Abstract
We use a form of “freeze-trap, kinetic-crystallography” to explore the migration of Xe atoms away from the dinuclear heme-a3/CuB center in Thermus thermophilus cytochrome ba3 oxidase. This enzyme is a member of the heme-copper oxidase super-family, and is thus crucial for dioxygen dependent life. The mechanisms involved in the migration of oxygen, water, electrons, and protons into and/or out of the specialized channels of the heme-copper oxidases are generally not well understood. Pressurization of crystals with Xe gas previously revealed a O2 diffusion channel in cytochrome ba3 oxidase that is continuous, Y-shaped, 18–20 Å in length and comprised of hydrophobic residues, connecting the protein surface within the bilayer to the a3-CuB center in the active site. To understand movement of gas molecules within the O2 channel, we performed crystallographic analysis of 19 Xe laden crystals freeze-trapped in liquid nitrogen at selected times between 0 and 480 seconds while undergoing out-gassing at room temperature. Variation in Xe crystallographic occupancy at five discrete sites as a function of time leads to a kinetic model revealing relative degrees of mobility of Xe atoms within the channel. Xe egress occurs primarily through the channel formed by the Xe1 → Xe5 → Xe3 → Xe4 sites, suggesting that ingress of O2 is likely to occur by the reverse of this process. The channel itself appears not to undergo significant structural changes during Xe migration, thereby indicating a passive role in this important physiological function.
Cytochrome c oxidases are terminal enzymes of respiration that comprise the heme-copper oxidase superfamily (1–3). They are found in the plasma membrane of prokaryotes and the mitochondrial inner membrane of eukaryotes, wherein they catalyze the reduction of molecular oxygen to water with concomitant oxidation of cytochrome c and generation of a proton gradient across the membrane. Accordingly, these enzymes contribute to the energy needs of a cell and are crucial for aerobic life.
Cytochrome ba3 oxidase (ba3) is a B-type cytochrome c oxidase from the plasma membrane of Thermus thermophilus and is the preferred terminal enzyme of cellular respiration at low oxygen tensions (4). The enzyme contains the four redox cofactors: heme a3, heme b, binuclear CuA, and CuB as illustrated in Fig. 1A (4–6). Electrons enter the enzyme at the CuA site and tunnel from there to the low-spin heme b and move to the heme a3-CuB dinuclear center (dnc) depending on its chemical state (3, 7–8). Protons enter from the cytoplasm and exit to the periplasm via specially adapted pathways (9).
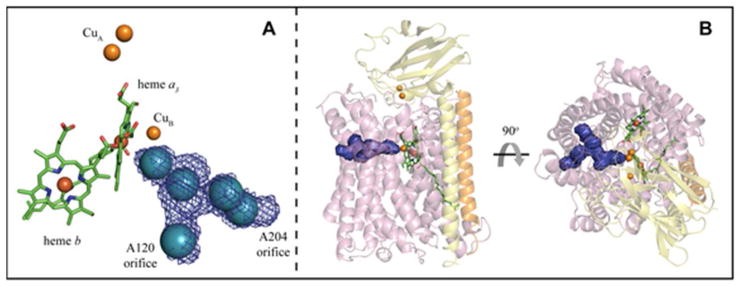
Figure 1.
A Metal centers of ba3 with Xe sites (blue spheres). The Y-shaped hydrophobic channel of ba3 (blue mesh) is overlaid on the Xe sites; the orifice to each fork of the channel is labeled by an adjacent residue. B Transmembrane (left) and overhead (right) views of ba3 illustrating the shape and location of the hydrophobic channel (blue surface). Subunit I is lavender, subunit II, which contains the soluble CuA domain, is yellow, and subunit IIa is orange; heme a3 and heme b are green.
Previously, we described a channel in ba3 using X-ray crystallography (10) using static Xe and Kr pressurization (11). This Y-shaped channel was defined by five major and two minor discrete gaseous atom binding sites that extend from Xe1 near the buried dnc to two possible entry/exit points at the membrane-facing surface of the protein (Fig. 1A, B). To the extent that the Xe atom serves as a mimic for the O2 molecule (see below and ref. 10), the previous work demonstrated the existence of a structurally defined, bifurcated channel within ba3 whose function is to attract O2 from the lipid bilayer into the dnc. The relative affinity for Xe and Kr at discrete sites within the channel could be determined in terms of relative occupancy under different pressurization conditions (10), but no information concerning movements of Xe atoms within the channel was obtained from the static structures (Protein Data Bank code 3BVD).
Xe pressurization of crystals has been used to identify a putative O2 diffusion channel in Rhodobacter sphaeroides cytochrome c oxidase (12), similar to that in ba3, and to define hydrophobic sites other non-heme containing proteins (13). For integral membrane proteins, Xe has been used to probe O2 channels in photosystem II (14), the NH3 channel in the Amt-1 transporter (15), and N2O sites in the NMDA receptor (16). Xe pressurization has also been used to perturb the conformation and activity in bacteriorhodopsin (17), and to delimit detergent micelles in crystals (18). In soluble enzymes, Xe has been used to define the pathways for O2 (19), CH4 (20), CO (21) and NO (22) into active sites. In heme nitric oxide/oxygen binding (H-NOX) domains, redistribution of Xe atoms within a hydrophobic channel is implied by anisotropic electron density and increasing occupancy closer to the heme (22). Aside from the latter observation, no crystallographic study of Xe binding sites has extracted direct experimental evidence concerning the relative movements of Xe atoms within hydrophobic channels or cavities.
