Abstract
Data from the Encyclopedia of DNA Elements (ENCODE) project show over 9640 human genome loci classified as long noncoding RNAs (lncRNAs), yet only ∼100 have been deeply characterized to determine their role in the cell. To measure the protein-coding output from these RNAs, we jointly analyzed two recent data sets produced in the ENCODE project: tandem mass spectrometry (MS/MS) data mapping expressed peptides to their encoding genomic loci, and RNA-seq data generated by ENCODE in long polyA+ and polyA− fractions in the cell lines K562 and GM12878. We used the machine-learning algorithm RuleFit3 to regress the peptide data against RNA expression data. The most important covariate for predicting translation was, surprisingly, the Cytosol polyA− fraction in both cell lines. LncRNAs are ∼13-fold less likely to produce detectable peptides than similar mRNAs, indicating that ∼92% of GENCODE v7 lncRNAs are not translated in these two ENCODE cell lines. Intersecting 9640 lncRNA loci with 79,333 peptides yielded 85 unique peptides matching 69 lncRNAs. Most cases were due to a coding transcript misannotated as lncRNA. Two exceptions were an unprocessed pseudogene and a bona fide lncRNA gene, both with open reading frames (ORFs) compromised by upstream stop codons. All potentially translatable lncRNA ORFs had only a single peptide match, indicating low protein abundance and/or false-positive peptide matches. We conclude that with very few exceptions, ribosomes are able to distinguish coding from noncoding transcripts and, hence, that ectopic translation and cryptic mRNAs are rare in the human lncRNAome.
In addition to over 20,000 protein-coding genes and known small-RNA, including microRNA host genes, the human genome includes at least 9640 loci transcribed solely into long, non-protein-coding RNAs (long noncoding RNAs; lncRNAs), often with multiple transcript isoforms (Derrien et al. 2012). Of these, only a minority (under 100) have been functionally characterized at an individual level by forward and reverse genetic approaches in organismal and cell culture models. The remainder are known purely via high-throughput discovery and expression analysis. Well-known examples of lncRNAs that have been functionally characterized in-depth include the imprinted Myc target H19 (Gabory et al. 2009), the epigenetic homeobox gene regulator HOTAIR, which promotes cancer metastasis (Gupta et al. 2010), and Xist, the lncRNA that is responsible for inactivation of the mammalian X-chromosome (Jeon and Lee 2011). While these few examples already attest to the diversity of lncRNA functions in chromatin remodeling and imprinting, the diversity of heretofore-uncharacterized lncRNAs hints at numerous additional lncRNA-dependent regulatory mechanisms in mammalian systems. Miat is another example of a recently discovered lncRNA that takes part in a direct network feedback loop with the Pou5f1 pluripotency factor in stem cells (Pou5f1 is also known as Oct4); Miat is both a direct target of and a direct regulator of Pou5f1 (Lipovich et al. 2010; Sheik Mohamed et al. 2010). Hence, lncRNAs can be both regulated by and regulators of key transcription factors. LncRNA genes are transcribed in a diverse range of human tissues and cell lines, and show highly specific spatial and temporal expression profiles, which, in conjunction with detailed molecular characterization of the lncRNAs, attest to numerous distinct functions. These functions include, but are not limited to, epigenetic and post-transcriptional gene expression regulation, sense-antisense interactions with known protein-coding genes, direct binding and regulation of transcription factor proteins, nuclear pore gatekeeping, and enhancer function by transcriptional initiation of lncRNAs that cause chromatin remodeling (Lipovich et al. 2010). Mammalian lncRNAs have epigenetic signatures comparable to those of protein-coding genes, frequently associate with the polycomb repressor complex PRC2 which renders them capable of regulating numerous target genes through histone modifications suppressing gene expression, and mediate global transcriptional programs of cancer transcription factors (Guttman et al. 2009; Khalil et al. 2009; Huarte et al. 2010; Derrien et al. 2012).
A particularly intriguing property of mammalian lncRNAs is their lack of evolutionary conservation, relative to protein-coding genes. Primate-specific lncRNAs in the human genome are increasingly well-documented in the literature (for a review citing multiple pertinent recent reports, see Lipovich et al. 2010). Previously, Tay et al. (2009) screened the human genome for primate-specific single-copy genomic sequences, uncovering 131 primate-specific transcriptional units supported by transcriptome data. The brain-derived neurotrophic factor (BDNF) gene, a key contributor to synaptic plasticity, learning, memory, and multiple neurological diseases, is overlapped by a cis-encoded primate-specific lncRNA (Pruunsild et al. 2007). Most recently, Derrien et al. (2012) found that ∼30% of human lncRNA transcripts in GENCODE, many of which are expressed in the brain, are primate specific. The resulting relevance of lncRNAs to species-specific phenotypes, including primate and human uniqueness, highlights the importance of using empirical methodologies to document whether lncRNAs are actually non-protein-coding.
The majority of definitively known lncRNAs have been annotated using empirical evidence such as cDNA and EST alignments to genome assemblies (Carninci et al. 2005; Katayama et al. 2005; Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009). Yet, despite the attention that they have received, the noncoding status of most lncRNA genes and transcripts has been established mostly through computational means including: examining the size of open reading frames (ORFs), assessing conservation of ORFs that are shorter than known proteins, and looking for conserved translation initiation and termination codons. However, a recent flurry of literature suggests that there may exist a class of bifunctional RNAs encoding both mRNAs and functional noncoding transcripts: Indeed, there is direct evidence for rare members of this transcript class in human, mouse, and fly (Hube et al. 2006; Kondo et al. 2010; Dinger et al. 2011; Ingolia et al. 2011; Ulveling et al. 2011). Hence, identifying the fraction of ostensibly noncoding RNAs that may encode polypeptides is a compelling and open question. In this report, we utilize empirical evidence to estimate, in two ENCODE cell lines, the fraction of annotated lncRNAs that may encode, and therefore possibly function through, polypeptides.
