Abstract
Bacterial biofilms are defined as a surface attached community of bacteria embedded in a matrix of extracellular polymeric substances that they have produced. When in the biofilm state, bacteria are more resistant to antibiotics and the host immune response than are their planktonic counterparts. Biofilms are increasingly recognized as being significant in human disease, accounting for 80% of bacterial infections in the body and diseases associated with bacterial biofilms include: lung infections of cystic fibrosis, colitis, urethritis, conjunctivitis, otitis, endocarditis and periodontitis. Additionally, biofilm infections of indwelling medical devices are of particular concern, as once the device is colonized infection is virtually impossible to eradicate. Given the prominence of biofilms in infectious diseases, there has been an increased effort toward the development of small molecules that will modulate bacterial biofilm development and maintenance. In this review, we highlight the development of small molecules that inhibit and/or disperse bacterial biofilms through non-microbicidal mechanisms. The review discuses the numerous approaches that have been applied to the discovery of lead small molecules that mediate biofilm development. These approaches are grouped into: 1) the identification and development of small molecules that target one of the bacterial signaling pathways involved in biofilm regulation, 2) chemical library screening for compounds with anti-biofilm activity, and 3) the identification of natural products that possess anti-biofilm activity, and the chemical manipulation of these natural products to obtain analogues with increased activity.
Introduction
A significant factor contributing to the pathogenesis, and antibiotic/host immune resistance to a number of medically important bacterial strains is the ability of the bacteria to form a biofilm. Bacterial biofilms are highly organized surface-associated communities of bacteria encased within an extracellular matrix. Bacteria within a biofilm exhibit distinct phenotypes from planktonic cells, particularly with respect to growth and gene expression.1 Bacterial biofilms have become recognized as a serious threat to both the medical and industrial sectors of society within the past 20 years.2 On a global scale, biofilm-related costs incur billions of dollars to the agricultural, engineering, and medical sectors of the economy.3 The correlation between biofilms and infectious disease is a connection that is becoming well documented in the medical community and The National Institutes of Health (NIH) estimates that 80% of all bacterial infections occurring in the human body are biofilm related.3 An estimated 17 million new biofilm infections arise each year in the U.S., which result in up to 550,000 fatalities annually. Common ailments that are driven and are perpetuated by bacterial biofilms include, but are not limited to: lung infections of cystic fibrosis (CF) patients, burn wound infections, ear infections, catheter infections, bacterial endocarditis, chronic wound infections, and tooth decay.3,4 Longer hospital stays, chronic infection, and increased fatalities as a result of biofilm-mediated infections place a significant economic burden on healthcare systems worldwide.5 Bacterial biofilms also underlie the persistent colonization of hospital facilities, both driving and sustaining nosocomial infections.
Biofilms are inherently insensitive to antiseptics and microbicides that would typically eliminate their planktonic brethren and are known to be upwards of 1000-times more resistant to conventional antibiotics6 and bacteria within a biofilm reach a much higher cell density (1011 CFU/mL) than do planktonic bacteria (108 CFU/mL).7 Multi-drug resistant (MDR) bacteria are becoming commonplace in the global healthcare setting and antibiotics which have previously been of last resort are being used with increased frequency in attempts to alleviate particularly aggressive infections.8 Compounding this problem, only two new classes of antibiotics (oxazolidinones and lipopeptides) have been introduced into the clinic over the last 40 years.9 Biofilms also underlie importunate infections of indwelling medical devices (IMDs), and it has been demonstrated that the presence of such a foreign body decreases the minimal infecting dose of Staphylococcus aureus by 100,000-fold.7 Eradication of these infections is virtually impossible, requiring aggressive antibiotic therapy, removal of the indwelling device, and surgical debridement.10
Phenotypic changes brought about by the formation of a biofilm contribute to bacterial resistance to antibiotics. These changes include production of the extracellular polymeric substance (EPS) and upregulation of genes responsible for porin proteins or specialized efflux pumps to purge antibiotics from the cell. While the 3-dimensional morphology of biofilms lends itself to nutrient distribution and waste disposal, it also provides a fertile environment for the efficient transfer of genetic material.11 Gene transfer rates in biofilms facilitated through the conjugation process have been reported to be up to 1000-fold higher than those found in planktonic cells.12 Another problem exacerbated by bacteria occupying a biofilm state is hypermutation. This occurs when bacteria, in response to environmental pressures (such as antibiotics), mutate at higher rates to evolve resistance and alleviate the particular stress on the community.13 Approximately 20% of Psuedomonas isolates from the lungs of cystic fibrosis patients’ display this phenomenon,14 making the task of treatment even more challenging.
The slow growth rates of biofilm communities have also been posited as a factor in increased resistance to eradication.15 Within a biofilm, there exists a heterogeneous population of cells which differ in growth rates dependant upon the area within the biofilm in which the cells are located. Bacterial cells embedded deep within the biofilm matrix grow more slowly due to lack of nutrients and oxygen.16 Cells with reduced metabolic activity are also inherently more recalcitrant to antimicrobial therapies17 as almost all antibiotics target five biosynthetic processes, all of which occur in actively growing bacteria: the biosynthesis of proteins, RNA, DNA, peptidoglycan and folic acid.18 Therefore most antibiotics that are capable of killing growing and dividing bacterial cells tend to be very inefficient at killing non-multiplying bacteria.19 This, coupled with the differential penetration distances into a biofilm that antibiotics display, explains how some drugs fail to completely eradicate biofilm bacteria.15
Our understanding of the diverse mechanisms that govern bacterial biofilm formation and maintenance is continually increasing. Recent research into the environmental cues and genetic elements that play a part in biofilm regulation, from two-component signal transduction systems, to indole-signaling, and the role of the second messenger c-di-GMP, have led to novel targets and compound scaffolds for the development of anti-biofilm agents. These new strategies, coupled with the development of antagonists of the more well studied quorum sensing pathways, have led to the development of a number of anti-biofilm molecules that have been explored for their potential to be developed into effective treatments for biofilm-mediated infections.20
As most pathogenic bacteria exist as biofilm communities, the discovery and development of agents with the ability to limit biofilm formation, or even eradicate established biofilms, will have the potential to enhance the efficacy of antibiotics that are ineffective against biofilm bacteria. Such therapeutic entities should have a profound impact on human medicine.2,7 It is important to distinguish molecular scaffolds that have the ability to affect biofilm development via non-microbicidal mechanisms from their microbicidal counterparts, as the pressure on bacteria to evolve resistance to agents which are not microbicidal will be significantly reduced or even eliminated as compared to the selective pressures exerted by conventional bactericidal entities. Therefore, this review seeks to present compounds that have been shown to explicitly demonstrate anti-biofilm behavior in a nonbactericidal manner and cause phenotype shifts among bacteria commonly known to form biofilms. This is significant, as there exists the potential for co-dosing newly developed anti-biofilm compounds that possess the above-mentioned characteristics with conventional antibiotics to eliminate established biofilm infections. This review provides an overview of the numerous approaches that have been applied to the discovery of small molecules able to mediate the formation and maintenance of bacterial biofilms. These approaches are grouped into: 1) the identification and development of small molecules that target one of the bacterial signaling pathways involved in biofilm regulation, 2) chemical library screening for compounds with anti-biofilm activity, and 3) the identification of natural products that possess anti-biofilm activity, and the chemical manipulation of these natural products to obtain analogues with increased activity.
Compounds that modulate biofilms by targeting bacterial signaling pathways
A significant amount of the current information we have regarding biofilm development is tied to research involving the phenomenon of bacterial quorum sensing (QS). QS is the term used to describe intercellular communication between bacteria that allows the community to make coordinated efforts at altering gene expression based upon population density.21 It is known that QS pathways heavily influence the formation of biofilms, in addition to the production of other virulence factors. A diverse range of biomolecules serve as the facilitators for QS systems in bacteria. Therefore, extensive research in this area has produced a number of analogues with the ability to modulate QS-dependent enzymes. These molecules compose the vast majority of compounds thus far investigated for biofilm control. The understanding of complex QS-based communication systems in Gram-negative organisms is based upon the study of a variety of bacteria, including the bioluminescent marine bacterium Vibrio fisheri and the human pathogens Vibrio cholerae and Pseudomonas aeruginosa.22-24 At its most basic level, QS in Gram-negative bacteria is effected by a set of two proteins. One protein is responsible for the production of a signaling molecule, which is typically referred to as an autoinducer (AI), while the other protein responds to the autoinducer. There are several classes of autoinducers, based upon on shared molecular features, these include acyl homoserine lactones (AHLs), autoinducing peptides (AIPs) and autoinducer-2 (AI-2).25
Acyl Homoserine Lactones (AHLs)
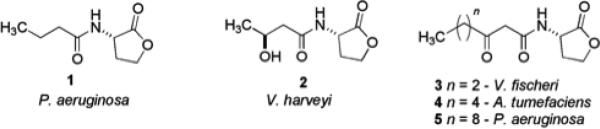
Gram-negative bacteria predominantly employ N-acyl homoserine lactones (AHLs) as autoinducers, and more than 70 species of bacteria communicate via AHL-mediated QS with specificity mediated via variation in the length and oxidation state of the acyl side chain (compounds 1-5) (Fig. 1).25 Native AHLs found in bacteria are L-isomers, and the D-isomers are known to be essentially devoid of biological activity. In V. fisheri, AHL synthesis occurs when the luxI gene is activated to produce the AHL synthase enzyme LuxI. When the newly produced AHL reaches a threshold intracellular concentration, binding to the transcriptional activator LuxR takes place, leading to activation of the luxR gene set. AHLs are able to freely diffuse in and out of bacterial cells, allowing the total AHL concentration to correlate to the total bacterial concentration, thus enabling population density-based control of gene expression. This cascade of events ultimately leads to the control of gene expression resulting in the control of virulence factor production and biofilm formation and maintenance.26 Thorough investigations of other Gram-negative QS pathways, most notably those belonging to P. aeruginosa, have shown that homologous systems to the LuxI and LuxR proteins (LasI and LasR) exist for the purpose of AHL synthesis and response.24 The most important AHLs responsible for QS in P. aeruginosa are N-butanoylhomoserine lactone 1 (C4-AHL, for the rhl system) and 3-oxo-C12-AHL 5 (for the las system).
Fig. 1.
AHLs utilized by Gram-negative bacteria for quorum sensing.
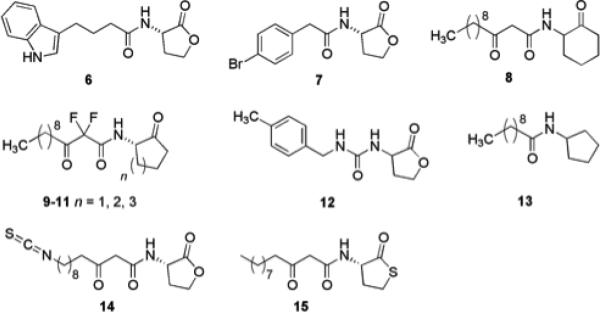
AHLs have served as one of the primary scaffolds studied over the past thirty years for the design of potential biofilm inhibitors.27,28 A considerable amount of work has been published involving the biological consequences of chemically modified AHL derivatives in a variety of QS systems. Work from the Blackwell group has documented the synthesis and identification of a number of natural and unnatural AHLs with the ability to modulate QS in P. aeruginosa and Agrobacterium tumefaciens.29 They also demonstrated that two of their most active synthetic AHLs could retard biofilm formation in P. aeruginosa PA01. Qualitative experiments were performed, in which an engineered P. aeruginosa strain capable of producing green fluorescent protein (GFP) for visualization was grown in the presence of synthetic ligands 6 and 7 (Fig. 2) at 50 μM. Significant inhibition of biofilm formation was observed for both derivatives. More recently, the Blackwell group has synthesized and presented the QS antagonist/agonist properties of diverse synthetic AHL-based libraries that were screened against P. aeruginosa, A. tumefaciens, and V. fischeri.30 Other research that includes the modification of AHLs to discern their effects on quorum sensing and biofilm formation in P. aeruginosa includes that of Suga and Spring. The work by Suga entailed the synthesis of a 96-member library constructed through solid phase protocols to mimic AHLs by replacing the homoserine lactone moiety with a variety of functionalities.31,32 A noteworthy compound indentified within this study was AHL derivative 8, which had no effect on biofilm growth, yet elicited a noticeable change in the biofilm morphology of P. aeruginosa PA01. Spring's research investigated analogues of P. aeruginosa AHLs in which the lactone functionality was replaced by a ketone, with additional difluorination between the β-keto amide positions (9-11).33 Other analogues have been reported involving ureas (12),34 thiolactones,35 sulfonamides,36 and N-acylated cyclopentylamides37 as viable modifications of AHLs to drive QS inhibition and thereby affect biofilm formation. One specific N-acyl cyclopentylamide, designated as C10-CPA (13), was shown to completely prevent biofilm formation by P. aeruginosa PA01:GFP at a concentration of 250 μM after seven days under flow conditions. These signaling molecule derivatives are particularly important because the biological activity of nearly every compound in this class is not driven by microbicidal properties.
Fig. 2.
Derivatives of the AHL scaffold that possess biological activity.