Ligand migration, however, as inferred from locations of static sites, has been extensively studied in the globins using Xe pressurization crystallography (13). The interpretation of data from these studies has benefitted from being carried out with crystals of relatively small proteins that diffract to high resolution, enabling detailed description of the binding sites. Petsko and coworkers (23) first reported crystal structures of myoglobin pressurized under several atmospheres of Xe gas, revealing four unique binding sites. Xe has been used, both experimentally and theoretically, to examine proteinaceous CO and O2 binding sites within sperm whale myoglobin (24), human hemoglobins (25), the hemoglobin, HbI, from the blood clam, Scapharca inequivalvis (26), and human cytoglobin (27). Theoretical work (28) suggested that the Xe binding sites in Mb may function as transient binding sites for ligands migrating from the solvent to the active site. However, as pointed out by Olson and colleagues (25), most experimental data, including Laue X-ray crystallography (29), do not support the theoretical conclusions but suggest, instead, that ligands escape and enter myoglobin by the His(E7) gate and are trapped in the distal pocket; this holds for the human hemoglobins as well ((25) and references therein). In the case of bacterial truncated hemoglobins and the neuronal mini-globin from the sea worm, ligands also enter the protein through an apolar tunnel between the C-terminal ends of the E and H helices (27) and are subsequently trapped in the distal pocket. Blocking this apolar tunnel with a Trp mutation or filling the tunnel with Xe markedly decreases both the fraction of ligands that escape to solvent and the bimolecular rate constant for entry of ligands to the active site (30). As noted, this behavior is to be distinguished from the other globins, in which ligands appear to enter and escape through the distal ligand gate.
Xe lacks several features of another oxygen mimic, carbon monoxide, whose spectroscopic properties have been widely used to examine its photoinduced release from and shorted ranged recombination with ferrous hemes within a variety of heme-containing proteins. Transient IR spectroscopies have also been used to follow recombination of CO in the heme-copper oxidases (31). Where CO falls short as a mimic of O2 is that its 14 X-ray scattering electrons require the availability of highly diffracting protein crystals to identify its position. A useful feature of Xe as a dioxygen mimic, however, in addition to its similar size and hydrophobicity, is that its 54 electrons, combined with significant anomalous X-ray scattering effects, allow facile identification of atomic positions and quantification of occupations even in crystals exhibiting only modest diffraction.
In this paper, we utilize an adaptation of traditional methods to trap intermediates using the apparatus developed by Soltis et al. (11, 32). During this operation, pressurized Xe binds to hydrophobic pockets within the protein matrix (23). For heavy atom derivatization, crystals are frozen immediately after depressurization to capture as much Xe as possible within the protein. In our approach, the crystals are pressurized for several minutes, quickly depressurized to 1 atm, and frozen by plunging into liquid nitrogen, but the time interval at 1 atm prior to freezing is varied from 0 to 480 seconds, allowing adsorbed Xe to diffuse out of the crystals. By crystallographically quantifying the positions and occupancies of Xe atoms as a function of time, a kinetic model can be derived describing their motions. To our knowledge, this is the first data to quantitate movement of gas molecules within a large integral membrane protein.
Experimental Procedures
Protein purification and crystallization
Protein preparation and purification was done following the methods described by Chen et al. (33). The modified crystallization procedure is detailed in the Supporting Information.
Xe derivatization and trapping
Xe derivatives of ba3 were produced following the procedure previously employed (10) using a single crystal pressurization cell developed at the Stanford Synchrotron Radiation Lightsource (SSRL) (11, 32). Crystals were pressurized to 100 psi (689.48 kPa) for 5 minutes followed by release of the pressure in the chamber. To capture the different stages of Xe diffusion out of the protein under the reduced Xe pressure, the crystals were removed from the chamber at select time intervals (0, 10, 30, 60, 120, 240, 360, and 480 s) and flash frozen into liquid nitrogen. Approximately 20 seconds are required to remove the crystal from the pressurization vessel and plunge it into liquid nitrogen.
Data collection and structure determination
X-ray diffraction experiments were conducted on SSRL beam lines 7-1 and 11-1. All data sets were collected using λ = 1.127 Å to simultaneously maximize the anomalous signal from the Fe, Cu, and Xe (f″ = 1.91, 2.74, and 4.37 electrons, respectively, at this wavelength) in the Xe-derivatized ba3 crystals. Each data set consisted of 360 frames collected with an oscillation angle of 1° with 10 s exposure per frame. Diffraction data were recorded at 100 K on an ADSC Q315 CCD detector, and no significant decay was observed at the resolution used in refinement (Table S1). The data sets were integrated and scaled using MOSFLM and SCALA (34). Detailed data collection and refinement statistics are presented in Table S1.
Using the wild-type recombinant structure of ba3 as a search model (PDB code 1XME), molecular replacement was done using MOLREP (35). All data sets were refined in the same fashion by rigid body and restrained refinement with REFMAC5 (36) using overall B factor refinement and a geometry weighting term of 0.3. 2|Fo|−|Fc| and |Fo|−|Fc| electron density maps, and anomalous difference Fourier maps, clearly revealed the positions of the Xe atoms as well as Cu and Fe atoms (Fig. 2) consistent with previous results (10). Unbiased coordinates and occupancies of the Fe, Cu, and Xe sites were refined using SHARP (37) against the highly redundant anomalous data sets (Table S1) and expected anomalous scattering factors (Table S2). Because all neighboring Xe-Xe distances are greater than the van der Waals contact distance (4.4 Ǻ) (Table S3; Fig. S2), the Xe sites were atomically resolved at the resolution of the data sets. Structures representing the highest resolution data set at each time point have been deposited with the RCSB (PDB codes 3S33, 3S38, 3S39, 3S3A, 3S3B, 3S3C, 3S3D, corresponding to time points 10, 30, 60, 120, 240, 360, 480 s, respectively). The 0 s time point structures are very similar to the previously deposited, 3.37 Ǻ resolution structure (PDB code 3BVD, ref. 10). However, because the previously deposited structure is not in the same space group, the newer but lower resolution data sets were used for the 0 s time point in the analysis (Tables S1, S2).
