Abstract
Statistical models have been used to quantify the relationship between gene expression and transcription factor (TF) binding signals. Here we apply the models to the large-scale data generated by the ENCODE project to study transcriptional regulation by TFs. Our results reveal a notable difference in the prediction accuracy of expression levels of transcription start sites (TSSs) captured by different technologies and RNA extraction protocols. In general, the expression levels of TSSs with high CpG content are more predictable than those with low CpG content. For genes with alternative TSSs, the expression levels of downstream TSSs are more predictable than those of the upstream ones. Different TF categories and specific TFs vary substantially in their contributions to predicting expression. Between two cell lines, the differential expression of TSS can be precisely reflected by the difference of TF-binding signals in a quantitative manner, arguing against the conventional on-and-off model of TF binding. Finally, we explore the relationships between TF-binding signals and other chromatin features such as histone modifications and DNase hypersensitivity for determining expression. The models imply that these features regulate transcription in a highly coordinated manner.
Transcription factors (TFs) are critical for the transcriptional regulation of gene expression (Takahashi and Yamanaka 2006; Vaquerizas et al. 2009). In humans, they represent the largest family of proteins, accounting for around 10% of genes (Babu et al. 2004). There are two types of TFs: general and sequence-specific. The former TFs act cooperatively with RNA polymerase II and are ubiquitously involved in the transcription of a large fraction of genes (Lee and Young 2000). The latter TFs bind specific subsets of target genes, leading to distinct spatiotemporal patterns of gene expression (Kadonaga 2004). Although systematic gene expression quantification has been available for a decade from microarray experiments (Schena et al. 1995), only recently has the genome-wide identification of TF-binding sites become possible, owing to the development of chromatin immunoprecipitation followed by microarray (ChIP-chip) and sequencing (ChIP-seq) technologies (Ren et al. 2000; Johnson et al. 2007).
In several previous studies, statistical models were constructed to study the regulatory functions of TF on gene expression based on the gene expression and TF-binding data (Ouyang et al. 2009; Cheng and Gerstein 2011). These studies showed that TF-binding signals around the transcription start sites (TSSs) of genes are predictive of gene expression levels with fairly high accuracy. But these studies have the following limitations: First, estimates of gene expression have relied on probes (microarray) or sequence reads (RNA-seq) spread across a gene, possibly across multiple unknown isoforms of that gene. It is often difficult to accurately determine the expression level of each transcript based on such data, which limits the predictive power of these models. Second, the numbers of TFs used in these models were quite limited and perhaps not representative (12 TFs in both studies). Third, the TF-binding data were available for only a single cell line, so it was not possible to investigate the specificity of the models by examining the degree to which differential TF binding between two conditions affects differential expression of genes in those conditions.
Fortunately, the ENCODE project has generated a large amount of data that enables us to overcome all of these limitations (The ENCODE Project Consortium 2012). In addition to expression quantification of transcripts from RNA-seq (Wang et al. 2009) and RNA–PET experiments (Ruan et al. 2007; JO Luo, JM Fullwood, YJ Koh, L Veeravalli, S Djebali, R Guigo, C Davis, T Gingeras, A Shahab, Y Ruan, et al., in prep.), the consortium has also used Cap Analysis of Gene Expression (CAGE) to quantify the expression levels of >130,000 TSSs (annotated by GENCODE). In contrast to RNA-seq, CAGE is a technology that directly measures the transcriptional signal at the TSS of genes (Shiraki et al. 2003; T Lassmann, P Carninci, in prep.). In total, the expression data include 267 expression profiles, representing RNA samples in multiple cell lines that are extracted from different cellular components using different RNA extraction protocols. Moreover, the ENCODE project has generated >400 TF-binding profiles for more than 120 human TFs or transcription related proteins, including both general and sequence-specific TFs (Gerstein et al. 2012). The completeness of the ENCODE data enable us to study the transcriptional regulation of TFs more accurately and comprehensively.
