Abstract
Neurogenesis in the developing neocortex relies on the ability of radial glial progenitor cells (RGCs) to switch from proliferative to differentiative neuron-generating divisions, but the molecular mechanisms that control this switch in a correct temporal manner are not well understood. Here, we show that DOCK7, a member of the DOCK180 family of proteins, plays an important role in the regulation of RGC proliferation versus differentiation. Silencing of DOCK7 in RGCs of developing mouse embryos impedes neuronal differentiation and maintains cells as cycling progenitors. In contrast, DOCK7 overexpression promotes RGC differentiation to basal progenitors and neurons. We further present evidence that DOCK7 influences neurogenesis by controlling apically directed interkinetic nuclear migration (INM) of RGCs. Importantly, DOCK7 exerts its effects by antagonizing the microtubule growth-promoting function of the centrosome-associated protein TACC3. Thus, DOCK7 interaction with TACC3 controls INM and the genesis of neurons from RGCs during cortical development.
The six-layered neocortex is formed by the orderly generation of postmitotic neurons through proliferative and differentiative divisions of neural progenitors during neurogenesis1–5. A major subtype of progenitors that give rise to most pyramidal neurons in the developing neocortex are radial glial cells (RGCs), which stem from neuroepithelial cells located at the apical-most region of the neuroepithelium, the ventricular zone (VZ)6–11. RGCs are highly polarized cells, extending an apical process into the ventricular lining and a long thin basal process to the pial surface2,3. A hallmark of these cells, responsible for the pseudostratified appearance of the VZ, is interkinetic nuclear migration (INM), where nuclei change position along the apical-basal axis during the course of the cell cycle12–14. Nuclei move away from the apical surface during G1, undergo S phase at a basal location in the VZ, and return to the apical surface during G2 to divide in the vicinity of the centrosomes, located in the apical endfeet of neuroepithelial and RGCs. Because of the apical location of their nuclei undergoing mitosis, these cells are also referred to as apical progenitors (APs).
Prior to the peak of neurogenesis, RGCs undergo predominantly symmetric proliferative divisions to amplify the progenitor pool. However, during the peak of neurogenesis (embryonic day 13–18 in mice) they largely divide asymmetrically to self-renew while simultaneously giving rise either to one neuron or to an intermediate/basal progenitor (BP), which subsequently divides symmetrically in the subventricular zone (SVZ) to produce two neurons7–9,15–17. Unlike APs, BPs do not display apical/basal polarity or INM9,17,18. While the differentiating progeny progressively migrate away to form the cortical plate (CP), renewing RGCs remain in the VZ for subsequent divisions, ensuring sufficient progenitors to produce later born neurons and glial cells2–4. Self-renewability of RGCs must be tightly balanced with differentiation for proper neocortical development19. Disruption of this balance has been associated with neurological and neuropsychiatric disorders, including microcephaly and schizophrenia20,21.
How APs precisely control self-renewal versus differentiation is not well understood. Besides cell cycle kinetics and asymmetric inheritance14,22–27, recent evidence suggests that INM can influence the balance between neurogenesis and progenitor pool maintenance by controlling the exposure time of AP nuclei to neurogenic versus proliferative signals along the basal-apical axis12,13,28–34. A high apical and low basal gradient of Notch activity, known to prevent APs from differentiating, has been reported in developing chick and zebrafish retina29,30. Furthermore, perturbations of INM in mouse neocortex have been linked to cell fate changes31–34, with impaired basal-to-apical (bl-to-ap) INM being associated with a decrease in APs, and a concomitant increase in BPs and/or neurons31,32,34. However, the consequences of the converse situation, accelerated bl-to-ap INM, on AP proliferation and neuronal differentiation remain unaddressed.
The cellular machinery that controls INM involves actomyosin- and microtubule-dependent systems12,13, with the former generally linked to the ap-to-bl leg of INM and the latter to the bl-to-ap leg in the developing rodent cortex32,33,35,36 (but see ref.37). Ample evidence indicates an important role for the centrosome, the main microtubule-organizing center in animal cells, in bl-to-ap INM13,31,32. Depletion of centrosomal proteins, such as Cep120 and TACCs, which associate with the microtubule lattice linking the centrosome and nucleus, disrupts this leg of INM and causes increased neuron production at the expense of progenitors32. Whereas these studies point to the importance of centrosomal proteins in INM and maintenance of the progenitor pool, little is known about their regulation in these events.
Here, we investigated the role of DOCK7, a member of the DOCK180-related protein superfamily, in cortical neurogenesis. The DOCK180 family emerged as a novel class of Rac/Cdc42 GTPase guanine nucleotide exchange factors (GEFs), which exhibit diverse cell type specific functions38–43. Among them, DOCK7 was initially identified as a Rac activator that is highly expressed in major regions of the developing rodent brain, including hippocampus and cortex44, and has been shown to control axon formation44 and myelination45 through activation of Rac and both Rac and Cdc42, respectively. Findings presented here demonstrate that DOCK7 plays a critical role in the generation of BPs and neurons from RGCs, and that it influences neurogenesis by controlling bl-to-ap INM of RGCs. Importantly, DOCK7’s role in INM and neurogenesis does not involve its GEF activity, but instead requires its interaction with the centrosome- and microtubule-associated protein TACC3. We found that DOCK7 exerts its effects by antagonizing the microtubule growth-promoting function of TACC3. Thus, our study identifies DOCK7 as a key regulator of INM and underscores the importance of this process in controlling RGC proliferation versus differentiation in the developing neocortex.
METHODS
DNA constructs
pCAGGS-FLAG-DOCK7 and pCAGGS-FLAG-DOCK7ΔDHR2 constructs have been described previously44. cDNAs encoding DOCK7Δ513–812 (DOCK7ΔDHR1), DOCK7Δ812–931, DOCK7Δ933–1164 (DOCK7ΔTACC3), DOCK6 and DOCK8 were cloned into pCAGGS with the addition of an N-terminal FLAG epitope tag. The EGFP-TACC3 plasmid was provided by L-H Tsai (MIT). pCAG-EGFP and pCAG-mKO2-F constructs were obtained by subcloning EGFP cDNA (Clontech) and mKO2-F cDNA (provided by F. Matsuzaki, RIKEN) into pCAGGS, respectively. The yeast two-hybrid (YTH) constructs pGBD-DOCK7-R1, -R2, and -R3 were generated by insertion of cDNAs encoding amino acids 1–664 (R1), 506–1164 (R2) and 1030–1644 (R3) of DOCK7 into pPC97. pGAD-TACC3 full-length (FL) and pGAD-TACC3-TACC were obtained in YTH screen. pGBD-Lamin, pGBD-H-Ras, pGAD-Lamin and pGAD-PI3KD were generated by subcloning cDNAs encoding Lamin, H-Ras and PI3-kinase delta into pPC86 and pPC97, respectively. The GST-TACC3 construct was obtained by subcloning full-length mTACC3 cDNA into pGEX-4T-3 (GE Healthcare Biosciences). For RNAi experiments, previously described DNA fragments encoding short hairpin RNAs directed against mouse DOCK7 mRNA (Dock7#2)44 or mTACC3 mRNA (Tacc3#1)32 were cloned into pSUPER (Oligoengine), a modified version of pSUPER in which an EGFP, EGFP-NLS (nuclear localization signal) or a DsRed-NLS encoding sequence driven from the chicken β-actin promoter was inserted into the XhoI site of pSUPER, and in the case of Dock7#2 shRNA also into the pTRIPΔU3-EF1α-EGFP lentiviral vector51.
Yeast two-hybrid screening
1 × 106 clones of a mouse fetal brain cDNA library in the pPC86 vector were screened using DOCK7-R1, -R2, or -R3 fragments cloned into pPC97 as baits in the PJ69a yeast reporter strain.
Cell culture and transfection/infection
HEK293, COS7, Neuro2A, N1E-115, and SKN-BE cells were cultured in DMEM containing 10% fetal bovine serum (HyClone), 4 mM L-glutamine (Gibco BRL), 100 I.U./ml penicillin (Gibco BRL), and 100 µg/ml streptomycin (Gibco BRL). Baf3 were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum and 10% WEHI-3 conditioned medium as a source of interleukin-3 (IL-3). Dissociated cortical progenitors were prepared from E12.5 or E13.5 cortices of CD1 mice and cultured in Neurobasal medium (Gibco BRL) containing 20 ng/ml FGF2 (Promega), 10 ng/ml EGF, 2% B27 (v/v, Gibco BRL), 4 mM L-glutamine, 100 I.U./ml penicillin, and 100 µg/ml streptomycin as described52. HEK293 cells were transfected using the calcium phosphate co-precipitation method. COS7 cells were transfected using the transfection reagent Fugene 6 (Roche), as outlined by the manufacturer. Neuro2A cells were infected with lentiviruses, which were generated as previously described51. Dissociated cortical progenitors were transfected using the Amaxa Nucleofection system (Amaxa, Lonza). 3–5 µg total of the indicated plasmids were used per electroporation.
In utero electroporation and BrdU injections
In utero electroporation was performed largely as described24. In brief, a timed-pregnant CD1 mouse (Charles River) at 13.5 d of gestation was anesthetized, the uterine horns were exposed, and approximately 1 to 2 µl of plasmid DNA was injected manually into the lateral ventricles of the embryos using a beveled and calibrated micropipette. Five 50 ms pulses of 40 V with a 950 ms interval were delivered across the uterus with two 5 mm electrode paddles positioned on either side of the head (BTX, ECM830). After electroporation, the uterus was placed back in the abdominal cavity and the wound was surgically sutured. For BrdU-labeling experiments, mouse embryos were electroporated at E13.5 and BrdU (150 mg per kg of body weight) was injected at E15.5. Animals were sacrificed at indicated time points following BrdU injection. All animal care protocols were approved by Cold Spring Harbor Laboratory.
Immunohistochemistry and immunocytochemistry
For immunostaining of tissue sections, whole embryos (E9.5 and E10.5) or brains of embryos were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 1 h at room temperature (RT; whole embryos) or overnight at 4°C (brains of embryos). Next, whole embryos and brains were either cryoprotected in 30 % sucrose in PBS and cut, after embedding in OCT compound, into 16 µm thick coronal sections on a Leica CM3050S cryostat (whole embryos and brains), or were sectioned into 40 µm thick coronal sections using a Vibratome (Leica VT1000S; brains). For immunostaining of COS7, SK-N-BE, and dissociated neocortical cells, the cells were fixed for 4 min with ice-cold 100% methanol at −20°C. Brain sections and cells were blocked and permeabilized with 10% normal goat serum (NGS) and 0.3% Triton X-100 (or 0.1 % for cells) in PBS for 1 h at RT, followed by incubation with primary antibodies diluted in 3% NGS and Triton X-100 in PBS at concentrations indicated below a t 4°C overnight. We used the following primary antibodies: anti-DOCK744 (rabbit, 1:500); anti-nestin (mouse, 1:200, Chemicon); anti-Tuj1 (mouse 1:1,500, Covance); anti-γ-tubulin (mouse, 1:2,000, Sigma); anti-GFP (chicken, 1:500, Aves Labs); anti-BrdU (rat, 1:500, AbD Serotec); anti-PH3 (rabbit, 1:1,000, Millipore); anti-Ki67 (rabbit, 1:250, Vector Laboratories); anti-Pax6 (rabbit, 1:500, Covance); anti-Tbr2 (rabbit, 1:300, Abcam); anti-RFP (rabbit, 1:500, Rockland); anti-pericentrin (mouse, 1:1,000, BD Biosciences); anti-TACC3 (mouse, 1:300, Santa Cruz); anti-α-tubulin (mouse, 1:2,000, Sigma); anti-ZO-1 (mouse, 1:100, Invitrogen); anti-GFAP (chicken, 1:500, Aves Labs). Of note, for BrdU staining, brain sections were incubated in 2N HCl solution for 30 min at 37°C to unmask the antigen, followed by a neutralization step with 0.1 M sodium tetraborate and 3 washes in PBS. The secondary antibodies used were: Alexa Fluor 488, 594, or 647 goat anti-mouse, rabbit, rat, or chicken (1:500 for immunohistochemistry and 1:1,000 for immunocytochemistry, Molecular Probes). Nuclei were counterstained with 0.3 µg/ml 4’,6-diamidino-2-phenylindole (DAPI, Sigma) in PBS for 10 min. Of note, all brain slices were counterstained with DAPI; images depicting DAPI staining were not always included for reasons of clarity. Images of brain sections and cells were acquired using a spinning disk confocal microscope (Perkin-Elmer).