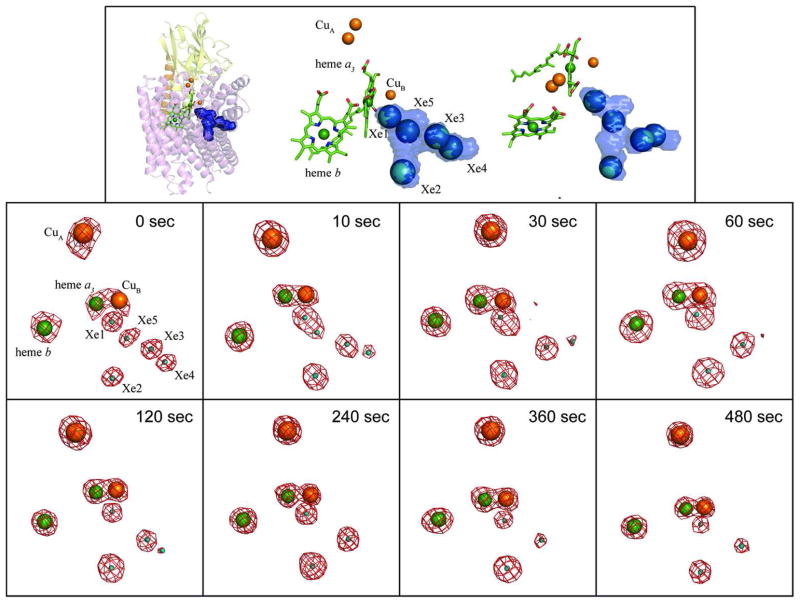
Figure 2.
The location and occupancies of Xe sites at eight time points during diffusion of Xe out of pressurized crystals. The top panel illustrates the location of the Xe sites lining the hydrophobic channel. The left and center figures in the top panel are in the same orientation as the other panels. The figure on the top right is an approximately 40° rotation towards the reader. The |F+|−|F−| anomalous difference Fourier map contoured at 6σ (red mesh) for the Cu, heme Fe, and Xe atoms of a Xe-pressurized crystal is shown for each time point. The 0 sec panel shows the relative location and naming convention of the Xe and metal sites (the binuclear CuA center was refined as a single atom given the resolution of the data). The average occupancy of Xe at each site at each time point (0 to 480 sec) was determined by occupancy normalization using a total of 19 data sets (Experimental Procedures).
Occupancy normalization
The accuracy of the anomalous signal in a given dataset depends upon diffraction quality, crystal orientation and absorption effects, site-specific radiation damage, and other systematic errors (38–39), which can lead to apparent differences in the occupancies of anomalous scattering centers. To compare Xe occupancies across multiple data sets, we used biochemical information to define an internal reference for normalization. Cytochrome ba3 oxidase contains four redox cofactors: heme b, heme a3, a single CuB site, and the binuclear CuA center. Analytical data and previous structures of ba3 showed the consistent full occupancy of these four metal centers, i.e. the occupancies of Feb, Fea3, and CuB are reasonably taken to be unity. However, as shown in Table S2 individual metal site occupancies vary significantly from 1.0. Utilizing the biochemical constraint, we normalized the occupancies of the metal sites as the mean of Feb, Fea3, and CuB relative to 1.0. For example, for data set 13 at 240 s (Table S2) the refined occupancies of the Feb, Fea3, and CuB sites are 0.978, 0.960, and 1.034, respectively, with mean 0.991. Multiplication by 1/mean normalizes the occupancies of all the metal and Xe sites for a given data set (FeFeCuB columns in Table S2). Normalized values per site are then averaged over the redundant data sets at each time point (AVG, DEV in Table S2).
The CuA center provides an internal control for this normalization procedure. For example, the normalized occupancy of CuA for data set 13 is 1.609/0.991 = 1.624 and the average for three data sets at 240 s is 1.704 (Table S2). The normalized average occupancy for CuA for all 19 data sets is 1.662 ± 0.048, or an error of less than 3%. The apparent occupancy of the CuA center, refined as a single site, is a non-integer value because the binuclear center (Cu-Cu distance 2.7 Ǻ) is not resolved at the ~4 Ǻ resolution of the data sets (Table S1). Hence, normalization based on the expected occupancy of the Feb, Fea3, and CuB sites in ba3 is validated by results for the CuA center, allowing for internal correction of Xe occupancies within each data set, and comparison of average Xe site occupancies at different time points.
Modeling kinetics
Using the Simbiology module of MATLAB (Natick, MA) and the normalized occupancies of Xe from our structures, we modeled the kinetics of Xe diffusion out of ba3 and obtained rate constants for each of the processes. Details of the model and results of the simulation are provided in the Results and Discussion section of the text, and the corresponding coupled differential equations are given in the Supporting Information.
Results and Discussion
Xe-pressurized crystals of recombinant, wild-type ba3 were flash frozen after release of pressure at times 0, 10, 30, 60, 120, 240, 360, and 480 s; 19 datasets having between 3.3 and 5.5 Å resolution were collected (Table S1). Following structure solution and refinement (Table S1), we monitored the presence of Xe at the five major binding sites using isomorphous and anomalous difference Fourier maps (Fig. 2). The refinement protocol uses the anomalous scattering from three metals sites in the native enzyme, CuB, Feb, and Fea3, to obtain internally normalized occupancies of the Xe atoms for each data set (Table S2). The normalized occupancy for the binuclear CuA center serves as an independent, internal indicator of error (see Experimental Procedures). The average occupancy, along with the calculated error bars, for each Xe site at each diffusion time point is plotted in Fig. 3. The estimated error of the quench times is 1–2 s (40).