In this study, we apply our previously developed model (Cheng and Gerstein 2011) to the ENCODE data to better understand transcriptional regulation. We quantify the relationship between TF-binding signals around TSS and the expression level of TSS measured by different technologies, and we study the relative contribution of different TF categories and of individual TFs. We compare the regulatory difference between different types of TSS. We also show that differential expression of genes can be determined largely by the differential binding of TFs. Finally, we explore how TFs coordinate with other chromatin features (e.g., chromatin modifications and DNase hypersensitivity) to regulate transcription.
Results
Relating TF-binding signals to gene expression levels
The ENCODE project has performed a large-scale analysis of gene expression and transcription factor (TF) binding in multiple human cell lines. In the gene expression data, the transcription levels of ∼130,000 GENCODE-annotated TSSs were quantified using three different technologies: cap analysis of gene expression (CAGE) RNA–PET, and RNA-seq in multiple cellular components, and with several different RNA extraction protocols. Meanwhile, the binding sites of ∼120 TFs in the human genome were determined by ChIP-seq experiments (Gerstein et al. 2012). These data sets enable us to investigate the relationship between TF binding and gene expression in a systematic and quantitative manner.
We have previously shown in mouse that the expression levels of transcripts can be accurately reflected by TF-binding signals in their TSS regions (Cheng and Gerstein 2011). In this study, we aim at validating this result using data from CAGE that directly measures the expression levels of TSSs, and to investigate the influences of different technologies and RNA extraction methods on TSS expression quantification. We constructed models to quantify the ability of TF-binding signals to statistically predict the expression levels of promoters. Unless stated otherwise, we represent the binding strength of a TF in a promoter by its average ChIP-seq signal in a 100-bp region centered on the TSS. We combined the TSS expression data with TF-binding data and then divided them into a training data set and a test data set. A model was trained on the training data set and then applied to the test data to predict the expression levels of TSSs (see Methods for details). The relationship between expression and TF binding was quantified by the correlation between predicted and actual expression levels (R), or by the coefficient of determination (R2), the percentage of variance of gene expression explained by the model. In order to evaluate the stability of our results, we built models using four different machine-learning methods: random forest (RF), support vector regression (SVR), multivariate adaptive regression splines (MARS), and multiple linear regression (MLR). Performance of the first three methods was roughly comparable, and was better than MLR, implying a nonlinear relationship between TF binding and TSS expression (Supplemental Fig. S1). In this article, to simplify presentation we focus on results from the RF method for models with multiple predictors and the SVR method for models with a single predictor (see Methods for details). Results from different methods are highly consistent and lead to the same conclusions, e.g., the relative importance of different TFs for predicting gene expression.
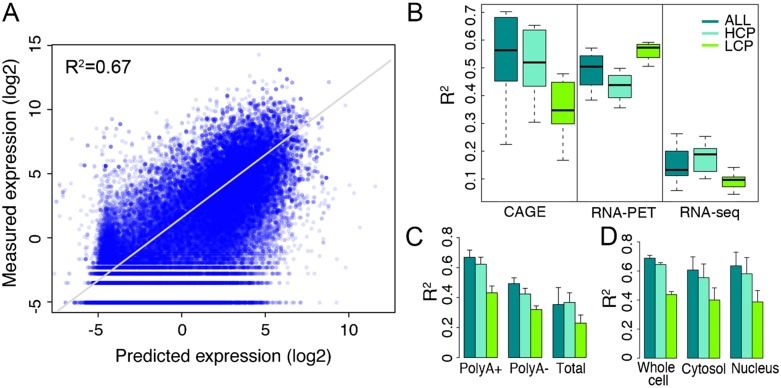
Our results indicate that TF-binding signals around the TSS are informative for “predicting” their expression levels. For example, Figure 1A shows the consistency between predicted and actual expression levels of TSSs measured by CAGE of whole-cell Poly A+ RNA in K562 cells. TF binding accounts for at least 67% of the variance of expression levels (R2 = 0.67). In total, there are 267 promoter expression profiles representing 12 different human cell lines in our data set. The performance of the model is not directly comparable between cell lines, because different numbers of TF-binding data sets are available for different cell lines. Since the most complete data were from K562, we chose this cell line for further analysis. The expression levels of a large fraction of TSSs (∼50% on average) are not detected (RPKM = 0) in any of these K562 data sets. Thus, we developed a more complicated model that first classifies TSSs into expressed and nonexpressed categories and then adopts a regression model to predict the expression levels for the expressed TSSs only (The ENCODE Project Consortium 2012). When applied to the TF data, this model achieves results very consistent with the methods without a classification step in terms of the R2 value and the relative importance of different TFs. We therefore focus on the classification-free models in the rest of this analysis.