As part of the Encyclopedia of DNA Elements (ENCODE) project, matched-sample long polyA+ and polyA− RNA-seq data were produced, along with tandem mass spectrometry (MS/MS) data for cellular proteins, for the Tier-1 “ENCODE-prioritized” human cell lines K562 and GM12878. The RNA-seq data provides measures of relative gene expression in various cellular compartments (Djebali et al. 2012); for both GM12878 and K562, nucleus, cytosol, and whole-cell samples were used to sequence both polyA+ and polyA− RNA populations. These data have been used to obtain measures of transcript abundance for all genes in GENCODE v7 annotation (the annotation generated for the ENCODE Consortium), based on ENCODE and other data (Harrow et al. 2012). The mass spec data were produced via a “shotgun” approach, wherein cells were cultured, subcellular fractionation performed, followed by protein separations, tryptic digestion, and MS/MS analysis. The resulting spectra were mapped directly to a 6-frame translation of the entire hg19 assembly to produce a “proteogenomic track” within the UCSC Genome Browser (Kent 2002; Karolchik et al. 2009), and were also mapped against the GENCODE gene annotation set (J Khatun, Y Yu, J Wrobel, BA Risk, HP Gunawardena, A Secrest, WJ Spitzer, L Xie, L Wang, X Chen, et al., in prep.). Integrative analysis of RNA and proteomics data has been explored in the literature and is examined in another ENCODE paper, highlighting translation of novel splice variants and expressed pseudogenes (Tian et al. 2004, Djebali et al. 2012). However, these data have not yet been applied to examine the empirical evidence for or against translation of computationally classified human long noncoding RNAs. A recent joint study of RNA and proteomic data in mouse revealed that protein levels and mRNA levels correlate such that RNA concentration is predictive of at least 40% of the variation in protein levels (Schwanhäusser et al. 2011). Since lncRNA genes are expressed, on average, at 4% of the level of protein-coding genes in the ENCODE cell lines (Derrien et al. 2012), we expect a similarly low level of expression for any putative protein(s) translated from lncRNAs. Therefore, to interrogate the translational competence of lncRNAs, we must account for the relative expression levels of these transcripts.
It has been shown that the quantity of detectable matches between MS/MS spectra and their corresponding peptides in a transcript correlate to protein abundance levels (Lu et al. 2007). This means that the number of detected peptide matches is an approximate surrogate for protein abundance (Liu et al. 2004; Vogel and Marcotte 2008). We used this characteristic to determine a calibration function that links mRNA expression abundance and protein expression abundance for the ENCODE data from K562 and GM12878. In our analysis, 21% of GENCODE v7 protein-coding genes are represented by at least one uniquely mapping peptide in any MS/MS sample, and the majority of those genes detected are expressed above 5 RPKMs in the whole-cell RNA-seq data (Harrow et al. 2012). We used these data, applying state-of-the-art machine-learning models to estimate the translational competence of transcripts as a function of RNA expression levels in various cellular compartments and RNA fractions. Using these models, we “regressed out” the expression-level effects to compare the translation competency of ostensibly noncoding transcripts to that of known mRNAs. We then manually examined each lncRNA for which we obtained empirical evidence of coding capacity. From these data, we determined the proportion of lncRNAs that appear to be truly “noncoding” in ENCODE Tier 1 cell lines, and we examined the exceptional cases where there was strong evidence of protein translation to determine whether these are indeed translated lncRNAs or simply misannotated mRNAs.
Results
The ENCODE Consortium has generated tandem mass spectrometry (MS/MS) data for the Tier-I ENCODE cell lines. This data has been mapped to the UCSC hg19 assembly in order to identify the best-fit genomic locus for each mass spectrum. The data comprised those peptides mapping within an estimated ≤10% FDR, based on decoy database searches. (See J Khatun, Y Yu, J Wrobel, BA Risk, HP Gunawardena, A Secrest, WJ Spitzer, L Xie, L Wang, X Chen, et al., in prep. for a detailed discussion of the proteogenomics data and mapping strategies.) Here, we further filtered the set to consider only peptides that mapped to unique genomic locations (i.e., unique peptide sequences). We queried each unique genomic peptide location for same-strand overlap with any exons of any of the 15,512 GENCODE lncRNA transcripts, all of which have been inferred from experimental (full-length clones or stranded RNA-seq) transcriptome data, and which summarily correspond to 9640 lncRNA genes.
Out of 350 distinct locations in which lncRNA-matching peptides were detected, there were 85 that uniquely mapped to exons of 111 GENCODE v7 lncRNA transcripts and nowhere else in any ORF in the human genome (see Methods). The 111 lncRNA transcripts are assignable to 69 distinct loci (GENCODE v7 genes) in the human genome. Of these uniquely mapping peptides, 26 peptides (from 10 loci) were “non-singleton hits,” in that either more than one peptide was detected mapping in-frame to the same lncRNA, or the same peptide was independently detected multiple times in independent MS/MS assays. The remaining 59 were singletons, detected only one time and in only one sample. We consider singleton hits to be potential evidence, but not confirmation of translation, and we consider only the non-singletons to correspond to detected instances of translation. This standard of at least technical replication is consistent with the data standards of the ENCODE Consortium (The ENCODE Project Consortium 2012).
RNA expression levels are correlated with the detectability of peptides in ENCODE MS/MS and RNA-seq data
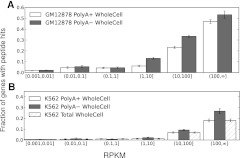
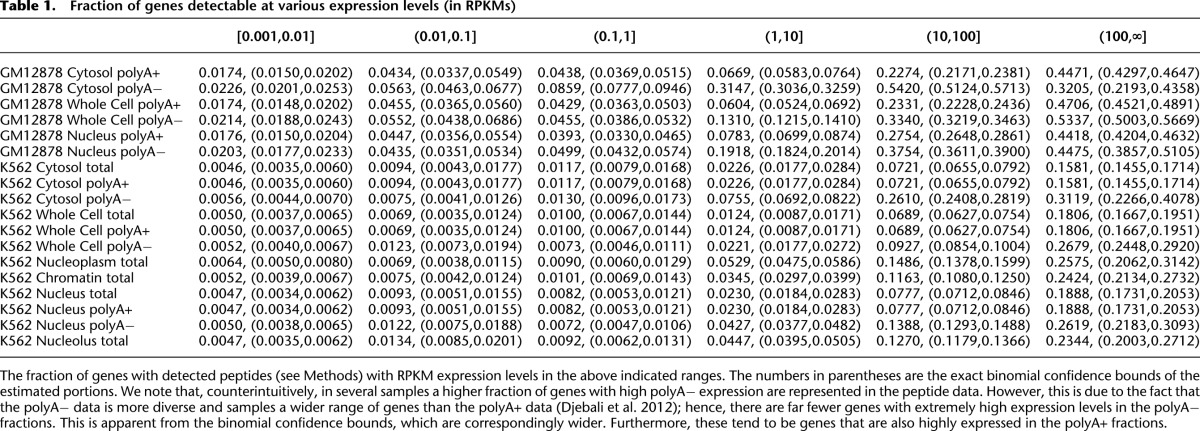
The MS/MS data generated by the ENCODE Consortium is nonquantitative, in the sense that the MS/MS protocol seeks only to detect the presence or absence of peptides, and does not attempt quantification of relative or absolute peptide levels (J Khatun, Y Yu, J Wrobel, BA Risk, HP Gunawardena, A Secrest, WJ Spitzer, L Xie, L Wang, X Chen, et al., in prep.). It was not clear a priori that previous results reporting the predictability of quantitative MS/MS data from mRNA levels would relate directly to this mode of MS/MS (Schwanhäusser et al. 2011). Furthermore, the RNA data collected by the ENCODE Consortium includes polyA± fractions in K562 and GM12878 nucleus, cytosol, and whole-cell samples, as well as total RNA in K562 nucleolus, nucleoplasm, and a sample purified from extracted chromatin, all in replicate, and all sequenced to a depth of more than 20 million reads (Djebali et al. 2012). In whole-cell RNA-seq data, lncRNA expression levels are on average 24-fold lower than mRNAs in polyA+ long RNA data, and 20-fold lower in polyA− long RNA-seq data. Figure 1, A and B shows that whole-cell RNA levels correlate with the rate of peptide detection (rank correlation, ρ ∼0.41 on average) (see also Table 1 and Supplemental Figs. 1, 2). Hence, it is clear that in order to understand the translational competency of RNAs, it is essential to normalize for expression level. Our ability to conduct this normalization is greatly enhanced by the richness of the RNA-seq data, with which we are able to study how differential RNA concentrations across cellular compartments and RNA fractions relate to the detectability of individual peptides by MS/MS.