The determination of the crystal structure of the P. aeruginosa transcriptional regulator LasR bound to its natural ligand (3-oxo-C12-AHL 5) by Bottomley et al. gives new insight into the binding features required for the design of potent inhibitors of LasR.25,38 This information was exploited in the design of covalent inhibitors of LasR in P. aeruginosa that were able to reduce biofilm formation. A series of inhibitors bearing electrophilic functional groups including isothiocyanates, bromoacetamides, and chloroacetamides, were designed to allow reaction with a nucleophilic cysteine residue present in the LasR binding pocket (Cys79). Isothiocyanate containing ligands were able to covalently and selectively bind Cys79 and using reporter strains were shown to inhibit quorum sensing. The lead compound, 14 (Fig. 2), was also shown to inhibit biofilm formation by wild type PA01 by almost 50% at a concentration of 50 μM.25
Despite the potential that these molecules may possess toward potentially unlocking the ability to modulate bacterial communication on demand, there exists differing opinions on how relevant AHL treatment is to the treatment of biofilms. On one hand, modulating AHL-driven processes have great promise as bacterial strains of P. aeruginosa deficient in AHL production grown as planktonic or biofilm populations have been found to be similarly susceptible to biocides as wild-type populations.39 However, homoserine lactones themselves are notoriously prone to hydrolysis at physiological pH and the ring-opened product is QS inactive, they have also been reported to possess immunomodulatory activity.40
Seeking to address the problem of lactone hydrolysis inherent to native AHLs, the potential of thiolactone AHL analogues as QS agonists and antagonists in Gram-negative bacteria has been investigated.41 A focused library of thiolactone analogues was examined against LuxR-type QS receptors in a number of bacteria: LasR (P. aeruginosa), LuxR (V. fischeri), and TraR (A. tumefaciens). Several highly active LuxR inhibitors were identified and the half-life of a representative active analogue 15 (Fig. 2), was shown to be almost double that of its parent AHL in bacterial growth medium. These analogues have not yet been tested for their activity as inhibitors of biofilm formation; however the thiolactone is a promising scaffold for the development of stable modulators of QS controlled biofilm formation.
Autoinducing peptides (AIPs)
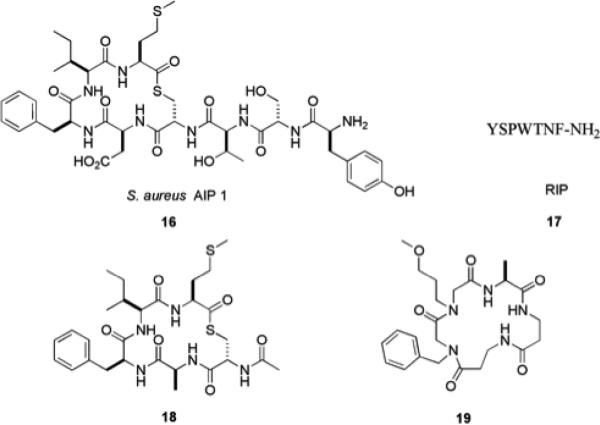
QS in Gram-positive bacteria is predominantly mediated by autoinducing peptides (AIPs) (Fig. 3),42 though it has been shown that Gram-positive bacteria may not exclusively utilize peptide-signaling molecules for communication and small molecules known as γ-butyrolactones have been identified as signaling molecules in some species of Streptomyces.43 Many AIPs possess hydrophobic domains, which have been shown to be crucial for activity. These motifs are postulated to play an important role in the promotion of hydrophobic interactions that influence receptor activation.44 The agr and TRAP QS systems in S. aureus regulate a number of virulence phenotypes, including biofilm formation. The accessory gene regulator (agr) operon contains the agrD gene, which encodes AgrD, the precursor of the S. aureus AIP 16. Upon reaching threshold levels, AIP will bind AgrC leading to the expression of a small non-coding RNA known as RNA-III, which subsequently down-regulates genes that encode adhesins required for biofilm formation.45 The RNA-III activating protein (RAP) activates TRAP (target of RNA-III activating peptide) via phosphorylation, leading to increased cell adhesion and biofilm formation, in addition to inducing expression of the agr operon.46
Fig. 3.
Peptide derived molecules with activity against S. aureus.
It has been demonstrated that the RNA-III inhibiting peptide (RIP) 17 inhibits phosphorylation of TRAP, leading to reduced biofilm formation. RIP has been studied in multiple animal models and has been shown to possess in vivo activity in preventing many types of infections including those caused by antibiotic resistant strains.47 More importantly, no toxicity has been observed and no RIP-resistant strains have yet been reported.48 Additional studies utilizing peptides and peptidomimetics as possible tools to probe quorum sensing have identified analogues of S. aureus AIP-1 that can also influence QS pathways in S. aureus. Truncated peptides such as 18 have been found to be potent AgrC-I QS antagonists,49 and the peptidomimetic 19 has been shown to promote biofilm formation in S. aureus.50
Autoinducer-2 (AI-2)
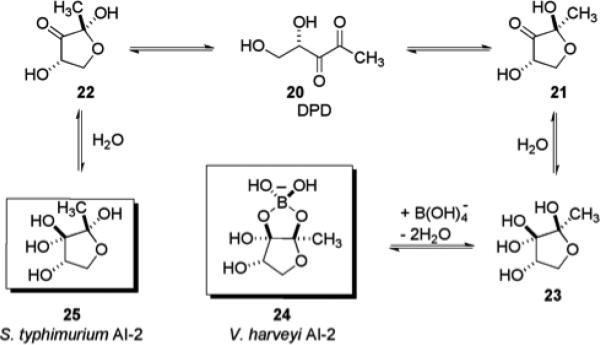
One of the putative universal quorum sensing mechanisms shared by both Gram-negative and Gram-positive bacteria involves the production of a small molecule termed autoinducer-2 (AI-2). All AI-2 molecules are derived from the precursor molecule (S)-4,5-dihydroxy-2,3-pentanedione (DPD) 20, and the synthase enzyme that drives DPD production has been found to be conserved in over 55 bacterial species.51 This finding has led many researchers to postulate that the AI-2 system is in fact the universal language of bacterial communication.52 Two AI-2 molecules that have been isolated and characterized are shown in Fig. 4, one from Vibrio harveyi 24 and another from Salmonella typhimurium 25. In the case of AI-2 isolated from V. harveyi, the DPD derivative exists as a borate diester 24, a result of the high concentrations of boron in the natural marine water habitat of this bacterium.53 DPD 20 is a highly reactive intermediate in the Activated Methyl Cycle (AMC), which recycles S-adenosyl-L-methionine (SAM), the primary methyl donor in bacterial and eukaryotic cells. SAM also plays a role as an intermediate in the pathway responsible for the synthesis of AHLs. LuxI synthase is known to catalyze the reaction of SAM with acylated acyl carrier proteins delivering AHLs.53
Fig. 4.
Role of DPD 20 in the synthesis of AI-2 molecules isolated from V. harveyi and S. typhimurium.
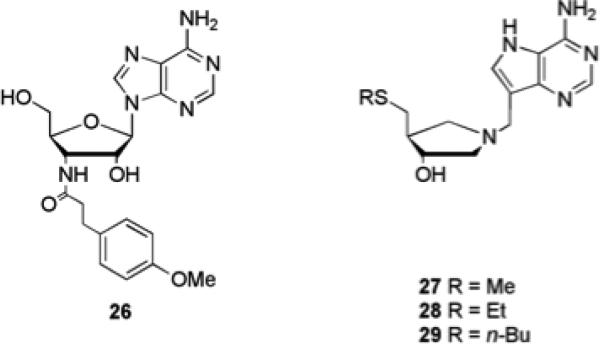
Recently Janda and co-workers as well as Sintim and co-workers have been able to synthesize and assay a small number of DPD analogues to delineate their effects on QS in V. harveyi and S. typhimurium.54-56 As the biosynthesis of DPD begins from SAM, the screening of a small library of nucleoside analogues for their ability to disturb AI-2-based QS in V. harveyi has also been investigated.57 The adenosine analogue 26 (Fig. 5) was found to block AI-2-based QS without interfering with bacterial growth. This compound was subsequently shown to affect biofilm formation in Vibrio anguillarum, Vibrio vulnificus and V. cholerae.57
Fig. 5.
Nucleoside analogues that interfere with AI-2 based QS pathways.
The 5’-methylthioadenosine nucleosidase (MTAN) enzymes are directly involved in the biosynthesis of AI-2 and AHLs, catalyzing the N-glycosyl hydrolysis of SAM and other adenosyl derivatives.58,59 Transition state analogues 27, 28, and 29 (Fig. 5) designed against MTANs were shown to inhibit MTAN activity with nanomolar IC50 values in cell lysates of a virulent V. cholerae strain. These transition state analogues were also shown to inhibit QS induction in V. harveyi reporter strains. Analogues 27 and 29 also inhibited MTAN in Escherichia coli and resulted in nontoxic inhibition of AI-2 production. Compound 29 also reduced biofilm formation in E. coli and V. cholerae by 18% and 71% respectively at a concentration of 1 μM without inhibiting planktonic growth.59
c-di-GMP
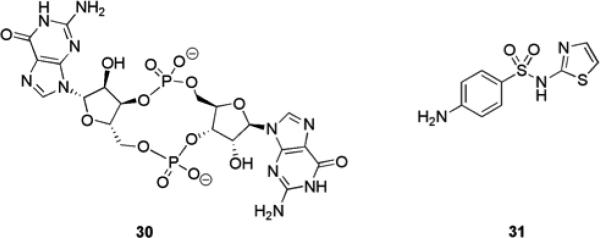
Bis-(3’5’)-cyclic di-guanylic acid (c-di-GMP) 30 (Fig. 6) is a second messenger signaling molecule that is thought to be ubiquitous in bacteria. Diguanylate cyclases (DGCs) and phosphodiesterases (PDEs) are responsible for the synthesis and breakdown of c-di-GMP, respectively.60 DGCs contain a conserved GGDEF (Gly–Gly–Asp–Glu–Phe) domain which has been observed in virtually all sequenced bacterial genomes61 and PDEs contain a conserved EAL domain (enriched in Glu–Ala–Leu).60 These proteins are highly regulated by various environmental and intracellular signals such as oxygen, light, and small molecules. The GGDEF domain-containing proteins effect changes in c-di-GMP levels in response to environmental and intracellular stimuli. Second messengers such as c-di-GMP can amplify the original signal into an intracellular signal capable of bringing about considerable biochemical changes within the cell.62 This is accomplished by the binding of c-di-GMP to downstream proteins, which results in conformational changes that lead to either positive or negative regulation of cellular functions. There is increasing evidence that the transition between the planktonic and biofilm lifestyle in P. aeruginosa is regulated via proteins with DGC or PDE activities through control of c-di-GMP levels.62 It has also been observed that exopolysaccharide synthesis (and thus the exopolysaccharide-dependent formation of biofilms) is regulated by c-di-GMP in various proteobacterial species such as V. cholera, P. aeruginosa, Pseudomonas fluorescens, A. tumefaciens, E. coli, and Salmonella enterica.61
Fig. 6.
c-di-GMP 30 and sulfathiazole 31 that inhibits DGC activity.
Dispersion of bacteria from a mature biofilm is another process that may be potentially regulated by c-di-GMP. A P. aeruginosa mutant that demonstrated elevated c-di-GMP levels exhibited defective detachment in response to environmental signals that typically elicit biofilm dispersion, such as changes in nutrient availability. It is postulated that c-di-GMP is maintained at a level high enough to promote the initial stages of biofilm formation and maturation (which require motility), but levels are then decreased to allow for full maturation of the biofilm.62 As QS and c-di-GMP signaling pathways likely regulate some of the same complex processes in bacteria, (e.g. biofilm formation), it is possible that the two signaling pathways are linked. A direct connection has not yet been reported though evidence for an indirect connection has been presented.63 For example, in V. cholerae, the QS–regulated transcription factor AphA influences the expression of genes encoding DGC and PDE. Additionally, during the QS-mediated formation of a symbiotic biofilm between the hyperthermophiles Thermotoga maritima and Methanococcus jannaschii, genes encoding DGC and PDE were upregulated and down-regulated respectively.63 One of the best-studied DGC is PleD, a signaling protein from the aquatic bacterium Caulobacter crescentus. Analysis of the crystal structure of the complex of PleD with c-di-GMP suggests that two PleD monomers dimerize and catalyze c-di-GMP synthesis from two molecules of GTP. The DGC activity of PleD is regulated by phosphorylation, which is thought to promote dimerization.63
Interfering with c-di-GMP signaling is an attractive target for control of biofilm formation, be it with c-di-GMP analogues, or with small molecules that interfere with the synthesis or degradation of c-di-GMP.64 The addition of exogenous c-di-GMP itself has been shown to inhibit biofilm formation in some bacterial species. S. aureus biofilm formation in vitro and adherence to HeLa cells was shown to be inhibited by c-di-GMP, while cyclic GMP (cGMP) and cyclic AMP (cAMP) had a much lower inhibitory effect on biofilm formation.65 Studies involving the dose-dependent effects of c-di-GMP on biofilm formation on abiotic surfaces in Streptococcus mutans revealed that biofilm formation could effectively be reduced in a dose dependent manner while simultaneously reducing adherence to tooth surfaces.66 In P. aeruginosa PA14, elevated intracellular c-di-GMP levels are known to be associated with increased biofilm formation and decreased swarming motility. Such behaviors can be influenced by the addition of exogenous amino acids that affect c-di-GMP levels. It has been shown that the addition of exogenous arginine results in an increase in c-di-GMP levels and concurrent stimulation of biofilm formation.67
Screening of a 1120 compound library with known biological activities against DGC identified sulfathiazole 31 (Fig. 6), as a hit, which inhibited DGC activity. Follow up biofilm inhibition assays confirmed that 31 was able to inhibit E. coli biofilm formation (IC50 = 5.8 μM) while not significantly inhibiting bacterial growth at concentrations up to 50-fold higher than those needed to inhibit biofilm formation.68
Indole signaling
Indole is another putative universal intercellular signal molecule amongst diverse bacteria69 that plays a direct role in the control of many bacterial behaviors in a QS-dependent fashion.70 Such behaviors include: antibiotic resistance,71 virulence,72 and biofilm formation73. Eighty-five species of bacteria have been documented to produce indole, with both indole-positive and indole-negative strains of bacteria altering various behaviors upon the extracellular presence of indole.69 High concentrations (>600 μM) of extracellular indole are produced by E. coli when cultured in rich medium and indole has been shown to decrease biofilm formation in E. coli in a non-toxic manner.74 The regulation of biofilm formation by indole in E. coli has been demonstrated to be temperature dependant, with a greater effect observed at lower temperatures (25 °C or 30 °C compared to 37 °C) and is mediated by the transcriptional regulator SdiA,75 which is a LuxR homologue76. The addition of exogenous indole to an E. coli strain unable to produce indole (lacking the tryptophanase (tnaA) gene) significantly affected the expression of 186 genes (more than two-fold) in biofilm cells at 30 °C, including the biofilm stress regulator bhsA, and bssR (yliH), which encodes a regulator of biofilm formation.75 Conversely, the presence of indole increases biofilm formation by P. aeruginosa and P. fluorescens despite the fact that these pseudomonads do not themselves produce indole.74
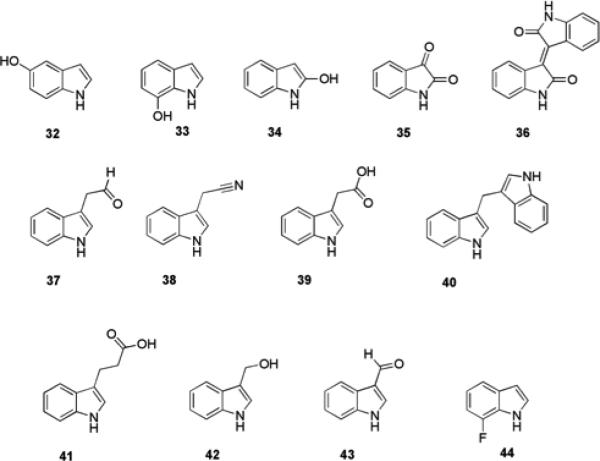
Indole is readily converted by oxygenases found in several bacterial species to a number of oxidized indole derivatives, such as hydroxyindoles 32-34, isatin 35, and isoindigo 36, (Fig. 7). It was therefore hypothesized that these oxidized indole metabolites may also play a role in biofilm formation.70 Screening of these compounds for the ability to inhibit biofilm formation by a strain of enterohemorrhagic E. coli (EHEC) revealed that 5-hydroxyindole 32 and 7-hydroxyindole 33 were able to inhibit biofilm formation by 11-fold and 27-fold respectively at a concentration of 1 mM. This is compared to 18-fold inhibition for indole itself at the same concentration.74 2-Hydroxyindole 34 at the same concentration had no effect on biofilm formation, while isoindigo 36 (at a concentration of 250 μM, which was the limit of its solubility) also had no effect. Isatin 35 increased biofilm formation by this strain of EHEC in a dose-dependant manner; with a four-fold increase effected at a concentration of 250 μM. None of the active indole derivatives significantly affected growth of EHEC, indicating that biofilm modulation is brought about via a non-toxic mode of action. Against PA01, 7-hydroxyindole 33 (500 μM) brought about a two-fold increase in biofilm formation while 5-hydroxyindole 32, isatin 35 and indole itself increased biofilm formation by 20-30% at the same concentration.74
Fig. 7.