Slice culture and time-lapse imaging
Time-lapse imaging of brain slices was largely performed as previously described53. In brief, mouse embryos at E13.5 were electroporated with plasmids expressing indicated shRNAs or cDNAs together with EGFP and mKO2-F expressing plasmids. The electroporated embryos were dissected 48 h later in ice-cold artificial cerebral spinal fluid (ACSF). Brains were isolated, embedded in 4% low-melting agarose in ACSF, and cut into 300 µm thick coronal slices using a Vibratome. Slices were transferred to a membrane insert (Millicell, 30 mm, 0.4 µm pore size) and incubated with slice culture medium (DMEM/F-12, 5% horse serum (v/v), 5% fetal bovine serum (v/v), N2 supplement (1:100, Invitrogen), B27 supplement without retinoic acid (1:50, Invitrogen), 100 U/ml penicillin/streptomycin, 10 ng/ml epidermal growth factor, and 10 ng/ml basic fibroblast growth factor) for about 3 h at 37°C. Subsequently, slices were immersed in 200 µl of type Ia collagen (Cellmatrix, Nitta Gelatin) for 1 h at 37°C, and inserts containing the slices were placed in a glass bottom microwell dish (MatTek). The samples were observed with an inverted spinning disk confocal microscope equipped with an environmental chamber (5% CO2, 37°C). Pictures were taken at 10 min intervals with 20–25 Z-sections (4 µm intervals). The time-lapse data were assembled and analyzed by ImageJ (NIH) and Volocity software (Improvision).
GST pull-down assays, coimmunoprecipitation and Western blot analysis
For GST pull-down assays, GST-TACC3 fusion protein and GST alone were immobilized onto Glutathione-Sepharose beads (GE Healthcare Biosciences). Mouse E13.5 cortices and HEK293 cells expressing FLAG-DOCK7, -DOCK7Δ513–812, -DOCK7Δ812–931, or -DOCK7Δ933–1164 fusion proteins, or empty control vector, were homogenized using Micropestles (Eppendorf) in lysis buffer (50 mM Tris pH 7.5, 1 % Triton X-100, 150 mM NaCl, 5% glycerol, 5 mM NaF, 1 mM Na3VO4 and protease inhibitors). Equal amounts of total lysates were incubated with GST-TACC3 fusion protein or GST. Immunoblots were probed with anti-DOCK7 or anti-FLAG polyclonal antibody (Sigma). For coimmunoprecipitations, HEK293 cells expressing EGFP-TACC3 and FLAG-DOCK7, or empty control vector, were homogenized in lysis buffer. Total lysates were incubated with anti-FLAG M2-agarose beads (Sigma) overnight at 4°C. Beads were washed 6 times with lysis buffer. Immunoprecipitates were then resolved by SDS-PAGE and immunoblotted with anti-EGFP (Invitrogen) and anti-FLAG (Sigma) polyclonal antibodies. For Western blots in Fig. 1a, mouse cortices (E11-P3) were homogenized using Micropestles (Eppendorf) in 75 mM Tris-HCl (pH 6.8), 3.8% SDS, 4 M urea, and 20% glycerol and subjected to Western blot analysis with anti-DOCK7 antibody, and anti-γ-tubulin as a loading control. Western blot analyses depicted in Supplementary Fig.3 were carried out using anti-DOCK7, anti-DOCK6 (MBL), or anti-DOCK8 (provided by P. Aspenström) antibody, and anti-γ-tubulin as a loading control.
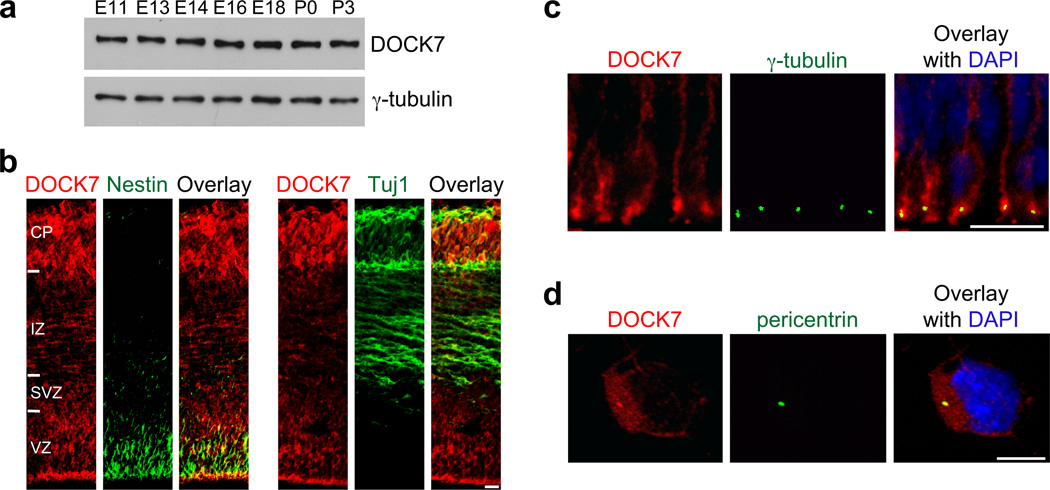
Figure 1.
Distribution and localization of DOCK7 in the developing mouse cortex. (a) DOCK7 levels in cortical lysates at different developmental stages. γ-tubulin was used as a loading control. Full-length Western blot is shown in Supplementary Figure 12. (b) Coronal cryosections of the neocortex at E13.5 immunostained for DOCK7 and neural stem/progenitor marker nestin or neuronal marker class III β-tubulin Tuj1. VZ, ventricular zone; SVZ, subventricular zone; IZ, intermediate zone; CP, cortical plate. (c) Coronal brain slices of the neocortex at E13.5 immunostained for DOCK7 and centrosomal marker γ-tubulin, and counterstained with DAPI. An enlarged view of the apical endfeet of VZ progenitors is shown. (d) Cultured cortical progenitors isolated from E13.5 neocortices immunostained for DOCK7 and centrosomal marker pericentrin, and counterstained with DAPI. Scale bars: 15 µm in b; 10 µm in c; 5 µm in d.
Analysis of microtubules
COS7 cells transfected with indicated plasmids were immunostained with antibodies against EGFP and FLAG (to identify transfected cells) and α-tubulin (to visualize microtubules), and counterstained with DAPI. Images were acquired using a Perkin-Elmer spinning disk confocal microscope. The area of the microtubule aster emanating from the centrosome was measured using ImageJ software. Dissociated neocortical cells were prepared from E13.5 neocortices 1 d post-electroporation with indicated plasmids as described32. Cells were plated on coverslips precoated with poly-D-lysine and laminin, and cultured for 2 d before fixation, immunostaining with antibodies against EGFP, RFP and α-tubulin, and counterstaining with DAPI. Confocal images were acquired and the area occupied by microtubule bundles around the nucleus was measured by ImageJ. Several Z-series confocal images of transfected cells were merged to show the entire microtubule “fork”-like structure.
Quantitative analysis of electroporated neocortices
All the quantification studies were carried out on transfected cells localized within the dorsolateral cortex. To score images, the GFP or RFP channel was first judged independently, followed by judgments of the other markers. A total of 3–6 brain sections were analyzed per animal by taking up to three 10–20 µm Z-stack images to cover the electroporated VZ, SVZ and/or IZ, CP of each coronal section with a 20× or a 40× objective and comparing them with equivalent sections in littermate counterparts. BrdU+, PH3+, Ki67+, Pax6+, Tbr2+ or Tuj1+ transfected cells were quantified using Volocity software. The number of BrdU+ and/or PH3+ or Ki67+ transfected (EGFP-NLS+ and/or RFP-NLS+) cells within the VZ, and of Pax6+ or Tbr2+ transfected cells within the VZ and SVZ, were quantified and calculated as the percentage of total transfected cells. Note, for quantification of Pax6+ and Tbr2+ transfected cells, fluorescence intensities of Pax6 and Tbr2 within the nuclear area were measured, and only transfected cells with Pax6 or Tbr2 fluorescence intensity above the mean plus 1.5 times the standard deviation of background intensity were counted as Pax6+ and Tbr2+ cells, respectively. Small puncta or signals that were not compliant with the EGFP-NLS+ and/or RFP-NLS+ nuclear labeling were ignored. Divisions at apical and basal positions were defined as those located less than and more than 20 µm, respectively, from the ventricular surface. Quantification of the distribution of transfected cells across the neocortical layers and of the number of transfected neurons were performed on single optical sections by quantifying all EGFP+, and EGFP+ Tuj1+ cells, respectively, and dividing by the number of EGFP+ cells in a radial stripe comprising all cortical layers. Transfected cells were scored as neurons only when Tuj1 immunostaining clearly surrounded a DAPI-positive nucleus in the EGFP+ and/or RFP+ cell. INM was measured by quantifying the number of BrdU+ transfected cells in each bin, 15 min, 2 h, 4 h and 6 h after a single BrdU injection (a bin was defined as a 20 µm-thick stripe parallel to the ventricle surface). The centrosome to nucleus distance in transfected RGCs was determined by measuring the distance from the centrosome to the lower edge of the nucleus. To measure the velocity of bl-to-ap nuclear migration, the change in the distance traveled by a transfected RGC (µm) was divided by the time (h) they traveled, using Volocity software.
Statistical analysis
Data were presented as mean ± s.e.m. from at least three independent experiments. Direct comparisons were made using Student’s t-test and multiple group comparisons were made using one-way analysis of variance (ANOVA). Statistical significance was defined as P < 0.05, 0.01 or 0.001 (indicated as *, ** or ***, respectively). P values ≥ 0.05 were considered not significant.
RESULTS
DOCK7 expression and localization in developing neocortex
To assess DOCK7’s role during cortical development, we began by examining its expression patterns in the developing cortex. Western blot analysis of cortical lysates showed DOCK7 is expressed at embryonic day 11 (E11), and expression persists until at least postnatal day 3 (P3), implying that DOCK7 is expressed during active neocortical neurogenesis (Fig. 1a). To determine the spatial expression pattern(s) of DOCK7 in the developing neocortex, we performed immunohistochemistry44 on cryosections of mouse brains around the onset of neocortical neurogenesis (E9.5, E10.5) and at E13.5. DOCK7 immunoreactivity was undetectable at E9.5 and weak at E10.5, but relatively high at E13.5 in Tuj1-positive neurons in the cortical plate (CP) and in nestin-positive VZ progenitors (Fig. 1b, and Supplementary Fig. 1), suggesting that onset of DOCK7 expression correlates with onset of neurogenesis.
