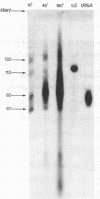
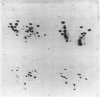
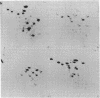
Abstract
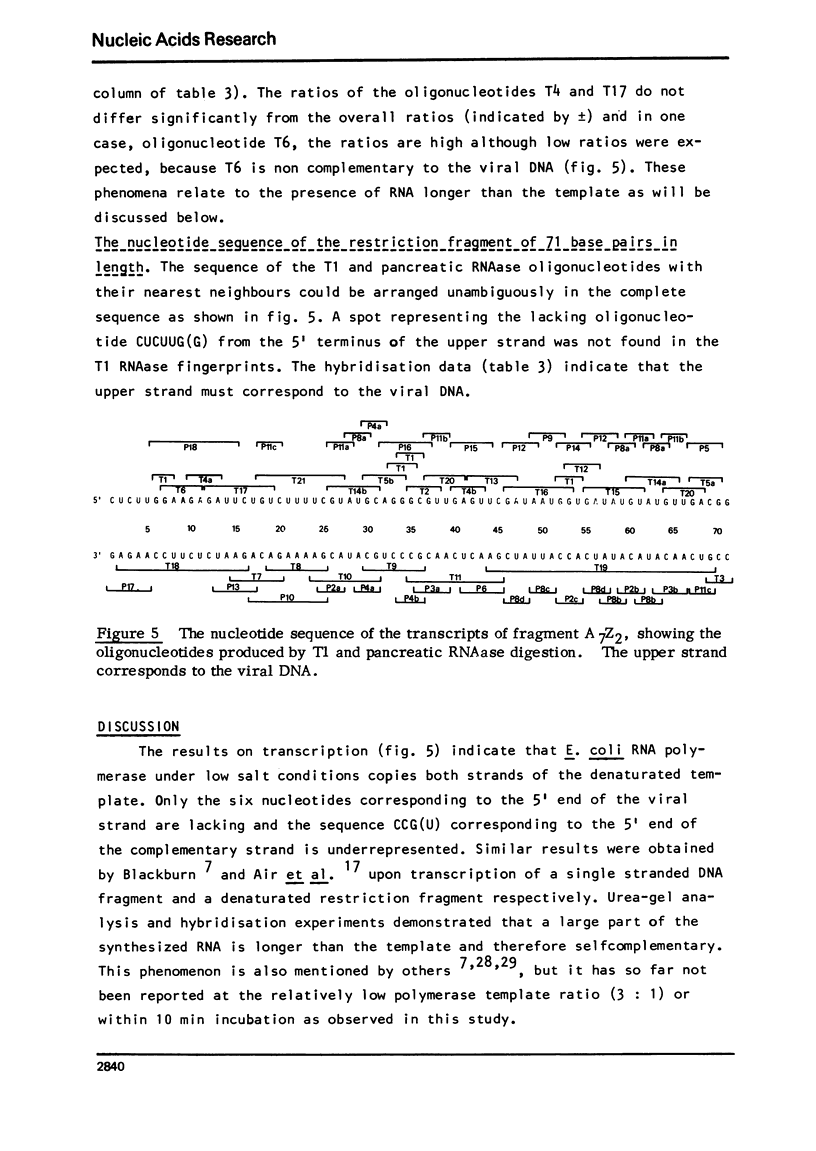
Part of the nucleotide sequence of a restriction fragment covering the origin of phiX174 DNA replication 1 has been determined. The fragment A7c was obtained by digestion of phiX174 RF DNA by the restriction enzyme from Arthrobacter luteus, Alu 1. It was further cleaved into two fragments, one large and one small, by the action of the restriction enzyme from Haemophilus aegyptius, Hae 111. The nucleotide sequence of the small fragment has been determined by analysis of the transcription products obtained by the action of Escherichia coli DNA-dependent RNA polymerase on denaturated template under conditions of low salt. Transcripts longer than the template were found. The whole sequence of 71 nucleotide pairs could be derived from complementary oligonucleotides, obtained after digestion of the transcripts with T1 or pancreatic RNAase. The sequence suggests that at least 4 of the 5 amber mutants 2 that have been mapped on this fragment are identical. On account of this and other evidence a reading frame is proposed.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D., Jansz H. S. Bacteriophage phiX174 DNA synthesis in a replication-deficient host: determination of the origin of phiX DNA replication. J Mol Biol. 1976 Apr 15;102(3):633–656. doi: 10.1016/0022-2836(76)90339-9. [DOI] [PubMed] [Google Scholar]
- Barrell B. G., Weith H. L., Donelson J. E., Robertson H. D. Sequence analysis of the ribosome-protected bacteriophase phiX174 DNA fragment containing the gene G initiation site. J Mol Biol. 1975 Mar 5;92(3):377–393. doi: 10.1016/0022-2836(75)90287-9. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H. Transcription by Escherichia coli RNA polymerase of a single-stranded fragment by bacteriophage phiX174 DNA 48 residues in length. J Mol Biol. 1975 Apr 15;93(3):367–374. doi: 10.1016/0022-2836(75)90283-1. [DOI] [PubMed] [Google Scholar]
- Borrias W. E., van de Pol J. H., van de Vate C., van Arkel G. A. Complementation experiments between conditional lethal mutants of bacteriophage psiX 174. Mol Gen Genet. 1969 Oct 13;105(2):152–163. doi: 10.1007/BF00445685. [DOI] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. The single-stranded DNA phages. CRC Crit Rev Microbiol. 1975 Dec;4(2):161–223. doi: 10.3109/10408417509111575. [DOI] [PubMed] [Google Scholar]
