Abstract
DNA double-strand breaks (DSBs) represent one of the most deleterious forms of DNA damage to a cell. In cancer therapy, induction of cell death by DNA DSBs by ionizing radiation (IR) and certain chemotherapies is thought to mediate the successful elimination of cancer cells. However, cancer cells often evolve to evade the cytotoxicity induced by DNA DSBs, thereby forming the basis for treatment resistance. As such, a better understanding of the DSB DNA damage response (DSB–DDR) pathway will facilitate the design of more effective strategies to overcome chemo- and radioresistance. To identify novel mechanisms that protect cells from the cytotoxic effects of DNA DSBs, we performed a forward genetic screen in zebrafish for recessive mutations that enhance the IR–induced apoptotic response. Here, we describe radiosensitizing mutation 7 (rs7), which causes a severe sensitivity of zebrafish embryonic neurons to IR–induced apoptosis and is required for the proper development of the central nervous system. The rs7 mutation disrupts the coding sequence of ccdc94, a highly conserved gene that has no previous links to the DSB–DDR pathway. We demonstrate that Ccdc94 is a functional member of the Prp19 complex and that genetic knockdown of core members of this complex causes increased sensitivity to IR–induced apoptosis. We further show that Ccdc94 and the Prp19 complex protect cells from IR–induced apoptosis by repressing the expression of p53 mRNA. In summary, we have identified a new gene regulating a dosage-sensitive response to DNA DSBs during embryonic development. Future studies in human cancer cells will determine whether pharmacological inactivation of CCDC94 reduces the threshold of the cancer cell apoptotic response.
Author Summary
Radiation therapy and most chemotherapies elicit cancer cell death through the induction of excessive DNA damage. However, cancer cells can harbor genetic defects that confer resistance to these therapies. To identify cellular components whose targeted therapeutic inactivation could potentially enhance the sensitivity of treatment-resistant cancer cells to DNA–damaging therapies, we have chosen an unbiased genetic approach in live whole zebrafish embryos to identify genes that normally protect cells from the lethal effects of DNA damage. This approach has yielded the discovery of a novel radioprotective gene called ccdc94. Upon inactivation of ccdc94, cells become more sensitive to radiation-induced cell death. Our further analysis revealed that the Ccdc94 protein functions in the Prp19 complex, which is known to regulate gene expression and repair of damaged DNA. We found that this complex normally represses radiation-induced cell death by inhibiting the expression of the p53 gene, a critical mediator of DNA damage–induced cell death. Future experiments that inactivate Ccdc94 and Prp19 complex proteins in human cancer cells will determine if inactivation of this complex represents a novel therapeutic strategy that could increase p53 expression to enhance sensitivity to DNA damaging therapies in chemo- and radio-resistant cancer cells.
Introduction
After cells undergo genotoxic stress, multiple DNA-damage response (DDR) pathways are essential for the faithful replication and transmission of chromosomes to subsequent generations. Depending on the type of lesion, different pathways are engaged to repair the DNA [1]. One of the most detrimental lesions to occur upon exposure to ionizing radiation (IR) and certain chemotherapies is the DNA double-stranded break (DSB). Immediate cell cycle arrest following DNA DSBs plays a critical role in promoting efficient DNA repair before cells enter mitosis. When exposed to excessive amounts of DNA DSBs that overwhelm their repair machinery, cells that are competent to do so will undergo p53-dependent apoptosis [2]. While the exact events that determine how this decision is made are not well understood, it is clear that p53-mediated transcriptional induction of the BH3-only protein Puma is critical for IR-induced apoptosis [3]–[5]. Puma induction triggers the activation of Bax and Bak [6] leading to mitochondrial outer membrane permeabilization, release of apoptotic factors including cytochrome C, and activation of the Caspase cascade of proteolytic degradation. Once Caspases are activated, an irreversible program of cellular destruction ensues. Anti-apoptotic members of the Bcl-2 family of proteins, like Bcl-2 and Bcl-xL, can inhibit this process by binding and sequestering Puma (and other BH3-only proteins) to prevent activation of Bax/Bak. Consequently, mutations that lead to the overexpression of Bcl-2 or to the impairment of the p53 pathway play pivotal roles not only in the development and progression of cancer, but also in the resistance to chemo- and radiotherapy that develops in established tumors [2], [7].
Interestingly, a number of genes with prominent functions in the DSB-DDR pathway are also required for normal development of the nervous system [8]. Ataxia-Telangiectasia (A–T) was one of the earliest recognized diseases that arise from defects in the DSB-DDR pathway and is characterized by severe ataxia, radiosensitivity, defective immune function, sterility and predisposition to cancer [9]. A–T is caused by homozygous recessive mutations in ataxia-telangiectasia mutated (ATM) [10], a gene encoding a kinase that plays pivotal roles in sensing DNA DSBs and coordinating a complex cellular signaling response that mediates the commitment to undergo cell cycle arrest, DNA repair and apoptosis [11]. Developing neurons are highly proliferative and the associated increase in oxidative stress likely exposes them to excessive DNA damage, thereby explaining their unique sensitivity to defects in the DSB-DDR pathway [8]. Since p53-dependent apoptosis is a common consequence of excessive DNA damage in this tissue [2], developing neurons are selectively dosage-sensitive to IR. Indeed, we and others have shown that the developing nervous system in zebrafish represents an excellent system to identify genes required for the DSB-DDR [3]–[4], [12].
The DSB-DDR pathway is complex and remains incompletely understood [13]. ENU-based genetic screens provide an unbiased method for identifying new components of the DSB-DDR pathway whose inactivation could kill cells that have become resistant to DNA-DSB-inducing therapies. To date, there are no published accounts of forward genetic screens performed in vertebrate in vivo models designed to identify novel radioprotective genes. Here we describe a rapid, thirty-hour zebrafish screen to identify mutations that enhance apoptosis after exposure to moderate levels of IR. One of the mutants we identified from this screen, which we named radiosensitizing mutation 7 (rs7), causes a severe sensitivity of zebrafish embryonic neurons to IR-induced apoptosis and is required for the proper development of the central nervous system. Hypersensitivity to DNA-DSBs by rs7 arises from an increase in p53 mRNA expression and activity. We have mapped the rs7 mutation to an early stop codon within ccdc94, a gene that encodes a protein with few known functions or informative domains but that is highly conserved from yeast to humans [14]. Using biochemical and genetic approaches, we demonstrate that Ccdc94 is a functional component of the Prp19 complex in vertebrate cells. Prp19 complex members have established roles in pre-mRNA splicing [15]–[16] and DNA repair [17]. We show that depleting components of this complex renders cells more sensitive to DNA damage because of inappropriately high p53 mRNA and protein levels, but this effect does not arise from a global splicing defect. In summary, by taking advantage of the powerful embryonic and genetic attributes of the zebrafish system, we have identified a new gene regulating a dosage-sensitive DSB-DDR pathway during development.
Results
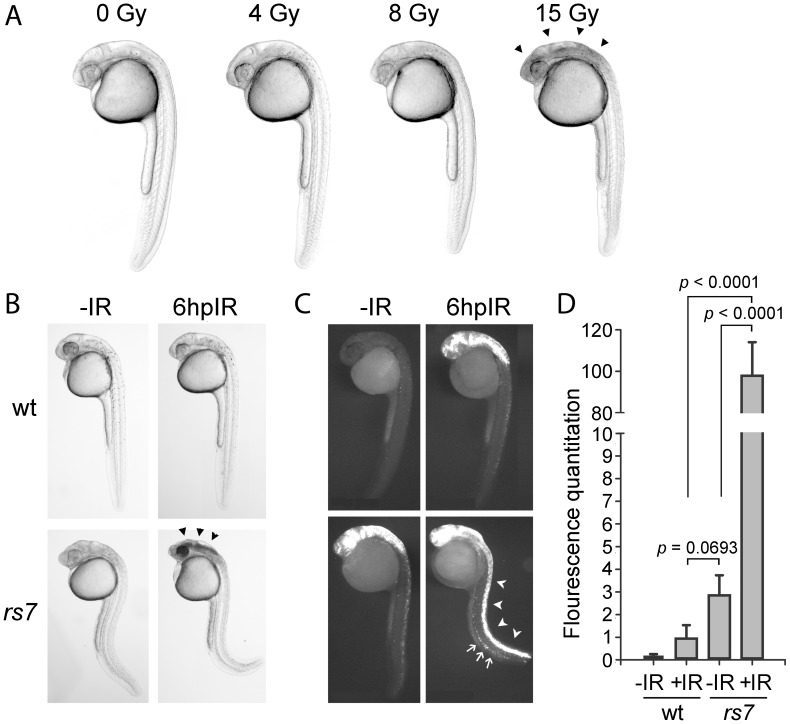
To discover novel radioprotective genes using the zebrafish genetic model, we first sought to identify an obvious bright-field phenotype in embryos that distinguishes different levels of apoptotic response to IR. High doses of IR, such as 15 Gy, administered to a transparent zebrafish embryo at 24 hours post-fertilization (hpf) cause extensive apoptosis in neural tissue resulting in the accumulation of opaque tissue in the head. This phenotype is very consistent and readily observable by bright-field microscopy by six hours post-IR (hpIR, Figure 1A, arrowheads). We treated wild-type zebrafish embryos with different levels of IR and found that the opaque tissue in the head was not observed using doses less than or equal to 8 Gy. These data show that a threshold of IR exposure exists between 8 and 15 Gy in wild-type embryos that gives rise to the obvious opaque neural tissue phenotype. We then reasoned that any mutation that inactivates a radioprotective gene should sensitize the embryonic neural tissue such that exposure of embryos to 8 Gy would cause a phenotype reminiscent of 15 Gy.
Figure 1. Identification of radiosensitizing mutation #7.