Figure 1.
Expression levels are correlated with peptide detectability via MS/MS. Peptide detectability (y-axis) as a function of RNA expression levels (RPKM, x-axis) in GM12878 (A) and K562 (B) whole-cell RNA samples. We identified peptides for only 1% of genes expressed at RPKM <0.1, whereas we detected peptides for ∼40% of genes expressed above RPKM 100. In general, the likelihood of detection rises as expression level rises.
Table 1.
Fraction of genes detectable at various expression levels (in RPKMs)
Lowly expressed RNAs rarely produce detectable peptides
Table 1 and Figure 1 illustrate that lowly expressed RNAs produce few detectable peptides. Hence, the marginal effect of increased expression levels in any given fraction is an increase in the likelihood that at least one peptide will be detected in MS/MS. However, there may also be complex joint effects between the various measurements, e.g., a marginal increase in polyA− nuclear transcription coupled with a decrease in polyA+ cytosol transcription may actually decrease the likelihood of peptide detection. To directly model these joint effects, we utilized a machine-learning algorithm based on Random Forests that incorporates both boosting and model-selection procedures to produce sparse, and therefore robust and interpretable classification models known as RuleFit3 (Friedman and Popescu 2008; see Methods for details).
Long RNA relative expression levels across cellular compartments and polyA+ and polyA− fractions accurately and robustly predict peptide detectability in MS/MS
We fit machine-learning models to the GM12878 and K562 data independently and on independent sets of replicates of RNA-seq data in order to assess the reproducibility of our conclusions (see Methods). Our classifiers distinguish between genes with at least one uniquely mapping peptide and those with no uniquely mapping peptides. We were able to construct models with misclassification rates of 21% in K562 and 23% in GM12878 computed on held-out test-sets in both cell lines and on either collection of independent replicates (see Methods). Furthermore, when the models are trained on one set of replicates and tested on the other, the average misclassification rate rises only slightly, to 22% in K562 and 25% in GM12878. Hence, our models are both biologically and technically robust.
Because the cellular RNA fractions sequenced in RNA-seq by ENCODE differ between the two cell lines, we had more information to build our predictor in K562 than in GM12878. In order to assess the similarity of the imputed models in either cell line, we fit a model in K562 that utilized only the compartments available in GM12878 (six fractions: polyA± in Cytosol, Nucleus, and Whole Cell). We then evaluated the performance of the K562 model on the GM12878 data and vice versa. The probability of misclassification increased modestly, from 25% within GM12878 and 22% within K562 to 26% for the GM12878 model tested on K562 and 24% for the K562 model tested on GM12878. The failure of the model to predict correctly in 21%–26% of the cases is not surprising, since we would expect that, for at least some proteins, transcript abundance is strongly dependent on the stability of the mRNA transcript as well as protein degradation rates.
An expression pattern indicative of translational competence
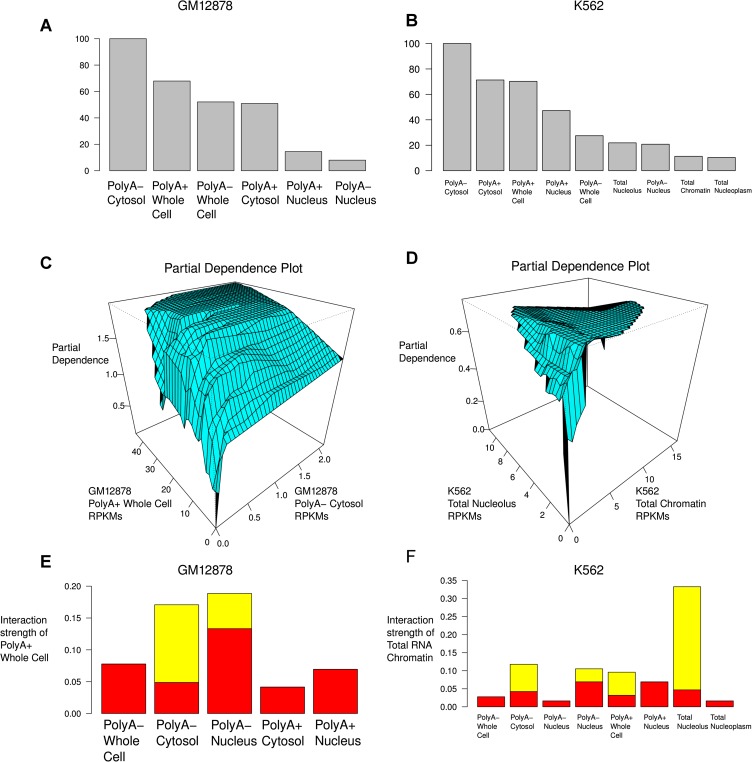
The most important predictor in either cell line (in both the K562 full model and the model using only GM12878 available data), is the polyA− Cytosol RNA fraction, and the direction of dependence is positive: higher polyA− Cytosol RNA levels correspond to an increased likelihood of detectable translation (Fig. 2; Supplemental Fig. 3). Although there is some substantial reordering of covariate importance down the rank-list, this has only a moderate effect on model performance between the two cell lines and, indeed, the precise order of covariates after polyA− Cytosol was unstable in K562 between biological replicates (see Supplemental Fig. 3). The usual interpretation of this sort of effect is colinearity between the variables: The various RNA fractions appear to provide some redundant information.
Figure 2.
Visualizing some of the properties of the model of RNA-seq and MS/MS data. (A,B) Relative importance of each of the covariates (RNA fractions). (C) Relative partial dependence of the likelihood of detecting at least one uniquely mapping peptide on the polyA+ Whole Cell and polyA− Cytosol fractions from GM12878. This is known as a “partial dependence plot.” We note that detectable polyA− Cytosol expression is nearly a prerequisite to detecting uniquely mapping peptides, even when polyA+ Whole Cell expression is extremely high. (D) Partial dependence plot for the total RNA nucleolus and chromatin fractions from K562. (E,F) “Interaction strength plots.” These show the relative importance of considering the dependence between pairs of covariates (fractions) in the overall predictive model. (Red bars) Standard deviation under the null of no association.