Figure 1.
Accuracy of the TF model for predicting TSS expression levels. (A) Consistency of predicted values with expression levels measured by CAGE in Poly A+ RNA samples extracted from whole cells. (B) Comparison of predictive accuracies of the TF model for expression data generated by three different technologies: CAGE, RNA–PET, and RNA-seq. (C) Comparison of predictive accuracies of the TF model for expression data from three different RNA extraction protocols: Poly A+, Poly A-, and total RNA. (D) Comparison of predictive accuracies of the TF model for expression data in different cellular components. In B–D, only data sets from K562 are used. The binding signals of 40 TFSSs are used as predictors. HCP and LCP are high and low CpG content promoters, respectively. Separate models are constructed for ALL, HCP, and LCP categories.
We compared the impact of different technologies, cellular components, and RNA extraction protocols on the “prediction accuracy” of models. We used the binding signals of 40 TFs to predict each of the 57 K562 expression profiles and compared the resulting accuracies in terms of R2 values. We found that the highest predictive accuracy was achieved for TSS expression data from CAGE (Fig. 1B). RNA-seq, as a method for quantifying expression at the transcript level, seems unable to precisely capture the expression levels of TSSs. Furthermore, prediction accuracies vary significantly among different RNA extraction protocols with Poly A+ > Poly A- > Total RNA (Fig. 1C). No obvious difference was observed between the prediction accuracies for expression data from different cellular components (Fig. 1D). It can also be seen that expression levels of promoters with high CpG content (HCP) are easier to predict than those with low CpG content (LCP). We will investigate the effect of CpG content on gene expression in more detail below.
Contribution of different TFs to the regulation of gene expression
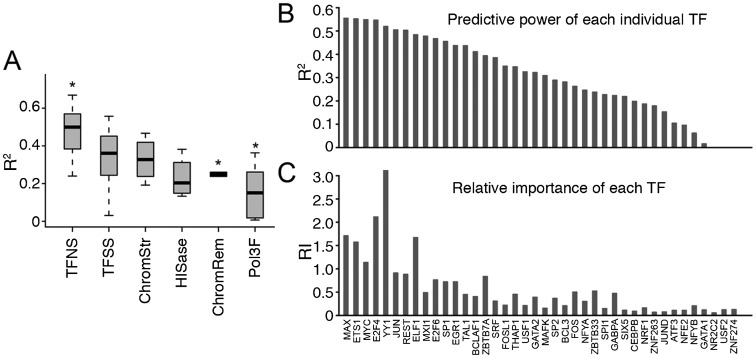
The ENCODE project has generated ChIP-seq data for a large number of DNA-binding proteins. These proteins can be roughly classified into six different categories, including sequence-specific TFs (TFSS), general or nonspecific TFs (TFNS), chromatin structure factors (ChromStr), chromatin remodeling factors (ChromRem), histone methyltransferases (HISase), and Pol3-associated factors (Pol3F) (Supplemental Table S1). For each TF, we constructed a model of expression prediction using it as the single predictor. We compared their capability for predicting expression levels of TSSs in K562 (e.g., whole cell poly A+ RNA). We found that individually, TFs in the TFNS category were significantly more predictive than proteins in other groups (P = 0.004, t-test), whereas proteins from the ChromRem and Pol3F categories were significantly less predictive (P = 0.0004 and P = 0.006, respectively, t-test) (Fig. 2A; Supplemental Table S1). TFs in the TFNS category are implicated in general transcriptional regulation. For instance, the TATA-binding protein (TBP) is a common subunit required by all three of the human RNA polymerases, I, II, and III (Kornberg 2007). Binding of these general TFs is essential for transcriptional initiation of most promoters, and therefore it makes sense that their binding signals have the highest predictive capabilities for gene expression. In contrast, it is expected that TFs in the Pol3F category are, in general, less predictive, because RNA Pol III is involved in initiating transcription of only a small fraction of promoters.