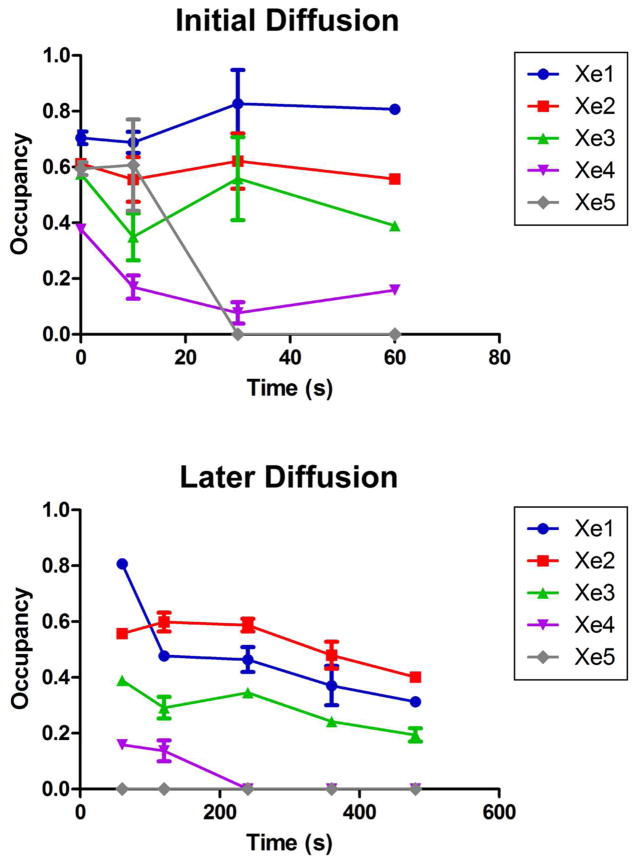
Figure 3.
Normalized occupancies at each Xe site plotted vs. time. Error bars represent standard error, s/√n, where s is the standard deviation and n is the number of observations, i.e. data sets at that time point. The top panel shows 0 – 60 sec data points, and the bottom panel shows 60 – 480 sec data points.
Protein structure and Xe locations
As shown in Table S1, the diffraction resolution of the 19 data sets ranges from 3.3 Å to 5.0 Å. Within this range, no side chain movements or backbone displacements relative to native structures of ba3 were observed upon Xe binding. This is also true for comparisons with ba3 structures at different redox states (41). Using isomorphous and anomalous difference maps, we located between three and five Xe atoms in each data set; early time points revealed the most Xe atoms while later time points showed fewer. The locations for each of the Xe sites in all of the structures are similar to the corresponding sites in the previously reported structures (10), and the same nomenclature is used to identify the individual sites (Fig. 2, top panel). Variations in position within the five discrete binding sites are small (Fig. S2); distances between neighboring Xe atoms, which are greater than the van der Waals contact distance of ~4.4 Ǻ, are given in Table S3. The amino acid residues surrounding each Xe site are primarily hydrophobic without propensity for a specific amino acid type, e.g., aromatics, to dominate (see Supporting Information of ref. (10)). The only noticeable change in a Xe position is a shift of Xe4 by ~2 Å away from the Xe3 site towards Val201 and Ala204, which occurs only at diffusion time points 30 and 120 s. The Xe4 site is the least occupied of these time points and is closest to the outside of the protein near Ala204 (footnote).
Temperature at the crystal after rapid freezing in liquid nitrogen
Clearly, passage of the crystal from a pressure of 100 psi (689.48 kPa) Xe at room temperature during the few seconds needed to move the crystal from ambient temperature to 77 K does not result in 100 % occupancy of the Xe binding sites, as shown in Table S2. While it is difficult to determine the temperature of each crystal when molecular motion ceases, other studies (40) have shown that the approximate time required to cool a macromolecular crystal from room temperature to 77 K is on the order of 1–2 s. Because the freezing time is ~1/10 of the shortest diffusion time, we assume that room temperature equilibrium distributions are trapped upon freezing following the 0 to 480 s intervals during which out-gassing is allowed to occur.
Xe egress
The cumulative occupancy of Xe sites 1, 5, 3, 2, and 4 decreased from 2.8 ± 0.38 Xe atoms to 0.9 ± 0.025 Xe atoms over time (Fig. S1), as would be expected as Xe escapes from the crystal. However, the distribution of occupancies at each time point displayed significant variation (Figs. 2, 3) thereby providing an opportunity to probe the mechanism of Xe egress and by reversal, Xe (O2) access. The Xe1 site, which is nearest to the dinuclear center, is likely the last transient stop prior for O2 prior to reduction (42). From 0 s to 10 s, all the Xe sites decrease in occupancy and maintain similar occupancy ranking. However, beginning at time point 30 s, the Xe5 site completely disappears and does not reappear at subsequent time points. Concurrently, the occupancies of the Xe1, Xe2, and Xe3 sites increase marginally while Xe4 continues to decrease (Fig. 3). Interestingly, before the Xe5 site disappears at time point 30 s, it begins to merge with the Xe1 site at time point 10 s (Fig. 2). Such merging of the density was observed only at time point 10 s. Anisotropy of Xe density in a hydrophobic channel has also been noted in H-NOX (22). By time point 120 s, the occupancy ranking among the Xe sites becomes switched such that the Xe2 site has the highest occupancy (Fig. 3). At 240 s and beyond, only the Xe1, Xe2, and Xe3, which are 8 – 12 Ǻ from each other, sites are occupied.