We studied the joint effect of the various cellular RNA fractions on our predictions (Fig. 2). In both cell lines, a number of pairwise interactions between the compartments were statistically significant (Fig. 2E,F; Supplemental Fig. 4). Here a statistically significant interaction is defined to be an interaction that is much stronger than would be expected if the expression levels were independent (Methods). Note that this does not mean that an interaction is important for predictive power. For instance, one of the most statistically significant interactions in K562 is total RNA Chromatin fraction with total RNA Nucleolus fraction, but the effect of increasing RPKM in the Nucleolus fraction is minimal (Fig. 2B,D,F): Very low values of expression in the Nucleolus imply a lack of detectable translation, and very high values slightly decrease the likelihood of translation, except when coupled with extremely high values in the Chromatin fraction.
The most stable interaction between both biological replicates and the two cell lines is observed between the polyA+ Whole Cell fractions and the polyA− Cytosol fractions in both cell lines (Fig. 2E,F). High polyA+ Whole Cell expression levels generally imply a high likelihood of detectable translation, except, interestingly, when little or no transcription is observed in the polyA− Cytosol fraction. The same dependence structure is observed between the polyA+ and polyA− Cytosol fractions, although it is not statistically significant in GM12878 (P > 0.01, permutation test). In contrast, neither the polyA+ nor polyA− nuclear samples (including, in K562, the total RNA Nucleoplasm sample) showed significant interactions.
An upper bound on the translational competency of lncRNAs in ENCODE cell lines GM12878 and K562
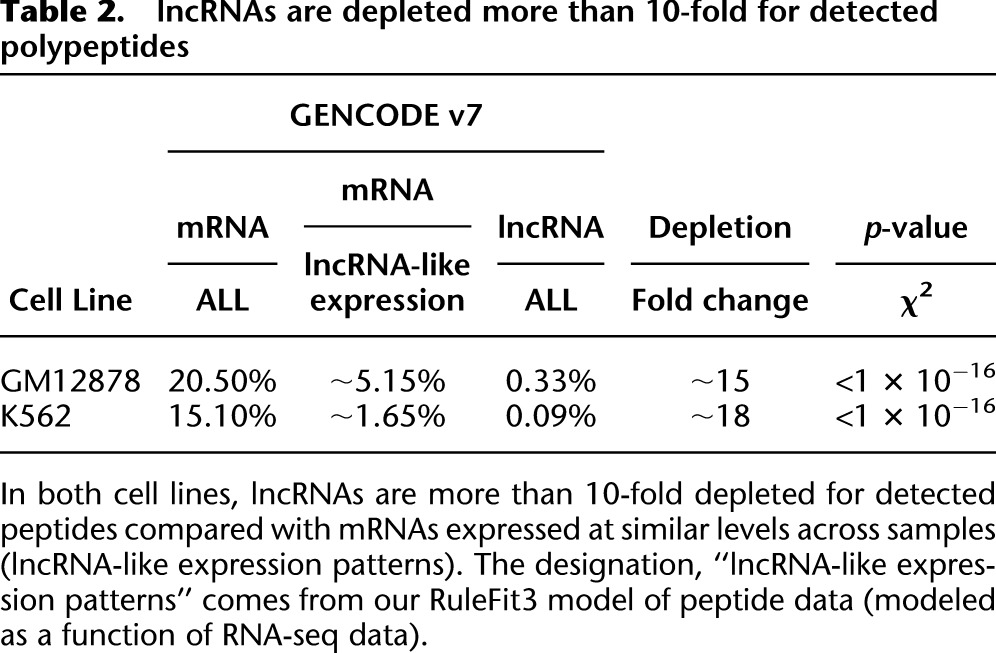
To estimate the fraction of lncRNAs that are translated in vivo, we compare the rate of detection of lncRNA translation with that for mRNAs expressed at similar levels. This is necessary, because otherwise any conclusions about the translational competency of lncRNAs would be subject to statistical confounding with levels and patterns of transcription. By interrogating our predictive models we can “regress out” transcriptional effects on the detectability of peptides (see Methods for details). For mRNAs with expression levels comparable to those of lncRNAs in GM12878, between 4.4% and 5.9% code for detected peptides (see Table 2). These numbers are directly comparable to the 0.33% of lncRNAs with detected translation in the same cell line (P < 10−16, two-sided χ2 test). For K562 we have detected translation for between 1.5% and 1.8% of mRNAs with lncRNA-consistent expression patterns and 0.09% of lncRNAs (P < 10−16, two-sided χ2 test). Hence, lncRNAs are likely between 13- and 20-fold depleted for detected translation given their expression patterns. We can obtain an upper bound for the fraction of GENCODE v7 lncRNAs translated in vivo by considering that we “should have detected” peptides corresponding to 100% of mRNAs. This is an upper bound because clearly not all mRNAs are expressed in these cells, and hence cannot produce peptides. Indeed, we have zero expression values across all compartments for 5.5% of GENCODE v7 mRNAs in GM12878 and 6.0% in K562, 7.1% are zero across all polyA+ samples in GM12878 and 9.2% in K562, and finally 60% are zero in at least one compartment in GM12878 and 51% in K562. Under the conservative model that all mRNAs were detectable, we infer that at least 92% of GENCODE v7 lncRNAs are untranslated in these cell lines.
Table 2.
lncRNAs are depleted more than 10-fold for detected polypeptides
The possibility of widespread translation of short polypeptides
We have demonstrated that lncRNAs are depleted for peptides that are detectable in our tandem MS/MS assay, but it remains possible that extremely short or rapidly degraded polypeptides exist that have gone undetected. The length of ORFs is largely uncorrelated with the number of peptides that we detected (ρ ∼0.08, r ∼0.005). In Supplemental Figure 5A we see that ORFs with detected peptides are enriched for long ORFs compared with the GENCODE v7 total ORF set (KS-2-sample test P < 10−16), but see that this effect is dominated by an enrichment for ORFs of more than 3K amino acids, rather than by a depletion of short ORFs (Supplemental Fig. 5B). However, the shortest GENCODE v7 ORF for which we have identified a peptide is 69 nucleotides, 23 amino acids; this implies an empirical size limit on detectability for our current data. Hence, it is possible that a population of short polypeptides has escaped the detection limits of our current MS/MS assay. Although we cannot rule out this possibility, we can provide an empirical bound: If the translation of short ORFs into stable polypeptides is widespread in the GENCODE v7 lncRNAs, then these likely encode polypeptides shorter than ∼23 amino acids in length.