Indole derivatives that affect biofilm formation.
Another indole derivative, indole-3-acetaldehyde, 37, produced by the plant pathogen Rhodococcus sp. BFI 332 inhibited biofilm formation by enterohemorrhagic E. coli O157:H7 without affecting planktonic growth.77 Compound 37 was shown to suppress expression of two curli operons, csgBAC and csgDEFG, and induce the expression of tryptophanase in E. coli O157:H7 biofilm cells. The spent medium of Rhodococcus sp. BFI 332, from which this compound was identified, was also shown to have an inhibitory effect on biofilm formation by two Staphylococcal species, S. aureus and Staphylococcus epidermidis.77
As many pathogenic bacteria rapidly degrade indole into some of its various metabolites depicted above, the ability of indole-derived plant secondary metabolites to modulate bacterial biofilm formation has also been investigated.78 3-Indolylacetonitrile (IAN) 38, indole 3-acetic acid (IAA) 39, 3,3′-methylene bisindole (MI) 40, indole-3-propioninc acid (I3PA) 41, indole-3-carbinol (I3C) 42, and indole-3-carboxyaldehyde (I3CA) 43 (Fig. 7) were screened at 100 μg/mL for their effects on biofilm formation by E. coli O157:H7 and PA01. The most active derivatives for suppression of biofilm formation by this E. coli strain were 38 and 43, which reduced biofilm formation by 11-fold and 24-fold respectively, compared to a three-fold reduction brought about by indole.78 In contrast to indole and the hydroxyindoles, 38 and 43 also weakly inhibited biofilm formation by PA01, effecting a 1.9-fold and 2.3-fold reduction in biofilm formation, respectively. IAN 38 was not degraded by either of these two bacterial strains, and did not affect planktonic growth of the bacteria. It was also shown that IAN increased indole production, providing a partial explanation for its ability to reduce biofilm formation.78
In a follow up study to identify indole derivatives with increased anti-virulence activity compared to IAN 38, 31 natural and synthetic indole derivatives were tested for the ability to inhibit biofilm formation and hemolytic activity of P. aeruginosa.79 The synthetic indole derivative, 7-fluoroindole (7FI) 44, was identified as the most active compound, and at a concentration of 1 mM reduced biofilm formation by four-fold and P. aeruginosa hemolytic activity by 14-fold. It was shown that 44 suppressed swarming motility, protease activity, and extracellular polymeric matrix production in P. aeruginosa. In addition, 44 reduced the production of QS-regulated virulence factors including 2-heptyl-3-hydroxy-4(1H)-quinolone, pyocyanin, rhamnolipid, and the two siderophores pyoverdine and pyochelin.79
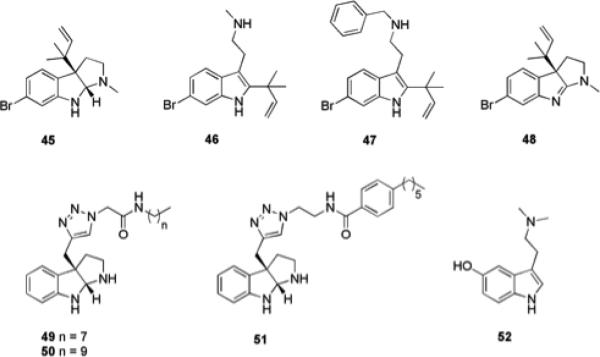
The flustramines are a collection of monobrominated secondary metabolites isolated from the bryozoan Flustra foliacea that contain a pyrroloindoline or indolic core (Fig. 8).80 Several of these compounds exhibit a host of biological activities, including antibacterial activity against several species of marine bacteria associated with the bryozoan, though they display no antibacterial activity against terrestrial bacterial strains.80 Dihydroflustramine C 45, also inhibited the production of reporter genes in an AHL-based QS reporter assay at concentrations below that at which it affected bacterial growth. It was therefore hypothesized that this class of natural products has the potential to act as a scaffold for the design of quorum sensing or indole signalling antagonists, which may have the ability to modulate biofilm formation.80
Fig. 8.
Flustramine derivatives and related compounds that inhibit biofilm formation.
To investigate the indolic flustramine scaffold as a potential modulator of biofilm formation, desformylflustrabromine (dFBr) 46 was tested for the ability to modulate S. aureus and E. coli biofilm formation. dFBr was able to inhibit biofilm formation by both strains, exhibiting IC50 values of 70 μM and 174 μM for E. coli and S. aureus, respectively. However, this compound demonstrated microbicidal effects on planktonic growth at these concentrations.81 Synthetic manipulation of four regions of the dFBr scaffold was performed with the aim of tuning the flustramine derivatives to modulate biofilm formation through a non-microbicidal mechanism. The most active compound identified, 47, inhibited biofilm formation by S. aureus and E. coli with IC50 values of 5.9 μM and 53 μM and was successful in eliciting its effects through a non-microbicidal mechanism.81 Mechanistic studies with 47 and indole itself in wild-type and knockout E. coli strains have shown that the activity of 47, like that of indole, is dependent on temperature, the transcriptional regulator SdiA, and tryptophanase; suggesting that the anti-biofilm activity of 47 may be occurring through modulation of indole-based signaling pathways.81
The potential of the flustramine pyrroloindoline scaffold to serve as a template for the design of compounds with anti-biofilm activity was assessed by synthesizing flustramine C, 48 and evaluating its activity against three diverse terrestrial bacterial pathogens: Acinetobacter baumannii, E. coli, and methicillin resistant S. aureus (MRSA). Flustramine C exhibited moderate anti-biofilm activity against the two Gram-negative strains studied, inhibiting biofilm formation by 20-30% at a concentration of 150 μM, but did not display any anti-biofilm activity (<5% inhibition) against MRSA at the same concentration.82 A library of flustramine C analogues was constructed in order to delineate the structural components required for anti-biofilm activity and identify more potent inhibitors. The library was designed to preserve the amphipathic nature of the molecule, with the reverse prenyl group of flustramine C replaced by a propargyl group from which structural diversity could be introduced through click chemistry with a series of azide amide moieties.82 Compounds from this library exhibited considerably increased activity compared to flustramine C. Compound 49 inhibited biofilm formation by A. baumannii with an IC50 value of 193 μM, while compound 50 inhibited biofilm formation by E. coli with an IC50 value of 36 μM. Furthermore, inhibitors of biofilm formation by Gram-positive bacteria were also identified from this library, with the most active compound 51 able to inhibit MRSA biofilm formation with a low micromolar IC50 value (3.4 μM). All three compounds were shown to be acting via a non-microbicidal mechanism.82
A number of structurally related brominated tryptamine derivatives have been isolated from the Mediterranean gorgonian Paramuricea clavata and assessed for their anti-biofouling activity against three marine biofilm bacteria: Pseudoalteromonas spp D41 and TC8 and Paracoccus sp. 4M6.83 Bufotenine 52 exhibited significant anti-adhesion activity against one of the tested strains (D41) and did not display microbicidal activity towards V. fischeri in the Microtox assay.83 This family of natural products may therefore provide a further indole-based scaffold that could be manipulated to obtain highly active inhibitors of biofilm formation by medically relevant bacteria.
Two-component systems
Two component signal transduction systems (TCS) are a class of regulatory systems found mainly in prokaryotes, though some have been identified in a small number of eukaryotic organisms. These regulatory systems are the predominant signaling mechanism in bacteria and allow the organism to sense and respond to changes in their environment. TCS consist of a histidine kinase and a response regulator. In response to an extracellular signal, the histidine kinase undergoes autophosphorylation at a conserved histidine residue. The histidine kinase then transfers the phosphate group to a conserved aspartate residue on the response regulator, resulting in activation of an effector domain, which then leads to the response.84 This typically involves the control of gene expression by DNA binding of the phosphorylated response regulator to an upstream regulatory region. Many histidine kinases can also act as phosphatases and dephosphorylate the phosphorylated response regulator, thus reversibly controlling gene expression.85 TCS are activated by a variety of environmental factors including pH, nutrient level, redox state, osmotic pressure, quorum signals, and the presence of antibiotics. TCS regulate the expression of genes that control behaviors such as cell growth, virulence, biofilm formation and maintenance, quorum sensing and antibiotic 85 resistance.85 Examples of TCS involved in biofilm regulation include, AgrAC (S. aureus)85 LytSR (S. aureus)86, BfmRS (A. baumannii)87-89 BfmRS (P. aeruginosa),90 BfiSR91 (P. aeruginosa) GacAS (P. aeruginosa)92 and VicRK (S. mutans)93.
The AgrAC TCS in S. aureus is part of the agr regulatory locus described earlier and controls virulence in response to AIPs. AIPs act upon the histidine kinase AgrC, leading to autophosphorylation and subsequent phosphorylation of AgrA. Once phosphorylated AgrA binds the DNA region between the RNA III and agrB genes, promoting transcription of RNA III.85 The LytSR TCS plays an important role in S. aureus biofilm development as a result of its effect on the expression of the lrgAB operon. The lrgAB operon regulates cell death and lysis, along with the cidABC operon. The gene product of cidA is an effector of murein hydrolase activity and cell lysis, while lrgA encodes an inhibitor of these processes. One function of the cid and lrg operons is to mediate cell death and lysis during biofilm development, providing a source of extracellular genomic DNA (eDNA) for use as a component of the biofilm matrix and inhibition of cell lysis by the lrg gene product inhibits DNA release in the biofilm.86 Disruption of lytSR results in altered murein (peptidoglycan) hydrolase activity in addition to spontaneous cell lysis.86 Inactivation of lytS and lrgAB does not result in a murein hydrolase or lysis phenotype during planktonic growth, but produces a pronounced phenotype during biofilm growth. LytS mutation results in a thicker, more adherent biofilm with increased levels of eDNA in the biofilm matrix.
The BfmRS TCS in A. baumannii has been shown to play an important role in biofilm formation. Inactivation of the response regulator of this TCS, BfmR, in the A. baumannii strain ATCC 19606 results in a decreased ability of this strain to form a biofilm on abiotic surfaces, and also results in a significant change in cell morphology. BfmR is required for the expression of the Csu pili chaperone–usher assembly system and the biofilm proficiency of this strain of A. baumannii is dependent upon this system, which plays a role in the initial steps of biofilm formation on abiotic surfaces by enabling adherence of the bacterial cells and formation of microcolonies. Inactivation of the sensor component of this TCS, BfmS, causes only a modest reduction in biofilm forming ability, suggesting that cross-talk between other sensors and BfmR may be important.87,88 It has been shown however, that mutation of bfmS results in a large reduction in motility (80% reduction compared to wild type).89
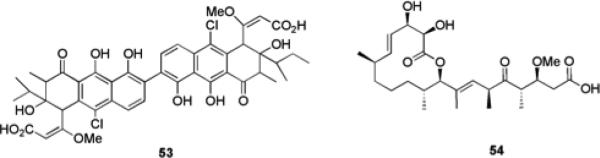
The role of TCS in the regulation of biofilm formation and maintenance makes them attractive potential targets for the development of anti-biofilm compounds, which has been underexploited to date. It has been shown that both the histidine kinase and response regulator of TCS are targetable by small molecules.94 Walkmycin C 53 (Fig. 9) is a known histidine kinase inhibitor that was isolated from a screen for inhibitors of the WalK histidine kinase in Bacillus subtilis, and has previously been shown to inhibit autophosphorylation of WalK in both B. subtilis and S. aureus.95 Walkmycin C targets the conserved catalytic domain of WalK and was therefore posited to potentially possess inhibitory activity against multiple histidine kinases. One such example is the VicRK TCS in S. mutans, an orthologue of WalRK that is involved in sucrose dependant biofilm formation. Walkmycin C was shown to inhibit the in vitro autophosphorylation activity of purified VicK with an IC50 of 2.87 μM (2.53 μg/mL).95 Addition of walkmycin C at levels below the minimum inhibitory concentration (MIC), resulted in the formation of abnormal biofilms and a reduction in biofilm mass (37.6% of the control at 0.63 μg/mL), demonstrating the potential of targeting TCS with small molecules as an anti-biofilm strategy.95
Fig. 9.