(A) Wild-type AB strain embryos were exposed to the indicated amount of IR at 24 hpf and visualized by brightfield microscopy 6 hours later. A threshold of IR exists between 8 and 15 Gy that gives rise to obvious cell death (seen as opaque tissue, marked by arrowheads) in the brain. (B) Wild-type embryos (derived from crossing wild-type parents) or rs7 mutant embryos (derived from crossing rs7 heterozygous parents) were exposed to 8 Gy IR at 24 hpf and visualized by brightfield microscopy 6 hours later. Arrowheads in the rs7 mutant mark cell death reminiscent of exposure of wild-type embryos to 15 Gy (as shown in A) (C) Embryos in (B) were sorted by phenotype, fixed at 6 hpIR and analyzed by immunofluorescence to detect activated Caspase-3. The “curly-up” tail phenotype and opaque tissue in the head were used to identify rs7 mutants. These phenotypes are present (to different degrees of severity) in both unirradiated and irradiated mutants such that they can be readily distinguished from siblings and wild-type at 30 hpf. After fixation of mutants, tails were clipped to distinguish them from wild-type embryos, which were analyzed in the same tube for Caspase-3 activity. Embryos were grouped for analysis according to whether embryos were irradiated or not, and rs7 mutants were identified based on the presence of tail-clips. Arrowheads point to the enhanced apoptosis in the spinal cord of an rs7 mutant, and arrows point to increased apoptosis in the ICM region of an rs7 mutant. (D) Immunofluorescence in the spinal cords from at least 10 embryos per group in (C) was quantified. wt; wild-type. See also Figure S1.
Based on this logic, we performed a recessive genetic screen using 8 Gy IR (Figure S1) and identified a number of mutations that sensitize embryos to IR. The first mutation we characterized was rs7 (Figure 1B) because these mutants showed a pronounced response to 8 Gy IR. The rs7 mutation gives rise to a homozygous recessive radiosensitive phenotype. Embryos that are heterozygous for the rs7 mutation are phenotypically identical to homozygous wild-type embryos with reference to all of the assays performed in this study. To verify that the observed neural cell death in irradiated rs7 mutants was due to apoptosis, we fixed the embryos at six hpIR and performed immunofluorescence with an antibody to detect activated Caspase-3 (Figure 1C). Wild-type embryos showed moderate levels of activated-Caspase-3-positive apoptotic cells in the brain and spinal cord in response to 8 Gy IR. By comparison, irradiated rs7 mutants showed a dramatic increase in cell death in the neural tissue when compared to wild-type irradiated embryos (Figure 1C, arrowheads). We also occasionally noticed an increase in IR-induced apoptosis in the intermediate cell mass (ICM) where primitive hematopoietic tissue resides (Figure 1C, arrows), but we remained focused on analyzing the consistent neural radiosensitive phenotype. Unirradiated rs7 mutant embryos exhibited a level of apoptosis in neural tissue that appeared similar to that of irradiated wild-type embryos indicating that the rs7 mutation also causes apoptosis in neural tissue independent from IR. For clarity, this will subsequently be referred to as “rs7-mediated neurodegeneration.”
Since radiosensitization is characterized by a multiplicative, rather than additive, effect on the response to IR, we questioned whether the enhanced apoptosis in the irradiated rs7 mutants represented a true radiosensitization or simply an addition of rs7-mediated neurodegeneration to the apoptosis that is normally induced by 8 Gy IR. To address this question, we quantified levels of fluorescence in each of the four experimental groups from Figure 1C and plotted the values in Figure 1D, normalizing the response of irradiated wild-type embryos to one. As suggested by Figure 1C, unirradiated rs7 mutants showed levels of fluorescence that were not significantly different from those of irradiated wild-type embryos. Notably, compared to irradiated wild-type embryos, irradiated rs7 mutants have a 95-fold increase in activated-Caspase-3 staining. These data show that rs7 is a bona fide radiosensitizing mutation.
While identification of mutations like rs7 that cause neurodegeneration is a relatively common event in zebrafish ENU-mutagenesis screens [18]–[19], we found that only 11% (4/36) of the neurodegenerative mutants we identified also gave rise to a radiosensitizing phenotype. Conversely, we have also identified radiosensitizing mutations that do not affect the survival of neural tissue at 30 hpf (our unpublished data). Indeed, morpholino knockdown of known components of the DSB-DDR pathway also leads to radiosensitization of neural tissue with or without associated developmental neurodegeneration ([20]–[21], our unpublished observations). This suggests that sensitivity to IR is not a frequent consequence of compromised survival in neural cells.
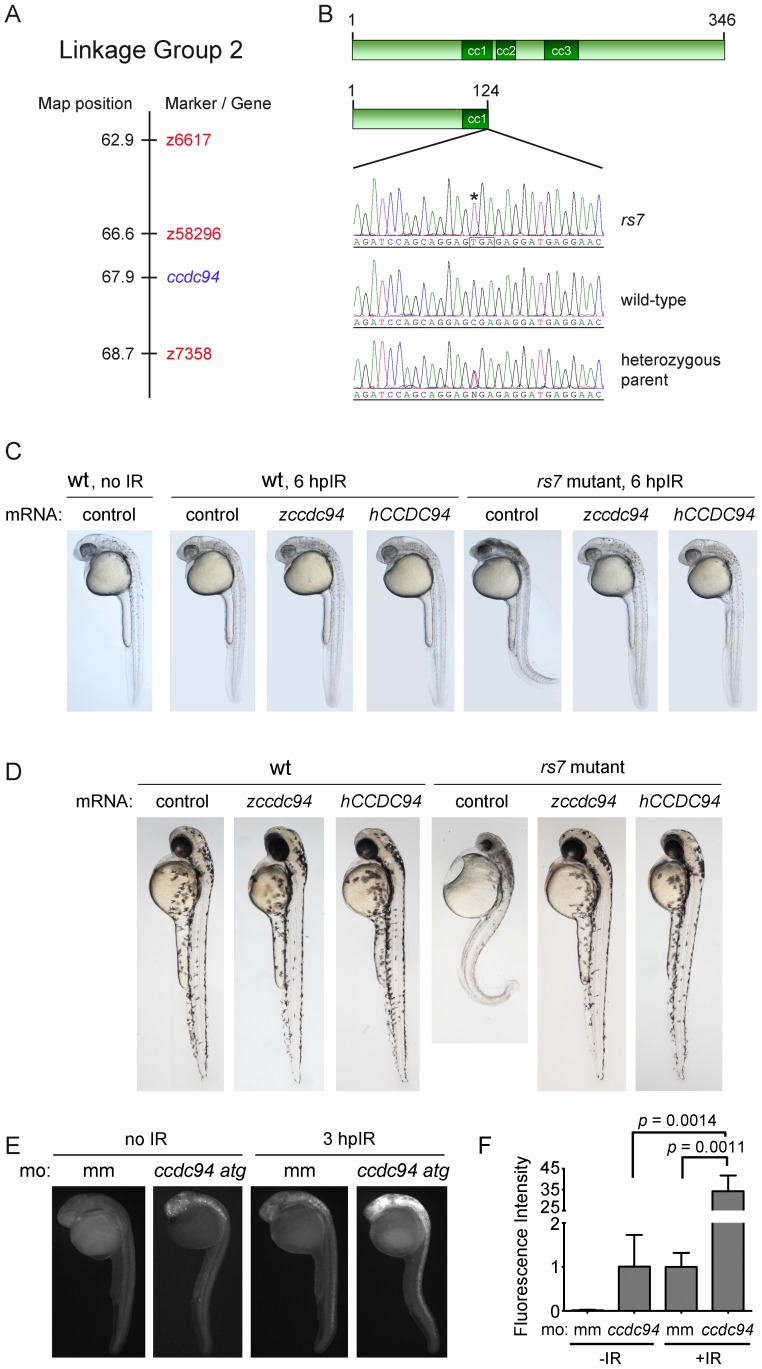
Genetic linkage analysis of the rs7 phenotype to 239 microsatellite markers revealed that markers z58296 and z7358 flanked a 2.1 centimorgan region on chromosome 2 that harbored the rs7 mutation (Figure 2A). Only 40 genes were annotated between these markers in the available zebrafish genome assembly Zv7 (http://www.ensembl.org/Danio_rerio), and none of these genes were implicated in the DSB-DDR response pathway. We reasoned that the gene harboring the rs7 mutation should be expressed in the brain and spinal cord because these tissues were selectively radiosensitized in rs7 mutants. Using published gene expression data for the 40 genes in the interval, we narrowed our list of candidates to six genes based on high levels of RNA expression in neural tissues (http://zfin.org/cgi-bin/webdriver?MIval=aa-pubview2.apg&OID=ZDB-PUB-040907-1). We sequenced five of these genes and found a premature stop codon in coiled-coil domain containing gene 94 (ccdc94, R125Stop, Figure 2B).
Figure 2. The rs7 phenotype is caused by a mutation in the ccdc94 gene.
(A) The rs7 mutation was localized to a 2.1 cM region (compared to the Massachusetts General Hospital genetic map for linkage group 2) as defined by flanking markers z58296 and z7358. (B) Sequencing of candidate genes (based on gene expression profiles) revealed a premature stop codon in the ccdc94 gene. Sequence from the ccdc94 gene exon 4 is shown for an rs7 mutant, an unrelated wild-type AB embryo, and a heterozygous parent of the rs7 mutant. Asterisk indicates the rs7 point mutation, and box illustrates mutation to a TGA nonsense codon. (C) Wild-type AB strain or rs7 heterozygous fish were incrossed, and the progeny were injected with mRNA encoding the indicated genes at the one-cell stage of development. At 24 hpf, embryos were exposed to 8 Gy IR and visualized by brightfield microscopy 6 hours later. (D) Embryos were injected similar to (C) but were not irradiated. Instead, they were left to develop until 48 hpf. (C and D) Because rs7 mutant and sibling embryos injected with wild-type ccdc94 mRNAs were morphologically indistinguishable, pictures were taken first, and the embryos were subsequently genotyped to identify mutants. (E) Wild-type embryos were injected with 400 µM mismatch morpholino (mm) as a negative control, or a translation-blocking ccdc94 morpholino (ccdc94 atg). Embryos were then irradiated at 24 hpf with 8 Gy IR and analyzed three hours later by immunofluorescence to detect activated Caspase-3. (F) Embryos in (E) were quantified similar to Figure 1D. Comparisons between mismatch morpholino-injected embryos plus and minus IR generated a p value of 0.0114. See also Figures S2 and S3.