Exhaustive manual reannotation of putatively translated lncRNAs
We performed in-depth visual manual annotation of each peptide uniquely mapping to a GENCODE v7 lncRNA by concurrent interpretation of the output from four tools: UCSC Genome Browser (Karolchik et al. 2009), UCSC BLAT (Kent 2002), NCBI BL2SEQ (Tatusova and Madden 1999) with TBLASTN functionality, and NCBI ORF Finder (Wheeler et al. 2003) (see Supplemental Material). We have stratified this annotation in three different ways: by locus, by individual transcript, and by peptide (see sheets 1 through 3, respectively, of Supplemental Data set 1). As a result of our annotation, we separated the 85 distinct peptides into three categories: 38 map to protein-coding genes that overlap GENCODE v7 lncRNA transcripts in the same orientation, 19 correspond to translatable lncRNA genes that do not overlap a coding transcript, and 28 are untranslatable in that they are located in-frame to, and downstream from, one or more stop codons in exons of lncRNA transcripts. All but 10 of these peptides were observed only once (see Supplemental Data set 1).
Protein-coding genes constitute the most abundant class of loci in this data set. In some cases, it is clear why the specific GENCODE transcript was annotated as an lncRNA, as the transcript's splicing was different from that of RefSeq isoforms of the same gene. However, in all cases, the genomic mapping of the peptide (see Methods) corresponds to a protein-coding exon shared by a RefSeq isoform and the lncRNA. Because MS/MS data provides only short peptides, not full-length protein sequences, we were unable in any of these cases to prove that the peptide was necessarily translated from the noncoding GENCODE transcript. Hence, we do not consider peptides that match exons shared by conventional coding and noncoding transcripts of the same protein-coding gene to be evidence of the translation of the differently spliced lncRNA. This conservative position is supported by the fact that 8 of 10 of the loci with shared coding exons give rise to non-singleton peptides. This means that 80% of the lncRNAs in our data set for which we have non-singleton peptides can be explained by shared coding exons and, hence, by translation of the canonical mRNA of the corresponding protein-coding gene.
We identified two lncRNA loci with non-singleton peptides (Fig. 3), which we refer to as putative cryptic mRNAs. In both cases, only one peptide per locus was discovered, and the peptide was identified in only one cell and compartment type, although on different runs of the machine (technical replication). The fact that only a single peptide would be detectable for a particular protein is not necessarily surprising; it may be that only one peptide is detectable by a mass spectrometer due to the dependence of the ENCODE protocol on enzymatic digestion (Rohrbough et al. 2006).
Figure 3.
Manual annotation of non-singleton peptides aligning to GENCODE lncRNA exons: case studies. (A–C) An untranslatable non-singleton peptide encoded downstream from an unprocessed pseudogene. (A) BL2SEQ TBLASTN peptide-to-lncRNA alignment. (B) NCBI ORF Finder view of the translation containing this peptide and upstream stops. (C) UCSC Genome Browser view of the peptide (red box). Direction is positive strand (data not shown). (D–F) An untranslatable non-singleton peptide encoded by a standalone lncRNA exon. (D) BL2SEQ TBLASTN peptide-to-lncRNA alignment. (E) NCBI ORF Finder view of the translation containing this peptide and upstream stops. (F) UCSC Genome Browser view of the peptide (red box) encoded on the negative strand of the genome.
The peptide SSLSILSCCAVIFSQAR (Fig. 3A–C) from the K562 nuclear fraction was exonically matched to the same-orientation GENCODE lncRNA transcript ENST00000454997.1. Encoded by the +1 ORF of this transcript, the peptide was untranslatable, owing to an in-frame upstream TAG and a lack of intervening start codons. This transcript is a part of a much larger transcriptional unit corresponding to the unprocessed non-protein-coding AFG3L2-like pseudogene, AFG3L1P. However, the peptide-matching exon is located downstream from the region containing the pseudogene's homology with its parental gene. We confirmed the lack of any matches to this peptide in human AFG3L2-family genes by BLASTP. There are no other protein-coding genes on the same (positive) genomic strand, in the region shown in the figure, that could produce transcripts accounting for this peptide's translation.
The peptide CIPLAFQRASK from the GM12878 nuclear fraction (Fig. 3D–F) was exonically matched to the same-orientation GENCODE lncRNA transcript ENST00000434292.1. Mapping to the +1 ORF of the lncRNA, this peptide was also untranslatable, as the cystein TGC codon is immediately preceded by a TGA stop. The peptide maps to the last exon of a standalone lncRNA gene with extremely limited cDNA and EST support. The splice acceptor site preceding the exon is conserved only in primates. Similarly to the previous case, there are no protein-coding genes in this region whose alternative or uncharacterized transcripts could account for this peptide.
These two cases represent the only data points in the lncRNA–proteogenomics intersection, where peptides matching bona fide lncRNAs rather than false GENCODE lncRNA annotations arising from GENCODE errors, were detected more than once. The remaining peptides, 59 singletons, were comprised of 27 untranslatable lncRNA genes, 18 translatable lncRNA genes, and 24 matches to overlapping protein-coding transcripts (Supplemental Data set 1). Figure 4, A–C highlights one of the 18 theoretically translatable lncRNA genes associated with a singleton peptide. The peptide TGLRSISQHLGERMR is clearly contained in-frame between an ATG start codon and a TGA stop codon (Fig. 4B). The peptide is entirely within exon 2 of the negative-strand GENCODE lncRNA shown, and cannot belong to either of the protein-coding genes in the locus as those genes are genomically on the positive strand. Figure 4, D and E, on the other hand, pinpoints one of the 24 matches to overlapping protein-coding transcripts. These matches are the result of rare GENCODE misannotations in release 7 (the GENCODE release used for this and all other ENCODE companion papers). Three in-frame translatable peptides (red, Fig. 4E) correspond to GENCODE transcripts that are given a GENCODE lncRNA biotype, but that are, logically, uncharacterized protein-coding splice variants of the protein-coding gene EMG1 that were missed by GENCODE's automated biotype assignment approach. These misannotations have been largely eliminated in subsequent releases of GENCODE.
Figure 4.
Manual annotation of translatable peptides aligning to GENCODE lncRNA exons: case studies. (A–C) A translatable singleton peptide encoded at a bona fide lncRNA locus. (A) BL2SEQ TBLASTN peptide-to-lncRNA alignment. (B) NCBI ORF Finder view of the translation containing this peptide (highlighted) including its furthest upstream ATG and its downstream stop (red rectangles). (C) UCSC Genome Browser view of the peptide (red box). Direction is negative strand. The singleton peptide is encoded by exon 2 of a GENCODE lncRNA that is divergently transcribed in the antisense orientation relative to the known gene CACNA1G. (D,E) A translatable non-singleton peptide traceable to a GENCODE misclassification of a protein-coding transcript of the EMG1 gene that had been assigned an lncRNA biotype. (D) Three peptides (red) that are in-frame to the EMG1 known protein (full-length shown) but are assigned to the GENCODE lncRNA ENST00000439543.2. (E) UCSC Genome Browser view of the peptide (red box). Direction is positive strand. The lncRNA is a noncoding transcript from the coding EMG1 locus. However, the peptides correspond to known coding exons of the EMG1 RefSeq, not solely to exons of the noncoding transcript. (E1) Peptide matches the common coding mRNA exon, not a unique exon of the lncRNA (this is true in all cases). (E2) GENCODE v7 lncRNA match.