A state model for the mechanism of gas diffusion
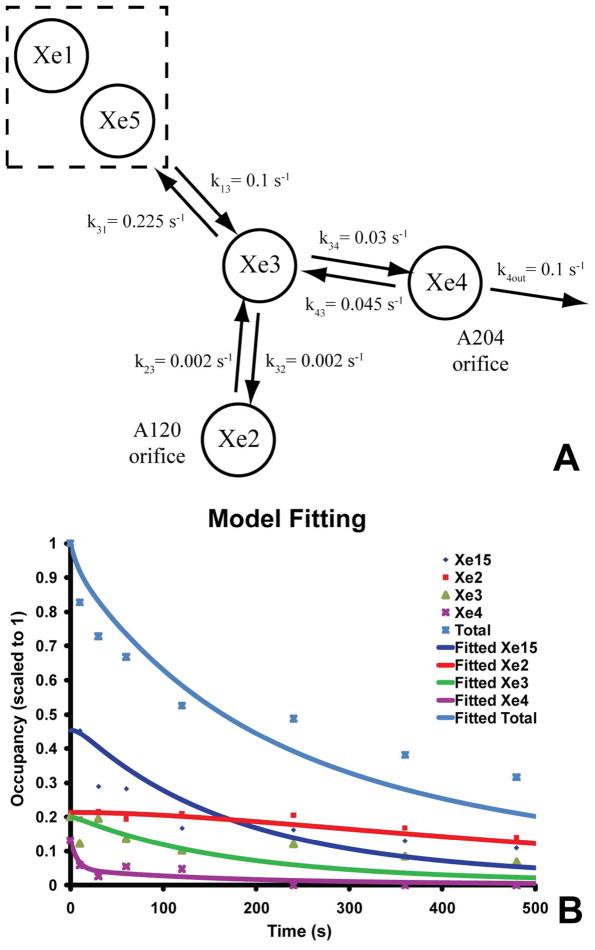
To infer a mechanism for migration of Xe out of the enzyme, we created a simple state model based on the occupancy data. The model (Fig. 4A) treats Xe migration as occurring via a series of reactions that move Xe from one site to the next, and in which Xe atoms do not interact with each other. The model involves four Xe binding locations in the protein (Xe2, Xe3, Xe4, and a combined site Xe15=Xe1+Xe5), plus an external bath. As noted above, the Xe1 and Xe5 sites merge at time point 10 s (Fig. 2); therefore, we modeled them as behaving as one by combining their occupancies, i.e., max = 2, min = 0.
Figure 4.
A A kinetic model for gas diffusion through the hydrophobic channel of ba3 showing the connectivity of the different Xe sites and their derived rate constants. The sites Xe1 and Xe5 were treated as a single site (see text). B Results of the simulation of rate constants after least squares minimization are plotted vs. the experimental occupancies using the model in A.
Due to spatial constraints within the hydrophobic channel, only those reactions that connect pairs of sites, as shown in Fig. 4A, are possible. It is also assumed that there is no reverse flux from Xe in the external bath into the Xe4 site, i.e. k4in [Xe]bath ≪ k4out [Xe4]. Based on the observed changes in Xe2, departure of Xe from this site via the A120 orifice to the external bath (footnote) does not occur. Hence, in the model, the Xe2 site communicates only with the protein interior (Fig. 4A). We then used the Sundials solver in the Simbiology module of MATLAB to numerically solve the set of coupled differential equations corresponding to the reaction model (see Supporting Information for the rate and equilibrium equations). The occupancies under the pressurized condition (equilibrium, time-zero) were used to define equilibrium constants for the various reversible reactions. With these constants, four independent rate constants (k13, k23, k34 and k4out) were utilized here as input to derive the time-dependent decay of the system and its individual components. The corresponding equilibrium constants, K13, K23, and K33, permitted calculation of the rate constants of the back reactions: k31, k32, and k43. To estimate the rate constants in the model, the trust-region-reflective algorithm in the MATLAB ‘lsqnonlin’ function was used to minimize the sum of squares of the residuals between the simulated data and the experimental occupancies at the various sites at different time-points.
K13 = (k13/k31) = 0.444, K23 = (k23/k32) = 0.935, and K34 = (k34/k43) = 0.658 (see Supporting Information). Due to errors in the occupancy data (Table S2) and other potential considerations, the noted value for k13 (0.1 s−1) is a lower bound on this rate constant. Further increases in this value result in only small changes in the residuals, without reaching convergence; hence k13 was fixed at this value during minimization.
Interesting relationships emerge from the estimated rate constants. For example, k23 and k32 (0.002 s−1) and k34, k43 (0.03–0.045 s−1) are separated by a factor of 15 – 22.5, meaning that exchange between the Xe3 and Xe4 sites is an order of magnitude faster than between the Xe3 and Xe2 sites. Similarly, exchange between the Xe3 and combined Xe1/Xe5 sites is about 3.5 times faster than between the Xe3 and Xe4 sites. The rate constant for exit from the Xe4 site of the protein, k4out, is approximately 0.1 s−1 (Fig. 4A). However, this is not the rate limiting step for the overall loss of Xe; rather slower internal steps significantly impact this global Xe diffusion out of the protein.
The Gibbs free energy for each equilibrium was obtained from ΔG° = −RTlnKi, where R = 1.986 x 10−3 kcal mol−1 K−1, T = 298 K, and Ki are equilibrium constants K13, K23, and K34. The corresponding reaction free energies are ΔG° = 0.48, 0.04, and 0.25 kcal/mol, respectively, being well below RT = 0.59 kcal mol−1 near room temperature. Nevertheless, the general picture indicates that diffusion of Xe away from the active site is thermodynamically unfavorable, though weakly. Future molecular dynamics studies may clarify the meaning of these small free energy differences (28), but they are beyond the scope of this work.
The nature of the channel may provide qualitative information about the rate at which a hydrophobic ligand may migrate from site-to-site. Thus, the channel is essentially uniform in diameter and continuous in its hydrophobic nature all the way from the intramembrane protein surface near the Xe4 site into the Xe1 site. This would suggest that the activation energies for passage between the sites are also small compared to RT, a condition, which suggests that properly defined molecular dynamics simulations of Xe motions in ba3 may facilitate further understanding of the mechanism shown here. Such studies have been done for O2, NO, CO, and Xe diffusion within sperm whale myoglobin (28) and for O2, NO in H-NOX domains (43).
