSC. Hybridized blots were imaged and analyzed using a PhosphoImager (Molecular Dynamics, Sunnyval, CA, USA).
Sequence analyses of pk1 and pk2 genes
Homologous sequences of both genes were first aligned in the server-based program MAFFT (http://align.bmr.kyushu-u.ac.jp/mafft/online/server/) using automatic settings. The resulting matrices were manually checked according to the predicted amino acid translation using BioEdit v7.0.9 [53]. MEGA5 software [54] was used to calculate nucleotide sequence divergence. For each locus, the GC content, the number of variable sites and the level of nucleotide diversity per site (Pi) were calculated. Ka/Ks likelihood analysis was also performed using the Selecton web server [55].
Recombination analysis was performed with RDP version v3.42 [56] using an alignment of non-redundant pk1 and pk2 nucleotide region encoding ANK-repeat domains. The parameters were set as follows: sequences were considered linear, the highest acceptable P value cut-off was 0.01, a Bonferroni correction was applied, consensus daughter sequences were found, gaps were included, different window sizes of variable sites were tested and 1,000 permutations were performed.
The best-fitted model of DNA evolution was estimated with jModelTest v0.1.1 [57] according to the corrected Akaike Information Criterion [58]. The selected model was TIM + G for pk1 and HKY + I for the pk2 locus encoding the ANK domain cluster. Gene genealogies were constructed using MrBayes v3.1.2 software [59,60] and supported by Bayesian and Maximum likelihood (ML) probabilities. Two Metropolis-coupled Markov chain Monte Carlo (MCMC) analyses were run for 5,000,000 generations and sampled every 250 generations. The first 25% of sampled trees were considered burn-in trees and were discarded before constructing a 50% majority rule consensus tree. ML analyses were carried out in PhyML 3.0 [61]. Node support came from 1,000 multiparametric bootstrap replicates. The networks were visualized with FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree). The network tree of the wsp gene was built following an identical Bayesian methodology (model: TPM3uf + I + G) ( Additional file 1: Figure S2).
Expression of ankyrin genes
Total RNAs were isolated from 20 to 50 gonads dissected from all species using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Ovaries were used in A. vulgare and A. nasatum where only females are infected. After a treatment with DNaseI (2U/μL, Ambion) at 37° for 30 min, 1 μg of RNA was used for reverse transcription using Superscript III kit (Invitrogen) as described by the manufacturer. To determine the expression of each gene, 1 μL of the reverse transcriptase reaction was used as template for the RT-PCR experiments. Control of the RT reactions was performed by omitting reverse transcriptase in the negative (RT-) controls and by testing the expression of the Wolbachia 16S rDNA gene ( Additional file 1: Table S1). Genomic DNA of all species was also used as a positive control of the PCR reactions as well as the one of the uninfected population (Nice, France) as negative control. Transcriptional analyses of pk2b2 and orf7 genes in several tissues of A. vulgare harbouring the feminizing wVulC Wolbachia strain were run as previously described [52]. The 16S rDNA gene expression was again used to check the quality of the extracted RNA in all these tissues.