Noteworthy, DOCK7 fluorescence in the VZ of E13.5 cortices was strong at the ventricular (apical) surface of the VZ (Fig. 1b), where AP centrosomes reside25,32. Closer examination of apical endfeet of nestin-positive progenitors revealed overlapping staining of DOCK7 with the centrosomal marker γ-tubulin, suggesting the presence of DOCK7 at the centrosome (Fig. 1c). To corroborate this, we examined the subcellular localization of DOCK7 in cultured cortical progenitors derived from E13.5 mouse cortices. While DOCK7 fluorescence was detectable throughout the cytoplasm, an intense signal was noticeable at the centrosome, where DOCK7 colocalized with the centrosomal marker pericentrin (Fig. 1d). These data confirm the presence of DOCK7 at the centrosome in cortical progenitors.
DOCK7 depletion enhances VZ progenitor proliferation in utero
Next, we asked whether DOCK7 affects progenitor proliferation in the embryonic mouse neocortex. To approach this, we generated a vector that expresses a short hairpin RNA (shRNA) targeting the 3’-untranslated region (UTR, Dock7#2) of mouse DOCK7 mRNA44. Dock7#2 shRNA significantly reduced DOCK7 levels in cortical progenitors, whereas control shRNA (scr#1) had no effect (Supplementary Fig. 2). Notably, DOCK6 levels were not affected upon DOCK7 knockdown and DOCK8 expression was not detectable in cortical progenitors; these proteins are most closely related to DOCK7 (Supplementary Fig. 3). The Dock7#2 or scr#1 shRNA construct, together with an EGFP-NLS (nuclear localization signal) expressing plasmid, was introduced into E13.5 mouse brains by in utero electroporation, BrdU was administered at E15.5, and brains were analyzed 2 h later. We found that the percentage of BrdU-positive progenitors in the VZ was significantly higher in the Dock7#2 shRNA group than in the scr#1 shRNA group (Fig. 2a,c). Rescue experiments using DOCK7 cDNA that lacks the 3’UTR and is therefore resistant to Dock7#2 shRNA-mediated RNAi (Supplementary Fig. 2) demonstrated that the DOCK7 RNAi effect is specific (Fig. 2a,c). We further performed immunostainings for phosphorylated histone H3 (PH3), a mitotic marker, and determined the mitotic index (i.e. the percentage of transfected cells PH3-positive in the VZ). This index was considerably higher in the Dock7#2 shRNA group compared to control and rescue groups (Fig. 2b,d). Thus, DOCK7 knockdown increases the percentage of proliferating VZ progenitors in the embryonic neocortex. When examining the impact of ectopic DOCK7 expression, we observed the opposite effects. The percentage of BrdU-positive transfected cells, as well as the mitotic index, was significantly reduced in the FLAG-DOCK7 expressing group compared to the control vector group (Fig. 2e–h). Notably, immunohistochemistry for cleaved caspase-3 at 2 d post-electroporation of FLAG-DOCK7 expressing vector showed that cell survival was unaffected (Supplementary Fig. 4), excluding the possibility that apoptosis was responsible for the decrease in the number of proliferating progenitors.
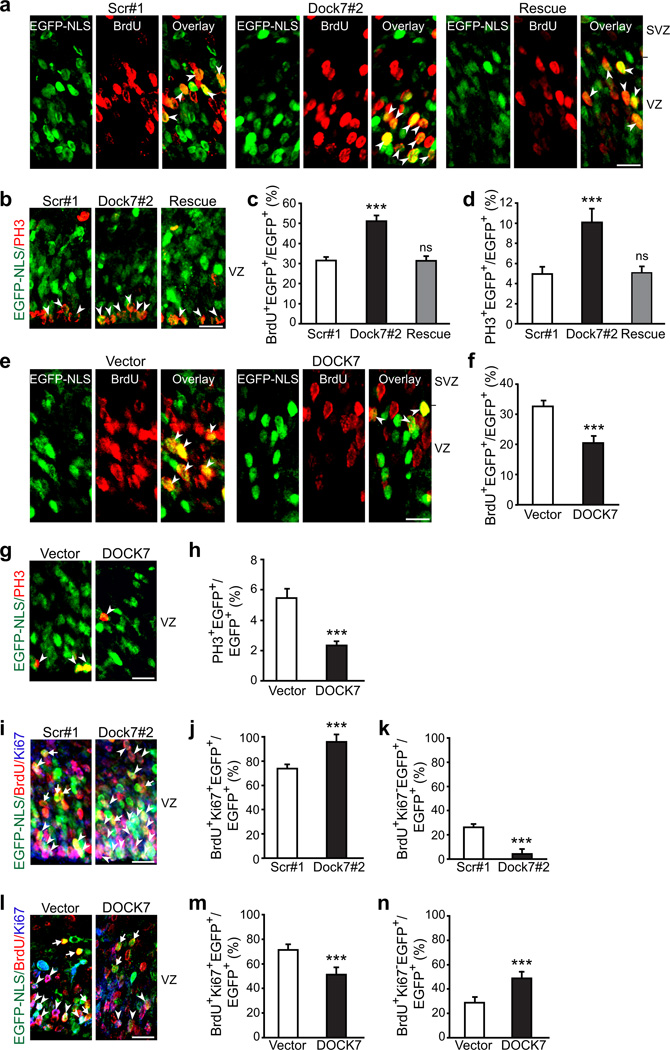
Figure 2.
DOCK7 modulates the VZ progenitor pool size. (a–h) Analysis of proliferating S-phase (BrdU+) and mitotic (PH3+) cells. Mouse embryos were co-electroporated at E13.5 with plasmids expressing EGFP-NLS and non-targeting shRNA (scr#1), Dock7 targeting shRNA (Dock7#2) or Dock7#2 shRNA and FLAG-DOCK7 (rescue) (a–d), or control vector (vector) or FLAG-DOCK7 (DOCK7) (e–h), pulse-labeled with BrdU for 2 h at E15.5 (a,c,e,f), or unlabeled (b,d,g,h). Brain slices were co-immunostained with anti-EGFP and anti-BrdU (a,c,e,f) or anti-PH3 (b,d,g,h) antibodies. (a,b,e,g) Confocal images of the VZ (including SVZ in a,e). Arrowheads indicate transfected cells that are BrdU+ (a,e) or PH3+ (b,g). (c,d,f,h) Quantification of BrdU+ (c,f) or PH3+ (d,h) transfected cells in VZ. Data are mean ± s.e.m.; n = 921–1738 cells for each condition. ***P < 0.001; ns, not significant; one-way ANOVA (c,d) and Student’s t-test (f,h). (i–n) Analysis of cell cycle re-entry and exit. Mouse embryos were electroporated at E13.5 and pulse-labeled with BrdU at E15.5 for 24 h. Brain slices were co-immunostained for EGFP, BrdU and Ki67. (i,l) Confocal images of the VZ. Cycling Ki67+ BrdU+ EGFP-NLS+ cells and Ki67− BrdU+ EGFP-NLS+ cells withdrawn from the cell cycle are indicated with arrowheads and arrows, respectively. (j,k,m,n) Quantification of transfected cells in VZ that continued cycling (BrdU+ Ki67+; j,m) and those that exited the cell cycle (BrdU+ Ki67−; k,n). Data are mean ± s.e.m.; n = 867–1292 cells for each condition.. ***P < 0.001; Student’s t-test. Scale bars, 20 µm. For details on quantifications, see supplemental data.
To gain further insight into how altered DOCK7 expression could affect the progenitor pool size, we analyzed the proliferative status of VZ progenitors in more detail, by performing BrdU pulse-labeling, to visualize S-phase cells, combined with Ki67 staining to reveal cells in the cell cycle. First, embryos were electroporated at E13.5, 2 d later a 24 h pulse of BrdU was administered, and the fractions of transfected cells that entered the cell cycle (BrdU+, Ki67+) or withdrew from the cell cycle (BrdU+, Ki67−) were quantified. The percentage of BrdU and Ki67 double-positive cells increased in the Dock7#2 shRNA group (Fig. 2i,j), and decreased in the FLAG-DOCK7 group (Fig. 2l,m). Conversely, the percentage of BrdU-positive and Ki67-negative cells decreased in the Dock7#2 shRNA group (Fig. 2 i,k), and increased in the FLAG-DOCK7 group (Fig. 2 l,n). These data indicate that DOCK7 knockdown favors maintenance of cells as cycling progenitors, whereas ectopic DOCK7 expression promotes cell cycle exit. In a second set of experiments, a short BrdU pulse (30 min) was delivered, and the ratio of cycling Ki67-positive progenitors in S phase (BrdU+) over Ki67-positive progenitors in the VZ was measured. This ratio was not significantly different between Dock7#2 shRNA, FLAG-DOCK7, and control transfected groups (Supplementary Fig. 5a,b), implying that altering DOCK7 levels does not appreciably affect cell cycle duration. In concurrence, DOCK7 knockdown in Neuro-2A cells did not alter cell cycle distribution, as revealed by flow cytometric analysis (Supplementary Fig. 5c). Thus, DOCK7 levels likely affect progenitor pool size by influencing cell cycle exit/re-entry, but not cell cycle duration of VZ progenitors.
DOCK7 is required for the genesis of neurons
Based on our findings that DOCK7 knockdown expands the VZ progenitor pool and ectopic DOCK7 expression reduces it, we reasoned that DOCK7 could play a role in controlling the switch of RGCs from proliferation to differentiation/neurogenesis. To assess this, we electroporated E13.5 embryos with Dock7#2 shRNA or FLAG-DOCK7 expressing vector, together with an EGFP-NLS or EGFP expression plasmid, and 2 d later determined the number of transfected RGCs, BPs, and neurons. To identify RGCs and BPs, brain slices were immunostained for Pax6 and Tbr2, markers of RGCs and BPs, respectively. DOCK7 knockdown resulted in more Pax6-positive cells, but fewer Tbr2-positive cells, compared to control scr#1 shRNA electroporated cortices (Fig. 3a–d). Conversely, ectopic DOCK7 expression caused a decrease in Pax6-positive cells, but increase in Tbr2-positive cells, compared to the control vector condition (Fig. 3e–h). Thus, DOCK7 depletion promotes amplification of the RGC pool, while DOCK7 overexpression diminishes this pool and promotes the transition from RGCs to BPs.