TCS inhibitors walkmycin C 53 and carolacton 54.
The bacterial secondary metabolite carolacton 54, affects the viability of S. mutans biofilms at nanomolar concentrations.93,96 This compound had only minor effects on the growth of planktonic bacteria but killed bacterial cells within a biofilm state. Carolacton was subsequently shown to affect the expression of six different TCS in S. mutans biofilm cells: CiaRH, VicKRX, RelRS, SMU.1037c/1038c, SMU.659/660, and ComDE.93 Microarray analysis identified VicKRX and ComDE as playing essential roles in the carolacton response, and known VicR controlled genes were downregulated upon treatment with carolacton. It was shown that this response is mediated through the serine/threonine protein kinase PknB, which the authors posited could be due to phosphorylation of VicR by PknB.93 Although the ability of carolacton to modulate S. mutans biofilms is not through a strictly non-microbicidal mechanism, the fact that this compound does not affect planktonic growth represents an alternative strategy for the small molecule control of biofilms and further validates targeting of TCS as a means of controlling bacterial biofilms.
Other signaling molecules
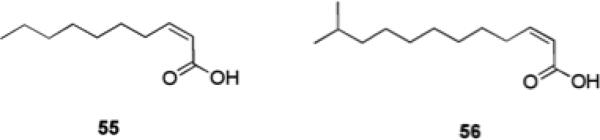
Davies et al. identified a fatty acid signaling molecule, cis-2-decenoic acid 55 (Fig. 10), produced by, P. aeruginosa during growth which was shown to inhibit biofilm development.97 Another fatty acid, cis-11-methyl-2-dodecenoic acid 56, known as diffusible signal factor (DSF) which was recovered from Xanthomonas campestris, is able to disaggregate cell flocs formed by X. campestris.98cis-2-Decenoic acid 55, was also shown to induce the dispersion of established biofilms formed by a number of bacteria including E. coli, Klebsiella pneumoniae, B. subtilis, and S. aureus at low nanomolar concentrations, as evidenced by microcolony disaggregation and measurement of the number of cells released into the bulk biofilm culture medium.97 The ability of cis-2-decenoic acid 55 to disperse pre-formed biofilms of A. baumannii and S. aureus, when investigated using a crystal violet reporter assay, which measures the biofilm mass remaining, was determined to be much lower and <10% dispersion was observed at a concentration of 400 μM, while the compound exhibited IC50 values of 121 μM and 220 μM against two S. aureus strains.99
Fig. 10.
Fatty acid signaling molecules that affect biofilm formation.
Certain D-amino acids, which are produced by bacteria during stationary phase, are thought to be a native signal for biofilm disassembly in B. subtilis and were shown to inhibit biofilm formation in fresh cultures of B. subtilis, S. aureus and P. aeruginosa.100,101 The inhibitory activity of D-amino acids is thought to be a consequence of disruption of the connection between an extracellular matrix protein and the bacterial cell leading to blocking of the growth of initial foci into larger assemblies of cells. Against B. subtilis, D-tyrosine, D-leucine, D-tryptophan, and D-methionine inhibited biofilm formation while the analogous L-amino acids, and other D-amino acids including D-alanine and D-phenylalanine were inactive. The individual amino acids showed inhibitory activity at concentrations from low micromolar (D-tyrosine) to millimolar (D-leucine); however combination of the four amino acids resulted in markedly increased activity compared to any alone, inhibiting biofilm formation at a concentration of 10 nM.100 Against S. aureus, the combination of D-phenylalanine, D-proline, and D-tyrosine exhibited greater biofilm inhibitory activity than the combination of D-tyrosine, D-leucine, D-tryptophan, and D-methionine.101 D-amino acids did not affect planktonic bacterial growth.101
Chemical Library Screening
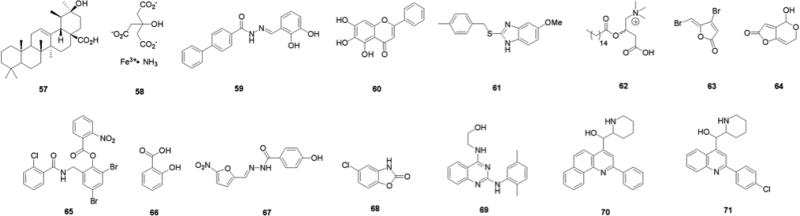
Due to the scarcity of known molecular scaffolds that inhibit/disperse bacterial biofilms, high throughput screening (HTS) has been employed in attempts to discover leads for new anti-biofilm modulators (Fig. 11). One of the first reports detailing the screening of a large library of compounds with the objective of identifying novel small molecules that possessed anti-biofilm activity was disclosed in 2005 by the Wood group.102 The screening of 13,000 compounds revealed a hit (0.08%), identified as ursolic acid 57, which effectively inhibited E. coli biofilm formation at concentrations as low as 22 μM without affecting growth. Microarray analysis of E. coli samples treated with compound 57 showed upregulation of genes responsible for chemotaxis, motility, and heat shock, all genes closely tied to biofilm formation.
Fig. 11.
Compounds with anti-biofilm activity identified through HTS.
In 2005, the Hergenrother group reported the identification of iron salts as effective non-antibiotic inhibitors and disruptors of P. aeruginosa biofilms from a screen of over 4,500 compounds which belonged to the University of Illinois Marvel Library Compound Collection (MLCC).103 Six compounds (0.13%) were found to inhibit biofilm development by at least 30% at a concentration of 50 μM in the preliminary assays. Ferric ammonium citrate (FAC) 58 was found to be a non-toxic inhibitor of P. aeruginosa PA14 biofilms, exhibiting an IC50 value of approximately 60 μM. No toxicity was observed even when FAC concentrations were increased upwards of 500 μM. Further investigation revealed that neither the ammonium or citrate ions were responsible for the observed anti-biofilm activity. The analysis of other iron salts (ferric chloride, ferric sulfate, ferrous sulfate) revealed biofilm inhibitory profiles that were comparable to that of FAC. They also demonstrated that by switching to an iron-rich growth media, established P. aeruginosa biofilms could be disrupted and cleared in continuous flow experiments. Additionally, analysis of various clinically isolated sputum samples from patients with cystic fibrosis showed that each genetically different strain exhibited varying degrees of susceptibility to elevated iron levels. Iron is known to be an important chemical in quorum sensing and bacterial virulence, high concentrations of iron are believed to suppress the expression of genes that encode for enzymes that scavenge excess iron, thus making the absence of these products detrimental to biofilm growth and maintenance.104 This same group has also shown that various iron chelates can suppress biofilm formation in P. aeruginosa.105
Clardy and Junker reported a comprehensive HTS in 2007, in which they sought to identify novel small molecules that inhibited and dispersed PA01 biofilms.106 A total of 66,095 unique molecules derived from libraries containing known bioactive compounds, natural products, and commercially available entities were screened using a luminescence-based assay. Of the compounds analyzed, 61 (0.09%) were shown to possess notable activities in bacterial anti-attachment assays and of these, 30 compounds were determined to exhibit IC50 values of less than 20 μM. The screens also identified a number of molecules that elicited biofilm antibiotic-enhancing effects. The most active compound discovered, 59, was shown to possess an IC50 value of 530 nM for biofilm inhibition. This makes 59 one of the most active biofilm modulators ever disclosed against either Gram-positive or Gram-negative bacteria. Compound 59 was also reported to exhibit an EC50 value for biofilm dispersion of 230 μM. The active compounds and the general scaffold classes they represent do not have any known bacterial targets or mechanisms of action. The authors, however, state that the chemical nature of some of the compounds may lead them to be metal chelators. The chemical composition of the scaffolds identified from the screens included aromatic based hydrazines and hydrazides, quinolines, and aryl substituted pyrazoles.
Computer-aided drug design (CADD) techniques have also been employed in the search for new biofilm modulators. One such study involved the investigation of 51 active compounds derived from traditional Chinese medicines whose antibacterial properties had been previously documented.107 By virtually determining if any of the target compounds exhibited similar docking characteristics in a 3-D environment to a known inhibitor of the A. tumefaciens quorum sensing transcriptional regulator protein TraR, the authors hypothesized that hits in the virtual screen would translate into compounds that would also display anti-biofilm activity against P. aeruginosa. Five compounds that performed well in the virtual screens were able to inhibit biofilm development and completely disperse P. aeruginosa biofilms at 200 μM, however no IC50 values were determined. The most active compound, baicalein 60, had no noticeable effect on bacterial growth. Baicalein is a flavonoid that is speculated to play a role in apoptosis and to also inhibit lipid peroxidation. The highlight of this article was the finding that ampicillin-resistant P. aeruginosa biofilms could be sensitized to the antibiotic in the presence of baicalein. No effects were observed when just the resistant cells were treated with only antibiotic or the anti-biofilm agent, yet when dosed together a significant synergistic effect was seen.
Screening of approximately 66,000 compounds and natural-product extracts from the Center for Chemical Genomics at the University of Michigan to identify compounds that affected induction of a V. cholerae c-di-GMP-inducible transcriptional fusion led to the discovery of a novel benzimidazole 61.108 This compound was examined for the ability to inhibit biofilm formation by a number of pathogenic bacterial strains. Compound 61 was shown to be a broad spectrum inhibitor of biofilm formation, significantly inhibiting biofilm formation by P. aeruginosa (CF-145), K. pneumoniae, Erwinia amylovora, and Shigella boydii, at 100 μM and by MRSA USA300, and S. aureus Newman at 25 μM, using the minimum biofilm eradication concentration (MBEC) static assay, without affecting bacterial growth. IC50 values, as calculated from the MBEC assay for inhibition of biofilm formation by P. aeruginosa and V. cholerae were determined to be 45.9 nM and 32.3 nM, respectively. The compound was, however, unable to disperse pre-formed biofilms.108
Examples of more focused libraries include the screening of 80 known eukaryotic protein kinase inhibitors for effects on biofilm formation.109 These compounds were selected as they have the potential to interact with broad range of targets due to their ATP-like scaffolds and the effects of such compounds had not yet been explored in microbial systems. Palmitoyl-DL-carnitine 62 was identified as an inhibitor of both PA01 and E. coli biofilm formation, and exhibited IC50 values of 13 μM and 3.0 μM, respectively.109 This compound was also subsequently shown to inhibit biofilm formation by Listeria monocytogenes with an IC50 value of 5.9 μM110 yet was unable to disperse pre-formed biofilms. The compound was observed to inhibit expression of the P. aeruginosa LasR/LasI quorum sensing system, though it is thought that this system is not the main target for these compounds as evidenced by the fact that palmitoyl-DL-carnitine exhibited almost comparable biofilm inhibitory effects on P. aeruginosa MW1, a lasI/rhlI double mutant, which is unable to produce autoinducers. The exact mechanism of action of palmitoyl-DL-carnitine therefore remains to be determined.109
A structure-based virtual screen (SB-VS) for the identification of putative quorum sensing inhibitors was carried out using a focused database comprising compounds that possess structural similarities to the known quorum-sensing inhibitors furanone C30 63, patulin 64, the P. aeruginosa LasR natural ligand (3-oxo-C12-AHL 5) and a known quorum-sensing receptor agonist TP-1, 65.111 This screen led to the discovery of three compounds, all recognized drugs: salicylic acid 66, nifuroxazide 67, and chlorzoxazone 68, which were subsequently shown to significantly inhibit quorum sensing-regulated gene expression at concentrations at which they did not affect bacterial growth. In addition to affecting quorum sensing regulated virulence factor production, these compounds were shown to affect biofilm formation by PA01. Biofilms treated with these compounds appeared thinner and less structured than control biofilms, and total biofilm mass was reduced in the presence of these quorum sensing inhibitors.111
A HTS of an 8,000-compound structurally diverse chemical library for small-molecule inhibitors of bacterial motility using V. cholerae identified several compounds that inhibited motility without exhibiting toxicity.112 The lead compound, a quinazoline-2,4-diamino analogue 69, completely suppressed motility in swarm agar plates without affecting the planktonic growth rate and was subsequently shown to inhibit biofilm formation by 1.6 and 1.9-fold at 18 h and 30 h respectively, at a concentration of 10 μg/mL, using a crystal violet staining assay.112
A high-throughput epifluorescence microscopy imaging based HTS has been developed for the identification of small molecule inhibitors of biofilm formation in V. cholerae.113 A number of compounds that reduced biofilm formation without altering bacterial cell viability were identified, with the two lead compounds 70 and 71, which are analogues of the antimalarial drug mefloquine, resulting in biofilm coverage of 0.5 and 1.8% respectively (compared to coverage of 20% in untreated wells) with comparable bacterial growth to control wells. Mefloquine has known antibiotic activity against Gram-positive bacteria but has not been shown to have any antibiotic activity against Gram-negative strains.113
Natural Products and Natural Product Analogues
Natural products provide a diverse array of chemical structures and possess a plethora of biological activities. A number of natural products that possess the ability to inhibit or disperse bacterial biofilms have been used as the starting points for medicinal chemistry programs in which synthetic manipulation of the natural product scaffold has allowed for the design of more efficacious compounds. Much of the natural product inspiration for these programs has come from compounds isolated from plants and marine organisms.
Plant extracts
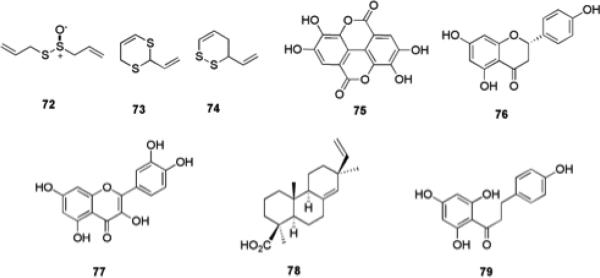
Interest in studying natural products derived from plant sources for the discovery of new biologically active compounds is not uncommon as many traditional medicines have been rooted heavily in the use of plant extracts for the treatment and cure of disease symptoms.114 Garlic possesses antibiotic properties which have been traced to the compound allicin 72 (Fig. 12).115 Renewed interest in garlic as a source for novel small molecules that modulate biofilms was generated with the finding that garlic extracts have shown the ability to clear pulmonary P. aeruginosa infections in mouse models.116 This particular study also demonstrated that P. aeruginosa biofilms grown in vitro and treated with garlic extracts were more susceptible to treatment with the antibiotic tobramycin and to grazing by polymorphonuclear (PMN) leukocytes. Isolation and analysis of the compounds responsible for these activities lead to the identification of a number of new compounds that possess antibiotic properties, while two particular dithianes 73 and 74, have shown the ability to modulate QS in LuxR systems without microbicidal activity.117
Fig. 12.