Discussion
The integration of transcriptome and proteome data provides an empirical route to validate existing annotations of specific transcripts as endowed with, or devoid of, protein-coding capacity. Though mapping genomic loci against lncRNA sequences has provided new insights, we understand that whole-genome proteogenomic mapping—the method by which the loci were identified—has some limitations. Due to the target size, mapping against the entire human genomic sequence decreased the sensitivity of identifications at a constant FDR. Also, we did not consider peptides encoded by multiple exons across splice junctions, or post-translationally modified (PTM) peptides, in the mapping process. Approximately 25% of tryptic-digested peptides span exon boundaries, so not including them in a search can miss crucial identifications (Tanner et al. 2007; Castellana et al. 2008). Nor did we consider known SNPs or RNA-editing events as mapped by RNA-seq (Djebali et al. 2012). Though inclusion of trans-exons, PTM, SNPs, and RNA-editing events would increase sensitivity, their inclusion would also greatly increase the database size (>1000 times larger) and thereby decrease specificity. At present, searching a database of this magnitude is not feasible. This motivated our comprehensive manual annotation of the lncRNA hits. Because this analysis only utilized contiguous whole-genome six-frame translation, peptides that provide compelling evidence of lncRNA translation in our results could not be explained by splice junctions, SNPs, or RNA-editing events in protein-coding transcripts.
However, we determined that 38 of the 85 peptides we detected map to lncRNAs that entirely contain well-known coding ORFs, meaning that we have identified several misannotations in GENCODE. This highlights the need for continued HAVANA manual annotation. Our machine-learning approach enabled us to extrapolate an upper bound on the fraction of translated lncRNAs, and since that was around 8%, we conclude that the GENCODE effort has already produced an over 90% accurate disaggregation of coding from noncoding genes. Additional targeted proteomics data, especially data at higher sensitivities in these and other human cell lines and tissues, will be necessary to fully vet future annotations of the human genome. While the machine-learning approach predicts 8% of bona fide lncRNAs to be translated, our manual annotation of this particular peptide data set only identified translation of ∼0.4% of lncRNAs, including the few cases where the lncRNAs were mRNAs misannotated by GENCODE. Our model indicates that this discrepancy is a result of the depletion of low-abundance proteins in the mass spec data.
Our machine-learning approach also revealed a previously unknown positive correlation between translation and polyA− Cytosol RNA. The marginal positive effect of increased polyA− Cytosol expression is actually greater than the marginal effect of increased polyA+ Cytosol expression (Fig. 2). Indeed, polyA− Cytosol RNA level is the single most important covariate for prediction, although there are minor differences between the two cell lines that may be due to underlying differences in RNA processing and degradation efficiency. We note that K562 is a chronic myeloid leukemia cell line, while GM12878 is a normal but EBV-immortalized LCL. We hypothesize that this fraction may be measuring post-translational RNA processing, by which we mean the degradation and metabolism of transcripts after translation, resulting in polyA− fragments localized in the cytosol. This illustrates the importance of considering the direction of causality in statistically predictive models. Although the natural biological temptation is to think of RNA levels as “causing” or “influencing” protein levels (and therefore peptide detectability) the opposite may be true: Abundant proteins may be translated from high-abundance transcripts with correspondingly abundant degradation products. That the presence of such degradation products, if our hypothesis is correct, is a better indicator of translational competence than the polyA+ Cytosol fraction remains an intriguing subject for future study. Future experiments should investigate whether these degraded polyA− sequences, derived from previously translated RNAs in the cytosol, are nonfunctional, or whether they are stable due to post-cleavage 5′-capping (Otsuka et al. 2009) and may carry out additional roles in the cell, attesting to multifunctionality and interrelatedness of long and short RNAs.
We found two putative cryptic mRNAs in the GENCODE manually annotated lncRNAs, and this observation coupled with our machine-learning approach leads us to conclude that translations of cryptic mRNAs are rare in the proteome of the ENCODE cell lines K562 and GM12878. Moreover, our confidence in these two transcripts as cryptic mRNAs is tempered by the fact that the matched peptides were preceded by upstream stop codons, meaning that upstream splicing, ribosomal frameshifting, or RNA editing, not captured in public cDNA/EST data from these loci, would be necessary to explain the incidence and structure of any translatable transcripts from either locus.
We did not observe any evidence of pervasive translation, i.e., a process conceptually analogous to the accepted paradigm of pervasive transcription (Clark et al. 2011). This result stands in contrast to the work of Ingolia et al. (2011) in mouse ES cells. We note that MS/MS is dependent on the steady-state abundance of proteins, whereas Ribo-seq measures the instantaneous rate of translation, and tells us nothing about the longevity of any resulting polypeptides. It could be that the majority of the ubiquitous translation identified in Ingolia et al. (2011) corresponds to rapidly degraded molecules. Furthermore, high-throughput immunogenomic analysis of human peptides indicates that they can function as novel autoantigens (Larman et al. 2011). Ectopic lncRNAs may represent one source of ORFs giving rise to such autoantigenic peptides. Accordingly, disease specificity and immunogenicity of lncRNA-encoded peptides may warrant future investigation, and may enhance ENCODE's impact on clinical and translational medicine. Since we already have MS/MS in these cell lines, conducting Ribo-seq on matched samples will be a priority for future work.
In at least one case, we may have observed an example of Gouldian exaptation (Brosius and Gould 1992): AFG3L1P is a transcribed pseudogene that appears translated. By manual annotation, we conclusively mapped a repeatedly detected peptide to a novel downstream exon of the pseudogene transcriptional unit, an exon beyond the 3′end of the parental gene similarity region and absent in the parental gene locus. Hence, this may represent an instance of the exaptation of a transcribed pseudogene into novel protein-coding function, distinct from the parental gene (Fig. 3).