The Ccdc94 gene is present in genomes from yeast to humans (http://www.ensembl.org/Danio_rerio/Gene/Summary?g=ENSDARG00000026185r=2:52746237-52760438) and has previously been shown to regulate pre-mRNA splicing [14]. While the zebrafish and human protein sequences share 67% overall identity (Figure S2), the first 175 amino acids are nearly an exact match (94% identity). The Ccdc94 protein contains three predicted coiled-coil domains (http://www.ensembl.org/Danio_rerio/Transcript/Summary?g=ENSDARG00000026185r=2:52746237-52760438t=ENSDART00000036813), all of which would be either eliminated or disrupted by the R125Stop mutation, suggesting that the R125Stop mutation interrupts a highly conserved function of Ccdc94.
To determine if the R125Stop mutation caused the rs7 radiosensitivity phenotype, we tested whether wild-type ccdc94 mRNA could rescue the excessive IR-induced apoptosis in rs7 mutants. We injected one-cell stage wild-type embryos, or embryos derived from a cross between rs7 heterozygotes, with mRNAs encoding either zebrafish ccdc94, human CCDC94 or egfp (control). We irradiated the clutches with 8 Gy and found that both zebrafish ccdc94 and human CCDC94 mRNA rescued the rs7 bright-field radiosensitization phenotype (Figure 2C). We also followed the development of rs7 mutant embryos in the absence of IR and found that the rs7 mutation causes major deterioration of neural tissue resulting in a small head and curled tail phenotype by day 2 (Figure 2D, Figure S3) and death by the end of day 3. To prove that this phenotype was also caused by the mutation in ccdc94, we performed the rescue experiment described above, but instead of irradiating the embryos at 24 hpf, we allowed them to develop unperturbed until 48 hpf. Figure 2D demonstrates that both zebrafish ccdc94 and human CCDC94 mRNA rescued the rs7 morphological phenotype at 48 hpf. Ultimately, however, this transient rescue (injected mRNA generally lasts for up to three days) did not rescue these embryos from lethality, as they eventually succumbed to complications from developing edema and overall dysmorphic effects by day 6 (data not shown). Nonetheless, these data demonstrate that the early embryonic radiosensitizing and neurodegenerative phenotypes of rs7 mutants are specifically due to the loss of a highly conserved function of Ccdc94.
To independently show that loss of the ccdc94 gene causes the rs7 phenotype, we knocked-down the endogenous ccdc94 in wild-type animals with an anti-sense translation-blocking morpholino. Figure 2E–2F demonstrates that ccdc94 knockdown radiosensitizes embryos as measured by whole-mount immunofluorescence to detect activated Caspase-3. Since the ccdc94 knockdown only increases Caspase-3 activity by 34-fold (compared to 95-fold in rs7 mutants, Figure 1D), the morpholino likely induces a partial knockdown of Ccdc94. While the “curly-up” phenotype seen in the rs7 mutants (Figure 1B, Figure 2C–2D) is not obvious at 27 hpf in ccdc94 morphants, it becomes prominent by 2 days-post-fertilization (data not shown). As such, we have confirmed by three independent assays that the rs7 phenotype is due to the effects of a recessive loss-of-function mutation in the ccdc94 gene.
The anti-apoptotic oncoprotein Bcl-2 has been shown in a number of studies to confer cancer-cell resistance to IR and chemotherapy [22]–[24]. To determine whether the rs7 mutation could overcome bcl-2 overexpression and restore apoptosis after exposure to IR, we injected one-cell stage wild-type or rs7 mutant embryos with 5 pg of bcl-2 mRNA. Figure S4 shows that while 5 pg of bcl-2 mRNA completely abolishes all apoptosis in irradiated wild-type embryos, it cannot achieve the same response in rs7 mutant animals, which exhibit typical levels of IR-induced apoptosis despite overexpression of bcl-2 mRNA. However, since rs7 mutants injected with 5 pg of bcl-2 mRNA exhibit less apoptosis than control-injected rs7 mutants (Figure S4), we hypothesized that higher levels of bcl-2 expression would fully block rs7-mediated radiosensitivity. Indeed, injection of 50 pg of bcl-2 mRNA (or mRNA encoding bcl-xL, another anti-apoptotic member of the Bcl-2 family) was able to completely block the rs7-mediated radiosensitization. These experiments indicate that Ccdc94 is a dose-dependent modifier of the anti-apoptotic function of bcl-2 in a manner that is genetically upstream of bcl-2 in the DSB-DDR pathway.
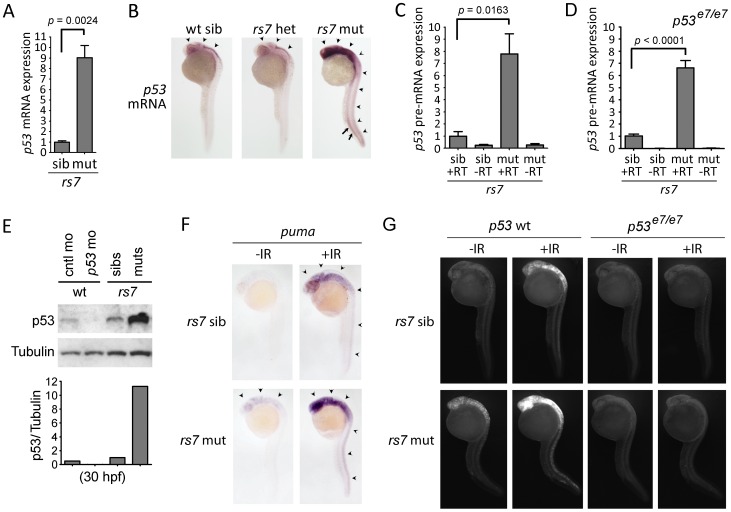
Since the only described function for Ccdc94 involved the regulation of splicing in yeast [14], we investigated if it played a similar role in vertebrates. We sequenced the transcriptome of 30 hpf wild-type, rs7 siblings and rs7 mutants by Illumina RNA-Seq analysis and compared gene expression profiles. We found no obvious global differences in splicing since greater than 96% of genes showed no significant differences (i.e., p>0.05) in mRNA expression, and there was no evidence of changes to alternative splicing. Instead we found a remarkable increase in p53 mRNA levels that were 3.4 and 5.1-fold higher in rs7 mutants compared to rs7 siblings and wild-type embryos, respectively. These results were confirmed by quantitative real-time reverse transcriptase PCR (qPCR) and whole-mount in situ hybridization with a probe complementary to p53 mRNA (Figure 3A, 3B). Interestingly, the high expression of p53 in both the neural and hematopoietic tissue in rs7 mutants (Figure 3B) mirrors the high expression of ccdc94 in these same tissues in wild-type embryos (http://zfin.org/cgi-bin/webdriver?MIval=aa-pubview2.apg&OID=ZDB-PUB-040907-1 and data not shown). To determine if elevated p53 mRNA levels were likely due to either increased p53 transcription or enhanced mRNA stabilization, we measured levels of intronic p53 sequence as a representation of pre-mRNA. Indeed, rs7 mutants show a similar increase in p53 pre-mRNA (Figure 3C) suggesting that the elevated levels of p53 are likely due to increased transcription.
Figure 3. The rs7-mediated radiosensitizing phenotype is caused by an increase in p53 mRNA expression.
(A) Thirty hpf rs7 siblings and mutants were identified based on morphology (similar to 1C). RNA was isolated, reverse transcribed using oligo-dT primers and analyzed for the expression of p53 mRNA by qPCR. Expression of the gapdh gene was also analyzed to normalize p53 mRNA levels. All data was then compared to sibling data, which was adjusted to a value of one. (B) rs7 siblings and mutants were grown to 24 hpf and analyzed by whole-mount in situ hybridization with a probe complementary to p53 mRNA. High levels of p53 mRNA expression are evident in neural tissue (arrowheads) and the ICM (arrows). Pictures were taken first, and the embryos were subsequently genotyped to identify mutants and heterozygous or wild-type siblings. (C) RNA from (A) was reverse transcribed with random hexamers, and intron 9 of the p53 gene was analyzed by qPCR to determine levels of p53 pre-mRNA. Expression of 28S RNA was also analyzed to normalize p53 pre-mRNA levels. Similar results were obtained from an analysis of intron 4 (data not shown). All data was then compared to sibling data, which was adjusted to a value of one. (D) rs7+/−;p53e7/e7 fish were incrossed to analyze rs7 siblings and mutants in a p53 homozygous mutant background. Rs7 mutants and siblings were distinguished by morphology since loss of p53 does not prevent the rs7-mediated “curly-up” tail phenotype. RNA was harvested at 30 hpf and analyzed as in (C). (E) Protein was harvested from rs7 siblings and mutants, and p53 and control morphants (injected at 400 µM) at 30 hpf and analyzed for p53 and Tubulin (as a loading control). ImageJ software was used to quantify band intensity from film. Shown below the blot are values for p53 divided by values for Tubulin (corresponding to above lanes) with rs7 siblings normalized to one. (F) rs7 sibling or mutant embryos were irradiated at 24 hpf with 8 Gy IR and analyzed 2 h later by whole-mount in situ hybridization with a probe complementary to puma mRNA. Arrowheads point to puma expression in neural tissue. (G) rs7 sibling or mutant embryos in the p53 wild-type or homozygous mutant background were irradiated at 24 hpf with 8 Gy IR and analyzed three hours later by immunofluorescence to detect activated Caspase-3. For panels (A), (C), and (D), error bars represent the standard error of the mean from at least three independent experiments. Panels (B) and (F–G) show representative data from at least three independent experiments. RT; reverse transcriptase. See also Figures S4, S5, S6.
Robu et al [25] have demonstrated the activation of a p53-dependent “general stress response” driven by morpholino off-target effects. While this pathway likely required the post-translational activation of p53, rather than upregulation of p53 transcription, we were concerned that induction of full-length p53 mRNA in rs7 mutants could be a downstream feed-forward mechanism of the p53-dependent general stress response. We tested this possibility by performing the same experiments in a p53 mutant background (p53e7/ e7, [26]) to eliminate the transcriptional activity of the p53 protein. Analysis of rs7;p53 double mutants showed that p53 mRNA remains highly elevated in this context and is not simply an indirect consequence of a general p53-dependent stress response (Figure 3D).