Molecules isolated from plant extracts and their derivatives that possess anti-biofilm activity.
Extracts from Rubus ulmifolius Schott., Rosaceae (Elmleaf blackberry), which are rich in the polyphenol ellagic acid 75 and glycosylated derivatives, have been shown to have an inhibitory effect on biofilm formation by a number of diverse S. aureus strains at concentrations well below those required to limit bacterial growth.7 Ellagic acid alone was also shown to possess anti-biofilm properties.7 Meanwhile, a number of flavonoids found in citrus species, including naringenin 76, and quercetin 77, which are antagonists of homoserine lactone and AI-2-mediated cell–cell signaling in V. harveyi, were able to inhibit biofilm formation by V. harveyi BB120 and E. coli O157:H7 in a dose-dependent manner.118
Another plant derived natural product that has recently been identified as a potential inhibitor of biofilm formation by oral pathogens is the resin acid 4-epi-pimaric acid 78, which was isolated from Aralia cachemirica L. (Araliaceae). 4-epi-Pimaric inhibited biofilm formation by S. mutans on a saliva-coated surface at sub-MIC concentrations, and was shown by confocal microscopy to inhibit clumping and attachment.119
The antioxidant phloretin 79 (found in apples) has been shown to markedly reduce biofilm formation by enterohemorrhagic E. coli O157:H7 without affecting the growth of planktonic cells, and also without affecting commensal E. coli K-12 biofilms.120 This is significant as the eradication of commensal bacteria by conventional antibiotics is a major problem and can result in increased susceptibility to infections. Therefore, antibacterial strategies that selectively eliminate pathogenic bacteria are much sought after. Phloretin repressed several genes including those encoding toxins, AI-2 importer genes, and curli genes in E. coli O157:H7 biofilm cells and it was also shown that phloretin resulted in reduced fimbria production, suggesting possible modes of action for this compound.120
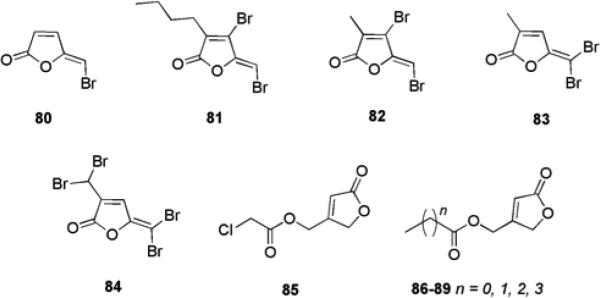
Halogenated Furanones
Comprehensive research reports have detailed the biological activities and effects that halogenated furanones (HFs) have on bacterial QS.121-124 These small molecules are natural products native to the marine red algae Delisea pulchra and primarily serve as a defense mechanism by acting as natural chemical deterrents (Fig. 13).125 It has been proposed that these small molecules are an evolutionary response to the adverse effects of AHL driven colonization of the host by marine bacterial species.126 Studies have shown that natural HFs isolated from the algae possess the ability to prevent biofilm formation and swarming in E. coli and B. subtilis.123,124 These findings have led to an extensive effort to chemically modify the furanone scaffold in hopes of tuning biological activity. Structurally, the similarity between HFs and AHLs is apparent when considering how each possesses a non-polar aliphatic carbon “tail” attached to a relatively polar “head”. As seen in AHLs, the length of the carbon chain attached to the furanone ring is varied in addition to the presence of hydroxyl or acyl functionalities. The presence of bromine atoms in these small molecules is one of the most significant structural differences between AHLs and HFs.
Fig. 13.
Halogenated furanones that possess anti-biofilm activity.
The activity of HFs in bioassays designed to assess AHL inhibitory activity has been well documented and includes the ability to impede luminescence in V. fischeri, and to affect the expression of 93 genes in P. aeruginosa studied through DNA microarray analysis.127,128 Most of the affected genes contribute to the production of virulence factors, notably biofilm formation. Giskov and co-workers have shown that specific furanones have the ability to affect these AHL-dependent activities by binding to LuxR protein homologues (the target receptors of AHLs) and promoting their degradation through proteolytic cleavage.129,130 Furthermore, the concentration of furanone needed to elicit the biological effect is consistent with concentrations of furanones found in D. pulchra.131 These observations have been thought to be a consequence of the strong binding affinity that comes from attack by nucleophilic residues in the binding pocket of the receptor to the electrophilic enone moiety of the HF, which results in structural changes that make the protein more prone to degradation.132
Studies involving the explicit use of HFs as tools to modulate bacterial biofilm growth and maintenance have revealed that synthetic furanones possess the ability to penetrate P. aeruginosa biofilm matrices and interfere with bacterial QS without any associated microbicidal properties.121 HF 80, which lacks a side chain and possesses a vinyl bromide, has been reported to display such activity. When biofilms were grown in the presence of HF 80, noticeable changes in biofilm maturation and structure were seen, similar to those observed in some P. aeruginosa mutants lacking the lasI system. While the furanone had no effect on initial bacterial attachment, bacterial detachment was more rapid resulting in a loss of more biomass from the treated sample in comparison to the untreated. Other experiments in which biofilms were grown in the presence or absence of 80 showed that there was no noticeable difference in biofilm morphology, yet the furanone treated biofilm was sensitized to antibiotic treatment with tobramycin.86
The activity of furanones against QS pathways in Gram-positive bacteria is less known, however, the natural furanone (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone 81, from D. pulchra has been shown to enhance biofilm formation by S. epidermidis and S. aureus at concentrations of 10 to 20% of the MIC.133 This increase in biofilm formation correlated with an increase in polysaccharide intercellular adhesin (PIA) and was shown to be a result of interference with luxS.133 Additionally, furanone 80 has also been shown to exhibit activity against a number of Gram-positive oral Streptococcus biofilms.134 Furanone 80 at a concentration of 60 μM resulted in a 63% and 76% reduction in biofilm growth in polystyrene wells of S. mutans and Streptococcus intermedius respectively.
The importance of the degree and pattern of bromination in furanones on the effects of biofilm formation in E. coli has been investigated.135 Compounds 82, 83, and 84 were shown to reduce biofilm formation at concentrations of 224 μM (82 and 83) and 141 μM (84) by 75%, 63%, and 80%, respectively. In contrast, another set of five chemically synthesized furanones (85-89), which contained an exocyclic ester functionality attached to the furanone ring and were completely devoid of bromination, were shown to be effective inhibitors of P. aeruginosa biofilm formation in flow cell confocal microscopy experiments.136 Molecular modeling was then employed to estimate the binding energies of the synthetic furanones to the LasR receptor in comparison to the known P. aeruginosa AHL signaling molecule 3-oxo-C12-AHL 5. Docking studies indicated that the furanone portion of the molecule overlapped with the lactone moiety of the AHL in the binding pocket, leading to docking scores that corresponded well with one another and provided a plausible explanation for the observed activity.
There are a number of issues that still need to be addressed to allow the use of halogenated furanones for human treatment therapies. These include toxicity, carcinogenic properties, and instability under aqueous conditions.137 Future research should address these issues to help solidify this class of small molecules as important players in the development of new therapeutics to combat bacterial biofilms.
Other Marine Natural Products and Analogues
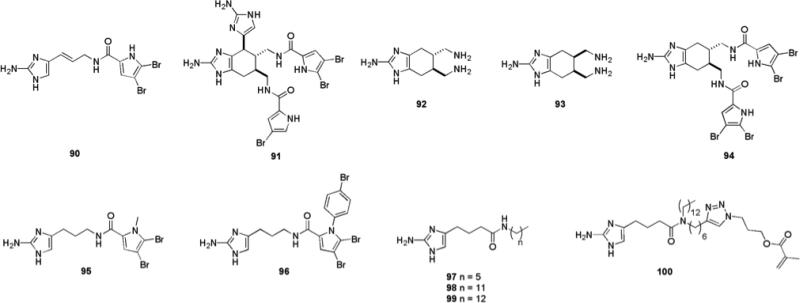
Some of the most active anti-biofilm compounds discovered to date have been based upon the molecular scaffolds of natural products isolated from marine natural products. The oroidin family of alkaloids possess nitrogen-dense architectures characterized by the incorporation of one or more 2-aminoimidazole (2-AI) sub-units.138 These molecules are found in marine sponges of the family Agelasidae, which reside in tropical water environments.139 Their role is believed to be that of a chemical anti-feeding defense mechanism against predators.140 The first report of oroidin family members inhibiting the formation of biofilms occurred in 1997 with the discovery that both oroidin 90 and bromoageliferin 91 (Fig. 14) displayed anti-biofouling activity against the Gram-negative marine α-proteobacterium Rhodospirillum salexigens.141 Bromoageliferin was reported to have an IC50 value of 2.43 nM while oroidin exhibited an IC50 value of 169 μM. Oroidin has also been reported to possess anti-attachment activity against V. vulnificus.142
Fig. 14.
2-Aminoimidazole alkaloids from sponges of the family Agelasaide: oroidin 90, bromoageliferin 91 and structurally simplified analogues that retain anti-biofilm activity.
Based upon these findings, it was posited that the architecture embedded within these marine alkaloids could provide a basis for the development of novel small molecules with the ability to inhibit and disperse bacterial biofilms. The ubiquity of the 2-aminoimidazole motif in these compounds led to the hypothesis that it was the key pharmacophore responsible for biological activity. Indeed, initial studies with two simplified analogues of bromoageliferin, TAGE (trans-bromoageliferin) 92 and CAGE (cis-bromoageliferin) 93 (Fig. 14), showed that these simple analogues inhibited biofilm formation by PA01 and PA14 under static conditions with IC50 values of 100 μM and 190 μM against PA01 and PA14 respectively for TAGE and IC50 values 100 μM and 180 μM against PA01 and PA14 respectively for CAGE. Both compounds were shown to be acting via a non-microbicidal mechanism.143 TAGE was also shown to possess the ability to disperse established P. aeruginosa biofilms (EC50 of 82 μM and 114 μM against PA01 and PA14 respectively). Confocal microscopy revealed a significant reduction in PA01 biofilm biomass after 48 hours in the presence of 100 μM TAGE. The ratio of live vs. dead cells in TAGE treated vs. untreated cells was similar, validating that the molecule was not selectively killing biofilm bacteria. Analysis of established PA01:GFP biofilms under flow condition confirmed the results obtained under static conditions.144 Installation of di-brominated acylpyrrole moieties similar to those present in bromoageliferin onto the TAGE scaffold result in compound 94, which exhibited increased biofilm inhibitory activity against P. aeruginosa, including against a mucoid variant (PD0300) representative of bacteria isolated from patients with cystic fibrosis. IC50 values for this compound were 1.77 μM, 12.0 μM and 2.47 μM against PA01, PA14 and PD0300 respectively. However, none of the acylated analogues were able to disperse pre-formed biofilms with the same efficiency as TAGE.144
Oroidin 90 exhibited similar activity to TAGE and CAGE against P. aeruginosa, with IC50 values of 190 μM and 166 μM against PA01 and PA14 respectively, confirming that a more simple architecture based upon the 2-AI scaffold was viable as an anti-biofilm agent.145 Structure-activity-relationships (SARs) for anti-biofilm activity were investigated thorough the construction of a 50-member library of analogues based upon the oroidin template.145 Three regions within the oroidin template were systematically varied in order to delineate which structural features of the molecule were required for biological activity in each section, these regions were: the pyrrole tail group, the linker chain, and the 2-AI head group. These studies revealed that any modifications to the 2-AI head of the molecule completely abolished activity. The most active molecule discovered was dihydrosventrin (DHS) 95 (Fig. 14), an analogue of the oroidin family alkaloid sventrin, which has also been isolated from marine sponges. DHS inhibited the formation of biofilms produced by PA01, PA14, PD0300, A. baumannii, and Bordatella bronchiseptica strain RB50 with IC50 values of 51 μM, 111 μM, 115 μM, 110 μM, and 238 μM, respectively.146 DHS was also able to disperse preformed biofilms of PA01, PA14, A. baumannii, and RB50. A library of second generation analogues designed to probe the effect of the N-alkyl substituent on the pyrrole ring on anti-biofilm activity in comparison to DHS147 led to the identification of compound 96, which possesses a para-bromo phenyl substituent, and exhibited greater non-microbicidal anti-biofilm activity against A. baumannii than DHS (IC50 = 27 μM, EC50 = 41 μM).