Authors’ contributions
SP carried out the molecular genetic studies, participated in the data acquisition and performed all analyses and drafted the manuscript. CL and LC participated in the data acquisition. RAG was involved in project conception and critical revision of the manuscript. PG and DB coordinated the study, participated in its design, in the data acquisition and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Southern blotting analyses. Reconstituted Southern blots of EcoRI or BamHI digested DNA from 8 Wolbachia-infected terrestrial isopod species hybridized with three different probes (see text for details). White triangles highlight positions of the hybridized fragments. Lanes were loaded with DNA from Wolbachia strain endosymbionts of PDP as P. dilatatus petiti; PDD as P. dilatatus dilatatus; CC as C. convexus; AVC as A. vulgare strain wVulC; AVM as A. vulgare strain wVulM; AN as A. nasatum; OA as O. asellus; PP as P. pruinosus strain wPruIII. The number of bands in some lanes is higher than the number of copies presented in Table 2 due to EcoRI and/or BamHI restriction site(s) in these copies, as confirmed by sequencing. Upper light bands correspond to partially digested DNA fragments. Figure S2. Phylogenetic tree ofWolbachiastrains based on thewspgene.Wolbachia strains of isopods are shown in bold (wAlbum: Armadillidium album; wAse: Oniscus asellus; wConV: Cylisticus convexus; wDil: Porcellio dilatatus dilatatus; wElo: Chaetophiloscia elongata; wHoo: Sphaeroma hookeri; wMus: Philoscia muscorum; wNas: Armadillidium nasatum; wOce: Ligia oceanica; wPet: Porcellio dilatatus petiti; wPruIII: Porcellionides pruinosus; wRug: Sphaeroma rugicauda; wScaber: Porcellio scaber; wVulC, wVulM, wVulP: Armadillidium vulgare). The additional B-supergroup Wolbachia strains and the host phenotypes they induce are based on previously published information (wAlbB: Aedes albopictus; wAlt: Chelymorpha alternans; wAu, wMa, wNo, wRi: Drosophila simulans; wBol: Hypolimnas bolina; wCauB: Cadra cautella; wCon: Tribolium confusum; wDei: Trichogramma deion; wEnc: Acraea encedon; wFor: Encarsia formosa; wFir: Gryllus firmus; wKue: Ephestia kuehniella; wMel: Drosophila melanogaster; wOri: Tagosodes orizicolus; wPip-JHB, wPip-Pel: Culex pipiens quinquefasciatus; wScap: Ostrinia scapulalis; wSn: Drosophila sechellia; wStri: Laodelphax striatellus; wTai: Teleogryllus taiwanemma; wVitA: Nasonia vitripennis). Confirmed or suspected induced-phenotypes of Wolbachia strains of isopods are drawn from Bouchon et al. (2008). The red colour of strains corresponds to the feminizing induced-phenotype, blue to CI, green to male killing, light grey to parthenogenesis, and black to suspected feminization. Node supports are shown by posterior probabilities from Bayesian inferences. Figure S3. SMART outputs representing the number of ANK motifs found in Pk1 translated sequences. Figure S4. SMART outputs representing the number of ANK motifs found in Pk2 translated sequences. Table S1. List of primers used in this study for sequencing (PCR), for expression analyses (RT-PCR), or for Southern blots (SB). Expected PCR product size in base pair (bp) was calculated relative to the wVulC reference sequences. Table S2. List ofpk1andpk2sequences used for Figure 1, Additional file 1: Figure S3 and Additional file 1: Figure S4. Accession numbers from this study are in bold.
Contributor Information
Samuel Pichon, Email: samuel.pichon@unibas.ch.
Didier Bouchon, Email: didier.bouchon@univ-poitiers.fr.
Chao Liu, Email: chaoliu@bio.ku.dk.
Lanming Chen, Email: lanmingchen@yahoo.com.cn.
Roger A Garrett, Email: garrett@bio.ku.dk.
Pierre Grève, Email: pierre.greve@univ-poitiers.fr.
Acknowledgements
We are grateful to C. Debenest, C. Delaunay, D. Guyonnet, A. Lafitte, J. Lesobre and M. Raimond for technical assistance. We thank I. Mazerie and H. de Verdal for their contributions to the experiments. The authors acknowledge four anonymous reviewers whose comments helped improve the final manuscript. This research was funded by the Centre National de la Recherche Scientifique (CNRS), the French Ministère de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche, the French embassy in Denmark, by an EU grant (EuWol, QLRT-2000-01079) and the Agence Nationale de la Recherche (ANR-06-BLAN-0316). SP was supported by a Ph.D. fellowship from Région Poitou-Charentes.