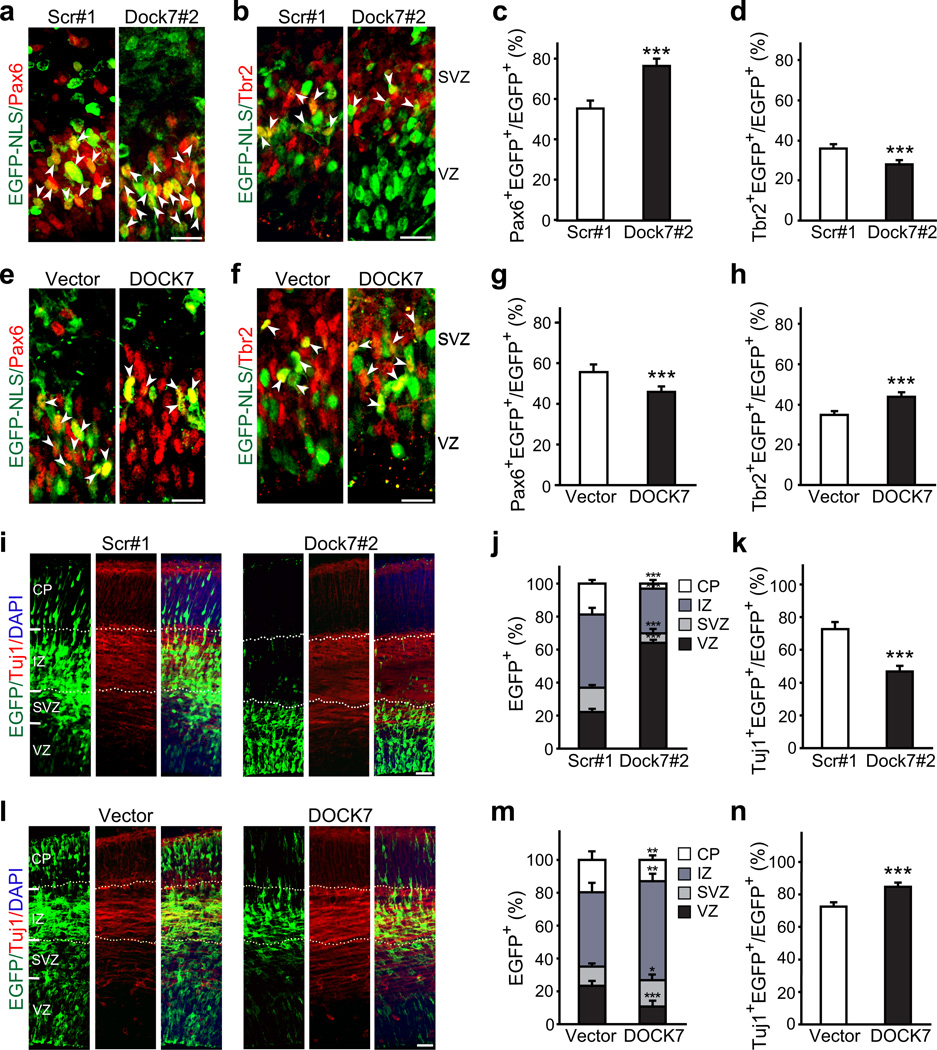
Figure 3.
DOCK7 is required for the transition of RGCs to BPs and genesis of neurons. (a–h) Analysis of RGCs and BPs. Mouse embryos were co-electroporated at E13.5 with plasmids expressing EGFP-NLS and non-targeting shRNA (scr#1) or Dock7 targeting shRNA (Dock7#2) (a–d), or empty control vector (vector) or FLAG-DOCK7 (DOCK7) (e–h), and sacrificed at E15.5. Brain slices were co-immunostained for EGFP and Pax6 (a,e,c,g) or Tbr2 (b,f,d,h), markers for RGCs and BPs, respectively. (a,b,e,f) Confocal images of the VZ/SVZ. Arrowheads indicate transfected cells that are Pax6+ (a,e) or Tbr2+ (b,f). Scale bars, 20 µm. (c,d,g,h) Quantification of transfected cells that are Pax6+ (c,g) or Tbr2+ (d,h) in the VZ/SVZ. Data are mean ± s.e.m.; n = 853–1268 cells for each condition. ***P < 0.001; Student’s t-test. (i–n) Analysis of neurons. Mouse embryos were electroporated as described above, except that EGFP, instead of EGFP-NLS, was included as marker protein. Brain slices were co-immunostained for EGFP and Tuj1, and counterstained with DAPI. (i,l) Confocal images of neocortices showing distribution of EGFP+ transfected cells across neocortical layers. Scale bars, 40 µm. (j,m) Quantification of the distribution of EGFP+ transfected cells in entire neocortex. Data are shown as mean ± s.e.m.; n = 1983–2453 cells for each condition. *P < 0.05; **P < 0.01; ***P < 0.001; Student’s t-test. (k,n) Quantification of transfected cells that are Tuj1+. Data are mean ± s.e.m.; n = 1318–2331 cells for each condition. ***P < 0.001, Student’s t-test. For details on quantifications, see supplemental data.
We further found that the number of EGFP-positive neurons was decreased in cortices of embryos electroporated with Dock7#2 shRNA expressing vector. This was first revealed by altered cell distribution across the neocortical layers. Newly generated neurons at E15.5 typically migrate towards the CP through the intermediate zone (IZ), while self-renewing RGCs remain in the VZ. Compared to the control group, Dock7#2 shRNA transfected cells were more abundant in the VZ and less abundant in the CP and IZ (Fig 3i,j). Also, a decrease in the percentage of transfected cells in the SVZ, where BPs reside, was observed. Furthermore, the percentage of Tuj1-positive transfected cells was significantly decreased in the Dock7#2 shRNA group (Fig. 3k). Importantly, when E13.5 or E15.5 cortices were electroporated and analyzed 8 or 9 d later at P1.5 and P4.5, respectively, we found that these perturbations persisted. While in the control groups (at P1.5 and P4.5) the majority of transfected cells were located in the cortical mantle, with only a small fraction (< 10%) residing in the proliferative VZ/SVZ, in the Dock7#2 shRNA groups ≥ 40% of the transfected cells still resided in the VZ/SVZ and the percentage of transfected cells in the cortical mantle was correspondingly reduced (Supplementary Fig. 6a.d,g,j). Consistently, the percentage of Tuj1-positive transfected cells was significantly decreased in the Dock7#2 shRNA groups (Supplementary Fig. 6b,e,h,k). We did not detect a significant difference in the percentage of GFAP-positive transfected cells between the Dock7#2 shRNA and scr#1 shRNA groups (Supplementary Fig. 6c,f,i,l), implying that DOCK7 knockdown does not induce premature astrocyte differentiation. These data indicate that silencing DOCK7 impairs the genesis of neurons.
When examining the effects of DOCK7 overexpression, we observed a decrease in the percentage of transfected cells in the VZ, an increase in the SVZ and IZ, but, notably, a decrease in the CP (Fig. 3l,m). The number of Tuj1-positive transfected cells was significantly increased upon DOCK7 expression (Fig. 3n); however, the majority of these cells resided in the IZ, and relatively few were detectable in the CP (Fig. 3l,m). Noteworthy, most of the cells located in the IZ displayed a multipolar phenotype. These data suggest that ectopic DOCK7 expression promotes the genesis of neurons, which are able to reach the IZ, but are defective or delayed in migrating toward the CP, possibly due to a defect in the transition from multipolar to bipolar phase. Thus, in addition to controlling the genesis of neurons, DOCK7 may also play a role in the polarization and/or migration of IZ neurons.
DOCK7 is important for interkinetic nuclear migration
We next asked how DOCK7 affects neurogenesis and controls RGC proliferation versus differentiation. Cellular parameters that influence RGC progenitor output include cell cycle duration, cell polarity and asymmetric inheritance, and, more recently, INM12–14,19,23–26. Altering DOCK7 levels did not affect cell cycle duration (Supplementary Fig. 5). Also, DOCK7 did not appear to directly affect RGC polarity and adhesion, revealed by RGCs coupling by adherens junctions (AJs) to the apical side of the neuroepithelium (Supplementary Fig. 7). However, a hint for a role for DOCK7 in INM came from our BrdU labeling experiments, where we noted a difference in the location of BrdU-labeled nuclei of Dock7#2 shRNA and FLAG-DOCK7 expressing cells (Fig. 2a,e; arrowheads). Compared to the control group, the distribution of the BrdU-positive nuclei was shifted toward the apical surface in the Dock7#2 shRNA group, whereas in the FLAG-DOCK7 group the nuclei were positioned more basally in the upper VZ. To further establish whether and how altered DOCK7 expression affects INM, we examined in more detail the migration of RGC nuclei concurrent with cell progression from S to M phase (see Supplementary Fig. 8). To this end, embryos were electroporated at E13.5, 2 d later BrdU was administrated, and the positions of BrdU-labeled nuclei of transfected cells were determined 15 min, 2 h, 4 h, and 6 h after BrdU injection (Fig. 4).
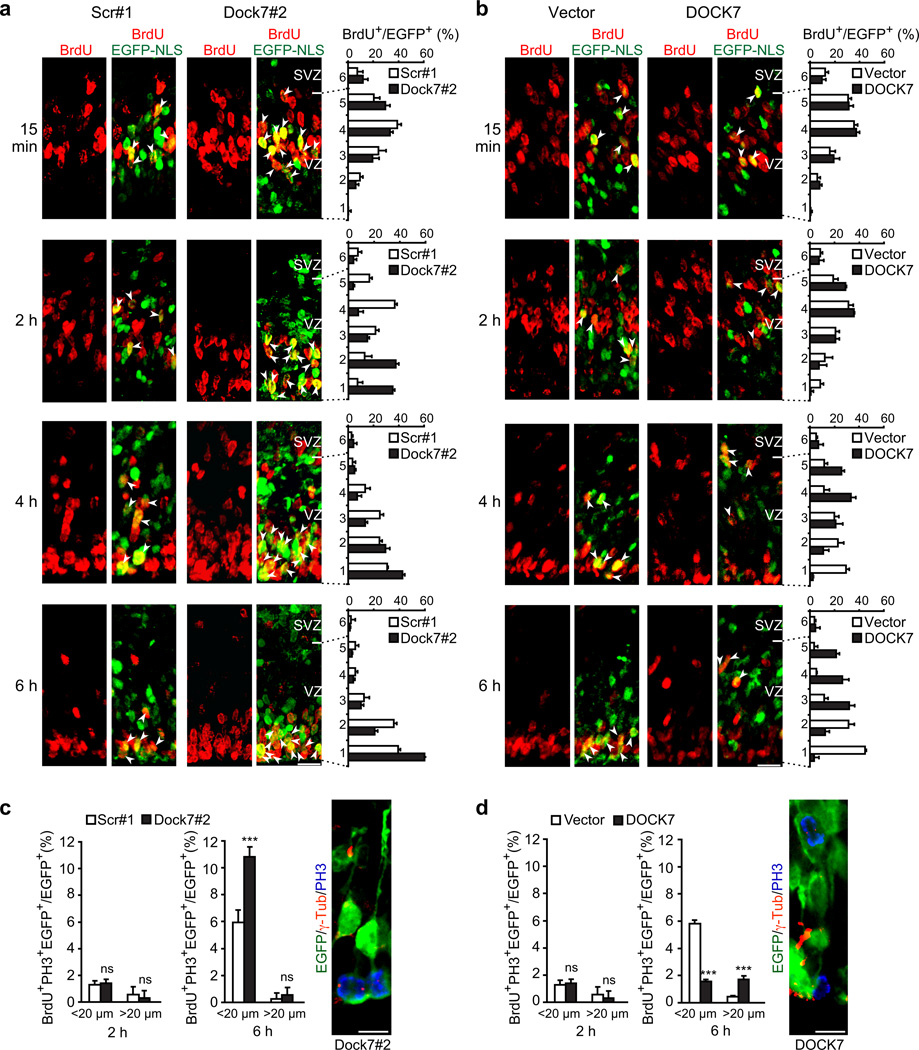
Figure 4.