The vast majority of the peptides we detected were singletons and, hence, likely correspond to either false positives or rare proteins (Greenbaum et al. 2003; Lu et al. 2007). A possible explanation for some of the singleton lncRNA-encoded peptides may be the pioneer round of translation by ribosomes at the nuclear periphery. Many lncRNAs, including an abundant subset of endogenous antisense lncRNAs, are nuclear (Kiyosawa et al. 2005). Export of an lncRNA into the cytoplasm could subject it to the same nonsense-mediated decay machinery as a mRNA. This machinery entails ribosomal proofreading, with a pioneer round of translation by ribosomes located just outside of the nucleus, not Golgi/ER. RNAs with multiple post-stop splice junctions are targeted for degradation (Hwang et al. 2010). However, it may also be that these proofreading transcripts are insufficiently abundant to be detected by MS/MS. Another intriguing possibility is that we are observing the outcomes of ribosomal frame shifting, RNA editing, or splicing not yet identified by ENCODE or GENCODE. This possibility is supported by the observation that many of our singleton peptides are preceded by an in-frame upstream stop codon and lack an in-frame ATG codon. Of course, these may also simply be false positives in the MS/MS data, and/or the products of ectopic translation. Exploring these possibilities will be the topic of future work.
This is the first study to empirically evaluate the translational competence of human lncRNAs. An important caveat of proteogenomic interrogations of ENCODE cell line data sets is that lncRNA translation may be different in vivo in organismal tissues, and may be highly atypical (either suppressed or unusually frequent) in the two cancer cell lines that we studied here. Hence, we expect that many additional tissues will be needed in order to understand the protein-coding capacity of human transcripts. A secondary concern is the incomplete nature of the GENCODE lncRNA data set. Mining raw cDNA and EST data for lncRNA genes lacking GENCODE counterparts should create a more inclusive lncRNA reference data set for future proteogenomic studies of the complete human lncRNAome. Mass spectrometric approaches need to be substantially improved in order to accurately detect and quantify peptide abundance from human tissue samples and primary cell cultures in addition to cancer cell lines.
Methods
Quantification of transcription rates at the level of whole genes
RNA-seq data was obtained from the ENCODE RNA Dashboard (http://genome.crg.es/encode_RNA_dashboard/). Quantifications for GENCODE v7 genes were derived using the Flux Capacitor algorithm (http://flux.sammeth.net/) and these quantities were obtained from Supplemental Material in Djebali et al. (2012). In total, we utilized gene-by-gene quantifications for biological replicates of samples GM12878: poly(A)− Whole Cell, Cytosol, Nuclear; and poly(A)+ Whole Cell, Cytosol, Nuclear; and K562: Total RNA Nucleoplasm, Nucleolus, Chromatin; poly(A)− Whole Cell, Cytosol, Nuclear; and poly(A)+ Whole Cell, Cytosol, Nuclear.
Proteomic analysis
We performed shotgun proteomic analyses for two ENCODE Tier1 cell lines K562 and GM12878. Subcellular fractionation was performed on both cell lines following a common protocol, producing four fractions for each cell line: nuclear, mitochondrial, cytosolic, and membrane fractions (Cox and Emili 2006). SDS-PAGE separation and in-gel digestion was then performed as described by Shevchenko et al. (2006). The collected protein fractions were further processed using filter-aided sample preparation (FASP) (Wisniewski et al. 2009) or the GOFAST method (Y Yu, L Xie, HP Gunawardena, J Khatun, C Maier, M Leerkes, M Giddings, X Chen, in prep.). Tandem mass spectrometry data were produced using an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) (J Khatun, Y Yu, J Wrobel, BA Risk, HP Gunawardena, A Secrest, WJ Spitzer, L Xie, L Wang, X Chen, et al., in prep.). We obtained eight different sets of data, totaling 998,570 high-resolution MS/MS spectra. To perform proteogenomic mapping, we used two proteogenomic mapping software packages, Peppy (http://geneffects.com/Peppy) and Genome Fingerprint Scanning (GFS). HMM_Score was used in both programs to match and score each MS/MS spectrum to the best-fit peptide sequence. A 6-frame translation and proteolytic digestion of the whole human genome (UCSC hg19, 2009) was used for proteogenomic mapping. See also Supplemental Methods and Legends.
Integrating RNA-seq expression data with the proteogenomic mappings—a machine learning approach
Machine learning was conducted using the RuleFit3 package (Friedman and Popescu 2008; http://www-stat.stanford.edu/∼jhf/r-rulefit/rulefit3/R_RuleFit3.html). RuleFit3 classification models are composed of several decision trees that make weighted contributions to the final classification rule applied to each observation. The package was used in classification mode with the option tree.size=6, and all other options at default values. We fit our predictors to distinguish between genes with at least one uniquely mapping peptide and genes with none; however, using a threshold of two uniquely mapping peptides did not qualitatively change any results. Due to the sparsity of the MS/MS data, to increase the sensitivity of our classifier, we sampled training sets such that 50% of observations corresponded to RNAs with at least one peptide match observed in the uniquely mapping peptide data, and the other 50% without any uniquely mapping peptides. Predictions were then tested on held-out test data in the usual way. Additionally, the RuleFit3 package gives the predicted values as a numeric score whose absolute value reflects confidence that its sign is the same as that of the response. We used a fivefold cross-validation to select the optimal cutoff for these scores to further improve the error rates (maximizing the sum of true positive and true negative classification rates).
We further validated the robustness of our classification approach by fitting on one set of replicates of the RNA-seq experiments, and then validating on a held-out test set in a nonoverlapping collection of replicates. Because we do not have biological replicates of the MS/MS data at this time, we could not adopt the same approach with our response variable.
Obtaining plots of the relative importance of the RNA fractions for predictive power
Variable importance (the contribution of each covariate to the overall predictive power of the fitted model) is computed as in Friedman and Popescu (2008), (with an overview in Gerstein et al. 2012). In general, the relative importance of a covariate quantifies its usefulness for the prediction problem at hand. We assessed the importance of each RNA fraction in both cell lines and in nonoverlapping collections of biological replicates. Only results that were stable under both biological replication and between cell lines were reported. The function “varimp” was used in its default settings to produce the variable importance bar plots in Figure 2, A and B.
Assessing the interactions between RNA fractions
We assessed the statistical significance of pairwise covariate interactions using the RuleFit3 function “twovarint,” which utilizes a bootstrapped null interaction model to impute the usefulness of the covariates for prediction when only their marginal distributions are known. This is described in detail in Friedman and Popescu (2008). For statistically significant interactions, as in Figure 2, we used the function “pairplot” to visualize the nature of the dependence of our classifications on pairs of covariates. That is, pairplot cannot be used to estimate the marginal increase in likelihood of classification as a function of a small increase in either covariate (that is beyond the capabilities of the software), but it can provide a visualization of the nature of the dependence of predictions on the covariates; hence, these plots can be considered “scale free,” only the shape of the 2D curve is relevant. For instance, a pairplot following the plane z = x + y would indicate linear dependence on the covariates. In the current study, most dependencies are approximately monotonic, but none are linear.