Other oroidin analogues, which could be accessed by more facile synthetic routes, included those in which the native amide bond directionality was reversed,148 those in which the amide bond was removed, moved along the linker chain of the molecule, and substituted with additional aliphatic side chains.149 Compounds possessing an aliphatic tail group exhibited low micromolar inhibition of PA01 and PA14 biofilm development, with even the least active compound (97) (Fig. 14) possessing activity four to five times greater than that of oroidin 90. The most active inhibitor of this first generation of reverse amide analogues (98) was also an extremely effective dispersion agent (IC50/EC50 = 2.84 μM/33 μM against PA01, IC50/EC50 = 2.26 μM/21 μM against PA14). A clear trend in which potency of the analogue was directly proportional to the length of the aliphatic chain was observed, with a break point against PA14 of a chain length of 13 carbons (99), for which an IC50 value of 729 nM was recorded. Removal of the amide bond entirely indicated that the presence of the amide bond or its respective directionality in this class of analogues is not entirely crucial for anti-biofilm activity, though these analogues exhibited lower activities than the reverse amide series.149
Covalent incorporation of one simple oroidin analogue, 99, into a methacrylate polymer in efforts to develop new biofilm-resistant material for use on indwelling medical devices (IMD) produced a polymeric composite of 4% monomer 100, which was observed to be resistant to biofilm colonization by A. baumannii.150
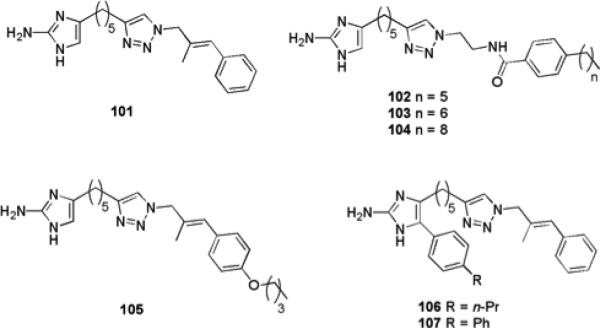
The incorporation of a triazole moiety to the oroidin analogues resulted in a number of highly active compounds. A number of 2-aminoimidazole/triazole (2-AIT) conjugates were generated,151 with the most active compounds possessing a substituted unsaturated aryl pendant group. This series yielded a highly active compound (101) (Fig. 15), which possessed broad spectrum inhibitory and dispersal properties against a variety of bacterial strains. Compound 101 was active against both Gram-positive and Gram-negative bacteria with IC50 values of 5.6 μM, 530 nM, 980 nM, and 810 nM and against PA01, PA14, A. baumannii, and S. aureus, respectively. This compound exhibited a synergistic effect with several antibiotics in the dispersion of pre-established biofilms of several bacterial strains, and was also shown to re-sensitize planktonic bacteria of drug-resistant strains of S. aureus and A. baumannii to the effects of conventional antibiotics.152
Fig. 15.
Highly active oroidin analogue 115 and derivatives that possess biofilm inhibition and dispersal activity.
Replacing the unsaturated aryl appendage of 101 with a diverse array of alkyl linked amides153,154 resulted in the discovery of several compounds with low micromolar biofilm inhibition and dispersal activity against a number of Gram-positive and Gram-negative bacterial strains (in addition to fungal anti-biofilm activity and activity against mixed species biofilms). The most active compounds possessed para-alkyl phenyl substituents and activity could again be tuned by adjusting the alkyl chain length. Compound 102, which has a six carbon alkyl chain, exhibited IC50/EC50 values of 21.4 μM/36.2 μM against V. vulnificus and an IC50 of 8.9 μM against R. salexigens, making this compound significantly more active than oroidin. Compound 103, which has a chain length of seven carbons, exhibited IC50 values of 11.2 μM and 1.9 μM against E. coli and a multi-drug resistant strain of A. baumannii respectively, and respective EC50 values of 23.4 μM and 7.9 μM against the same two strains. Compound 103 was highly active against Gram-positive bacteria, exhibiting IC50/EC50 values of 0.5 μM/0.7 μM against vancomycin-resistant enterococci (VRE) and 0.6 μM/28.1 μM against S. epidermidis. The most active compound against MRSA was compound 102, which exhibited IC50/EC50 values of 0.7 μM/0.9 μM. Compound 104, which has a nine carbon chain, exhibited increased biofilm inhibition activity against PA01 compared to 101, with an IC50 of 0.5 μM compared to 5.6 μM, and increased biofilm dispersion activity against PA14, with an EC50 of 2.5 μM compared to 22 μM for 101. All compounds were shown to be acting via a non-microbicidal mechanism.
Further efforts to increase the activity of 101 through modification of the aryl appendage have been carried out through the use of Suzuki–Miyaura coupling.155 A number of these compounds were found to be effective biofilm dispersion agents against A. baumannii, with the most active compound 105, exhibiting an EC50 of 44.7 μM, compared to 120 μM for the parent compound 101.
The effect of addition of substituents to the 2-AI moiety upon the ability of 101 to modulate biofilms has been investigated through a synthetic approach involving Grignard addition to α-amino Weinreb amides.99 The most potent compound identified from this library (106) exhibited an IC50 value of 1.42 μM against MRSA. Further substitution of the newly introduced phenyl ring resulted in the identification of a compound (107) with improved biofilm inhibition activity against MRSA (IC50 1.34 μM). The most potent non-microbicidal inhibitor of A. baumannii biofilm formation from this series (106) was not as active than the parent compound 101, exhibiting an IC50 value of 11.28 μM, though it was able to disperse pre-formed A. baumannii biofilms with an EC50 value of 44.6 μM.
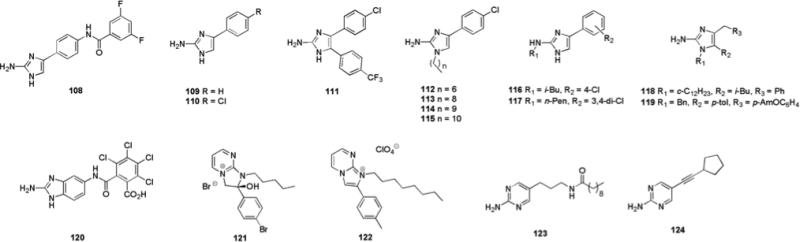
Construction of a 30-member library based upon an aryl 2-AI scaffold (Fig. 16) lead to the identification of compound 108, which was able to inhibit E. coli biofilm formation via a non-microbicidal mechanism, exhibiting an IC value of 5.2 μM.156
Fig. 16.
Aryl 2-AI, 2-ABI, and 2-AP compounds that possess anti-biofilm activity
Steenackers and co-workers have subsequently reported the anti-biofilm activity of a related series of aryl 2-AI compounds.157 The parent compound 109, which possess a phenyl group at the 4-position of the 2-AI ring, displayed moderate biofilm inhibitory activity, inhibiting biofilm formation by S. typhimurium with an IC50 of 130 μM at 25 °C and an IC50 of 146 μM at 37 °C, and P. aeruginosa biofilm formation with IC50 values of 72.6 and 22.7 μM at 25 °C and 37 °C, respectively. Substitution of the phenyl ring at the para-position resulted in increased biofilm inhibition activity, with the most active compound, 110, which possesses a chlorine substituent, inhibiting biofilm formation by S. typhimurium with IC50 values of 16.0 and 30.1 μM at 25 °C and 37 °C respectively, and P. aeruginosa biofilm formation with IC50 values of 3.5 and 29.3 μM at 25 °C and 37 °C respectively. Introduction of a number of aryl substituents at the 5-position of the 2-AI ring resulted in a slight increase in activity relative to 110, with the most active analogue 111 inhibiting biofilm formation by S. typhimurium at 25 °C with an IC50 value of 10.8 μM. The introduction of alkyl substituents at the 1-position of the 2-AI ring also modulated activity, short chain substituents in general resulted in a decrease in activity, while the most active analogues against P. aeruginosa biofilm formation were those possessing an alkyl chain of intermediate length, with 112, which has a heptyl substituent, exhibiting an IC value of 2.6 μM.157 Longer alkyl chain substituents resulted in the highest activity against S. typhimurium, with compounds possessing nonyl (113), decyl (114) and undecyl (115) chains exhibiting IC50 values of ~4 μM at 25 °C. The synthesis of a related series of 2,4-disubstituted 2-aminoimidazoles also identified a number of inhibitors of biofilm formation with the most active compounds, 116 and 117, able to inhibit biofilm formation by S. typhimurium, with IC50 values of 2.0 and 2.2 μM respectively, and P. aeruginosa biofilm formation, with IC50 values of 0.9 and 0.7 μM respectively at 25 °C.158
The generation of a series of 1,4,5-trisubstituted naamine alkaloids via a silver (I) mediated synthesis from secondary propargylamines produced several compounds that could inhibit biofilm formation by S. typhimurium and P. aeruginosa without significantly affecting planktonic growth. The lead compounds, 118 and 119, exhibited IC50 values of 10.3 μM (118) and 18.3 μM (119) against S. typhimurium and 27.4 μM (118) and 17.4 μM (119) against P. aeruginosa.159
Scaffolds related to the 2-AI scaffold that have been investigated for anti-biofilm properties include 2-aminobenzimidazole (2-ABI)160 and 2-aminopyrimidine (2-AP)161 (Fig. 16). The most active non-microbicidal 2-ABI identified was compound 120, with IC50 and EC50 values of 890 nM and 2.9 μM (MRSA), 1.4 μM and 75 μM (VRE), and 570 nM and 7.3 μM (S. epidermidis), respectively. The ability of compound 120 to prevent biofilm formation by these Gram-positive bacterial strains was inhibited by the presence of Zn (II), and the compound was subsequently shown to bind Zn (II) directly, providing insight into the potential mode of action of this compound.160
A series of 2-hydroxy-2-aryl-2,3-dihydroimidazopyrimidinium salts, which are intermediates in the synthesis of 2,4,5-trisubstituted 2-aminoimidazoles, included a number of compounds with low micromolar biofilm inhibition activity against S. typhimurium and P. aeruginosa at 25 °C, with the most active compound, 121, exhibiting IC50 values of 1.7 μM and 4.2 μM.231 Similarly, a series of imidazo[1,2-α]pyrimidinium salts produced several active compounds, with the most active compound, 122, exhibiting IC50 values of 1.5 μM and 1.0 μM against S. typhimurium and P. aeruginosa at 25 °C.157 However, it is believed that both of these pyrimidinium scaffolds are cleaved in vivo to the corresponding substituted 2-aminoimidazoles, which may be responsible for the observed activity.157,158
A number of 2-aminopyrimidine derivatives that could not be cleaved to a 2-aminoimidazole were synthesized to unequivocally evaluate the 2-AP scaffold for the ability to modulate bacterial biofilms. 2-AP derived compounds were found to be less active as inhibitors of biofilm formation than their 2-AI counterparts, particularly against Gram-negative bacteria. None were able to disperse pre-formed biofilms. The most active, non-microbicidal 2-APs discovered were compounds 123, which inhibited MRSA biofilm formation with an IC50 of 72 μM, and compound 124, which inhibited biofilm formation by a methicillin sensitive strain of S. aureus with an IC50 of 67 μM.161
Two diterpene alkaloids, (-)-agelasine D 125, and the corresponding oxime derivative (-)-ageloxime D 126 (Fig. 17), were isolated from the marine sponge Agelas nakamurai.162 (-)-Agelasine D 125 inhibited planktonic growth of S. epidermidis (MIC < 0.0877 μM) but did not inhibit biofilm formation, while the converse was true for (-)-ageloxime D 126, which inhibited biofilm formation without affecting bacterial growth. A synthetic route to (+)-agelasine D has been developed163 and this scaffold has the potential to be exploited for the development of compounds with anti-biofilm activity via synthetic manipulation, as has been achieved for the oroidin and bromoageliferin scaffolds.162
Fig. 17.
(-)-Agelasine D 125, (-)-ageloxime D 126 and marine derived carbamates that possess anti-biofilm activity
The structurally simple bacterial metabolite ethyl N-(2-phenethyl) carbamate 127 (Fig. 17), which was isolated from the culture medium of the marine bacterium SCRC3P79 (Cytophaga sp.) has been reported to possess moderate anti-biofilm activity against R. salexigens.141 This compound was also shown to be moderately active in inhibiting biofilm formation by medically relevant strains of bacteria including S. epidermidis, MRSA, VRE, multi-drug resistant A. baumannii (MDRAB), and E. coli.164 A library of analogues based upon compound 127 was constructed, leading to the identification of a number of compounds with increased activity. The menthyl derived compounds 128-130 were the most active, inhibiting biofilm formation by several strains of S. aureus with IC50 values in the mid to low micromolar range.164
Conclusion
Bacterial biofilms pose a serious threat to society and are responsible for the morbidity and mortality of a myriad of diseases as they are inherently insensitive to antibiotics and the host immune response. The increase in antibiotic resistance among many bacterial strains has put a tremendous pressure on the medical community to find alternative approaches for the treatment of biofilm-mediated diseases. This review describes the current standing of research into the control of bacterial biofilms using small molecules that do not act as antibiotics or microbicides, and instead elicit anti-biofilm activity through alternative mechanisms, such as interfering with the complex regulatory systems that control biofilm formation and maintenance. Molecules with such characteristics should offer the unique ability to mitigate bacterial selection pressures while simultaneously providing insight into how bacteria communicate with one another. Agents with the ability to prevent the formation of, or even eradicate, biofilms have the potential to restore the efficacy of antibiotics that are ineffective against biofilm bacteria, and should have a profound impact upon human medicine.