We next sought to clarify whether the increased p53 mRNA in rs7 mutants was due to specific regulation of p53 transcription or a general activation of the DSB-DDR pathway. To answer this question, we first tested wild-type embryos for IR-mediated induction of p53 mRNA. We found that IR exposure in wild-type embryos leads to an increase in p53 mRNA expression (Figure S5A) that is modest compared to unirradiated rs7 mutants (Figure 3A). We next analyzed the rs7 mutants for upregulation of genes that are known to be induced by IR-induced activation of E2F1 in a p53-independent manner, such as apaf1, caspase7, and p73 [27]. To measure p53-independent gene expression, we performed the experiment in the p53e7/ e7 background. Figure S5B demonstrates that none of these genes are significantly upregulated by the rs7 mutation. These experiments suggest that the rs7 mutation specifically induces p53 expression through a selective mechanism that is not simply due to general activation of the DSB-DDR pathway.
To determine whether the increased p53 mRNA in rs7 mutants translated to increased p53 protein, we analyzed protein levels in rs7 siblings and mutants by western analysis using a previously described antibody to zebrafish p53 (ZFp53-9.1, [28]). Indeed, we found that the rs7 mutation caused a dramatic increase in p53 protein compared to siblings (Figure 3E). A previously characterized p53 morpholino [26] was included to demonstrate antibody specificity. RNA-Seq analysis showed that a reduction in Mdm2 expression was likely not contributing to increased p53 protein levels since mdm2 mRNA levels were 1.5- and 1.2-fold higher in rs7 mutants than in rs7 siblings and wild-type embryos, respectively. These experiments define a new role for Ccdc94 in embryonic development as a negative regulator of p53 mRNA and protein expression.
We next questioned whether an increase in p53 protein activity accounts for the extreme radiosensitivity of the rs7 mutants to IR. Since p53-dependent puma induction is essential for IR-induced apoptosis in mammals and zebrafish [4]–[5], [29], we analyzed expression of puma mRNA as a measure of p53 transcriptional activity. As expected, after exposure to IR, puma expression is much stronger in rs7 mutants than siblings (Figure 3F), and the expression of puma in irradiated mutants and siblings requires wild-type p53 (data not shown). To validate the requirement for p53 in rs7-mediated radiosensitization, we tested the rs7 mutation in the p53 mutant background. We irradiated embryos at 24 hpf, analyzed them three hours later for apoptosis, and found that wild-type p53 is required to execute the rs7-mediated radiosensitivity (Figure 3G). These experiments suggest that the rs7-dependent increase in p53 protein and IR-induced activity accounts for the rs7-mediated radiosensitivity of developing neural tissue.
In the absence of IR, rs7 mutants show increased puma expression in neural tissue (Figure 3F, arrowheads) compared to siblings, a sign that increased p53 mRNA expression translates to an increase in p53 pro-apoptotic activity even in the absence of an exogenous DNA-damage signal. Overexpression of puma has been shown to have a potent pro-apoptotic effect in zebrafish embryos [3], [12], so we questioned whether this might contribute to the developing neurodegenerative phenotype that becomes strikingly evident by day 2 in rs7 mutants (Figure 2D, Figure S3 (arrowheads)). We found that expression levels of puma were extremely high in rs7 mutants as measured at 32 hpf and 48 hpf by qPCR, and this aberrant expression was entirely dependent on the presence of wild-type p53 (Figure S6A). In p53 mutants, where puma expression was absent, there was a marked reduction in neural cell death and an obvious improvement in brain development in rs7 mutants by brightfield microscopy (Figure S6B, arrows). While loss of wild-type p53 prolonged the life of rs7 mutants by 2–3 days, it ultimately failed to rescue the developmental lethality (data not shown) indicating that Ccdc94 also has essential p53-independent roles in development. However, these data show that inactivation of p53 significantly rescues the developmental neurodegeneration in rs7 mutants, and the pro-apoptotic activity of Puma likely contributes to rs7-mediated neurodegeneration.
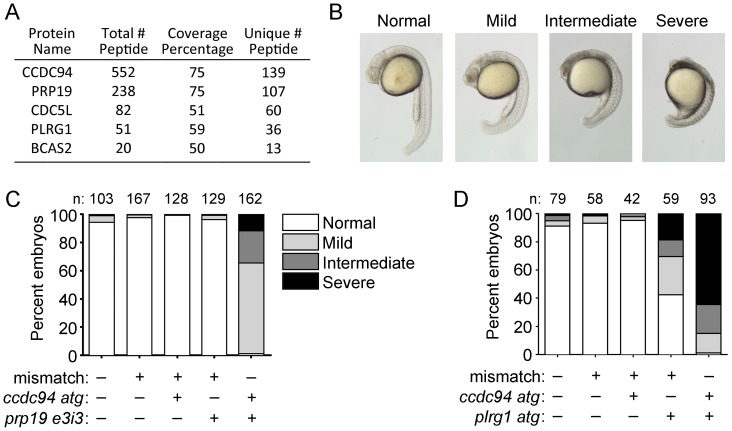
To gain further insight into the molecular function of Ccdc94 in the DSB-DDR pathway, we performed a tandem-affinity purification followed by mass spectrometry (TAP/MS) of human CCDC94 in mammalian cells. Specifically, CCDC94 cDNA was cloned into the pGlue vector encoding a dual-affinity tag containing streptavidin-binding protein, calmodulin-binding protein, and the hemagglutinin epitope. Lines of 293T cells expressing low levels of the tagged-bait fusion proteins were generated, detergent-solubilized, subjected to two rounds of affinity purification, trypsinized and analyzed by liquid chromatography–tandem mass spectrometry. The top hit was CCDC94 (Figure 4A) which serves as a positive control for the analysis. Interestingly, some of the other highly significant hits (PRP19, CDC5L, PLRG1, BCAS2) are core members of the PRP19 complex [30]–[32] which has previously been shown to be required for pre-mRNA splicing in yeast [15]–[16] and for DNA repair in both yeast and human cells [33]–[37].
Figure 4. Ccdc94 interacts with core members of the Prp19 complex.
(A) TAP/MS analysis shows that human CCDC94 binds to core members of the PRP19 complex. “Total Peptide #” indicates the number of peptides present in the analysis that are derived from the indicated protein. “Coverage Percentage” refers to the percent of protein sequence for a given protein that is represented by the peptides. “Unique # Peptide” refers to the number of unique peptides found for the indicated protein. (B, C) Wild-type embryos were injected with the ccdc94 atg morpholino (400 µM), prp19 e3i3 morpholino (200 µM), or mismatch morpholino (as a negative control and to keep the total concentration of morpholino injections at 600 µM) and grown to 22 hpf. A range of phenotypes were observed in the embryos as shown in (B). Embryos showing the different phenotypes from (B) were quantified in (C). (D) Wild-type embryos were injected with the ccdc94 atg morpholino (400 µM), plrg1 atg morpholino (20 µM), or mismatch morpholino (as a negative control and to keep the total concentration of morpholino injections at 420 µM), grown to 22 hpf, and analyzed as in (C). Data in (C) and (D) is derived from one representative experiment; however, three independent experiments were qualitatively similar for each panel. Number (n) of embryos analyzed for each group is indicated. See also Figure S7.
As an independent validation that Ccdc94 interacts with Prp19 complex members, we analyzed whether ccdc94 could genetically interact with prp19 or plrg1 in vivo. We reasoned that targeting each gene by morpholino knockdown would allow us to titrate levels of each protein to best reveal a potential genetic interaction. We designed a splice-blocking morpholino targeting prp19 (called prp19 e3i3, Figure S7) and made use of a previously characterized translation-blocking plrg1 morpholino [38]. We injected the highest dose of each morpholino that gave rise to minimal apoptosis or developmental abnormalities. At 22 hpf, injection with ccdc94 or prp19 morpholino alone gave rise to mostly normally developing embryos whereas the plrg1 morpholino alone led to an accumulation of cell death in the central nervous system in about half of injected embryos with varying severity (represented in Figure 4B and quantified in Figure 4D), similar to plrg1 mutants [39] and previously characterized morphants [38]. Strikingly, these same developmental abnormalities were highly abundant among embryos co-injected with ccdc94 morpholino and either prp19 or plrg1 morpholinos (Figure 4B–4D). These experiments indicate that ccdc94 genetically interacts with both prp19 and plrg1 in vivo.
To determine whether the radioprotective function of Ccdc94 is derived from its interaction with the Prp19 complex, we questioned whether loss of prp19 or plrg1 could phenocopy the rs7 mutation. We injected the prp19 morpholino into wild-type embryos and analyzed expression of p53 mRNA by qPCR. To ensure that we were analyzing full-length p53 mRNA and not a truncated isoform that has been shown to be non-specifically induced by morpholino off-target effects [25], we used primers that were designed to amplify across exons 1 and 2 of the p53 mRNA. The ccdc94 morpholino and a mismatch morpholino were injected as positive and negative controls for induction of p53 mRNA, respectively. The prp19 morpholino independently caused a significant increase in p53 mRNA and pre-mRNA expression, similar to knockdown of ccdc94 (Figure 5A–5B). Of note, upregulation of p53 transcripts in the ccdc94 and prp19 morphants was reduced compared to that observed in the rs7 and plrg1 mutants, likely due to incomplete knockdown by the morpholinos.
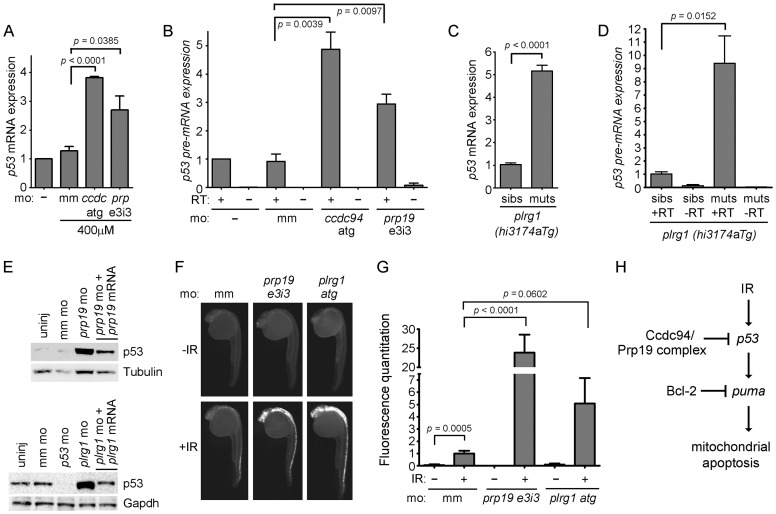
Figure 5. Loss of prp19 or plrg1 phenocopies, loss of ccdc94.