In spite of these approximations and caveats, the ba3, Xe occupancy data provide a first experimental view on the nature of gas movement within the protein. Our simple model provides good agreement with most features of the data (Fig. 4B), but the rapid partial initial decay of Xe15 (Fig. 3) remains to be better understood. While it is possible that other pathways of Xe leakage might exist for the Xe5 site, merging of the anomalous difference electron density between the Xe1 and Xe5 sites (Fig. 2) suggests a very low energy barrier between them.
Reversal of ligand diffusion may suggest a mechanism for O2 ingress
The results of the modeling suggest that diffusion of oxygen into the dnc of ba3 may occur via Xe4 → Xe3 → Xe5 → Xe1 sites. Accordingly, it is of interest to compare the hydrophobic channels in other cytochrome c oxidases of known structure (see Fig. 2 of (10)). In particular, the Rhodobacter sphaeroides cytochrome aa3 oxidase channel, also determined by Xe pressurization (12), has only one entry point, analogous to the A120 (footnote) orifice (Fig. 1A), and roughly overlaps with the Xe2 → Xe3 → Xe5 → Xe1 branch of the ba3 channel. Due to homology in the structures and hence an expected passive role of the channel in R. sphaeroides, diffusion rates may be similar in the two proteins. A recent study of the reaction between fully reduced ba3 and O2 indicates a second-order, diffusion-controlled process by which O2 binds to Fea3 to form compound A, an oxy complex (44). Therefore, O2 likely enters the channel by diffusion, followed by a faster, first-order process in which O2 moves from the mouth of the channel at A204 to Fea3.
Examination of structures of ba3 crystallized from detergents suggested that the hydrophobic channel is devoid of crystallographic waters or other small molecules (PDB Codes, 1XME, 1EHK). Recently, a 1.8 Å resolution crystal structure of ba3 in the lipidic cubic phase has identified one water molecule in the hydrophobic channel, that being a water in the Xe1 site hydrogen bonded to a water in the a3-CuB center (42); the rest of the channel contains no observable electron density. The higher affinity of the Xe1 site for a hydrophobic ligand, even at longer times (Fig. 3), is consistent with the idea that O2 bound in the Xe1 site has a functional relationship to the active site, being the last “stop” before being reduced to water.
The Xe2 sites may function a storage site
The small changes in occupancy of the Xe2 site over time as Xe diffuses out of ba3 indicate that very little to no Xe exits the protein through this pathway; hence the absence of a k2out in our model. This finding is consistent with studies of the ba3 I-A120F mutant: even though the phenylalanine replacement physically blocks access to the Xe2 site via the A120 orifice, the mutant retains oxidase activity similar to recombinant, wild-type ba3 (42). Hence, the Xe2 arm of the bifurcated channel may not be utilized for primary oxygen input to the enzyme. The much slower rate between the Xe2 to Xe3 sites suggests that once the Xe2 site is filled by pathway Xe4 → Xe3 → Xe2, very little Xe returns to the main input channel, i.e., pathway Xe4 → Xe3 → Xe5 → Xe1, unless the Xe3 site is empty. Therefore, when O2 is abundant, the Xe2 site may function as a reservoir for O2, which continues to enter through the main input channel during turnover. Under low oxygen tensions, however, the main input channel would become depleted via the Xe4 site, allowing transiently stored O2 in the Xe2 site to enter the Xe3 site and from there move exergonically to the Xe1 site. This transient supply site would bridge the time when the enzyme consumes all the O2 in the main input channel during turnover and when the channel gets refilled again. This mechanism is in agreement with the utilization of ba3 by T. thermophilus as the preferred cytochrome c oxidase when O2 is scarce (22). Using internal cavity networks as storage for small molecules in proteins is not without precedence. For example, the truncated hemoglobin family of proteins has been implicated in both O2 and CO storage (45), while hydrophobic channels in H-NOX proteins modulate the flux of O2 and NO, thereby affecting signaling (22).
Conclusions
An extensive Xe-pressurized, crystallographic study supports the presence of a large, hydrophobic channel within which gaseous ligands such as Xe, O2, CO and NO are able to migrate into and out of the active site of the cytochrome ba3 oxidase. One of the Xe binding sites may also function as a temporary storage site for O2. Because the ba3 structure in Xe pressurized crystals is isomorphous with the native protein [including structures in different redox states (41)], and because the data can be adequately fit with a kinetic model, it appears that the hydrophobic channel functions in a passive manner, and it is unnecessary to consider significant rearrangements within the protein itself. In contrast, in H-NOX proteins dynamics controls O2 and NO diffusion through ‘transiently formed pathways’, affecting ligand concentration within an interior cavity proximal to the heme, which in turn affects heme binding and signaling (22). This behavior is functionally opposite to that in ba3 where O2 is a substrate and rapid acquisition from the membrane is necessary for activity and proton pumping (22). Hence, in ba3 the populations of Xe in five discrete binding sites, and the rates of exchange among the sites, are determined only by the relative thermodynamic affinity of Xe for the sites and the inherent activation barriers between them. Our observations contribute to a general understanding of how hydrophobic gas molecules gain access to deeply buried active sites while revealing details of their relative mobility within a large integral membrane protein.
Supplementary Material
Acknowledgments
We thank Dr. David Goodin for helpful discussions on the development of the diffusion-trapping technique and Ying Chen for help with protein preparation. We also thank Dr. Ana Gonzalez at SSRL for generously loaning us the Xe-pressurization apparatus. Portions of this research were carried out at the Stanford Synchrotron Radiation Light Source, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209), and the National Institute of General Medical Sciences.
We dedicate this paper to our colleague and mentor, J. A. Fee, who passed away shortly after the manuscript was submitted for review.
Abbreviations
- ba3
Thermus thermophilus cytochrome ba3 oxidase
- dnc
dinuclear center
Footnotes
This research was supported by National Institutes of Health grant GM035342 (JAF) and National Science Foundation grants MCB1121959 and PHY0750049 (AAD).