DOCK7 controls basal-to-apical INM of RGCs. (a–d) Mouse embryos were electroporated at E13.5 with a plasmid expressing EGFP-NLS together with one of the indicated plasmids, pulse-labeled with BrdU at E15.5, and sacrificed 15 min, 2 h, 4 h or 6 h after BrdU injection. Brain slices were co-immunostained for EGFP and BrdU (a,b) and EGFP, BrdU and PH3 (c,d, left). (a,b) Left panels: Confocal images showing position of BrdU+ and EGFP-NLS+ nuclei in neocortices transfected with plasmids expressing scr#1 shRNA (scr#1) or Dock7#2 shRNA (Dock7#2) (a) or control vector (vector) or FLAG-DOCK7 (DOCK7) (b), at indicated times after BrdU injection. Scale bars, 20 µm. Arrowheads indicate BrdU+ nuclei of transfected cells. Right panels: Distribution of BrdU+ transfected cells quantified as percentage of all transfected cells per bin (one bin: 20 µm high) across VZ and part of SVZ, which was divided into 6 bins. Data are mean ± s.e.m.; n = 383–601 cells for each condition. (c,d) Quantification of BrdU+ and PH3+ transfected cells at apical (< 20 µm) or more basal (> 20 µm) locations relative to ventricular surface in VZ at 2 and 6 h after BrdU injection. Data are mean ± s.e.m.; n = 754–1009 cells for each condition. ***P < 0.001; ns, not-significant; Student’s t-test. Right panels: Representative images of RGCs expressing EGFP and Dock7#2 shRNA (c) or FLAG-DOCK7 (d) immunostained for EGFP (green), PH3 (blue) and centrosomal marker γ-tubulin (red). Scale bars, 10 µm. For details on quantifications, see supplemental data.
At the 15 min time point, the majority of BrdU-labeled nuclei of control, Dock7#2 shRNA, and FLAG-DOCK7 expressing VZ cells were located in the upper half of the VZ, consistent with the known location of RGC nuclei in S phase33 (Fig. 4a,b). BrdU-labeled control nuclei began to migrate toward the ventricle surface (VS) within 2 h, and continued to do so for about another 4 h (Fig 4a,b; scr#1, vector), in concurrence with an S phase duration of ≈ 4 h and G2 phase duration of ≈ 2 h at this developmental stage33. After reaching the VS, they entered apical mitosis (< 20 µm from VS), as judged by their PH3 immunoreactivity (Fig. 4c,d). DOCK7 knockdown accelerated the bl-to-ap INM (Fig. 4a, Dock7#2). Whereas only a few control BrdU-labeled nuclei reached the apical region of the VZ after 2h, a much larger fraction of BrdU-labeled nuclei of Dock7#2 shRNA group transfected cells already reached this location (Fig. 4a), where most of them stayed for another ≈ 4 h, only to then enter mitosis at the apical surface (Fig. 4c). Indeed, an increase in the number of BrdU+, PH3+ VZ cells in the Dock7#2 shRNA group compared to the control group was observed at 6 h, but not 2 h, following BrdU injection (Fig. 4c). Conversely, when examining the nuclei position of FLAG-DOCK7 expressing VZ cells, we noted that the bl-to-ap migration of BrdU-labeled nuclei was delayed, with many of the nuclei remaining in the upper half of the VZ 6 h after BrdU injection (Fig. 4b). This extended stay at basal locations was associated with an increased number of mitoses away from the apical surface, as judged by the higher percentage of BrdU+, PH3+ cells at basal positions (> 20 µm from VS) in the DOCK7 group compared to the control vector group (Fig. 4d).
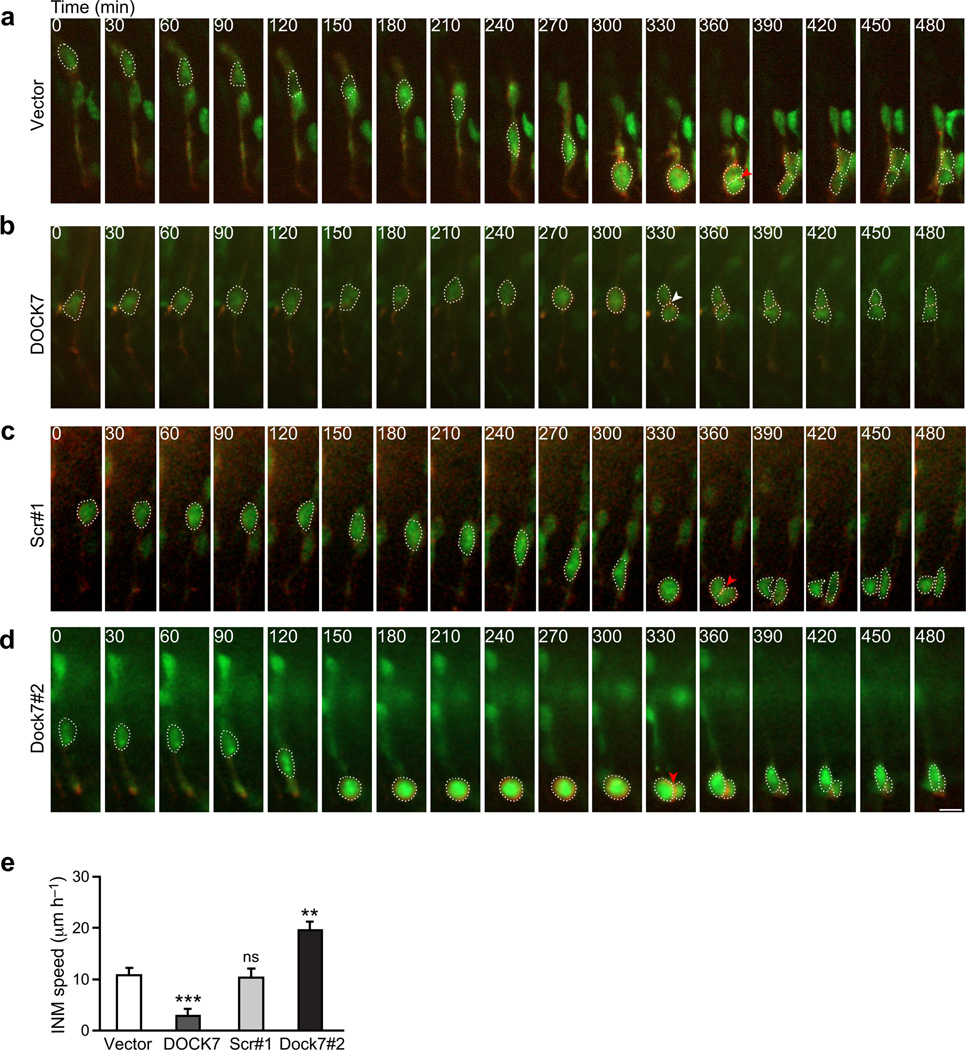
To corroborate and extend the above findings, we carried out time-lapse imaging on acute cortical slices 2 d after in utero electroporation of plasmids expressing empty control vector, FLAG-DOCK7, scr#1 shRNA, or Dock7#2 shRNA. EGFP and mKO2-F expressing plasmids were co-electroporated to fluorescently label both the cytoplasm (EGFP) and plasma membrane (mKO2-F contains a CAAX farnesylation motif) of the RGCs, and slices were imaged over 8 to 10 h. We found that nuclei-surrounding cell bodies of most control vector expressing cells migrated steadily toward the ventricle and cells divided at the apical surface (Fig. 5a,e, Supplementary Video 1). In contrast, cell bodies of a significant fraction of DOCK7 overexpressing cells remained at basal locations and the cells divided away from the ventricular surface (Fig. 5b,e, Supplementary Video 2). Some cell bodies of DOCK7 overexpressing cells were observed to move slowly toward the ventricle. However, the distance traveled was short, and those cells divided away from the ventricular surface (Supplementary Video 3). These findings support that DOCK7 overexpression impedes bl-to-ap INM, leading to extended residence time of RGC nuclei at basal locations and ectopic mitoses. On the other hand, cell bodies of most Dock7#2 shRNA expressing cells migrated considerably faster to the ventricular surface, than those of control scr#1 shRNA expressing cells, where, importantly, they then resided for several hours before undergoing apical mitoses (Fig. 5c–e, Supplementary Videos 4–6). These findings corroborate that DOCK7 knockdown accelerates bl-to-ap INM, leading to extended residence of RGC nuclei at apical locations and apical mitoses. Taken together, we conclude that DOCK7 plays a critical role in the regulation of the bl-to-ap INM process.
Figure 5.
Altered DOCK7 expression affects apically directed INM of RGCs in acute cortical slices. (a–e) Mouse embryos were co-electroporated with plasmids expressing EGFP and mKO2-F marker proteins, and empty control vector (vector) (a), FLAG-DOCK7 (DOCK7) (b), non-targeting shRNA (scr#1) (c) or Dock7 targeting shRNA (Dock7#2) (d) at E13.5, and time-lapse imaging was performed 2 d later. (a–d) Time-lapse video sequences (over an 8 h time period) of EGFP and mKO2-F positive transfected cells undergoing INM. Ventricular surface is located at the bottom of the images. Time is denoted in the upper left corner. Cell bodies are delineated by dashed lines. Red arrowheads indicate mitosis at apical surface; white arrowhead indicates ectopic mitosis. Scale bar, 10 µm. (e) Quantification of bl-to-ap INM velocity. Data are mean ± s.e.m.; n = 19–43 cells from at least 3 experiments for each condition (see supplemental data for details). **P < 0.01; ***P < 0.001; ns, not significant; one-way ANOVA.
DOCK7 interacts with TACC3
To gain insight into the molecular mechanism(s) by which DOCK7 controls INM and neurogenesis, we first explored whether DOCK7’s GEF activity is involved. DOCK7, like other DOCK180 members, contains a conserved DHR2 domain that catalyzes the exchange of GDP for GTP on Rac and/or Cdc4238,44,45. This domain was shown to be required for DOCK7’s role in axon formation44. To determine whether it is important for DOCK7’s role in INM and genesis of neurons, we tested whether a DOCK7ΔDHR2 mutant44 can rescue DOCK7 RNAi-evoked effects on INM, RGC expansion and neuron production. Intriguingly, DOCK7ΔDHR2 rescued all phenotypes to a similar degree as seen with DOCK7 WT (Fig. 7k–o), implying that DOCK7’s DHR2 domain and GEF activity toward Rac/Cdc42 are dispensable for its role in INM and cortical neurogenesis.
Figure 7.
DOCK7 antagonizes TACC3 function during cortical neurogenesis. (a–e) DOCK7 and TACC3 have opposing functions. E13.5 mouse embryos were co-electroporated with plasmids expressing EGFP-NLS (a–d) or EGFP (e) and scr#2 or Tacc3#1 shRNA, pulse-labeled with BrdU for 2 h (a,c) or 6 h (b) at E15.5, or not labeled (d,e). Brain slices were co-immunostained for EGFP and BrdU (a,c), BrdU and PH3 (b), PH3 (d), or Tuj1 (e). (a) Confocal images of VZ/SVZ co-immunostained for EGFP and BrdU. Arrowheads indicate BrdU+ transfected cells. (b–e) Quantification of transfected cells that are BrdU+ and PH3+ < 20 µm or > 20 µm away from ventricular surface in VZ (b); BrdU+ in VZ (c); PH3+ in VZ (d); or Tuj1+ in entire neocortex (e). (f–j) Simultaneous knockdown of TACC3 rescues DOCK7 knockdown phenotypes. E13.5 embryos were co-electroporated with plasmids co-expressing EGFP-NLS (or EGFP in j) and scr#1 or Dock7#2 shRNA and/or plasmids co-expressing RFP-NLS (or RFP in j) and scr#2 or Tacc3#1 shRNA, and processed as in a–e. (f) Confocal images of VZ/SVZ co-immunostained for EGFP, RFP and BrdU. Arrowheads indicate double-transfected (EGFP-NLS+ and RFP-NLS+) cells that are BrdU+. (g–j) Quantification of double-transfected cells as in b–e. (k–o) DOCK7ΔTACC3 (ΔTACC3), but not DOCK7ΔDHR2 (ΔDHR2), fails to rescue DOCK7 knockdown phenotypes. E13.5 embryos were co-electroporated with EGFP-NLS-expressing plasmid and indicated constructs, and processed as in a–e. (k) Confocal images of VZ/SVZ co-immunostained for EGFP and BrdU. Arrowheads indicate BrdU+ transfected cells. (l–o) Quantification of transfected cells as in b–e. Data are mean ± s.e.m.; n = 739–2498 cells for each condition (see supplemental data for details). **P < 0.01, ***P < 0.001; ns, not significant; Student’s t-test (b–e) and one-way ANOVA (g–j, l–o); Scale bars, 20 µm.