(A) Wild-type embryos were injected with the indicated morpholinos. RNA was harvested at 30 hpf, reverse transcribed using oligo-dT primers and analyzed for the expression of p53 mRNA by qPCR. Expression of the gapdh gene was also analyzed to normalize p53 mRNA levels. All values were then compared to the average value from uninjected embryos, which was adjusted to one. (B) RNA from (A) was reverse transcribed with random hexamers, and intron 9 of the p53 gene was analyzed by qPCR to determine levels of p53 pre-mRNA. Expression of 28S RNA was also analyzed to normalize p53 pre-mRNA levels. All values were then compared to the average value from uninjected embryos, which was adjusted to one. (C, D) Siblings and mutants from the plrg1(hi3174aTg) line were distinguished by morphology at 30 hpf and collected for analysis. RNA was harvested and analyzed by qPCR as in (A, B). For panels (A–D), error bars represent the standard error of the mean from at least three independent experiments. (E) Wild-type embryos were injected with the indicated morpholinos and analyzed at 30 hpf for p53 protein similar to Figure 3E. (F) Wild-type embryos were injected with the indicated morpholinos, irradiated (or not) at 24 hpf with 8 Gy and analyzed three hours later by whole-mount immunofluorescence to detect activated Caspase-3. Three independent experiments showed that knockdown of prp19 or plrg1 radiosensitized the embryos. (G) Activated-Caspase-3-specific immunofluorescence from (F) was quantified as in Figure 1D. (H) Genetic diagram showing that the Ccdc94 and core members of the Prp19 complex inhibit the transcription of p53, and therefore p53-mediated induction of puma expression, and normally restrict IR-induced mitochondrial apoptosis, upstream of Bcl-2. For all relevant panels in this figure, morpholinos were injected at the same concentrations as described in Figure 4B-4D. RT; reverse transcriptase, mm; mismatch, ccdc; ccdc94, prp; prp19. See also Figure S8.
We next took advantage of a plrg1 mutant zebrafish line [plrg1(hi3174aTg), [39]]. This line contains a retroviral insertion in intron 1 of the plrg1 gene that leads to severely reduced mRNA levels (Figure S8A). Embryos that are homozygous for this retroviral insertion exhibit major developmental cell death in the central nervous system and usually die by the end of day two [39]. Injection of wild-type plrg1 mRNA completely rescued this developmental phenotype at 24 hpf (Figure S8B). We analyzed p53 mRNA and pre-mRNA levels in the plrg1 mutants and found that, similar to the rs7 mutants, they express abnormally high levels of p53 mRNA and pre-mRNA (Figure 5C–5D).
We reasoned that the increase in p53 expression in response to knockdown or loss of prp19 and plrg1, respectively, should lead to an increase in IR-induced apoptosis, similar to knockdown or loss of ccdc94 (Figure 2E–2F and Figure 1C, respectively). Since the plrg1(hi3174aTg) mutants have severe neurodegeneration that interfered with our ability to evaluate irradiated embryos for an increase in apoptosis, we elected to use the plrg1 morpholino [38] to titrate plrg1 expression levels. We first analyzed whether knockdown of prp19 and plrg1 would phenocopy the rs7-mediated increase in p53 protein expression seen in Figure 3E. Figure 5E shows that knockdown with either the prp19 e3i3 or plrg1 atg morpholinos caused an increase in p53 expression that was specifically rescued by overexpression of the respective wild-type mRNAs. Knockdown of either prp19 or plrg1 also sensitized the embryos to IR-induced apoptosis (Figure 5F–5G). Together these results suggest that Ccdc94 is a component of the Prp19 complex which functions to protect proliferating embryonic neural cells from genotoxic stresses such as IR by modulating the levels of p53 mRNA expression (Figure 5H).
Discussion
We have previously shown that zebrafish embryonic neural tissue is an excellent model system to dissect the DSB-DDR pathway since it faithfully recapitulates many of the complex molecular signaling events elucidated in mammalian systems [4], [12]. With the notion that conserved embryonic pathways are exploited by cancer cells to promote survival, we embarked on a unique approach to use an unbiased genetic screen in zebrafish to identify novel radioprotective genes in the DSB-DDR pathway. The goal of these studies is to better understand the role of the DSB-DDR pathway in development, knowledge that can be ultimately translated to improved therapies for cancer treatment. Our efforts led us to identify a novel radioprotective gene called ccdc94. The Ccdc94 S. cerevisiae ortholog (named Yju2) has previously been shown to interact with the Prp19 complex [14], [30], [32] and is required for the first catalytic step in pre-mRNA splicing in yeast [14]. Our experiments confirm that the Ccdc94/Prp19 complex interaction is conserved in vertebrate cells; however, we have identified a new function for this complex in repressing p53 mRNA expression during development.
The Prp19 complex has a well-established role in pre-mRNA splicing in yeast [15]–[16], but whether the complex has an equivalent role in vertebrates is not clear [17]. Our studies show that the Prp19 complex normally represses p53 mRNA expression in neural cells to promote cell survival. There is a large body of evidence documenting the regulation of p53 activity by post-translational mechanisms [40] but a relative dearth of studies addressing potential mechanisms by which p53 is regulated at the transcriptional level despite the fact that potent transcriptional up-regulation could conceivably overcome post-translational mechanisms that restrict p53 activity. Indeed, we found that the 9-fold up-regulation of p53 mRNA transcripts in rs7 mutants translates to increased p53 activity. To date, there is no evidence to suggest that the Prp19 complex directly regulates transcription. An analysis of down-regulated genes in the rs7 mutants could yield novel, direct mechanisms by which the p53 gene is transcriptionally regulated.
The Prp19 complex has also been linked to DNA repair [17]. Indeed, the prp19 gene was originally identified in S. cerevisiae in a screen for radiosensitizing mutations [33]. The prp19 mutant identified in this screen, initially called xs9, was subsequently renamed pso4-1 since it was shown to have much greater sensitivity to the DNA interstrand cross-linking (ICL) agent PAVA (psoralen plus UVA light therapy) than to IR [34]. Further analysis of this thermoconditional mutant revealed that Prp19 functions in repair of DNA ICLs in a pathway that appears to be genetically separable from its role in pre-mRNA splicing [33]–[34], [36]. Studies of human PRP19 confirmed the role of the PRP19 complex in ICL repair [37] and also suggested a role for PRP19 in the repair of DNA DSBs [35]. Therefore, the increase in p53 expression resulting from loss of Prp19 complex gene expression could ultimately stem from a lack of DNA repair.
The neural tissue is one of the most radiosensitive tissues in the early developing zebrafish embryo. One likely explanation arises from the supposition that developing neurons are already poised to undergo apoptosis if they fail to receive neurotrophic survival factors from their synaptic targets [41]. As such, readiness to undergo DNA-damage-induced apoptosis is a feature that embryonic neural tissue shares with many cancers [42]. The neural tissue is also one of the most highly proliferative tissues in the developing embryo (our unpublished observations), another feature in common with cancer [43]. By exploiting the similarities between embryonic neural tissue and cancer cells, we have been able to use an unbiased genetic approach to identify novel and conserved genes involved in the DSB-DDR that represent potentially important targets in both neurodegenerative disease and radio-resistant cancers.
Sensitivity to IR-induced apoptosis by the rs7 mutation appears to be restricted to neural tissue and possibly primitive hematopoietic tissue in the ICM (Figure 1C). These areas are consistent with disproportionately high levels of ccdc94 mRNA expression during zebrafish embryonic development (http://zfin.org/cgi-bin/webdriver?MIval=aa-pubview2.apg&OID=ZDB-PUB-040907-1 and data not shown) as well as regions of high p53 mRNA expression (Figure 3B) in rs7 mutants. In general, tissues other than neural or ICM have modest expression of ccdc94 by comparison suggesting that Ccdc94-independent mechanisms evolved to regulate p53 expression in these other tissues. However, CCDC94 appears to be ubiquitously expressed in tissues of adult humans (http://www.ncbi.nlm.nih.gov/UniGene/clust.cgi?ORG=Hs&CID=21811) suggesting that it may have a broader role in the regulation of p53 expression beyond development.
In conclusion, we have identified and analyzed the role of a novel radioprotective gene, ccdc94, in the first genetic screen in a vertebrate system designed to identify radiosensitizing mutations in vivo. We found that in vertebrate cells Ccdc94 interacts with core members of the Prp19 complex both biochemically and genetically and that protection from IR-induced apoptosis by this complex is mediated by inhibition of p53 expression. Upregulation of p53 expression could potentially overcome mechanisms that evolve during cancer progression to restrict its protein activity. Future experiments will determine whether CCDC94 and PRP19 complex components will be useful targets for sensitizing p53 wild-type cancer cells to IR therapy.
Materials and Methods
Ethics statement
All experiments involving zebrafish conformed to the regulatory standards and guidelines of the Dana-Farber Cancer Institute and University of Utah Institutional Animal Care and Use Committee.
Zebrafish lines
Zebrafish were maintained, mutagenized and bred as described [44]. Wild-type embryos and the rs7 mutant line were derived from the AB strain. The rs7 mutant line was outcrossed to wild-type AB fish seven times and all phenotypes described herein are representative of crosses between at least seventh generation rs7 heterozygous fish. The plrg1(hi3174aTg) line was obtained from the Zebrafish International Resource Center (http://zebrafish.org/zirc/home/guide.php). We have previously described the p53e7/e7 zebrafish line that carries a homozygous M214K mutation in the p53 coding sequence [26].
Irradiator usage
IR was administered with either a Cs-137 gamma irradiator (Gammacell 1000) or an X-ray irradiator (RadSource RS2000). Irradiators were completely interchangeable such that 8 Gy of X-rays gave rise to identical embryonic phenotypes described in this study as 8 Gy of gamma rays.