Acknowledgements
Financial support is gratefully acknowledged from the V Foundation, Agile Sciences, the National Institutes of Health (GM055769), The Department of Defense DMRDP program (W81XWH-11-2-0115), The North Carolina Biotechnology Center, the Kennan Foundation, and the University of North Carolina Competiveness Research Fund. The DMRDP program is administered by the Department of Army; The U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, Fort Detrick, MD 21702-5014 is the awarding and administering acquisition office. The content of this manuscript does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
References
- 1.Donlan RM, Costerton JW. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musk DJ, Hergenrother PJ. Curr. Med. Chem. 2006;13:2163–2177. doi: 10.2174/092986706777935212. [DOI] [PubMed] [Google Scholar]
- 3.Davies D. Nat. Rev. Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 4.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 5.Smith K, Hunter IS. J. Med. Microbiol. 2008;57:966–973. doi: 10.1099/jmm.0.47668-0. [DOI] [PubMed] [Google Scholar]
- 6.Rasmussen TB, Givskov M. Int. J. Med. Microbiol. 2006;296:149–61. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 7.Quave CL, Estevez-Carmona M, Compadre CM, Hobby G, Hendrickson H, Beenken KE, Smeltzer MS. PLoS ONE. 2012;7:e28737. doi: 10.1371/journal.pone.0028737. doi:10.1371/journal.pone.0028737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neu HC. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 9.Conly JM, Johnston BL. Can. J. Infect. Dis. Med. Microbiol. 2005;16:159–160. doi: 10.1155/2005/892058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart PS, Costerton JW. Lancet. 2001;358:135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 11.Davey ME, O'Toole GA. Microbiol. Mol. Biol. Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hausner M, Wuertz S. Appl. Environ. Microbiol. 1999;65:3710–3713. doi: 10.1128/aem.65.8.3710-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blazquez J. Clin. Infect. Dis. 2003;37:1201–1209. doi: 10.1086/378810. [DOI] [PubMed] [Google Scholar]
- 14.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. Science. 2000;288:1251–1253. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 15.Mah TF, O'Toole GA. Trends Microbiol. 2001;9:34–39. doi: 10.1016/s0966-842x(00)01913-2. [DOI] [PubMed] [Google Scholar]
- 16.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 17.Spoering AL, Lewis K. Journal of Bacteriology. 2001;183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurdle JG, O'Neill AJ, Chopra I, Lee RE. Nat. Rev. Microbiol. 2011;9:62–75. doi: 10.1038/nrmicro2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coates ARM, Hu Y. Trends Pharmacol. Sci. 2008;29:143–150. doi: 10.1016/j.tips.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Harmsen M, Yang L, Pampand SJ, Tolker-Nielsen T. FEMS Immunol. Med. Microbiol. 2010;59:253–268. doi: 10.1111/j.1574-695X.2010.00690.x. [DOI] [PubMed] [Google Scholar]; Bacteriol. 2001;183:1047–57. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camilli A, Bassler BL. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henke JM, Bassler BL. J. Bacteriol. 2004;186:6902–6914. doi: 10.1128/JB.186.20.6902-6914.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jayaraman A, Wood TK. Annu. Rev. Biomed. Eng. 2008;10:145–167. doi: 10.1146/annurev.bioeng.10.061807.160536. [DOI] [PubMed] [Google Scholar]
- 24.Parsek MR, Greenberg EP. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8789–8793. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amara N, Mashiach R, Amar D, Krief P, Spieser SAH, Bottomley MJ, Aharoni A, Meijler MM. J. Am. Chem. Soc. 2009;131:10610–10619. doi: 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]
- 26.Finch RG, Pritchard DI, Bycroft BW, Williams P, Stewart GS. J. Antimicrob. Chemoth. 1998;42:569–571. doi: 10.1093/jac/42.5.569. [DOI] [PubMed] [Google Scholar]
- 27.Manefield M, Kjelleberg S, Givskov M. Curr. Med. Chem. Anti Infect. Agents. 2003;2:213–218. [Google Scholar]
- 28.Geske GD, O'Neill JC, Blackwell HE. Chem. Soc. Rev. 2008;37:1432–1447. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. J. Am. Chem. Soc. 2005;127:12762–12763. doi: 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]
- 30.Geske GD, O'Neill JC, Miller DM, Mattmann ME, Blackwell HE. J. Am. Chem. Soc. 2007;129:13613–13625. doi: 10.1021/ja074135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith KM, Bu YG, Suga H. Chem. Biol. 2003;10:563–571. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 32.Smith KM, Bu YG, Suga H. Chem. Biol. 2003;10:81–89. doi: 10.1016/s1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 33.Glansdorp FG, Thomas GL, Lee JJ, Dutton JM, Salmond GP, Welch M, Spring DR. Org. Biomol. Chem. 2004;2:3329–3336. doi: 10.1039/B412802H. [DOI] [PubMed] [Google Scholar]
- 34.Frezza M, Castang S, Estephane J, Soulere L, Deshayes C, Chantegrel B, Nasser W, Queneau Y, Reverchon S, Doutheau A. Bioorg. Med. Chem. 2006;14:4781–4791. doi: 10.1016/j.bmc.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Schaefer AL, Hanzelka BL, Eberhard A, Greenberg EP. J. Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castang S, Chantegrel B, Deshayes C, Dolmazon R, Gouet P, Haser R, Reverchon S, Nasser W, Hugouvieux-Cotte-Pattat N, Doutheau A. Bioorg. Med. Chem. Lett. 2004;14:5145–5149. doi: 10.1016/j.bmcl.2004.07.088. [DOI] [PubMed] [Google Scholar]
- 37.Ishida T, Ikeda T, Takiguchi N, Kuroda A, Ohtake H, Kato J. Appl. Environ. Microbiol. 2007;73:3183–3188. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bottomley MJ, Muraglia E, Bazzo R, Carfi A. J. Biol. Chem. 2007;282:13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 39.MacLehose HG, Gilbert P, Allison DG. J. Antimicrob. Chemoth. 2004;53:180–184. doi: 10.1093/jac/dkh090. [DOI] [PubMed] [Google Scholar]
- 40.Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, Goldner M, Dessaux Y, Camara M, Smith H, Williams P. Infect. Immun. 2002;70:5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McInnis CE, Blackwell HE. Bioorg. Med. Chem. 2011;19:4820–4828. doi: 10.1016/j.bmc.2011.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abraham WR. Curr. Med. Chem. 2006;13:1509–1524. doi: 10.2174/092986706777442039. [DOI] [PubMed] [Google Scholar]
- 43.Takano E, Chakraburtty R, Nihira T, Yamada Y, Bibb MJ. Mol. Microbiol. 2001;41:1015–1028. doi: 10.1046/j.1365-2958.2001.02562.x. [DOI] [PubMed] [Google Scholar]
- 44.Lyon GJ, Muir TW. Chem. Biol. 2003;10:1007–1021. doi: 10.1016/j.chembiol.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 45.Bordi C, de Bentzmann S. Annals of Intensive Care. 2011;1:19. doi: 10.1186/2110-5820-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fux CA, Stoodley P, Hall-Stoodley L, Costerton JW. Expert Rev. Anti-infect. Ther. 2003;1:667–683. doi: 10.1586/14787210.1.4.667. [DOI] [PubMed] [Google Scholar]
- 47.Giacometti A, Cirioni O, Gov Y, Ghiselli R, Del Prete MS, Mocchegiani F, Saba V, Orlando F, Scalise G, Balaban N, Dell'Acqua G. Antimicrob. Agents Chemother. 2003;47:1979–1983. doi: 10.1128/AAC.47.6.1979-1983.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balaban N, Cirioni O, Giacometti A, Ghiselli R, Braunstein JB, Silvestri C, Mocchegiani F, Saba V, Scalise G. Antimicrob. Agents Chemother. 2007;51:2226–2229. doi: 10.1128/AAC.01097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan WC, Coyle BJ, Williams P. J. Med. Chem. 2004;47:4633–4641. doi: 10.1021/jm0400754. [DOI] [PubMed] [Google Scholar]
- 50.Fowler SA, Stacy DM, Blackwell HE. Org. Lett. 2008;10:2329–2332. doi: 10.1021/ol800908h. [DOI] [PubMed] [Google Scholar]
- 51.Waters CM, Bassler BL. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 52.Hardie KR, Heurlier K. Nat. Rev. Microbiol. 2008;6:635–643. doi: 10.1038/nrmicro1916. [DOI] [PubMed] [Google Scholar]
- 53.Vendeville A, Winzer K, Heurlier K, Tang CM, Nat KR. Rev. Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- 54.Lowery CA, Park J, Kaufmann GF, Janda KD. J. Am. Chem. Soc. 2008;130:9200–9201. doi: 10.1021/ja802353j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meijler MM, Hom LG, Kaufmann GF, McKenzie KM, Sun CZ, Moss JA, Matsushita M, Janda KD. Angew. Chem. Int. Ed. 2004;43:2106–2108. doi: 10.1002/anie.200353150. [DOI] [PubMed] [Google Scholar]
- 56.Smith JAI, Wang J, Nguyen-Mau S-M, Leeb V, Sintim HO. Chem. Commun. 2009:7033–7035. doi: 10.1039/b909666c. [DOI] [PubMed] [Google Scholar]
- 57.Brackman G, Celen S, Baruah K, Bossier P, Van Calenbergh S, Nelis HJ, Coenye T. Microbiology. 2009;155:4114–4122. doi: 10.1099/mic.0.032474-0. [DOI] [PubMed] [Google Scholar]
- 58.Ronning DR, Iacopelli NM, Mishra V. Protein Science. 2010;19:2498–2510. doi: 10.1002/pro.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutierrez JA, Crowder T, Rinaldo-Matthis A, Ho M-C, Almo SC, Schramm VL. Nat. Chem. Biol. 2009;5:251–257. doi: 10.1038/nchembio.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan H, Chen W. Chem. Soc. Rev. 2010;39:2914–2924. doi: 10.1039/b914942m. [DOI] [PubMed] [Google Scholar]
- 61.Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. J. Bacteriol. 2005;187:1792–1798. doi: 10.1128/JB.187.5.1792-1798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tamayo R, Pratt JT, Camilli A. An. Rev. Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camilli A, Bassler BL. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sintim HO, Smith JA, Wang J, Nakayama S, Yan L. Future Med. Chem. 2010;2:1005–1035. doi: 10.4155/fmc.10.185. [DOI] [PubMed] [Google Scholar]
- 65.Karaolis DK, Rashid MH, Chythanya R, Luo W, Hyodo M, Hayakawa Y. Antimicrob. Agents Chemother. 2005;49:1029–1038. doi: 10.1128/AAC.49.3.1029-1038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan W, Qu T, Zhao H, Su L, Yu Q, Gao J, Wu B. Microbiol. Res. 2010;165:87–96. doi: 10.1016/j.micres.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 67.Bernier SP, Ha DG, Khan W, Merritt JH, O'Toole GA. Res. Microbiol. 2011;162:680–688. doi: 10.1016/j.resmic.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Antoniani D, Bocci P, Maciag A, Raffaelli N, Landini P. Appl Microbiol Biotechnol. 2010;85:1095–1104. doi: 10.1007/s00253-009-2199-x. [DOI] [PubMed] [Google Scholar]
- 69.Lee J, Lee J-H. FEMS Microbiol. Rev. 2010;34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 70.Wood TK. Environ. Microbiol. 2009;11:1–15. doi: 10.1111/j.1462-2920.2008.01768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nishino K, Nikaido E, Yamaguchi A. J. Bacteriol. 2007;189:9066–9075. doi: 10.1128/JB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bartlett DH, Mueller RS, Beyhan S, Saini SG, Yildiz FH. J. Bacteriol. 2009;191:3504–3516. doi: 10.1128/JB.01240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee J, Jayaraman A, Wood TK. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK. Appl. Environ. Microbiol. 2007;73:4100–4109. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wood TK, Lee J, Zhang XS, Hegde M, Bentley WE, Jayaraman A. ISME J. 2008;2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
- 76.Kanamaru K, Kanamaru K, Tatsuno I, Tobe T, Sasakawa C. Mol. Microbiol. 2000;38:805–816. doi: 10.1046/j.1365-2958.2000.02171.x. [DOI] [PubMed] [Google Scholar]
- 77.Lee J-H, Kim Y-G, Kim C-J, Lee J-C, Cho MH, Lee J. Appl. Microbiol. Biotechnol. 2012 DOI 10.1007/s00253-012-3881-y. [Google Scholar]
- 78.Lee J-H, Cho MH, Lee J. Environ. Microbiol. 2011;13:62–73. doi: 10.1111/j.1462-2920.2010.02308.x. [DOI] [PubMed] [Google Scholar]
- 79.Lee J-H, Kim Y-G, Cho MH, Kim J-A, Lee J. FEMS Microbiol. Lett. 2012 DOI 10.1111/j.1574-6968.2012.02500.x. [Google Scholar]
- 80.Peters L, Konig GM, Wright AD, Pukall R, Stackebrandt E, Eberl L, Riedel K. Appl. Environ. Microbiol. 2003;69:3469–3475. doi: 10.1128/AEM.69.6.3469-3475.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bunders CA, Minvielle MJ, Worthington RJ, Ortiz M, Cavanagh J, Melander C. J. Am. Chem. Soc. 2011;133:20160–20163. doi: 10.1021/ja209836z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bunders C, Cavanagh J, Melander C. Org. Biomol. Chem. 2011;9:5476–5481. doi: 10.1039/c1ob05605k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Penez N, Culioli G, Perez T, Briand J-F, Thomas OP, Blache Y. J. Nat. Prod. 2011;74:2304–2308. doi: 10.1021/np200537v. [DOI] [PubMed] [Google Scholar]