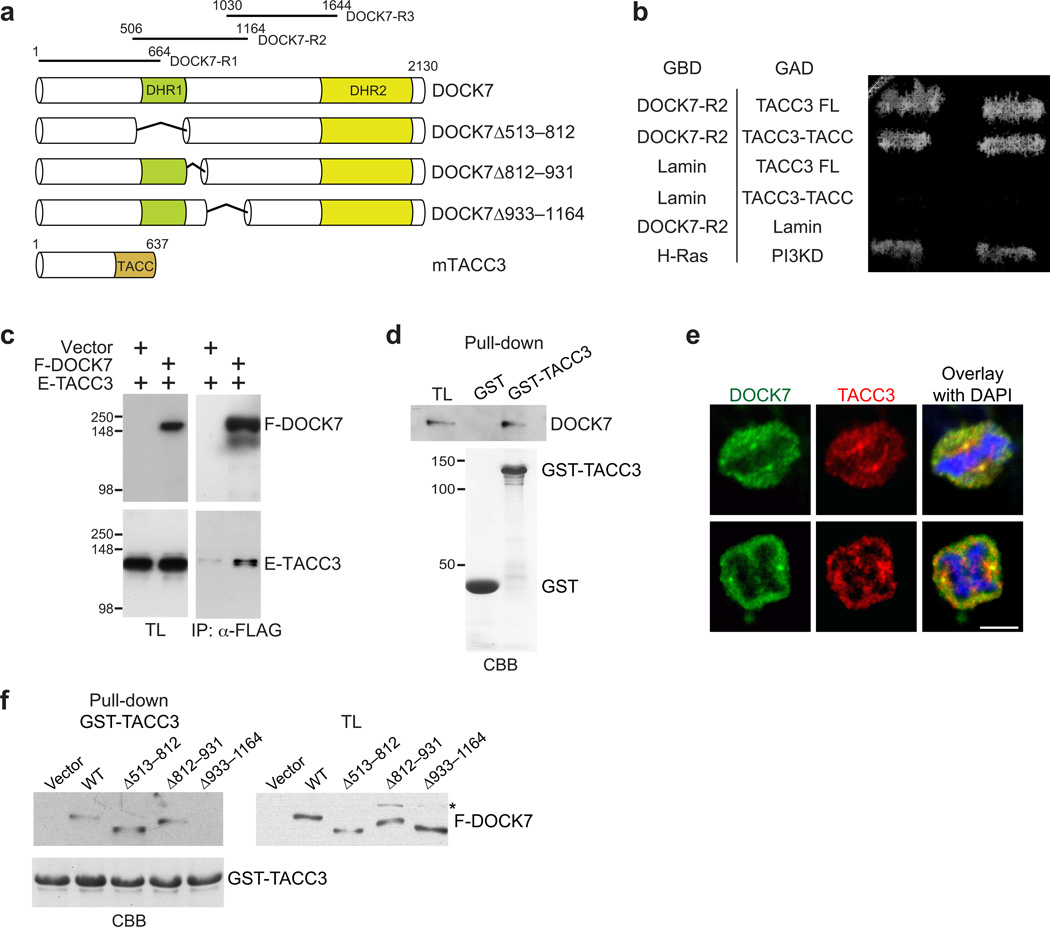
To identify other potentially relevant DOCK7 functions, we decided to search for DOCK7-interacting proteins other than small GTPases. To this end, the yeast two-hybrid approach was used to screen an embryonic mouse brain cDNA library with three different DOCK7 fragments as bait (Fig. 6a). Two of the positive clones contained identical cDNAs matching the TACC domain-encoding sequence of TACC3, and another contained a cDNA encoding full-length TACC3. These clones were obtained using DOCK7-R2 (aa 506–1164) as bait (Fig. 6a). TACC3 is a member of the TACC family, centrosome- and microtubule-associated proteins implicated in centrosome-directed microtubule growth, nuclear migration, and, significantly, in maintaining the neural progenitor pool during mouse neocortical development32,46,47.
Figure 6.
DOCK7 interacts with TACC3. (a) DOCK7 and TACC3 domain structure and deletion constructs. DOCK7 fragments used as baits in YTH screens are indicated at the top. (b) YTH interaction between DOCK7-R2 and TACC3. Yeast transformed with plasmids expressing DOCK7-R2 fused to GAL4 DNA-binding domain (GBD), and full-length (TACC3-FL) or TACC-motif (TACC3-TACC) of TACC3 fused to GAL4 activation domain (GAD), were grown on medium lacking histidine. Lamin served as negative, and H-Ras and PI3-kinase delta (PI3KD), as positive control. (c) DOCK7-TACC3 interaction in mammalian cells. Lysates from HEK293 cells transiently expressing FLAG-DOCK7 (F-DOCK7) and/or EGFP-TACC3 (E-TACC3) were immunoprecipitated (IP) with anti-FLAG antibody and analyzed by immunoblotting with anti-FLAG and anti-EGFP antibodies. TL, total lysate. (d) GST-TACC3 fusion protein, or GST alone (lower panel, Coomassie Brilliant Blue/CBB staining), immobilized on beads was incubated with lysates from E13.5 mouse cortices. Bound DOCK7 was detected by immunoblotting with anti-DOCK7 antibody. (e) DOCK7 colocalizes with TACC3 at centrosome of mitotic human neuroblastoma SK-N-BE cells. Cells were co-immunostained for DOCK7 (green) and TACC3 (red), and counterstained with DAPI (blue). Confocal images of cells in metaphase (top) and in anaphase (bottom) are shown. Scale bar, 2 µm. (f) Mapping TACC3-binding region in DOCK7. GST-TACC3 protein (lower panel, CBB) immobilized on beads was incubated with lysates from HEK293 cells expressing control vector (vector), FLAG-DOCK7 wild-type (WT) or one of the indicated FLAG-DOCK7 deletion constructs. Bound proteins were detected by immunoblotting with anti-FLAG antibody. Asterisk indicates non-specific band. Full-length Western blots are shown in Supplementary Figure 12.
The interaction between DOCK7 and TACC3 was validated by several approaches. First, the YTH interaction (Fig. 6b) was confirmed by coimmunoprecipitation experiments, using lysates from HEK293 cells co-expressing FLAG-DOCK7 and EGFP-TACC3 (Fig. 6c). Second, the association between DOCK7 and TACC3 was demonstrated by a GST-fusion protein pull-down assay. Beads loaded with GST-TACC3 efficiently pulled-down DOCK7 from lysates of E13.5 mouse cortices (Fig. 6d). Finally, immunocytochemistry studies in human SK-N-BE neuroblastoma cells revealed that DOCK7 and TACC3 colocalize, particularly, at the centrosome (Fig. 6e). Thus, these data demonstrate that the two proteins interact, with the R2 fragment of DOCK7 sufficient for binding. To further delineate the TACC3-binding domain in DOCK7, mutants containing deletions within the R2 fragment were generated (Fig. 6a) and tested for their ability to interact with TACC3 in GST pull-down assays. We found that DOCK7Δ933–1164, henceforth called DOCK7ΔTACC3, failed to bind TACC3 (Fig. 6f).
DOCK7 controls INM and neurogenesis by antagonizing TACC3
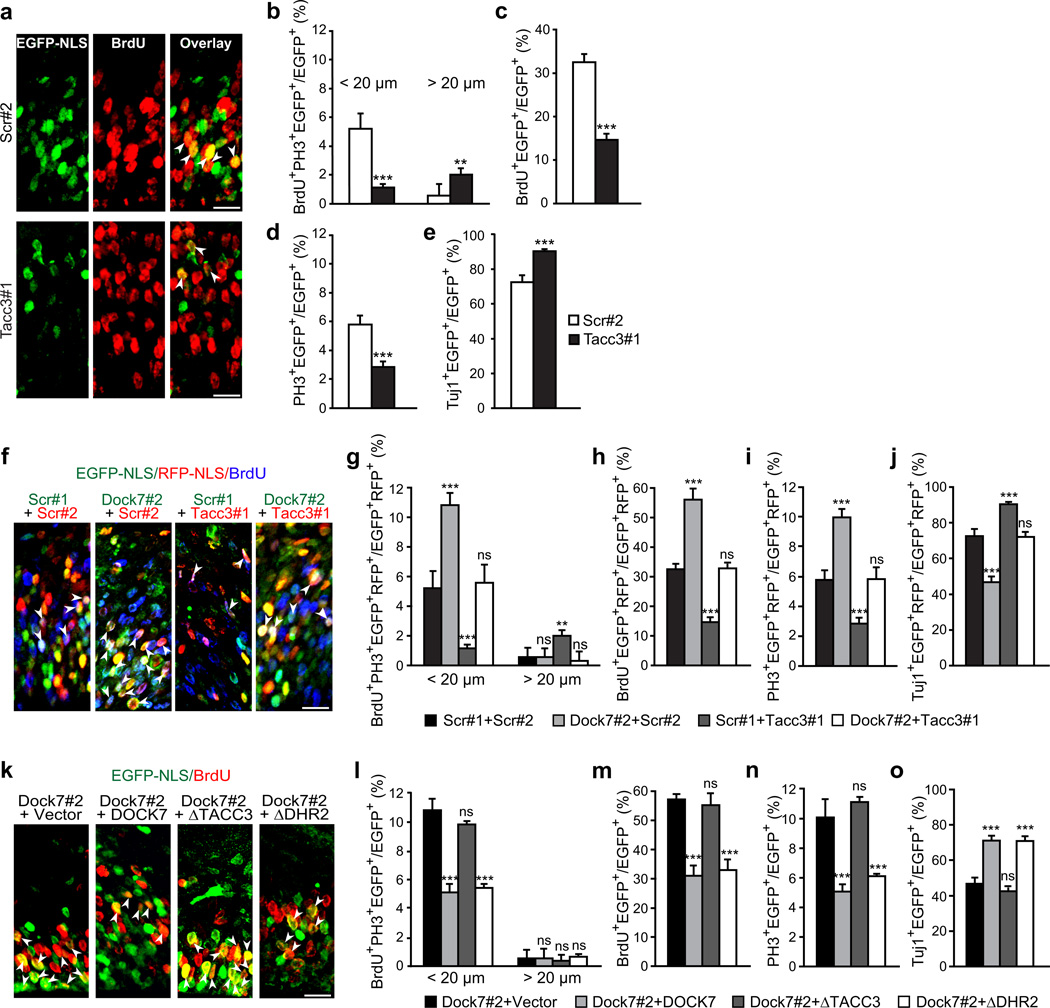
We next investigated the relevance of the DOCK7-TACC3 interaction in INM and neurogenesis. A previous study showed that knockdown of TACCs halted bl-to-ap INM, decreased the number of proliferating progenitors, and increased neuron production32, all opposite to the effects of DOCK7 knockdown. Since in the latter study a mixture of siRNAs targeting all three TACC family members was used, we first investigated whether knockdown of TACC3 alone is sufficient to produce the above phenotypes. We found that this is indeed the case; knockdown of TACC3 impaired bl-to-ap INM, decreased BrdU labeling and the mitotic index, and increased the percentage of Tuj1+ cells (Fig. 7a–e). These data indicated that DOCK7 and TACC3 have opposing functions during cortical neurogenesis. Therefore, we asked whether DOCK7 exerts its effect on INM and neurogenesis by antagonizing TACC3 function.