Figure 2.
The capabilities of different TFs to predict TSS expression level. (A) Comparison of the predictive accuracies of individual DNA-binding proteins in six different categories. (*) Indicates that the predictive powers of TFs in a corresponding category are significantly different from those of the other TFs. (B) The predictive accuracy of using each individual TFSS as the single predictor. (C) The relative importance of each TFSS in the Random Forest model. The calculation is based on the CAGE expression data in Poly A+ RNA samples extracted from K562 whole cells. Note that TFSS labels are shared by B and C.
For each of the 40 TFSSs assayed in K562, we investigated its individual predictive power in a degenerate model that uses this TF as a single predictor (Fig. 2B). Strikingly, each TF alone can predict TSS expression levels of all genes with fairly high accuracy. As shown, the binding signal of MAX alone can explain 55% of the variance in expression of all TSS, which is only ∼12% lower than the variance explained by the full model (67%). The R2 in a degenerate model indicates the power of a TF for predicting expression individually. In the full model, the relative importance of TFs for predicting the expression levels of promoters is roughly reflected by their Relative Importance score (RI score, see Methods) (Fig. 2C). We use the standard RI metrics of different machine learning methods, which indicate the contribution of TFs after considering their intercorrelations in a model, and thus provide complementary information to the individual predictive power. Specifically, in a random forest model the RI of a TF is calculated as the increase of prediction error (%IncMSE) when binding data for this TF is permuted. In general, highly predictive TFs have more binding peaks, particularly in the TSS proximal regions. We found in the full model that the top five most important TFs in K562 are YY1, E2F4, MYC, MAX, and ELF1. We also examined the effect of TF–TF interaction on the predictive accuracy. Our results indicated that including interaction terms in the model did not lead to further improvement.
In principle, we would expect the binding of transcriptional activators to positively correlate with gene expression levels, and a negative correlation for transcriptional repressors. Surprisingly, we observe a positive correlation between the expression level of TSSs and the binding signal of most ENCODE TFs (Supplemental Table S2). For instance, the binding of REST, which represses neuronal genes in non-neuronal tissues (Schoenherr and Anderson 1995), is positively correlated with gene expression (ρ = 0.70). This implies that TF occupancy alone may not be sufficient to determine the function of a TF at a locus, as has been demonstrated in a recent study (Lickwar et al. 2012). For many TFs, their binding signal in a DNA region may simply reflect the accessibility of the local chromatin structure.
The effect of promoter CpG content on gene expression
The CpG content of promoters in eukaryotes has been shaped by DNA methylation (Deaton and Bird 2011). Cytosines in CpG dinucleotides can be methylated to form 5-methylcytosine, which undergoes a high rate of mutation into uracil. Meanwhile, methylation of CpG sites within the promoter is a critical regulatory mechanism to inactivate a gene (Pai et al. 2011). As a consequence, genes repressed in germ-line cells or early developmental stages tend to have lower CpG content in their promoters (Deaton and Bird 2011). When genes are repressed by methylation of CpG cytosines in their promoters, those cytosines tend to mutate to uracil, so there is a sort of “evolutionary arms race” between CpG-based repression and mutation to uracil that lowers CpG content.