References
- Baldo L, Dunning Hotopp JC, Jolley KA. et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchon D, Cordaux R, Grève P. In: Insect Symbiosis. Bourtzis K, Miller TA, editor. Taylor & Francis Group, Boca Raton; 2008. Feminizing Wolbachia and the evolution of sex determination in isopods; pp. 273–294. [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf A. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 2005;60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- Bourtzis K. Miller TA: Insect Symbiosis. Taylor & Francis Group, Boca Raton; 2003. [Google Scholar]
- Werren JH. Biology of Wolbachia. Annu Rev Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- Foster J, Ganatra M, Kamal I. et al. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Walker T, Sebaihia M. et al. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol. 2008;25:1877–1887. doi: 10.1093/molbev/msn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Westberg J, Sapountzis P. et al. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci USA. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg SL, Dunning Hotopp JC, Delcher AL. et al. Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 2005;6:R23. doi: 10.1186/gb-2005-6-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J. et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:e69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- Li J, Mahajan A, Tsai M-D. Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry. 2006;45:15168–15178. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng Z-Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kim KJ, Choi K-S, Grab DJ, Dumler JS. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell Microbiol. 2004;6:743–751. doi: 10.1111/j.1462-5822.2004.00400.x. [DOI] [PubMed] [Google Scholar]
- Zhu B, Nethery KA, Kuriakose JA. et al. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect Immun. 2009;77:4243–4255. doi: 10.1128/IAI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, den Dulk-Ras A, Hooykaas PJJ, Rikihisa Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007;9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- Fenn K, Blaxter M. Wolbachia genomes: revealing the biology of parasitism and mutualism. Trends Parasitol. 2006;22:60–65. doi: 10.1016/j.pt.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, O’Neill SL. Wolbachia-host interactions: connecting phenotype to genotype. Curr Opin Microbiol. 2007;10:221–224. doi: 10.1016/j.mib.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Duron O, Boureux A, Echaubard P. et al. Variability and expression of ankyrin domain genes in Wolbachia variants infecting the mosquito Culex pipiens. J Bacteriol. 2007;189:4442–4448. doi: 10.1128/JB.00142-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I, Burke GR, Riegler M, O’Neill SL. Distribution, Expression, and Motif Variability of Ankyrin Domain Genes in Wolbachia pipientis. J Bacteriol. 2005;187:5136–5145. doi: 10.1128/JB.187.15.5136-5145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkins SP, Walker T, Lynd AR. et al. Wolbachia variability and host effects on crossing type in Culex mosquitoes. Nature. 2005;436:257–260. doi: 10.1038/nature03629. [DOI] [PubMed] [Google Scholar]
- Walker T, Klasson L, Sebaihia M. et al. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol. 2007;5:39. doi: 10.1186/1741-7007-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atyame CM, Delsuc F, Pasteur N, Weill M, Duron O. Diversification of Wolbachia Endosymbiont in the Culex pipiens Mosquito. Mol Biol Evol. 2011;28:2761–2772. doi: 10.1093/molbev/msr083. [DOI] [PubMed] [Google Scholar]
- Atyame CM, Duron O, Tortosa P. et al. Multiple Wolbachia determinants control the evolution of cytoplasmic incompatibilities in Culex pipiens mosquito populations. Mol Ecol. 2011;20:286–298. doi: 10.1111/j.1365-294X.2010.04937.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Furukawa S, Nikoh N, Sasaki T, Fukatsu T. Complete WO phage sequences reveal their dynamic evolutionary trajectories and putative functional elements required for integration into the Wolbachia genome. Appl Environ Microbiol. 2009;75:5676–5686. doi: 10.1128/AEM.01172-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent BN, Funkhouser LJ, Setia S, Bordenstein SR. Evolutionary Genomics of a Temperate Bacteriophage in an Obligate Intracellular Bacteria (Wolbachia) PLoS One. 2011;6:e24984. doi: 10.1371/journal.pone.0024984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braquart-Varnier C, Grève P, Félix C, Martin G. Bacteriophage WO in Wolbachia infecting terrestrial isopods. Biochem Biophys Res Commun. 2005;337:580–585. doi: 10.1016/j.bbrc.2005.09.091. [DOI] [PubMed] [Google Scholar]