The coordinates and observed structure factor amplitudes for the structures in this paper have been deposited in the Protein Data Bank (PDB codes: 3S33, 3S38, 3S39, 3S3A, 3S3B, 3S3C, 3S3D).
The two arms of the Y-shaped channel are defined as follows: orient the molecule to place the CuA domain at the top (Fig. 1A). The fork of the Y facing downward terminates near Ala120; the fork to the right terminates near Ala204. The two orifices of the channel are designated with respect to these residues on the protein surface.
Supporting Information Available
Protein preparation, purification, and crystallization procedures can be found in the Supporting Information. Table S1 contains crystallographic statistics for all 19 data sets. The rest of the tables and figures found in this section contain further detail on the occupancies of each of the metal sites as well as calculations used in both occupancy normalization and kinetic modeling. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Michel H, Behr J, Harrenga A, Kannt A. Cytochrome c oxidase: structure and spectroscopy. Annual review of biophysics and biomolecular structure. 1998;27:329–356. doi: 10.1146/annurev.biophys.27.1.329. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson-Miller S, Babcock GT. Heme/Copper Terminal Oxidases. Chemical reviews. 1996;96:2889–2908. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 3.Babcock GT, Wikstrom M. Oxygen activation and the conservation of energy in cell respiration. Nature. 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 4.Keightley JA, Sanders D, Todaro TR, Pastuszyn A, Fee JA. Cloning and expression in Escherichia coli of the cytochrome c552 gene from Thermus thermophilus HB8. Evidence for genetic linkage to an ATP-binding cassette protein and initial characterization of the cycA gene products. The Journal of biological chemistry. 1998;273:12006–12016. doi: 10.1074/jbc.273.20.12006. [DOI] [PubMed] [Google Scholar]
- 5.Zimmermann BH, Nitsche CI, Fee JA, Rusnak F, Munck E. Properties of a copper-containing cytochrome ba3: a second terminal oxidase from the extreme thermophile Thermus thermophilus. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:5779–5783. doi: 10.1073/pnas.85.16.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soulimane T, Buse G, Bourenkov GP, Bartunik HD, Huber R, Than ME. Structure and mechanism of the aberrant ba3-cytochrome c oxidase from Thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fee JA, Case DA, Noodleman L. Toward a Chemical Mechanism of Proton Pumping by the B-Type Cytochrome c Oxidases: Application of Density Functional Theory to Cytochrome ba3 of Thermus thermophilus. Journal of the American Chemical Society. 2008;130:15002–15021. doi: 10.1021/ja803112w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farver O, Wherland S, Antholine WE, Gemmen GJ, Chen Y, Pecht I, Fee JA. Pulse Radiolysis Studies of Temperature Dependent Electron Transfers among Redox Centers in ba3-Cytochrome c Oxidase from Thermus thermophilus: Comparison of A- and B-Type Enzymes. Biochemistry ASAP. 2010 doi: 10.1021/bi100548n. [DOI] [PubMed] [Google Scholar]
- 9.Chang HY, Hemp J, Chen Y, Fee JA, Gennis RB. The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16169–16173. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luna VM, Chen Y, Fee JA, Stout CD. Crystallographic Studies of Xe and Kr Binding within the Large Internal Cavity of Cytochrome ba3 from Thermus thermophilus: Structural Analysis and Role of Oxygen Transport Channels in the Heme-Cu Oxidases†,‡. Biochemistry. 2008;47:4657–4665. doi: 10.1021/bi800045y. [DOI] [PubMed] [Google Scholar]
- 11.Soltis SM, Stowell MHB, Wiener MC, Phillips GN, Rees DC. Successful flash-cooling of xenon-derivatized myoglobin crystals. J Appl Crystallogr. 1997;30:190–194. [Google Scholar]
- 12.Svensson-Ek M, Abramson J, Larsson G, Tornroth S, Brzezinski P, Iwata S. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 13.Prange T, Schiltz M, Pernot L, Colloc’h N, Longhi S, Bourguet W, Fourme R. Exploring hydrophobic sites in proteins with xenon or krypton. Proteins. 1998;30:61–73. [PubMed] [Google Scholar]
- 14.Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W. Cyanobacterial photosystem II at 2.9-angstrom resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol. 2009;16:334–342. doi: 10.1038/nsmb.1559. [DOI] [PubMed] [Google Scholar]
- 15.Andrade SLA, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colloc’h N, Santos JSD, Retailleau P, Vivares D, Bonnete F, d’Estainto BL, Gallois B, Brisson A, Risso JJ, Lemaire M, Prange T, Abraini JH. Protein crystallography under xenon and nitrous oxide pressure: Comparison with in vivo pharmacology studies and implications for the mechanism of inhaled anesthetic action. Biophysical journal. 2007;92:217–224. doi: 10.1529/biophysj.106.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayakawa N, Kasahara T, Hasegawa D, Yoshimura K, Murakami M, Kouyama T. Effect of Xenon Binding to a Hydrophobic Cavity on the Proton Pumping Cycle in Bacteriorhodopsin. J Mol Biol. 2008;384:812–823. doi: 10.1016/j.jmb.2008.09.075. [DOI] [PubMed] [Google Scholar]
- 18.Sauer O, Roth M, Schirmer T, Rummel G, Kratky C. Low-resolution detergent tracing in protein crystals using xenon or krypton to enhance X-ray contrast. Acta Crystallogr D. 2002;58:60–69. doi: 10.1107/s0907444901017292. [DOI] [PubMed] [Google Scholar]
- 19.Duff AP, Trambaiolo DM, Cohen AE, Ellis PJ, Juda GA, Shepard EM, Langley DB, Dooley DM, Freeman HC, Guss JM. Using xenon as a probe for dioxygen-binding sites in copper amine oxidases. J Mol Biol. 2004;344:599–607. doi: 10.1016/j.jmb.2004.09.075. [DOI] [PubMed] [Google Scholar]
- 20.Whittington DA, Rosenzweig AC, Frederick CA, Lippard SJ. Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase. Biochemistry. 2001;40:3476–3482. doi: 10.1021/bi0022487. [DOI] [PubMed] [Google Scholar]