- 84.Stock AM, Robinson VL, Goudreau PN. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 85.Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. Curr. Opin. Microbiol. 2010;13:232–239. doi: 10.1016/j.mib.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 86.Sharma-Kuinkel BK, Mann EE, Ahn J, Kuechenmeister LJ, Dunman PM, Bayles KW. J. Bacteriol. 2009;191:4767–4775. doi: 10.1128/JB.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaddy JA, Actis LA. Future Microbiol. 2009;4:273–278. doi: 10.2217/fmb.09.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tomaras AP, Flagler MJ, Dorsey CW, Gaddy JA, Actis LA. Microbiol. 2008;154:3398–3409. doi: 10.1099/mic.0.2008/019471-0. [DOI] [PubMed] [Google Scholar]
- 89.Clemmer KM, Bonomo RA, Rather PN. Microbiol. 2011;157:2534–2544. doi: 10.1099/mic.0.049791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Petrova OE, Schurr JR, Schurr MJ, Sauer K. Mol. Microbiol. 2011;81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Petrova OE, Sauer K. J. Bacteriol. 2010;192:5275–5288. doi: 10.1128/JB.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Parkins MD, Ceri H, Storey DG. Mol. Microbiol. 2001;40:1215–1226. doi: 10.1046/j.1365-2958.2001.02469.x. [DOI] [PubMed] [Google Scholar]
- 93.Reck M, Rutz K, Kunze B, Tomasch J, Surapaneni SK, Schulz S, Wagner-Döbler I. J. Bacteriol. 2011;193:5692–5706. doi: 10.1128/JB.05424-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Belcheva A, Golemi-Kotra D. J. Biol. Chem. 2008;238:12354–12364. doi: 10.1074/jbc.M710010200. [DOI] [PubMed] [Google Scholar]
- 95.Eguchi Y, Kubo N, Matsunaga H, Igarashi M, Utsumi R. Antimicrob. Agents Chemother. 2011;55:1475–1484. doi: 10.1128/AAC.01646-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kunze B, Reck M, Dötsch A, Lemme A, Schummer S, Irschik H, Steinmetz H, Wagner-Döbler I. BMC Microbiol. 2010;10:199. doi: 10.1186/1471-2180-10-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davies DG, Marques CNH. J. Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dow JM, Crossman L, Findlay K, He Y-Q, Feng J-X, Tang J-L. Proc. Natl. Acad. Sci. 2003;100:10995–11000. doi: 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Su Z, Peng L, Worthington RJ, Melander C. ChemMedChem. 2011;6:2243–2251. doi: 10.1002/cmdc.201100316. [DOI] [PubMed] [Google Scholar]
- 100.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hochbaum AI, Kolodkin-Gal I, Foulston L, Kolter R, Aizenberg J, Losick R. J. Bacteriol. 2011;193:5616–5622. doi: 10.1128/JB.05534-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ren DC, Zuo RJ, Barrios AF, Bedzyk LA, Eldridge GR, Pasmore ME, Wood TK. Appl. Environ. Microbiol. 2005;71:4022–4034. doi: 10.1128/AEM.71.7.4022-4034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Musk DJ, Banko DA, Hergenrother PJ. Chem. Biol. 2005;12:789–796. doi: 10.1016/j.chembiol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 104.Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. Microbiol.-Sgm. 2007;153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- 105.Musk DJ, Hergenrother PJ. J. Appl. Microbiol. 2008;105:380–388. doi: 10.1111/j.1365-2672.2008.03751.x. [DOI] [PubMed] [Google Scholar]
- 106.Junker LM, Clardy J. Antimicrob. Agents Chemother. 2007;51:3582–3590. doi: 10.1128/AAC.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zeng Z, Qian L, Cao L, Tan H, Huang Y, Xue X, Shen Y, Zhou S. Appl. Microbiol. Biotechnol. 2008;79:119–126. doi: 10.1007/s00253-008-1406-5. [DOI] [PubMed] [Google Scholar]
- 108.Sambanthamoorthy K, Gokhale AA, Lao W, Parashar V, Neiditch MB, Semmelhack MF, Lee I, Waters CM. Antimicrob. Agents Chemother. 2011;55:4369–4378. doi: 10.1128/AAC.00583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wenderska IB, Chong M, McNulty J, Wright GD, Burrows LL. ChemBioChem. 2011;12:2759–2766. doi: 10.1002/cbic.201100500. [DOI] [PubMed] [Google Scholar]
- 110.Nguyen UT, Wenderska IB, Chong MA, Koteva K, Wright GD, Burrows LL. Appl. Environ. Microbiol. 2011 doi: 10.1128/AEM.07227-11. doi:10.1128/AEM.07227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang L, Rybtke MT, Jakobsen TH, Hentzer M, Bjarnsholt T, Givskov M, Tolker-Nielsen T. Antimicrob. Agents Chemother. 2009;53:2432–2443. doi: 10.1128/AAC.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rasmussen L, White EL, Pathak A, Ayala JC, Wang H, Wu J-H, Benitez JA, Silva AJ. Antimicrob. Agents Chemother. 2011;55:4134–4143. doi: 10.1128/AAC.00482-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peach KC, Bray WM, Shikuma NJ, Gassner NC, Lokey RS, Yildiz FH, Linington RG. Mol. BioSyst. 2011;7:1176–1184. doi: 10.1039/c0mb00276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lai PK, Roy J. Curr. Med. Chem. 2004;11:1451–60. doi: 10.2174/0929867043365107. [DOI] [PubMed] [Google Scholar]
- 115.Slusarenko AJ, Patel A, Portz D. Euro. J. Plant Pathol. 2008;121:313–322. [Google Scholar]
- 116.Bjarnsholt T, Jensen PO, Rasmussen TB, Christophersen L, Calum H, Hentzer M, Hougen HP, Rygaard J, Moser C, Eberl L, Hoiby N, Givskov M. Microbiol.-Sgm. 2005;151:3873–3880. doi: 10.1099/mic.0.27955-0. [DOI] [PubMed] [Google Scholar]
- 117.Persson T, Hansen TH, Rasmussen TB, Skinderso ME, Givskov M, Nielsen J. Org. Biomol. Chem. 2005;3:253–262. doi: 10.1039/b415761c. [DOI] [PubMed] [Google Scholar]
- 118.Vikram A, Jayaprakasha GK, Jesudhasan PR, Pillai1 SD, Patil BS. J. Appl. Microbiol. 2010;109:515–527. doi: 10.1111/j.1365-2672.2010.04677.x. [DOI] [PubMed] [Google Scholar]
- 119.Ali F, Sangwan PL, Koul S, Pandey A, Bani S, Abdullah ST, Sharma PR, Kitchlu S, Khan IA. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:149–159. doi: 10.1007/s10096-011-1287-x. [DOI] [PubMed] [Google Scholar]
- 120.Lee J-H, Regmi SC, Kim JA, Cho MH, Yun H, Lee C-S, Lee J. Infect. Immun. 2011;79:4819–4827. doi: 10.1128/IAI.05580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Hoiby N, Kjelleberg S, Givskov M. Microbiol.-Sgm. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- 122.Manefield M, Welch M, Givskov M, Salmond GP, Kjelleberg S. FEMS Microbiol. Lett. 2001;205:131–138. doi: 10.1111/j.1574-6968.2001.tb10936.x. [DOI] [PubMed] [Google Scholar]
- 123.Ren D, Sims JJ, Wood TK. Lett. Appl. Microbiol. 2002;34:293–299. doi: 10.1046/j.1472-765x.2002.01087.x. [DOI] [PubMed] [Google Scholar]
- 124.Ren DC, Sims JJ, Wood TK. Environ. Microbiol. 2001;3:731–736. doi: 10.1046/j.1462-2920.2001.00249.x. [DOI] [PubMed] [Google Scholar]
- 125.Denys R, Wright AD, Konig GM, Sticher O. Tetrahedron. 1993;49:11213–11220. [Google Scholar]
- 126.Kjelleberg S, Steinberg P, Givskov M, Gram L, Manefield M, De Nys R. Aquat. Microb. Ecol. 1997;13:85–93. [Google Scholar]
- 127.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song ZJ, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Hoiby N, Givskov M. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rasmussen TB, Manefield M, Andersen JB, Eberl L, Anthoni U, Christophersen C, Steinberg P, Kjelleberg S, Givskov M. Microbiol.-Uk. 2000;146:3237–3244. doi: 10.1099/00221287-146-12-3237. [DOI] [PubMed] [Google Scholar]
- 129.Manefield M, De Nys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg SA. Microbiol.-Uk. 1999;145:283–291. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- 130.Manefield M, Rasmussen TB, Henzter M, Andersen JB, Steinberg P, Kjelleberg S, Givskov M. Microbiol.-Sgm. 2002;148:1119–1127. doi: 10.1099/00221287-148-4-1119. [DOI] [PubMed] [Google Scholar]
- 131.Khmel IA, Metlitskaya AZ. Mol. Biol. 2006;40:195–210. [PubMed] [Google Scholar]
- 132.Konaklieva MI, Plotkin BJ. Mini-Rev. Med. Chem. 2006;6:817–825. doi: 10.2174/138955706777698589. [DOI] [PubMed] [Google Scholar]
- 133.Kuehl R, Al-Bataineh S, Gordon O, Luginbuehl R, Otto M, Textor M, Landmann R. Antimicrob. Agents Chemother. 2009;53:4159–4166. doi: 10.1128/AAC.01704-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lonn-Stensrud J, Petersen FC, Benneche T, Aamdal SA. Oral Microbiol. Immunol. 2007;22:340–346. doi: 10.1111/j.1399-302X.2007.00367.x. [DOI] [PubMed] [Google Scholar]
- 135.Han Y, Hou S, Simon KA, Ren D, Luk YY. Bioorg. Med. Chem. Lett. 2008;18:1006–1010. doi: 10.1016/j.bmcl.2007.12.032. [DOI] [PubMed] [Google Scholar]
- 136.Kim C, Kim J, Park HY, Park HJ, Lee JH, Kim CK, Yoon J. Appl. Microbiol. Biotechnol. 2008;80:37–47. doi: 10.1007/s00253-008-1474-6. [DOI] [PubMed] [Google Scholar]
- 137.Hentzer M, Givskov M. J. Clin. Invest. 2003;112:1300–1307. doi: 10.1172/JCI20074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hoffmann H, Lindel T. Synthesis-Stuttgart. 2003:1753–1783. [Google Scholar]
- 139.Braekman JC, Daloze D, Stoller C, Vansoest RWM. Biochem. Syst. Ecol. 1992;20:417–431. [Google Scholar]
- 140.Chanas B, Pawlik JR, Lindel T, Fenical W. J. Exp. Mar. Biol. Ecol. 1997;208:185–196. [Google Scholar]
- 141.Yamada A, Kitamura H, Yamaguchi K, Fukuzawa S, Kamijima C, Yazawa K, Kuramoto M, Wang GYS, Fujitani Y, Uemura D. Bull. Chem. Soc. Jpn. 1997;70:3061–3069. [Google Scholar]
- 142.Kelly SR, Jensen PR, Henkel TP, Fenical W, Pawlik JR. Aquat. Microb. Ecol. 2003;31:175–182. [Google Scholar]
- 143.Huigens RW, Richards JJ, Parise G, Ballard TE, Zeng W, Deora R, Melander C. J. Am. Chem. Soc. 2007;129:6966–6967. doi: 10.1021/ja069017t. [DOI] [PubMed] [Google Scholar]
- 144.Huigens RW, Ma LY, Gambino C, Moeller PD, Basso A, Cavanagh J, Wozniak D, Melander C. Mol. Biosys. 2008;4:614–621. doi: 10.1039/b719989a. [DOI] [PubMed] [Google Scholar]
- 145.Richards JJ, Ballard TE, Huigens RW, Melander C. Chembiochem. 2008;9:1267–1279. doi: 10.1002/cbic.200700774. [DOI] [PubMed] [Google Scholar]
- 146.Richards JJ, Huigens RW, Ballard TE, Basso A, Cavanagh J, Melander C. C. Inhibition and dispersion of proteobacterial biofilms. Chem. Commun. 2008:1698–1700. doi: 10.1039/b719802g. [DOI] [PubMed] [Google Scholar]
- 147.Richards JJ, Reed CS, Melander C. Bioorg. Med. Chem. Lett. 2008;18:4325–4327. doi: 10.1016/j.bmcl.2008.06.089. [DOI] [PubMed] [Google Scholar]
- 148.Richards JJ, Ballard TE, Melander C. Org. Biomol. Chem. 2008;6:1356–1363. doi: 10.1039/b719082d. [DOI] [PubMed] [Google Scholar]
- 149.Ballard TE, Richards JJ, Wolfe AL, Melander C. Chem. Euro. J. 2008;14:10745–10761. doi: 10.1002/chem.200801419. [DOI] [PubMed] [Google Scholar]
- 150.Peng L, DeSousa J, Su Z, Novak BM, Nevzorov AA, Garland E, Melander C. Chem. Commun. 2011;47:4896–4898. doi: 10.1039/c1cc10691k. [DOI] [PubMed] [Google Scholar]
- 151.Rogers SA, Melander C. Angew. Chem. Int. Ed. 2008;47:5229–5231. doi: 10.1002/anie.200800862. [DOI] [PubMed] [Google Scholar]
- 152.Rogers SA, Huigens RW, Cavanagh J, Melander C. Antimicrob. Agents Chemother. 2010;54:2112–2118. doi: 10.1128/AAC.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rogers SA, Bero JD, Melander C. ChemBioChem. 2010;11:396–410. doi: 10.1002/cbic.200900617. [DOI] [PubMed] [Google Scholar]
- 154.Reed CS, Huigens RW, Rogers SA, Melander C. Bioorg. Med. Chem. Lett. 2010;20:6310–6312. doi: 10.1016/j.bmcl.2010.08.075. [DOI] [PubMed] [Google Scholar]
- 155.Reyes S, Huigens RW, Su Z, Simon ML, Melander C. Org. Biomol. Chem. 2011;9:3041–3049. doi: 10.1039/c0ob00925c. [DOI] [PubMed] [Google Scholar]
- 156.Bunders C, Richards JJ, Melander C. Bioorg. Med. Chem. Lett. 2010;20:3797–3800. doi: 10.1016/j.bmcl.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 157.Steenackers HPL, Ermolat'ev DS, Savaliya B, Weerdt AD, De Coster D, Shah A, Van der Eycken EV, De Vos DE, Vanderleyden J, De Keersmaecker SCJ. J. Med. Chem. 2011;54:472–484. doi: 10.1021/jm1011148. [DOI] [PubMed] [Google Scholar]
- 158.Steenackers HP, Ermolat'ev DS, Savaliya B, De Weerdt A, De Coster D, Shah A, Van der Eycken EV, De Vos DE, Vanderleyden J, De Keersmaecker SCJ. Bioorg. Med. Chem. 2011;19:3462–3473. doi: 10.1016/j.bmc.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 159.Ermolat'ev DS, Bariwal JB, Steenackers HPL, De Keersmaecker SCJ, Van der Eycken EV. Angew. Chem. Int. Ed. 2010;49:9465–9468. doi: 10.1002/anie.201004256. [DOI] [PubMed] [Google Scholar]
- 160.Rogers SA, Huigens RW, Melander C. J. Am. Chem. Soc. 2009;131:9868–9869. doi: 10.1021/ja9024676. [DOI] [PubMed] [Google Scholar]
- 161.Lindsey EA, Worthington RJ, Alcaraz C, Melander C. Org. Biomol. Chem. 2012;10:2552–2561. doi: 10.1039/c2ob06871k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hertiani T, Edrada-Ebel R, Ortlepp S, van Soest RWM, de Voogd NJ, Wray V, Hentschel U, Kozytska S, Muller WEG, Proksch P. Bioorg. Med. Chem. 2010;18:1297–1311. doi: 10.1016/j.bmc.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 163.Vik A, Hedner E, Charnock C, Samuelson O, Larsson R, Gundersen L, Bohlin L. J. Nat. Prod. 2006;69:381–386. doi: 10.1021/np050424c. [DOI] [PubMed] [Google Scholar]
- 164.Rogers SA, Whitehead DC, Mullikin T, Melander C. Org. Biomol. Chem. 2010;8:3857–3859. doi: 10.1039/c0ob00063a. [DOI] [PubMed] [Google Scholar]