- Bordenstein SR, Wernegreen JJ. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 2004;21:1981–1991. doi: 10.1093/molbev/msh211. [DOI] [PubMed] [Google Scholar]
- Gavotte L, Henri H, Stouthamer R. et al. A Survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 2007;24:427–435. doi: 10.1093/molbev/msl171. [DOI] [PubMed] [Google Scholar]
- Masui S, Kamoda S, Sasaki T, Ishikawa H. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J Mol Evol. 2000;51:491–497. doi: 10.1007/s002390010112. [DOI] [PubMed] [Google Scholar]
- Masui S, Kuroiwa H, Sasaki T. et al. Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem Biophys Res Commun. 2001;283:1099–1104. doi: 10.1006/bbrc.2001.4906. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Pichon S, Ling A. et al. Intense transpositional activity of insertion sequences in an ancient obligate endosymbiont. Mol Biol Evol. 2008;25:1889–1896. doi: 10.1093/molbev/msn134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papafotiou G, Oehler S, Savakis C, Bourtzis K. Regulation of Wolbachia ankyrin domain encoding genes in Drosophila gonads. Res Microbiol. 2011;162:764–772. doi: 10.1016/j.resmic.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Yamada R, Iturbe-Ormaetxe I, Brownlie JC, O’Neill SL. Functional test of the influence of Wolbachia genes on cytoplasmic incompatibility expression in Drosophila melanogaster. Insect Mol Biol. 2011;20:75–85. doi: 10.1111/j.1365-2583.2010.01042.x. [DOI] [PubMed] [Google Scholar]
- Bu L, Bergthorsson U, Katju V. Local Synteny and Codon Usage Contribute to Asymmetric Sequence Divergence of Saccharomyces cerevisiae Gene Duplicates. BMC Evol Biol. 2011;11:279. doi: 10.1186/1471-2148-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Enkemann SA, Liang P. et al. Genome-wide gene expression profiling reveals aberrant MAPK and Wnt signaling pathways associated with early parthenogenesis. J Mol Cell Biol. 2010;2:333–344. doi: 10.1093/jmcb/mjq029. [DOI] [PubMed] [Google Scholar]
- Rigaud T, Moreau J, Juchault P. Wolbachia infection in the terrestrial isopod Oniscus asellus: sex ratio distortion and effect on fecundity. Heredity. 1999;83:469–475. doi: 10.1038/sj.hdy.6885990. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Bouchon D, Grève P. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet. 2011;27:332–341. doi: 10.1016/j.tig.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Negri I, Pellecchia M, Grève P. et al. Sex and stripping: the key to the intimate relationship between Wolbachia and host? Communicative & Integrative Biology. 2010;3:110–115. doi: 10.4161/cib.3.2.10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moret Y, Juchault P, Rigaud T. Wolbachia endosymbiont responsible for cytoplasmic incompatibility in a terrestrial crustacean: effects in natural and foreign hosts. Heredity. 2001;86:325–332. doi: 10.1046/j.1365-2540.2001.00831.x. [DOI] [PubMed] [Google Scholar]
- Ishmael N, Dunning Hotopp JC, Ioannidis P. et al. Extensive genomic diversity of closely related Wolbachia strains. Microbiology. 2009;155:2211–2222. doi: 10.1099/mic.0.027581-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand J-J, Martin G, Artault J-C. Correlation between the presence of a bacterial symbiont in oocytes of Porcellio dilatatus petiti and the sterility of the cross P. d. petiti male x P. d. dilatatus female. Arch Inst Pasteur Tunis. 1978;55:507–514. [PubMed] [Google Scholar]
- Cordaux R, Michel-Salzat A, Frelon-Raimond M, Rigaud T, Bouchon D. Evidence for a new feminizing Wolbachia strain in the isopod Armadillidium vulgare: evolutionary implications. Heredity. 2004;93:78–84. doi: 10.1038/sj.hdy.6800482. [DOI] [PubMed] [Google Scholar]
- Rigaud T, Souty-Grosset C, Raimond R, Mocquard J-P. Feminizing endocytobiosis in the terrestrial crustacean Armadillidium vulgare Latr. (Isopoda): recent acquisitions. Endocytobiosis & Cell Research. 1991;7:259–273. [Google Scholar]
- Rigaud T, Pennings PS, Juchault P. Wolbachia bacteria effects after experimental interspecific transfers in terrestrial isopods. J Invertebr Pathol. 2001;77:251–257. doi: 10.1006/jipa.2001.5026. [DOI] [PubMed] [Google Scholar]