- Donelson J. E., Barrell B. G., Weith H. L. The use of primed synthesis by DNA polymerase I to study an intercistronic sequence of phiX-174 DNA. Eur J Biochem. 1975 Oct 15;58(2):383–395. doi: 10.1111/j.1432-1033.1975.tb02385.x. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Min Jou W., Molemans F., Raeymaekers A., Van den Berghe A. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976 Apr 8;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Galibert F., Sedat J., Ziff E. Direct determination of DNA nucleotide sequences: structure of a fragment of bacteriophage phiX172 DNA. J Mol Biol. 1974 Aug 15;87(3):377–407. doi: 10.1016/0022-2836(74)90093-x. [DOI] [PubMed] [Google Scholar]
- Godson G. N. Origin and direction phiX174 double- and single-stranded DNA synthesis. J Mol Biol. 1974 Nov 25;90(1):127–141. doi: 10.1016/0022-2836(74)90261-7. [DOI] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. Fragment maps of phiX-174 replicative DNA produced by restriction enzymes from haemophilus aphirophilus and haemophilus influenzae H-I. J Virol. 1974 Nov;14(5):1142–1151. doi: 10.1128/jvi.14.5.1142-1151.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P. G., Sanders L., Slocombe P. M. A restriction cleavage map of phiX 174 DNA by pulse-chase labelling using E. coli DNA polymerase. Nucleic Acids Res. 1976 May;3(5):1323–1329. doi: 10.1093/nar/3.5.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. H., Sinsheimer R. L. Structure of an intermediate in the replication of bacteriophage phiX174 deoxyribonucleic acid: the initiation site for DNA replication. J Mol Biol. 1974 Feb 15;83(1):47–61. doi: 10.1016/0022-2836(74)90423-9. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Smith H. O. A restriction enzyme from Hemophilus influenzae. II. J Mol Biol. 1970 Jul 28;51(2):393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- Kleid D., Humayun Z., Jeffrey A., Ptashne M. Novel properties of a restriction endonuclease isolated from Haemophilus parahaemolyticus. Proc Natl Acad Sci U S A. 1976 Feb;73(2):293–297. doi: 10.1073/pnas.73.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. S., Sinsheimer R. L. A cleavage map of bacteriophage phiX174 genome. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2882–2886. doi: 10.1073/pnas.71.7.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. S., Sinsheimer R. L. A continuous electroelution method for the recovery of DNA restriction enzyme fragments. Anal Biochem. 1974 Aug;60(2):640–644. doi: 10.1016/0003-2697(74)90279-6. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. M. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raué H. A., Stoof T. J., Planta R. J. Nucleotide sequence of 5-S RNA from Bacillus licheniformis. Eur J Biochem. 1975 Nov 1;59(1):35–42. doi: 10.1111/j.1432-1033.1975.tb02421.x. [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Myers P. A., Morrison A., Murray K. A specific endonuclease from Arthrobacter luteus. J Mol Biol. 1976 Mar 25;102(1):157–165. doi: 10.1016/0022-2836(76)90079-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Nathans D. Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J Mol Biol. 1973 Dec 15;81(3):419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M., Mitchell A. R. Chromatography of nucleic acid digests on thin layers of cellulose impregnated with polyethyleneimine. Biochem J. 1971 Jul;123(4):613–617. doi: 10.1042/bj1230613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Sugisaki H., Okamoto T., Takanami M. Studies on bacteriophage fd DNA. III. Nucleotide sequence preceding the RNA start-site on a promoter-containing fragment. Nucleic Acids Res. 1975 Nov;2(11):2091–2100. doi: 10.1093/nar/2.11.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TESSMAN I., PODDAR R. K., KUMAR S. IDENTIFICATION OF THE ALTERED BASES IN MUTATED SINGLE-STRANDED DNA. I. IN VITRO MUTAGENESIS BY HYDROXYLAMINE, ETHYL METHANESULFONATE AND NITROUS ACID. J Mol Biol. 1964 Aug;9:352–363. doi: 10.1016/s0022-2836(64)80212-6. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Borst P. Formation of self-complementary RNA on a single strand of DNA by RNA polymerase from Escherichia coli. Biochim Biophys Acta. 1971 Sep 24;246(3):450–457. doi: 10.1016/0005-2787(71)90781-7. [DOI] [PubMed] [Google Scholar]
- Terao T., Dahlberg J. E., Khorana H. G. Studies on polynucleotides. CXX. On the transcription of a synthetic 29-unit long deoxyribopolynucleotide. J Biol Chem. 1972 Oct 10;247(19):6157–6166. [PubMed] [Google Scholar]
- VERWOERD D. W., KOHLHAGE H., ZILLIG W. Specific partial hydrolysis of nucleic acids in nucleotide sequence studies. Nature. 1961 Dec 16;192:1038–1040. doi: 10.1038/1921038a0. [DOI] [PubMed] [Google Scholar]
- Vereijken J. M., Van Mansfeld A. D., Baas P. D., Jansz H. S. Arthrobacter luteus restriction endonuclease cleavage map of phi chi 174 RF DNA. Virology. 1975 Nov;68(1):221–233. doi: 10.1016/0042-6822(75)90163-4. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Jou W. M., Fiers W. Analysis of 32P-labeled bacteriophage MS2 RNA by a mini-fingerprinting procedure. Anal Biochem. 1976 May 7;72:433–446. doi: 10.1016/0003-2697(76)90551-0. [DOI] [PubMed] [Google Scholar]
- Weisbeek P. J., Vereijken J. M., Baas P. D., Jansz H. S., Van Arkel G. A. The genetic map of bacteriophage phiX174 constructed with restriction enzyme fragments. Virology. 1976 Jul 1;72(1):61–71. doi: 10.1016/0042-6822(76)90311-1. [DOI] [PubMed] [Google Scholar]
- Zain B. S., Weissman S. M., Dhar R., Pan J. The nucleotide sequence preceding an RNA polymerase initiation site on SV40 DNA. Part 1. The sequence of the late strand transcript. Nucleic Acids Res. 1974 Apr;1(4):577–594. doi: 10.1093/nar/1.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]