Zebrafish microinjections
Zebrafish one-cell stage embryos were injected with 500 picoliters of mRNA or the indicated concentration of morpholino. In every experiment, total RNA or morpholino concentrations were kept constant through the addition of egfp mRNA or a mismatch morpholino, respectively. Morpholinos were designed and created by GeneTools, and sequences are listed in Text S1. For mRNA microinjection, zebrafish cDNAs were sub-cloned into pCS2+, and mRNA was made by 1) linearization of each construct with NotI, 2) SP6 Message Machine kit (Ambion, AM1340) and 3) purification for microinjection with NucAway Spin Columns (Ambion, AM10070). For morpholino rescue experiments, the plrg1 coding sequence was mutated to prevent direct binding of plrg1 mRNA to the translation-blocking plrg1 atg morpholino while creating only silent changes in regard to Plrg1 amino acid sequence (relevant primers are listed in Text S1).
Whole-mount in situ hybridization
For in situ hybridization, zebrafish cDNAs were sub-cloned into the pGEM-T-Easy (Promega). The pGEM-puma vector and generation of antisense probe was described previously [46]. The full-length zebrafish tp53 coding sequence was amplified with primers based on the published GenBank sequence (NM_131327 and Text S1). Full-length antisense and sense p53 RNA was generated by digesting with Apa1 and EcoRI, respectively, and transcribed in vitro with Sp6 and T3 polymerase, respectively. Embryos were dechorionated, staged, and fixed overnight in 4% paraformaldehyde at 4°C. Fixed embryos were washed in 1× PBST (1× PBS plus 0.1% Tween-20) at room temperature (RT, 3×10 minutes) and incubated in 100% methanol at −20°C for a minimum of two hours. Embryos were rehydrated in 1× PBST (3×10 minute washes at RT) and incubated in Hyb-minus (50% formamide, 5× SSC, and 0.1% Tween-20) for one hour at 68°C, then transferred to Hyb-plus (Hyb-minus, 5 mg/ml torula RNA type VI, 50 ug/ml heparin) for three hours at 68°C. RNA probe was added to embryos in Hyb-plus at 1 ng/uL and incubated overnight at 68°C. Embryos were then subjected to 20-minute washes at 68°C in the following order: twice with 2× SSCT-formamide (2× SSC, 0.1% Tween-20, 50% formamide), once with 2× SSCT, twice with 0.2× SSCT. At RT, embryos were then washed (3×10 minutes) with MABT (100 mM maleic acid, 150 mM sodium chloride, 100 mM Tris pH 9.5, 0.1% Tween-20), then incubated in block [MABT, 2% Blocking Reagent (Roche #11096176001), 1% fetal bovine serum] at RT for one hour. Embryos were incubated overnight in 1∶5000 anti-digoxigenin-fluorescein Fab fragments (Roche #11207741910) in block at 4°C. Embryos were then washed in 1× MABT (3×10 minutes at RT) and 0.1 M Tris pH 9.5 (3×10 minutes at RT) and stained with Vector BCIP/NBT alkaline phosphatase (Roche #11697471001). Upon completion of staining, embryos were washed in 1× PBST (3×10 minutes at RT) to stop the reaction. Embryos were imaged in either 80% glycerol or 3% methylcellulose (to allow for subsequent genotyping).
Whole-mount immunofluorescence and quantitation
Whole-mount activated Caspase-3 immunofluorescence was performed and quantified as described previously [12]. Quantitation was performed after immunofluorescence experiments by removing embryo tails at the level of the yolk. Tails were laid flat on a petri dish in 80% glycerol and activated Caspase-3 staining was documented by fluorescence microscopy using a Nikon Digital Sight DS-2MBWc black and white camera. All fluorescent pictures were taken at exactly the same exposure, gain, and magnification. Pictures were then cropped in Adobe Photoshop to include the same size region (using the ruler function on Photoshop) of the spinal cord using the end of the yolk extension as a reference point for all measurements, and to exclude any fluorescence arising outside of the spinal cord. Quantitation of fluorescence was performed with Volocity software by using the “Find Objects Using Intensity” option. The same exact parameters for eliminating the inclusion (and therefore quantitation) of background fluorescence were applied to all pictures equally. Six to twelve embryos from each group were included for all quantifications. GraphPad Prism software was used to plot the data, and error bars represent the standard error of averaged data from the embryos in a single experiment. Statistical analyses were performed using GraphPad Prism software using an unpaired student's T test. Representative quantitations from at least three experiments are shown for all data. Quantification represents measurements of fluorescence intensity which is directly related to Caspase-3 activity. However, fluorescence intensity is likely to fluctuate within cells. Therefore, changes in fluorescence intensity likely represent both increasing apoptotic cell number as well as increasing Caspase-3 activity within cells.
RNA–Seq analysis
The transcriptome of 30 hpf wild-type, rs7 siblings and rs7 mutants was analyzed by Illumina RNA–Seq analysis. Total RNA samples were prepared for sequencing using an Illumina mRNA Seq Sample Prep Kit and were sequenced using standard protocols on an Illumina Genome Analyzer IIx. Paired 36 base pair reads were obtained from the ends of each sequenced fragment. Reads were aligned to the Danio rerio Zv8 genome build (release date December 2008, obtained from UCSC Genome Bioinformatics, http://genome.ucsc.edu) with the SOAPAligner software (release 2.19, http://soap.genomics.org.cn/index.html). The SOAPAligner was run to allow an insert range of 100 to 140 bp, as the mean library insert size was 120 bp. Sequencing and alignment of the samples yielded the following in terms of sample, read pairs, aligned reads, and percent aligned: 1) rs7 muts, 19,034,491, 18,358,098, 96.4%, 2) rs7 sibs, 20,036,828, 19,206,760, 95.9%, 3) wild-type, 16,604,110, 12,630,153, 76.1%. RNA–Seq data from the rs7 mutant, sibling, and wild-type samples was analyzed for differential expression and differential splicing using the DefinedRegionScanSeqs method of the USeq software [45]. Transcript coordinates for this analysis were collected from the UCSC Genome Browser database [46].
Quantitative real-time PCR
RNA was isolated from embryos (10–25 embryos/sample) using the Qiagen RNeasy kit (74104). One microgram of purified RNA was used to generate cDNA using the Invitrogen Thermoscript RT-PCR kit (11146-024). For mRNA analysis, RNA was reverse transcribed using oligo-dT primers. For pre-mRNA analysis, RNA was reverse transcribed with random hexamers. cDNA was diluted 1∶20 in nuclease-free water and three technical replicates were analyzed using the LightCycler 480 Probes Master PCR mix (4707494001) and the Roche 480 Light Cycler or Eppendorf Realplex. Primers were designed by Roche to be used with their Universal Probe Library and are listed in Text S1. Primers were designed to cross exons 1 and 2 of the p53 gene to ensure specific amplification of full-length p53 mRNA, and expression of the gapdh gene was analyzed to normalize p53 mRNA levels. Introns 4 and 9 of the p53 gene were analyzed by qPCR to determine levels of p53 pre-mRNA, and expression of 28S RNA (which does not undergo splicing) was analyzed to normalize p53 pre-mRNA levels. All data was averaged from at least three independent experiments. GraphPad Prism software was used to plot the data, and error bars represent the standard error of averaged data. Statistical analyses were performed using GraphPad Prism software using an unpaired student's T test.
Tandem affinity purification/mass spectrometry analysis
CCDC94 cDNA was cloned into the pGlue vector encoding a dual-affinity tag containing streptavidin-binding protein, calmodulin-binding protein, and the hemagglutinin epitope. Lines of 293T cells expressing low levels of the tagged-bait fusion proteins were generated, detergent-solubilized, subjected to two rounds of affinity purification, trypsinized, and analyzed by liquid chromatography–tandem mass spectrometry. Tryptic peptides were separated by reverse phase nano-HPLC using a nanoAquity UPLC system (Waters Inc). Peptides were first trapped in a 2 cm trapping column (75 µm ID, C18 beads of 2.5 mm particle size, 200 Å pore size) and then separated on a self-packed 25 cm column (75 µm ID, C18 beads of 2.5 mm particle size, 100 Å pore size) at room temperature. The flow rate was 350 nl/min over a gradient of 5% buffer B (0.1% formic acid in acetonitrile) to 40% buffer B in 200 minutes. The identity of the eluted peptides was determined with a Velos-Orbitrap mass spectrometer (Thermo-Scientific). Specifically, following a FT full scan, MS2 spectral data were acquired by collision induced dissociation (CID) on the 9 most intense ions from the full scan, taking into account dynamic exclusion. The polysiloxane lock mass of 445.120030 was used throughout. All raw data were converted to mzXML format before a semi-tryptic search of the resultant spectra using Sequest and the Transproteomic Pipeline (TPP) on a Sorcerer 2.0 platform (Sage N Research, Milpitas, CA).
Western analysis
Approximately 100 30-hpf dechorionated embryos were rinsed three times at RT with 1× PBS. Ice-cold PBS was added, and embryos were incubated on ice for 5 minutes. Embryos were then de-yolked by pipetting 40 times through a thin bore plastic bulb pipet (Samco 235) in ice-cold PBS. Tubes were returned to ice for two minutes to let embryo bodies sink to the bottom of the tube. Supernatant was removed and embryo bodies were washed three times in ice-cold PBS, pelleted by centrifugation (1 second at top speed), and lysed with RIPA buffer (1% Nonide P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) containing 1% protease inhibitors (Sigma P8340) and 1% benzonase (Novagen 70746-3). BCA protein assay kit (Thermo Scientific 23227) was used to determine protein concentration. Ten micrograms of protein was loaded onto a denaturing gel (Novex NP0301BOX) and transferred to pre-hydrated PVDF membrane (GE PV4HYA0010). Membrane was blocked in 10% non-fat milk diluted in tris-buffered saline plus 1% tween-20 (TBST) for anti-p53 antibody. Zebrafish p53 antibody was kindly provided by Dr. Lane at A*STAR, Singapore and used at 1∶30 in 10% milk with overnight incubation at 4°C. The blot was washed (4×5 minutes each) in 1× TBST. Anti-mouse-horseradish peroxidase secondary antibody (Cell Signaling 7076S) was used at 1∶5000 in the same block for primary antibody. The blot was washed (4×5 minutes each) in 1× TBST before chemiluminescent horseradish peroxidase substrate (Millipore WBKLS0500) and film (Thermo Scientific 34090) were used to detect signal. Blot was stripped for 45 minutes rotating at 60°C (62.5 mM Tris, 2% sodium dodecyl sulfate, 100 mM 2-mercaptoethanol) and washed (8×15 minutes at RT) in TBST. The blot was then re-blocked in 3% bovine serum albumin (Amresco 0332) in TBST and probed using anti-GAPDH antibody (Abcam 9484) at 1∶2000 or anti-α-Tubulin antibody (Sigma T9026) at 1∶10,000. The mouse secondary antibody and detection was performed as outlined above. Developed films were electronically scanned at 600 dots per inch and quantified using ImageJ software.