- Michel-Salzat A, Cordaux R, Bouchon D. Wolbachia diversity in the Porcellionides pruinosus complex of species (Crustacea: Oniscidea): evidence for host-dependent patterns of infection. Heredity. 2001;87:428–434. doi: 10.1046/j.1365-2540.2001.00920.x. [DOI] [PubMed] [Google Scholar]
- Bouchon D, Rigaud T, Juchault P. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proceedings of the Royal Society B: Biological Sciences. 1998;265:1081–1090. doi: 10.1098/rspb.1998.0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill SL, Giordano R, Colbert AME, Karr TL, Robertson HM. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci USA. 1992;89:2699–2702. doi: 10.1073/pnas.89.7.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix C, Pichon S, Braquart-Varnier C. et al. Characterization and transcriptional analysis of two gene clusters for type IV secretion machinery in Wolbachia of Armadillidium vulgare. Res Microbiol. 2008;159:481–485. doi: 10.1016/j.resmic.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Tamura K, Peterson D, Peterson N. et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Doron-Faigenboim A, Erez E. et al. Selecton 2007: advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007;35:W506–W511. doi: 10.1093/nar/gkm382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M. et al. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Southern blotting analyses. Reconstituted Southern blots of EcoRI or BamHI digested DNA from 8 Wolbachia-infected terrestrial isopod species hybridized with three different probes (see text for details). White triangles highlight positions of the hybridized fragments. Lanes were loaded with DNA from Wolbachia strain endosymbionts of PDP as P. dilatatus petiti; PDD as P. dilatatus dilatatus; CC as C. convexus; AVC as A. vulgare strain wVulC; AVM as A. vulgare strain wVulM; AN as A. nasatum; OA as O. asellus; PP as P. pruinosus strain wPruIII. The number of bands in some lanes is higher than the number of copies presented in Table 2 due to EcoRI and/or BamHI restriction site(s) in these copies, as confirmed by sequencing. Upper light bands correspond to partially digested DNA fragments. Figure S2. Phylogenetic tree ofWolbachiastrains based on thewspgene.Wolbachia strains of isopods are shown in bold (wAlbum: Armadillidium album; wAse: Oniscus asellus; wConV: Cylisticus convexus; wDil: Porcellio dilatatus dilatatus; wElo: Chaetophiloscia elongata; wHoo: Sphaeroma hookeri; wMus: Philoscia muscorum; wNas: Armadillidium nasatum; wOce: Ligia oceanica; wPet: Porcellio dilatatus petiti; wPruIII: Porcellionides pruinosus; wRug: Sphaeroma rugicauda; wScaber: Porcellio scaber; wVulC, wVulM, wVulP: Armadillidium vulgare). The additional B-supergroup Wolbachia strains and the host phenotypes they induce are based on previously published information (wAlbB: Aedes albopictus; wAlt: Chelymorpha alternans; wAu, wMa, wNo, wRi: Drosophila simulans; wBol: Hypolimnas bolina; wCauB: Cadra cautella; wCon: Tribolium confusum; wDei: Trichogramma deion; wEnc: Acraea encedon; wFor: Encarsia formosa; wFir: Gryllus firmus; wKue: Ephestia kuehniella; wMel: Drosophila melanogaster; wOri: Tagosodes orizicolus; wPip-JHB, wPip-Pel: Culex pipiens quinquefasciatus; wScap: Ostrinia scapulalis; wSn: Drosophila sechellia; wStri: Laodelphax striatellus; wTai: Teleogryllus taiwanemma; wVitA: Nasonia vitripennis). Confirmed or suspected induced-phenotypes of Wolbachia strains of isopods are drawn from Bouchon et al. (2008). The red colour of strains corresponds to the feminizing induced-phenotype, blue to CI, green to male killing, light grey to parthenogenesis, and black to suspected feminization. Node supports are shown by posterior probabilities from Bayesian inferences. Figure S3. SMART outputs representing the number of ANK motifs found in Pk1 translated sequences. Figure S4. SMART outputs representing the number of ANK motifs found in Pk2 translated sequences. Table S1. List of primers used in this study for sequencing (PCR), for expression analyses (RT-PCR), or for Southern blots (SB). Expected PCR product size in base pair (bp) was calculated relative to the wVulC reference sequences. Table S2. List ofpk1andpk2sequences used for Figure 1, Additional file 1: Figure S3 and Additional file 1: Figure S4. Accession numbers from this study are in bold.