Estimating the total fraction of translated lncRNAs in K562 and GM12878—identifying mRNAs with expression patterns similar to lncRNAs
The RuleFit3 approach utilizes a combination of linear models and “rules” to conduct classification and prediction. Rules are simply decision trees. Hence, the program functions like a version of Random Forests (Breiman 2001) with the addendum of recent boosting, regularization, and dimension reduction techniques. Although in principle the models that we fit could have included linear terms, they did not, they included only decision trees. There is also an option in the software “rulefit,” model.type=“rules,” which we could have used to enforce this behavior. This is significant because it permitted us to take a Random Forests approach to clustering. That is, as in Breiman(2001), we collected protein-coding genes that fell into the same terminal nodes in decision trees as lncRNA genes. In particular, we defined the “signature” of a gene to be a binary string encoding every terminal node into which the gene fell in the fitted model. We called the expression pattern of two genes “similar” when the binary strings were identical. This clustered 90% of lncRNA genes, on average, with at least one coding gene. A complete fitted rule set for each cell line can be found in Supplemental Data File 2.
We also attempted to fit more standard regression models, such as logistic regression, but the fit to the data was poor. For comprehensive details and additional Methods see Supplemental Data File 3.
Data access
Bed files for all proteogenomic mapping results uploaded to the UCSC Data Coordination Center (DCC) can be accessed via http://genome.ucsc.edu/cgi-bin/hgTrackUi?db=hg19&g=wgEncodeUncBsuProt. All proteomic data uploaded to Proteome Commons in DTA format and their accession numbers are available in the Supplemental Material and at http://giddingslab.org/data/encode/proteome-commons. Raw RNAseq reads can be accessed from the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE30567. Additional detailed methods for RNA sequencing can be obtained in the Production Documents under “CSHL Long RNA-seq” at http://genome.ucsc.edu/ENCODE/downloads.html. The lncRNA annotations can be found on the Guigo group website: http://big.crg.cat/bioinformatics_and_genomics/lncrna. Finally, the GENCODE annotation is freely available at http://www.gencodegenes.org. All data is accessible via the ENCODE DCC at http://genome.ucsc.edu/ENCODE/. All code developed for the analyses presented here can be found in Supplemental Data File 4.
Acknowledgments
B.B. is grateful for the support of the Rosztoczy Foundation. J.B.B. thanks Roger Hoskins, Susan Celniker, Haiyan Huang, and Nathan Boley for useful conversations.
Author contributions: B.B. and J.B.B. conducted all machine-learning analyses and were advised by A.K.; H.J. handled general bioinformatics; L.L. led manual annotation efforts and was assisted by E.W. and W.G.; J.K. conducted all peptide mappings and MS analysis and was assisted by B.R. A.K. and P.B. provided statistical consulting, and M.C.G. directed the analysis of the MS data. B.D.S. and K.K. produced synthetic peptides used for MS/MS confirmation. Mass spectrometry data generation and verification was done by H.P.G., Y.Y., and X.C. Sample generation (cell culture work) was done by L.X. J.B.B. and L.L. conceived and organized the project.
Footnotes
[Supplemental material is available for this article.]
Article and supplemental material are at http://www.genome.org/cgi/doi/10.1101/gr.134767.111.
Freely available online through the Genome Research Open Access option.
References
- Affymetrix/Cold Spring Harbor Laboratory ENCODE Transcriptome Project 2009. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457: 1028–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L 2001. Random Forests. Mach Learn 45: 5–32 [Google Scholar]
- Brosius J, Gould SJ 1992. On “genomenclature”: A comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA.” Proc Natl Acad Sci 89: 10706–10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP 2008. Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci 105: 21034–21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. 2011. The reality of pervasive transcription. PLoS Biol 9: e1000625 doi: 10.1371/journal.pbio.1000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B, Emili A 2006. Tissue subcellular fractionation and protein extraction for use in mass-spectrometry-based proteomics. Nat Protoc 1: 1872–1878 [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res (this issue). doi: 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Gascoigne DK, Mattick JS 2011. The evolution of RNAs with multiple functions. Biochimie 93: 2013–2018 [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi AM, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JH, Popescu BE 2008. Predictive learning via rule ensembles. Ann Appl Stat. 2: 916–954 [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forné T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. 2009. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 136: 3413–3421 [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan K-K, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D, Colangelo C, Williams K, Gerstein M 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol 4: 117 doi: 10.1186/gb-2003-4-9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. 2010. Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res (this issue). doi: 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube F, Guo J, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA, Blanchard AA, Wang X, Deng G, Myal Y, et al. 2006. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol 25: 418–428 [DOI] [PubMed] [Google Scholar]
- Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE 2010. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell 39: 396–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT 2011. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146: 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Kent WJ 2009. The UCSC Genome Browser. Curr Protoc Bioinformatics 28: 1.4.1–1.4.26 [DOI] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT–the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K 2005. Disclosing hidden transcripts: Mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res 15: 463–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y 2010. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329: 336–339 [DOI] [PubMed] [Google Scholar]
- Larman HB, Zhao Z, Laserson U, Li MZ, Ciccia A, Gakidis MA, Church GM, Kesari S, Leproust EM, Solimini NL, et al. 2011. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol 29: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovich L, Johnson R, Lin C-Y 2010. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochim Biophys Acta 1799: 597–615 [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR 2004. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76: 4193–4201 [DOI] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM 2007. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol 25: 117–124 [DOI] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, Schoenberg DR 2009. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol 29: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T 2007. Dissecting the human BDNF locus: Bidirectional transcription, complex splicing, and multiple promoters. Genomics 90: 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrbough JG, Breci L, Merchant N, Miller S, Haynes PA 2006. Verification of single-peptide protein identifications by the application of complementary database search algorithms. J Biomol Tech 17: 327–332 [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L 2010. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 16: 324–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Tanner S, Shen Z, Ng J, Florea L, Guigó R, Briggs SP, Bafna V 2007. Improving gene annotation using peptide mass spectrometry. Genome Res 17: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL 1999. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174: 247–250 [DOI] [PubMed] [Google Scholar]
- Tay SK, Blythe J, Lipovich L 2009. Global discovery of primate-specific genes in the human genome. Proc Natl Acad Sci 106: 12019–12024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, et al. 2004. Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol Cell Proteomics 3: 960–969 [DOI] [PubMed] [Google Scholar]
- Ulveling D, Francastel C, Hubé F 2011. Identification of potentially new bifunctional RNA based on genome-wide data-mining of alternative splicing events. Biochimie 93: 2024–2027 [DOI] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM 2008. Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data. Nat Protoc 3: 1444–1451 [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, Schuler GD, Schriml LM, Sequeira E, Tatusova TA, et al. 2003. Database resources of the National Center for Biotechnology. Nucleic Acids Res 31: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M 2009. Universal sample preparation method for proteome analysis. Nat Methods 6: 359–362 [DOI] [PubMed] [Google Scholar]