Supporting Information
Schematic of genetic screen to identify recessive radiosensitizing mutations. Wild-type AB strain male zebrafish were treated with ENU giving rise to approximately 192 gene-inactivating mutations per sperm. ENU-mutagenized males were crossed with wild-type AB strain female fish to yield the F1 generation. Each fish in the F1 generation carried 192 unique mutations and were incrossed to create F2 families carrying 384 mutations per family. Up to eight random incrosses were performed within each F2 family, and the F3 clutches were subsequently analyzed for radiosensitizing phenotypes as described in the figure.
(TIF)
Alignment of zebrafish, mouse and human CCDC94 protein sequences. Protein sequences from zebrafish, mouse and human CCDC94 were aligned with the Jotun Hein algorithm using the default parameters in Lasergene MegAlign software. Zebrafish Ccdc94 is 67% identical to human CCDC94 and 69% identical to mouse CCDC94. An asterisk denotes R125, the residue that is mutated to a stop codon in rs7 mutants.
(TIF)
The rs7 mutation causes severe neurodegeneration that culminates in embryonic lethality. Wild-type embryos (derived from wild-type parents) and rs7 mutants were analyzed by brightfield microscopy on day 2 and day 3 of development. The rs7 mutation causes massive accumulation of cell death in the brain and spinal cord likely contributing to a small-head and “curly-up” tail phenotype. Rs7 mutant embryos usually fail to survive past day three.
(TIF)
Loss of ccdc94 compromises the anti-apoptotic function of Bcl-2. Wild-type (wt, derived from crossing wild-type parents) or rs7 mutant embryos (derived from crossing rs7 heterozygous parents) were injected at the one-cell stage of development with the indicated amounts of mRNA encoding zbcl-2, zbcl-xL, or egfp as a control (cntl). Embryos were then irradiated at 24 hpf with 8 Gy IR and analyzed three hours later by immunofluorescence to detect activated Caspase-3. Mutants were genotyped as in Figure 2C–2D. Representative data from three independent experiments is shown.
(TIF)
The rs7 mutation does not appear to cause a general activation of the DSB-DDR pathway. (A) Wild-type embryos were exposed (or not) to 8 Gy IR at 27 hpf and harvested for analysis three hours later. Quantitative PCR to analyze p53 mRNA expression was performed as in Figure 3A. (B) Rs7 siblings and mutants in the p53e7/e7 background [26] were analyzed at 30 hpf for expression of the indicated genes by qPCR similar to Figure 3A. For both (A) and (B), gapdh mRNA expression was measured to normalize gene expression levels. All data was then compared to rs7 sibling data, which was adjusted to a value of one. None of the values in (B) are significantly different from each other.
(TIF)
p53-mediated puma expression causes neurodegeneration in rs7 mutants. (A) rs7 sibling or mutant embryos in the p53 wild-type or mutant background were collected, and RNA was harvested at 30 hpf, reverse transcribed and analyzed for the expression of puma mRNA by qPCR. Rs7 mutants and siblings were distinguished by morphology since loss of p53 does not inhibit the rs7-mediated “curly-up” tail phenotype. (B) Pictures were taken of 48 hpf embryos from (A). Arrows point to neurodegeneration in rs7 mutants that is substantially rescued by loss of wild-type p53. For panel (A), error bars represent the standard error from three independent experiments.
(TIF)
The prp19 morpholino inhibits proper splicing of the prp19 pre-mRNA transcript. (A) Schematic of the zebrafish prp19 gene with exons shown as boxes and introns shown as lines between boxes. The e3i3 morpholino targets the splice donor site at the border of exon 3 and intron 3. In (B), F1 and R2 primer-based PCR will amplify from exon 2 to intron 3. Properly spliced mRNA should not give rise to PCR product. Inclusion of intron 3 should give rise to a 245-basepair product. (B) RNA from embryos injected with mismatch morpholino controls or prp19 e3i3 morpholino was harvested at 24 hpf, reverse transcribed with oligo-dT primers, and analyzed by PCR using primers shown in (A). Bands were excised from the gel, cloned into pGEM-T-Easy, and sequenced. Lower-case letters point to bands on the gel which correspond to regions of the prp19 gene shown below. When embryos were injected with the e3i3 morpholino, F1/R2 primers gave rise to a band representing inclusion of intron 3 (b). This should cause premature termination the Prp19 protein since a stop codon is present in the middle of the intron. Unexpectedly, all samples gave rise to a 330-basepair band (a) which is likely derived from the presence of contaminating genomic DNA but does not change interpretation of the data.
(TIF)
Loss of plrg1 mRNA expression in the plrg1(hi3174aTg) line leads to severe developmental abnormalities characterized by excessive cell death. (A) Siblings and mutants were collected from the plrg1(hi3174aTg) line based on morphology. RNA was harvested from each group at 30 hpf, reverse transcribed and analyzed by qPCR to determine levels of plrg1 mRNA. Error bars represent the standard error from three independent experiments. (B) plrg1(hi3174aTg) heterozygous fish were incrossed, and the progeny were either left uninjected or injected with 100 ng/uL of the indicated mRNAs. Right panels show higher magnification of embryos from adjacent left panels. Overexpression of plrg1 rescues the developmental abnormalities in the plrg1(hi3174aTg) mutants.
(TIF)
Supplemental Experimental Procedures. (A) Primers used for subcloning are listed and include the name of the target gene as well as restriction sites used for directional cloning into the pCS2+ expression plasmid. (B) The sequences of morpholinos used in this study are listed. The plrg1 morpholino was previously described [38]. (C) Primers used to examine specificity of the prp19 e3i3 morpholino are listed. See also Figure S7. (D) Primers used to sub-clone zp53 into pGEM-T-Easy for use in p53 in situ hybridization experiments are shown. See also Figure 3B. (E) Primer/probe sets used for qPCR analysis are grouped by presence or absence of shading.
(PDF)
Acknowledgments
We would like to thank Dr. David Lane (A*STAR, p53 Laboratory, Immunos, Singapore) for kindly providing us with the ZFp53-9.1 antibody. We also thank the University of Utah Microarray and Sequencing Core Facilities.
Funding Statement
Support for this work was provided by NIH grant NIH K01 DK074555 (CJ), the Huntsman Cancer Institute/Huntsman Cancer Foundation (CJ, RAS, SS, EH), P30 CA042014 (BM), NIH 3R01CA119066 (ATL, UP, SC), and NS058608 (RAS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lord CJ, Ashworth A (2012) The DNA damage response and cancer therapy. Nature 481: 287–294. [DOI] [PubMed] [Google Scholar]
- 2. Gudkov AV, Komarova EA (2003) The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer 3: 117–129. [DOI] [PubMed] [Google Scholar]
- 3. Kratz E, Eimon PM, Mukhyala K, Stern H, Zha J, et al. (2006) Functional characterization of the Bcl-2 gene family in the zebrafish. Cell Death Differ 13: 1631–1640. [DOI] [PubMed] [Google Scholar]
- 4. Sidi S, Sanda T, Kennedy RD, Hagen AT, Jette CA, et al. (2008) Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell 133: 864–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, et al. (2003) p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302: 1036–1038. [DOI] [PubMed] [Google Scholar]
- 6. Gallenne T, Gautier F, Oliver L, Hervouet E, Noel B, et al. (2009) Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J Cell Biol 185: 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kirkin V, Joos S, Zornig M (2004) The role of Bcl-2 family members in tumorigenesis. Biochim Biophys Acta 1644: 229–249. [DOI] [PubMed] [Google Scholar]
- 8. McKinnon PJ (2009) DNA repair deficiency and neurological disease. Nat Rev Neurosci 10: 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taylor AM, Harnden DG, Arlett CF, Harcourt SA, Lehmann AR, et al. (1975) Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature 258: 427–429. [DOI] [PubMed] [Google Scholar]
- 10. Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, et al. (1995) A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268: 1749–1753. [DOI] [PubMed] [Google Scholar]
- 11. Shiloh Y (2003) ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer 3: 155–168. [DOI] [PubMed] [Google Scholar]
- 12. Jette CA, Flanagan AM, Ryan J, Pyati UJ, Carbonneau S, et al. (2008) BIM and other BCL-2 family proteins exhibit cross-species conservation of function between zebrafish and mammals. Cell Death Differ 15: 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu YC, Chen HC, Wu NY, Cheng SC (2007) A novel splicing factor, Yju2, is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol 27: 5403–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan SP, Kao DI, Tsai WY, Cheng SC (2003) The Prp19p-associated complex in spliceosome activation. Science 302: 279–282. [DOI] [PubMed] [Google Scholar]
- 16. Cheng SC, Tarn WY, Tsao TY, Abelson J (1993) PRP19: a novel spliceosomal component. Mol Cell Biol 13: 1876–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Legerski RJ (2009) The Pso4 complex splices into the DNA damage response. Cell Cycle 8: 3448–3449. [DOI] [PubMed] [Google Scholar]
- 18. Abdelilah S, Mountcastle-Shah E, Harvey M, Solnica-Krezel L, Schier AF, et al. (1996) Mutations affecting neural survival in the zebrafish Danio rerio. Development 123: 217–227. [DOI] [PubMed] [Google Scholar]
- 19. Furutani-Seiki M, Jiang YJ, Brand M, Heisenberg CP, Houart C, et al. (1996) Neural degeneration mutants in the zebrafish, Danio rerio. Development 123: 229–239. [DOI] [PubMed] [Google Scholar]
- 20. Liu TX, Howlett NG, Deng M, Langenau DM, Hsu K, et al. (2003) Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell 5: 903–914. [DOI] [PubMed] [Google Scholar]
- 21. Bladen CL, Navarre S, Dynan WS, Kozlowski DJ (2007) Expression of the Ku70 subunit (XRCC6) and protection from low dose ionizing radiation during zebrafish embryogenesis. Neurosci Lett 422: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyashita T, Reed JC (1992) bcl-2 gene transfer increases relative resistance of S49.1 and WEHI7.2 lymphoid cells to cell death and DNA fragmentation induced by glucocorticoids and multiple chemotherapeutic drugs. Cancer Res 52: 5407–5411. [PubMed] [Google Scholar]