If this is the case, simultaneous knockdown of TACC3 and DOCK7 should counteract the phenotypes resulting from DOCK7 deficiency, whereas a DOCK7 mutant defective in TACC3 binding should fail to rescue them. To test the former, E13.5 cortices were electroporated with plasmids co-expressing Dock7#2 or scr#1 shRNA and EGFP-NLS (or EGFP) together with plasmids co-expressing Tacc3#1 or scr#2 shRNA and RFP-NLS (or RFP), and brains examined 2 d later. As expected, co-electroporation of Dock7#2 and scr#2 shRNA expressing vectors resulted in a higher percentage of cells dividing at apical positions, an increased number of proliferating VZ progenitors, and a decrease in neuron production. Importantly, simultaneous TACC3 knockdown completely rescued these phenotypes (Fig. 7f–j). To test whether a DOCK7 mutant defective in TACC3 binding can rescue the DOCK7 RNAi-evoked phenotypes, we coelectroporated plasmids expressing Dock7#2 shRNA and DOCK7ΔTACC3. In contrast to DOCK7 WT and DOCK7ΔDHR2, DOCK7ΔTACC3 failed to rescue the INM defect, the increase in dividing VZ progenitors and decrease in neuron production caused by DOCK7 RNAi (Fig. 7k–o). Together, these data indicate that DOCK7 controls INM and cortical neurogenesis by antagonizing TACC3 function.
DOCK7 antagonizes microtubule-associated function of TACC3
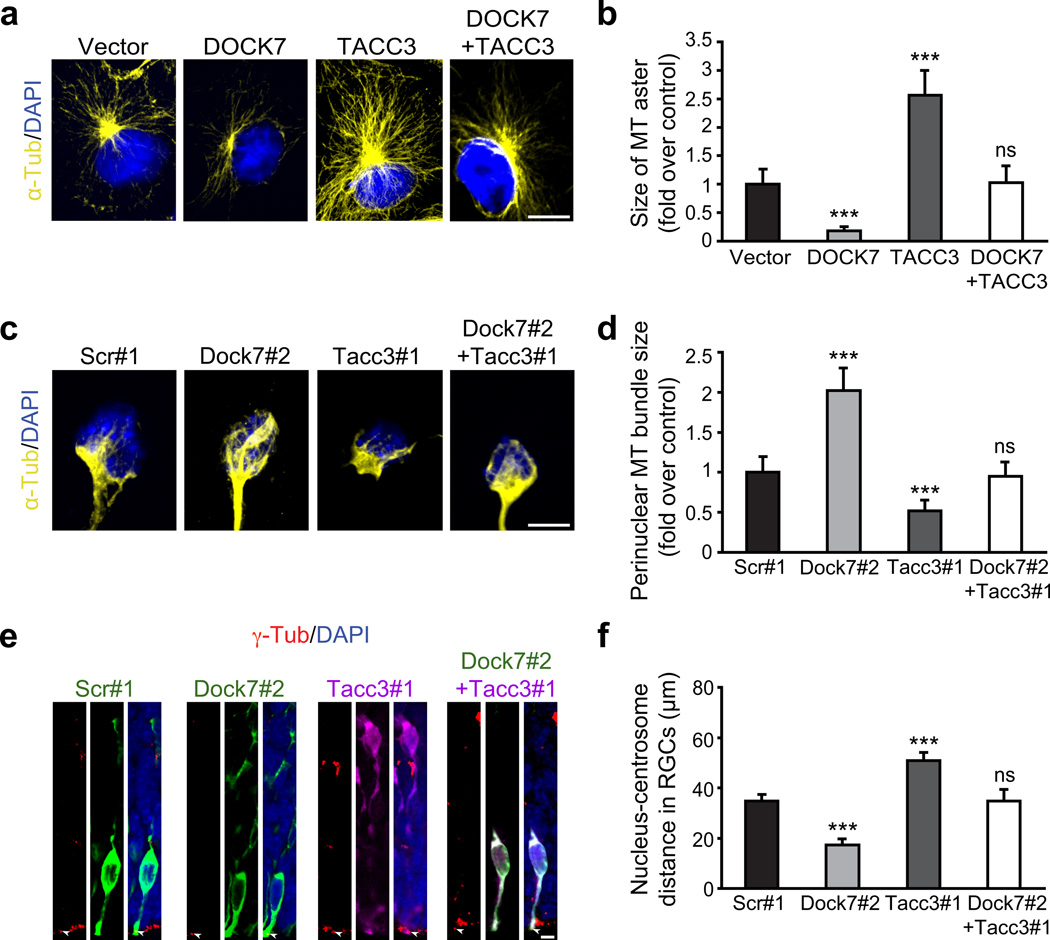
The TACC proteins were reported to control INM of cortical neural progenitors by regulating the growth and integrity of centrosome-associated microtubules coupling the centrosome and nucleus32. Hence, we assessed whether DOCK7 antagonizes the microtubule growth-promoting function of TACC3. We first examined this in COS7 cells, since TACC3 expression in these cells was shown to increase the size of the microtubule aster emanating from the centrosome32. In contrast to TACC3, DOCK7 expression decreased the size of the microtubule aster, and, importantly, co-expression of DOCK7 with TACC3 restored the microtubule aster size to that in control vector transfected cells (Fig. 8a,b). To determine whether these manipulations affected microtubule nucleation, COS7 cells were treated with nocodazole (which depolymerizes microtubules), and subsequently allowed to recover after drug washout. After a 5 min recovery period, most of the transfected cells in all groups showed a clear microtubule aster, indicating that microtubule nucleation was not affected. However, after 20 min of recovery, akin to the results in Fig. 8a, DOCK7 and TACC3 expressing cells showed a smaller and larger microtubule aster, respectively, compared to control cells and cells co-expressing DOCK7 and TACC3 (Supplementary Fig. 9).
Figure 8.
DOCK7 antagonizes microtubule growth-promoting/stabilizing function of TACC3. (a,b) DOCK7 antagonizes TACC3’s ability to increase microtubule aster size in COS7 cells. (a) α-tubulin immunostaining (yellow) and counterstaining with DAPI (blue) of COS7 cells expressing FLAG-DOCK7 (DOCK7) and EGFP-TACC3 (TACC3) alone, or in combination. (b) Measurement of microtubule aster size. The average aster size of control vector transfected cells was set as 1. Data are mean ± s.e.m.; n = 69–115 cells for each condition. ***P < 0.001; ns, not significant; one-way ANOVA. (c–d) Simultaneous silencing of TACC3 prevents enlargement of microtubule “fork”-like structure caused by DOCK7 knockdown in cultured neocortical cells. E12.5 mouse neocortices were transfected with plasmids co-expressing EGFP and scr#1 or Dock7#2 shRNA, RFP and Tacc3#1 shRNA, or both Tacc3#1 and Dock7#2 shRNA-expressing plasmids, and dissociated at E13.5. Cells were cultured for 2 d and immunostained for α-tubulin (yellow) and counterstained with DAPI (blue). (c) Representative images showing microtubule “fork”-like structure in transfected neocortical cells. (d) Measurement of perinuclear microtubule bundle size of “fork”-like structure. The average bundle size of scr#1 shRNA transfected cells was set as 1. Data are mean ± s.e.m.; n = 77–103 cells for each condition. ***P < 0.001; one-way ANOVA. (e–f) Silencing TACC3 rescues decrease in nucleus-centrosome distance caused by DOCK7 knockdown in RGCs in vivo. E13.5 embryos were electroporated as in c, and sacrificed at E15.5. Brain slices were immunostained for γ-tubulin (red), EGFP (green) and/or RFP (purple), and counterstained with DAPI (blue). (e) Examples of transfected RGCs. Arrowheads indicate centrosomes of transfected cells. (f) Quantification of nucleus-centrosome distance. Data are mean ± s.e.m.; n = 215–371 cells for each condition. ***P < 0.001; one-way ANOVA. For details on quantifications, see supplemental data. Scale bars, 10 µm (a) and 5 µm (c,e).
We then extended these studies to cortical neural progenitors. These cells display a microtubule “fork”-like structure consisting of two or more prominent microtubule bundles that couple the centrosome and the nucleus32. Whereas knockdown of TACC3 in cultured cortical progenitors decreased the size of the microtubule “fork”-like structure, knockdown of DOCK7 increased the overall size (Fig. 8c,d). Importantly, concurrent knockdown of TACC3 with DOCK7 restored the size of the microtubule “fork”-like structure to that of control cells (Fig. 8c,d).
Finally, we measured the nucleus-centrosome distance in RGCs expressing Dock7#2 shRNA, Tacc3#1 shRNA or both, as this distance is affected by alterations in microtubule growth and integrity32. Brain slices prepared from electroporated embryos were stained with γ-tubulin (centrosomal marker) and counterstained with DAPI (nuclear marker), and the nucleus-centrosome distance measured. In contrast to Tacc3#1 shRNA expressing RGCs, where the nucleus-centrosome distance was increased, this distance was significantly decreased in Dock7#2 shRNA expressing RGCs, and could be restored to that seen for scr#1 expressing RGCs by co-expression of Tacc3#1 shRNA (Fig. 8e,f). Combined with the above data, these findings indicate that DOCK7 antagonizes the microtubule growth-promoting/stabilizing function of TACC3. Notably, this effect of DOCK7 is independent of its DHR2 domain, as DOCK7ΔDHR2 was able to rescue the DOCK7 RNAi evoked decrease in nucleus-centrosome distance (Supplementary Fig. 10).
DISCUSSION
Fundamental to proper development of the mammalian neocortex is the ability of RGCs to balance self-renewal with neuronal differentiation in a correct temporal manner. Key determinants reported to influence RGC function during neurogenesis include cell polarity, cell cycle duration, and, more recently, INM2,12–14,19, which remains less understood. In this study, we present evidence that DOCK7, a member of the DOCK180 family, controls bl-to-ap INM by antagonizing the microtubule growth-promoting function of TACC3, and thereby controls the genesis of neurons from RGCs.
With the onset of neurogenesis, RGCs start switching from proliferative to differentiative neuron generating divisions9,15,16,18. Our data implicate DOCK7 in the regulation of this switch. Knockdown of DOCK7 expanded the radial glial progenitor pool by favoring maintenance of RGCs as cycling progenitors at the apical side of the VZ. Notably, with RGC expansion, generation of BPs and neurons was reduced, implying that neuronal differentiation was impaired at the RGC stage. Ectopic DOCK7 expression, on the other hand, increased BP and neuron production, and concomitantly reduced cycling RGCs, indicating their accelerated differentiation. These data indicate that DOCK7 plays a key role in regulating RGC proliferation versus differentiation and is required for proper genesis of neurons from RGCs. Our findings further imply that other DOCK180 family members do not compensate for this function of DOCK7, as knockdown of DOCK7 alone is sufficient to cause defects in neurogenesis. This is perhaps not surprising, since overall similarity among the DOCK180 members is largely restricted to the DHR2 and DHR1 domains; none of which appears to be required for DOCK7 function in neurogenesis (Fig. 7, Supplementary Fig. 11). Also, non-redundant functions for several DOCK180 family members have been previously reported41–43. While mice carrying a nonsense mutation in the Dock7 locus, originating from a chemical mutagenesis screen, were not reported to exhibit gross neurological or behavioral abnormalities in tests probing innate and/or anxiety/stress related behavior48, it will be interesting to assess them for specific cognitive or other impairments linked to cortical dysfunction.