- 21.Doukov TI, Blasiak LC, Seravalli J, Ragsdale SW, Drennan CL. Xenon in and at the end of the tunnel of bifunctional carbon monoxide dehydrogenase/acetyl-CoA synthase. Biochemistry. 2008;47:3474–3483. doi: 10.1021/bi702386t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter MB, Herzik MA, Kuriyan J, Marletta MA. Tunnels modulate ligand flux in a heme nitric oxide/oxygen binding (H-NOX) domain. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E881–E889. doi: 10.1073/pnas.1114038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tilton RF, Kuntz ID, Petsko GA. Cavities in Proteins - Structure of a Metmyoglobin-Xenon Complex Solved to 1.9-A. Biochemistry. 1984;23:2849–2857. doi: 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- 24.Ostermann A, Waschipky R, Parak FG, Nienhaus GU. Ligand binding and conformational motions in myoglobin. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 25.Birukou I, Maillett DH, Birukova A, Olson JS. Modulating distal cavities in the alpha and beta subunits of human HbA reveals the primary ligand migration pathway. Biochemistry. 2011;50:7361–7374. doi: 10.1021/bi200923k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knapp JE, Pahl R, Cohen J, Nichols JC, Schulten K, Gibson QH, Šrajer V, Royer WE., Jr Ligand Migration and Cavities within Scapharca Dimeric HbI: Studies by Time-Resolved Crystallography, Xe Binding, and Computational Analysis. Structure. 2009;17:1494–1504. doi: 10.1016/j.str.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Sanctis D, Dewilde S, Pesce A, Moens L, Ascenzi P, Hankeln T, Burmester T, Bolognesi M. Mapping protein matrix cavities in human cytoglobin through Xe atom binding. Biochem Bioph Res Co. 2004;316:1217–1221. doi: 10.1016/j.bbrc.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Cohen J, Arkhipov A, Braun R, Schulten K. Imaging the Migration Pathways for O2, CO, NO, and Xe Inside Myoglobin. Biophysical journal. 2006;91:1844–1857. doi: 10.1529/biophysj.106.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aranda Rt, Levin EJ, Schotte F, Anfinrud PA, Phillips GN., Jr Time-dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin. Acta crystallographica Section D, Biological crystallography. 2006;62:776–783. doi: 10.1107/S0907444906017318. [DOI] [PubMed] [Google Scholar]
- 30.Salter MD, Nienhaus K, Nienhaus GU, Dewilde S, Moens L, Pesce A, Nardini M, Bolognesi M, Olson JS. The Apolar Channel in Cerebratulus lacteus Hemoglobin Is the Route for O2 Entry and Exit. Journal of Biological Chemistry. 2008;283:35689–35702. doi: 10.1074/jbc.M805727200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koutsoupakis K, Stavrakis S, Pinakoulaki E, Soulimane T, Varotsis C. Observation of the equilibrium CuB-CO complex and functional implications of the transient heme a3 propionates in cytochrome ba3-CO from Thermus thermophilus. Fourier transform infrared (FTIR) and time-resolved step-scan FTIR studies. The Journal of biological chemistry. 2002;277:32860–32866. doi: 10.1074/jbc.M204943200. [DOI] [PubMed] [Google Scholar]
- 32.Cohen A, Ellis P, Kresge N, Soltis SM. MAD phasing with krypton. Acta crystallographica Section D, Biological crystallography. 2001;57:233–238. doi: 10.1107/s0907444900014670. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Hunsicker-Wang L, Pacoma RL, Luna E, Fee JA. A homologous expression system for obtaining engineered cytochrome ba3 from Thermus thermophilus HB8. Protein expression and purification. 2005;40:299–318. doi: 10.1016/j.pep.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Leslie AWG. The CCP4 suite: programs for protein crystallography. Acta crystallographica Section D, Biological crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.dela Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Method Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 38.Fourme R, Shepard W, Schiltz M, Prange T, Ramin M, Kahn R, de la Fortelle E, Bricogne G. Better structures from better data through better methods: a review of developments in de novo macromolecular phasing techniques and associated instrumentation at LURE. Journal of synchrotron radiation. 1999;6:834–844. [Google Scholar]
- 39.Schiltz M, Bricogne G. Modelling and refining site-specific radiation damage in SAD/MAD phasing. Journal of synchrotron radiation. 2007;14:34–42. doi: 10.1107/S0909049506038970. [DOI] [PubMed] [Google Scholar]
- 40.Teng TY, Moffat K. Cooling rates during flash cooling. J Appl Crystallogr. 1998;31:252–257. [Google Scholar]
- 41.Liu B, Chen Y, Doukov T, Soltis SM, Stout CD, Fee JA. Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme. Biochemistry (Mosc) 2009;48:820–826. doi: 10.1021/bi801759a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiefenbrunn T, Liu W, Chen Y, Katritch V, Stout CD, Fee JA, Cherezov V. High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment. PloS one. 2011;6:e22348. doi: 10.1371/journal.pone.0022348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Lu M, Cheng Y, Li Z. H-NOX domains display different tunnel systems for ligand migration. Journal of Molecular Graphics and Modelling. 2010;28:814–819. doi: 10.1016/j.jmgm.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 44.Szundi I, Funatogawa C, Fee JA, Soulimane T, Einarsdottir O. CO impedes superfast O2 binding in ba3 cytochrome oxidase from Thermus thermophilus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21010–21015. doi: 10.1073/pnas.1008603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittenberg JB, Bolognesi M, Wittenberg BA, Guertin M. Truncated hemoglobins: a new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes, and plants. The Journal of biological chemistry. 2002;277:871–874. doi: 10.1074/jbc.R100058200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.