- 23. Miyashita T, Reed JC (1993) Bcl-2 oncoprotein blocks chemotherapy-induced apoptosis in a human leukemia cell line. Blood 81: 151–157. [PubMed] [Google Scholar]
- 24. Sentman CL, Shutter JR, Hockenbery D, Kanagawa O, Korsmeyer SJ (1991) bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell 67: 879–888. [DOI] [PubMed] [Google Scholar]
- 25. Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, et al. (2007) p53 activation by knockdown technologies. PLoS Genet 3: e78 doi:10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, et al. (2005) tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A 102: 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Biswas AK, Johnson DG (2012) Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res 72: 13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee KC, Goh WL, Xu M, Kua N, Lunny D, et al. (2008) Detection of the p53 response in zebrafish embryos using new monoclonal antibodies. Oncogene 27: 629–640. [DOI] [PubMed] [Google Scholar]
- 29. Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, et al. (2003) Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4: 321–328. [DOI] [PubMed] [Google Scholar]
- 30. Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- 31. Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, et al. (2010) Molecular architecture of the human Prp19/CDC5L complex. Mol Cell Biol 30: 2105–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ren L, McLean JR, Hazbun TR, Fields S, Vander Kooi C, et al. (2011) Systematic two-hybrid and comparative proteomic analyses reveal novel yeast pre-mRNA splicing factors connected to Prp19. PLoS ONE 6: e16719 doi:10.1371/journal.pone.0016719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benathen IA, Beam CA (1977) The genetic control of X-ray resistance in budding yeast cells. Radiat Res 69: 99–116. [PubMed] [Google Scholar]
- 34. Henriques JA, Vicente EJ, Leandro da Silva KV, Schenberg AC (1989) PSO4: a novel gene involved in error-prone repair in Saccharomyces cerevisiae. Mutat Res 218: 111–124. [DOI] [PubMed] [Google Scholar]
- 35. Mahajan KN, Mitchell BS (2003) Role of human Pso4 in mammalian DNA repair and association with terminal deoxynucleotidyl transferase. Proc Natl Acad Sci U S A 100: 10746–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Revers LF, Cardone JM, Bonatto D, Saffi J, Grey M, et al. (2002) Thermoconditional modulation of the pleiotropic sensitivity phenotype by the Saccharomyces cerevisiae PRP19 mutant allele pso4-1. Nucleic Acids Res 30: 4993–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang N, Kaur R, Lu X, Shen X, Li L, et al. (2005) The Pso4 mRNA splicing and DNA repair complex interacts with WRN for processing of DNA interstrand cross-links. J Biol Chem 280: 40559–40567. [DOI] [PubMed] [Google Scholar]
- 38. Kleinridders A, Pogoda HM, Irlenbusch S, Smyth N, Koncz C, et al. (2009) PLRG1 is an essential regulator of cell proliferation and apoptosis during vertebrate development and tissue homeostasis. Mol Cell Biol 29: 3173–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, et al. (2004) Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A 101: 12792–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kruse JP, Gu W (2009) Modes of p53 regulation. Cell 137: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raff MC, Barres BA, Burne JF, Coles HS, Ishizaki Y, et al. (1993) Programmed cell death and the control of cell survival: lessons from the nervous system. Science 262: 695–700. [DOI] [PubMed] [Google Scholar]
- 42. Ni Chonghaile T, Sarosiek KA, Vo TT, Ryan JA, Tammareddi A, et al. (2011) Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science 334: 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 44.Westerfield M (1993) The Zebrafish Book.: University of Oregon Press, Eugene, OR.
- 45. Nix DA, Di Sera TL, Dalley BK, Milash BA, Cundick RM, et al. (2010) Next generation tools for genomic data generation, distribution, and visualization. BMC Bioinformatics 11: 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, et al. (2011) The UCSC Genome Browser database: update 2011. Nucleic Acids Res 39: D876–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic of genetic screen to identify recessive radiosensitizing mutations. Wild-type AB strain male zebrafish were treated with ENU giving rise to approximately 192 gene-inactivating mutations per sperm. ENU-mutagenized males were crossed with wild-type AB strain female fish to yield the F1 generation. Each fish in the F1 generation carried 192 unique mutations and were incrossed to create F2 families carrying 384 mutations per family. Up to eight random incrosses were performed within each F2 family, and the F3 clutches were subsequently analyzed for radiosensitizing phenotypes as described in the figure.
(TIF)
Alignment of zebrafish, mouse and human CCDC94 protein sequences. Protein sequences from zebrafish, mouse and human CCDC94 were aligned with the Jotun Hein algorithm using the default parameters in Lasergene MegAlign software. Zebrafish Ccdc94 is 67% identical to human CCDC94 and 69% identical to mouse CCDC94. An asterisk denotes R125, the residue that is mutated to a stop codon in rs7 mutants.
(TIF)
The rs7 mutation causes severe neurodegeneration that culminates in embryonic lethality. Wild-type embryos (derived from wild-type parents) and rs7 mutants were analyzed by brightfield microscopy on day 2 and day 3 of development. The rs7 mutation causes massive accumulation of cell death in the brain and spinal cord likely contributing to a small-head and “curly-up” tail phenotype. Rs7 mutant embryos usually fail to survive past day three.
(TIF)
Loss of ccdc94 compromises the anti-apoptotic function of Bcl-2. Wild-type (wt, derived from crossing wild-type parents) or rs7 mutant embryos (derived from crossing rs7 heterozygous parents) were injected at the one-cell stage of development with the indicated amounts of mRNA encoding zbcl-2, zbcl-xL, or egfp as a control (cntl). Embryos were then irradiated at 24 hpf with 8 Gy IR and analyzed three hours later by immunofluorescence to detect activated Caspase-3. Mutants were genotyped as in Figure 2C–2D. Representative data from three independent experiments is shown.
(TIF)
The rs7 mutation does not appear to cause a general activation of the DSB-DDR pathway. (A) Wild-type embryos were exposed (or not) to 8 Gy IR at 27 hpf and harvested for analysis three hours later. Quantitative PCR to analyze p53 mRNA expression was performed as in Figure 3A. (B) Rs7 siblings and mutants in the p53e7/e7 background [26] were analyzed at 30 hpf for expression of the indicated genes by qPCR similar to Figure 3A. For both (A) and (B), gapdh mRNA expression was measured to normalize gene expression levels. All data was then compared to rs7 sibling data, which was adjusted to a value of one. None of the values in (B) are significantly different from each other.
(TIF)
p53-mediated puma expression causes neurodegeneration in rs7 mutants. (A) rs7 sibling or mutant embryos in the p53 wild-type or mutant background were collected, and RNA was harvested at 30 hpf, reverse transcribed and analyzed for the expression of puma mRNA by qPCR. Rs7 mutants and siblings were distinguished by morphology since loss of p53 does not inhibit the rs7-mediated “curly-up” tail phenotype. (B) Pictures were taken of 48 hpf embryos from (A). Arrows point to neurodegeneration in rs7 mutants that is substantially rescued by loss of wild-type p53. For panel (A), error bars represent the standard error from three independent experiments.
(TIF)
The prp19 morpholino inhibits proper splicing of the prp19 pre-mRNA transcript. (A) Schematic of the zebrafish prp19 gene with exons shown as boxes and introns shown as lines between boxes. The e3i3 morpholino targets the splice donor site at the border of exon 3 and intron 3. In (B), F1 and R2 primer-based PCR will amplify from exon 2 to intron 3. Properly spliced mRNA should not give rise to PCR product. Inclusion of intron 3 should give rise to a 245-basepair product. (B) RNA from embryos injected with mismatch morpholino controls or prp19 e3i3 morpholino was harvested at 24 hpf, reverse transcribed with oligo-dT primers, and analyzed by PCR using primers shown in (A). Bands were excised from the gel, cloned into pGEM-T-Easy, and sequenced. Lower-case letters point to bands on the gel which correspond to regions of the prp19 gene shown below. When embryos were injected with the e3i3 morpholino, F1/R2 primers gave rise to a band representing inclusion of intron 3 (b). This should cause premature termination the Prp19 protein since a stop codon is present in the middle of the intron. Unexpectedly, all samples gave rise to a 330-basepair band (a) which is likely derived from the presence of contaminating genomic DNA but does not change interpretation of the data.
(TIF)
Loss of plrg1 mRNA expression in the plrg1(hi3174aTg) line leads to severe developmental abnormalities characterized by excessive cell death. (A) Siblings and mutants were collected from the plrg1(hi3174aTg) line based on morphology. RNA was harvested from each group at 30 hpf, reverse transcribed and analyzed by qPCR to determine levels of plrg1 mRNA. Error bars represent the standard error from three independent experiments. (B) plrg1(hi3174aTg) heterozygous fish were incrossed, and the progeny were either left uninjected or injected with 100 ng/uL of the indicated mRNAs. Right panels show higher magnification of embryos from adjacent left panels. Overexpression of plrg1 rescues the developmental abnormalities in the plrg1(hi3174aTg) mutants.
(TIF)
Supplemental Experimental Procedures. (A) Primers used for subcloning are listed and include the name of the target gene as well as restriction sites used for directional cloning into the pCS2+ expression plasmid. (B) The sequences of morpholinos used in this study are listed. The plrg1 morpholino was previously described [38]. (C) Primers used to examine specificity of the prp19 e3i3 morpholino are listed. See also Figure S7. (D) Primers used to sub-clone zp53 into pGEM-T-Easy for use in p53 in situ hybridization experiments are shown. See also Figure 3B. (E) Primer/probe sets used for qPCR analysis are grouped by presence or absence of shading.
(PDF)