Our data support a model in which DOCK7 influences the mode of RGC division and genesis of neurons through its regulatory effects on bl-to-ap INM. Ectopic DOCK7 expression impeded bl-to-ap INM of RGCs, without affecting cell cycle progression. This led to extended residence of RGC nuclei at basal locations and mitoses at ectopic sites away from the ventricular surface, producing daughter cells that likely differentiated into BPs and/or neurons, as the numbers of both were increased upon DOCK7 overexpression, while RGCs were decreased. In line with this, increased neurogenesis has been reported where INM was impaired upon Cep120 or Hook3 knockdown31,32. Conversely, DOCK7 knockdown accelerated the movement of RGC nuclei from bl-to-ap positions, resulting in extended apical residency of RGC nuclei and apical mitoses. This was associated with an increase in RGCs and reduction in BPs and neurons, implying that extended nuclear residence at the apical side favors RGCs to divide symmetrically, with two RGCs as progeny. In concurrence with our model are reports indicating enrichment of proliferative signals at the apical versus the basal axis of the neuroepithelium12,14,29,30. Thus, we posit that DOCK7 controls the bl-to-ap INM step and thereby influences RGC behavior and neurogenesis. Of note, our studies revealed that progenitors undergo S phase at their normal basal position regardless of the levels of DOCK7 expression, consistent with normal ap-to-bl basal migration of RGC nuclei during the G1 phase.
Although Rho GTPases regulate INM34,49,50, we found that DOCK7’s effects on INM and neurogenesis do not require its catalytic DHR2 domain, and, hence, are not mediated by DOCK7’s GEF activity toward Rac/Cdc42. Instead, evidence from our studies indicates that DOCK7 exerts its effects by antagonizing the centrosome-associated microtubule growth-promoting/stabilizing function of TACC3. Specifically, the two proteins exerted opposing effects on centrosomal microtubule growth, and on INM and neurogenesis. Additionally, the phenotypes associated with DOCK7 knockdown, including microtubule growth, INM and genesis of neurons, were rescued by reducing TACC3 levels. Finally, a DOCK7 mutant defective in TACC3 binding failed to rescue the DOCK7 RNAi-evoked phenotypes. We posit that fine-tuned regulation of the microtubule-associated function of TACC3 by DOCK7 controls the growth and dynamics of microtubules coupling the centrosome and nucleus. As such, gain or suppression of TACC3 function facilitates or impedes, respectively, the movement of the nucleus toward the centrosome. Translocation of the nucleus along the microtubules likely involves the minus end-directed motor dynein and associated proteins, as previously reported29,35,37. Future studies will be required to determine the precise mechanism(s) by which DOCK7 antagonizes TACC3’s microtubule-associated function, how DOCK7 is regulated, and whether DOCK7 acts on other microtubule-controlled cellular processes.
Besides controlling the genesis of neurons, our data suggest an additional role for DOCK7 in the polarization and/or migration of IZ neurons. This is shown by an increased number of Tuj1-positive neurons in the IZ, but not in the CP, upon ectopic DOCK7 expression. Since DOCK7’s GEF activity toward Rac/Cdc42 is required for axon formation of hippocampal neurons44 and migration of Schwann cells45, we envision DOCK7’s GEF activity also to be important for the polarization and/or migration of IZ neurons. Combined, these findings imply that DOCK7 executes several distinct functions in the developing brain by engaging in distinct protein-protein interactions. Here, we show that DOCK7 controls bl-to-ap INM, and, importantly, the genesis of cortical neurons from RGCs, via interaction with the centrosome- and microtubule-associated protein, TACC3. As such, our study not only offers new insight into DOCK7 function, but also sheds light on the INM process as it pertains to the regulation of RGC proliferation versus differentiation in the developing neocortex.
Supplementary Material
ACKNOWLEGDGMENTS
We thank members of the Van Aelst lab, E-E Govek and J. Skowronski for discussions and/or critical reading of the manuscript. We thank N. Gray and J.W. Tsai for technical advice regarding the in utero electroporation procedure, and K. John for yeast two-hybrid screening. We also thank F. Matsuzaki and L-H Tsai for reagents. This work was supported by US National Institutes of Health grant MH082808, and a New York STARR consortium grant to L.V.A. C-L.W. is supported by T32 CA 148056-1.
Footnotes
AUTHOR CONTRIBUTIONS
Y-T.Y., C-L.W. and L.V.A conceived and designed the project. Y-T.Y. and C-L.W. performed all the experiments and prepared the figures. L.V.A. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.McConnell SK. Constructing the cerebral cortex: neurogenesis and fate determination. Neuron. 1995;15:761–768. doi: 10.1016/0896-6273(95)90168-x. [DOI] [PubMed] [Google Scholar]
- 2.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 3.Farkas LM, Huttner WB. The cell biology of neural stem and progenitor cells and its significance for their proliferation versus differentiation during mammalian brain development. Curr Opin Cell Biol. 2008;20:707–715. doi: 10.1016/j.ceb.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Miyata T, Kawaguchi D, Kawaguchi A, Gotoh Y. Mechanisms that regulate the number of neurons during mouse neocortical development. Curr Opin Neurobiol. 2010;20:22–28. doi: 10.1016/j.conb.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Johansson PA, Cappello S, Gotz M. Stem cells niches during development--lessons from the cerebral cortex. Curr Opin Neurobiol. 2010;20:400–407. doi: 10.1016/j.conb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 7.Tamamaki N, Nakamura K, Okamoto K, Kaneko T. Radial glia is a progenitor of neocortical neurons in the developing cerebral cortex. Neurosci Res. 2001;41:51–60. doi: 10.1016/s0168-0102(01)00259-0. [DOI] [PubMed] [Google Scholar]
- 8.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 9.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 10.Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 11.Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 12.Latasa MJ, Cisneros E, Frade JM. Cell cycle control of Notch signaling and the functional regionalization of the neuroepithelium during vertebrate neurogenesis. Int J Dev Biol. 2009;53:895–908. doi: 10.1387/ijdb.082721ml. [DOI] [PubMed] [Google Scholar]
- 13.Taverna E, Huttner WB. Neural progenitor nuclei IN motion. Neuron. 2010;67:906–914. doi: 10.1016/j.neuron.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 14.Willardsen MI, Link BA. Cell biological regulation of division fate in vertebrate neuroepithelial cells. Dev Dyn. 2011;240:1865–1879. doi: 10.1002/dvdy.22684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi T, Nowakowski RS, Caviness VS., Jr The leaving or Q fraction of the murine cerebral proliferative epithelium: a general model of neocortical neuronogenesis. J Neurosci. 1996;16:6183–6196. doi: 10.1523/JNEUROSCI.16-19-06183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyata T, et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 19.Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- 20.Manzini MC, Walsh CA. What disorders of cortical development tell us about the cortex: one plus one does not always make two. Curr Opin Genet Dev. 2011;21:333–339. doi: 10.1016/j.gde.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barenz F, Mayilo D, Gruss OJ. Centriolar satellites: Busy orbits around the centrosome. Eur J Cell Biol. 2011 doi: 10.1016/j.ejcb.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8:438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 23.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 24.Bultje RS, et al. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gauthier-Fisher A, et al. Lfc and Tctex-1 regulate the genesis of neurons from cortical precursor cells. Nat Neurosci. 2009;12:735–744. doi: 10.1038/nn.2339. [DOI] [PubMed] [Google Scholar]
- 28.Baye LM, Link BA. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J Neurosci. 2007;27:10143–10152. doi: 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murciano A, Zamora J, Lopez-Sanchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- 31.Ge X, Frank CL, Calderon de Anda F, Tsai LH. Hook3 interacts with PCM1 to regulate pericentriolar material assembly and the timing of neurogenesis. Neuron. 2010;65:191–203. doi: 10.1016/j.neuron.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie Z, et al. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schenk J, Wilsch-Brauninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci U S A. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cappello S, et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- 35.Tsai JW, Chen Y, Kriegstein AR, Vallee RB. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosodo Y, et al. Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. Embo J. 2011;30:1690–1704. doi: 10.1038/emboj.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai JW, Lian WN, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci. 2010;13:1463–1471. doi: 10.1038/nn.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cote JF, Vuori K. Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J Cell Sci. 2002;115:4901–4913. doi: 10.1242/jcs.00219. [DOI] [PubMed] [Google Scholar]
- 39.Meller N, Merlot S, Guda C. CZH proteins: a new family of Rho-GEFs. J Cell Sci. 2005;118:4937–4946. doi: 10.1242/jcs.02671. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto Y, Yamauchi J. Cellular signaling of Dock family proteins in neural function. Cell Signal. 2010;22:175–182. doi: 10.1016/j.cellsig.2009.09.036. [DOI] [PubMed] [Google Scholar]
- 41.Laurin M, et al. The atypical Rac activator Dock180 (Dock1) regulates myoblast fusion in vivo. Proc Natl Acad Sci U S A. 2008;105:15446–15451. doi: 10.1073/pnas.0805546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukui Y, et al. Haematopoietic cell-specific CDM family protein DOCK2 is essential for lymphocyte migration. Nature. 2001;412:826–831. doi: 10.1038/35090591. [DOI] [PubMed] [Google Scholar]
- 43.Chen Q, et al. Loss of modifier of cell adhesion reveals a pathway leading to axonal degeneration. J Neurosci. 2009;29:118–130. doi: 10.1523/JNEUROSCI.3985-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watabe-Uchida M, John KA, Janas JA, Newey SE, Van Aelst L. The Rac activator DOCK7 regulates neuronal polarity through local phosphorylation of stathmin/Op18. Neuron. 2006;51:727–739. doi: 10.1016/j.neuron.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 45.Yamauchi J, Miyamoto Y, Chan JR, Tanoue A. ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. J Cell Biol. 2008;181:351–365. doi: 10.1083/jcb.200709033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gergely F, et al. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci U S A. 2000;97:14352–14357. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18:379–388. doi: 10.1016/j.tcb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Blasius AL, et al. Mice with mutations of Dock7 have generalized hypopigmentation and white-spotting but show normal neurological function. Proc Natl Acad Sci U S A. 2009;106:2706–2711. doi: 10.1073/pnas.0813208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minobe S, et al. Rac is involved in the interkinetic nuclear migration of cortical progenitor cells. Neurosci Res. 2009;63:294–301. doi: 10.1016/j.neures.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Hashimoto-Torii K, Torii M, Ding C, Rakic P. Gap junctions/hemichannels modulate interkinetic nuclear migration in the forebrain precursors. J Neurosci. 2010;30:4197–4209. doi: 10.1523/JNEUROSCI.4187-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janas J, Skowronski J, Van Aelst L. Lentiviral delivery of RNAi in hippocampal neurons. Methods Enzymol. 2006;406:593–605. doi: 10.1016/S0076-6879(06)06046-0. [DOI] [PubMed] [Google Scholar]
- 52.Kim WY, et al. GSK-3 is a master regulator of neural progenitor homeostasis. Nat Neurosci. 2009;12:1390–1397. doi: 10.1038/nn.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.