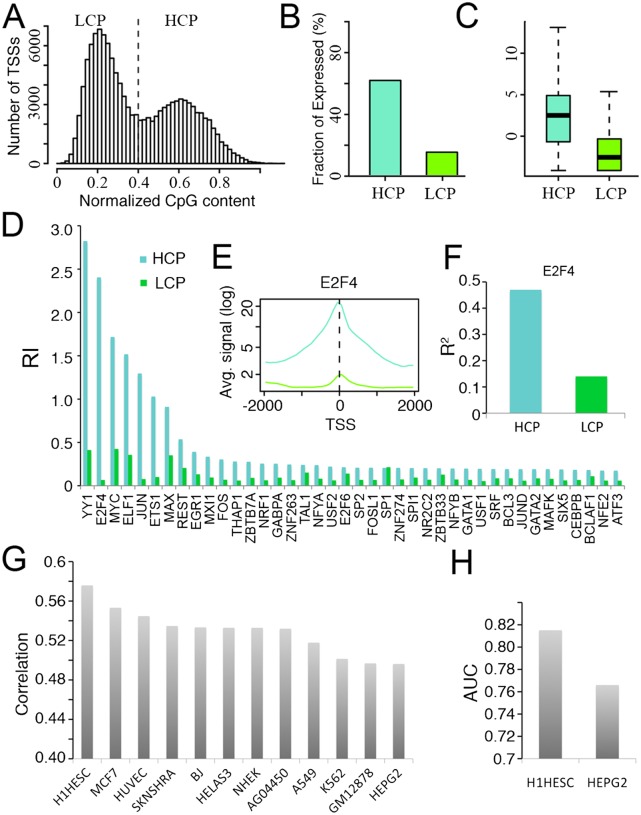
We calculated normalized CpG content for all GENCODE promoters (see Methods). As shown in Figure 3A, normalized CpG content follows a bimodal distribution, based on which we divided promoters into two classes: high CpG promoters (HCP) and low CpG promoters (LCP). HCP promoters are more highly expressed than LCP promoters as measured by CAGE experiments in all expression profiles. For example, in K562 whole-cell Poly A+ RNA, 62% of HCP promoters are expressed, while only 15.5% of LCP promoters are expressed (Fig. 3B). Furthermore, among the expressed TSSs, the expression level of HCP promoters is significantly higher than that of LCP promoters (Fig. 3C).
Figure 3.
The relationship between promoter CpG content and expression level. (A) The distribution of normalized CpG content for all human GENCODE TSSs. (B) The fraction of expressed TSSs in HCPs and LCPs. (C) The distributions of expression levels of expressed HCPs and LCPs. (D) The relative importance of each TF in the HCP- and LCP-specific models. (E) The aggregated binding signals of E2F4 around the TSS of HCPs and LCPs. (F) The predictive accuracies of HCP- and LCP-specific models using E2F4 as the single predictor. (G) The Spearman correlation coefficients between normalized CpG content and expression levels in different cell lines (CAGE data for Poly A+ RNA from whole cells). (H) The accuracies of using normalized CpG content to classify expressed and nonexpressed promoters in H1HESC and HEPG2. In B–F, the CAGE expression data for RNA extracted from K562 whole cells are used.
We have shown in Figure 1 that the expression levels of HCP promoters are easier to predict than those of LCP promoters. We further compared the relative importance of each TF for predicting the expression levels of HCP and LCP promoters. As shown in Figure 3D, the relative importance (RI) scores for the HCP model are generally greater than those for the LCP model, consistent with the higher predictive accuracy of the HCP model. The orders of the RI scores in the two models are roughly consistent, with the exception of E2F4. In the HCP model, E2F4 is the second most important TF, but in the LCP model its relative importance is very low. Consistently, the average binding signal of E2F4 at the TSS site is lower in LCP promoters than in HCP promoters (Fig. 3E). The binding signal of E2F4 alone accounts for 47% of the variance of expression levels for HCP promoters, but only 14% for LCP promoters (Fig. 3F). This finding implies that the regulation of E2F4 on gene expression might be affected by the status of CpG sites. In fact, it has been demonstrated that E2F binding can be regulated by CpG methylation (Campanero et al. 2000).
In promoters whose low expression level is mediated by CpG methylation, the methylated CpG dinucleotides have a relatively high chance to mutate into UpG. Especially for promoters repressed in germline cells or in early developmental stages, such mutations can be passed on to the next generation, resulting in a reduction in CpG content in that promoter region in future generations. We examined the correlation coefficient of normalized CpG content with expression levels of promoters in different cell lines. We found that the best correlation was obtained in H1HESC (H1 human embryonic stem cells), indicating that CpG content best reflects promoter expression status in this cell line. This indicates that gene expression and DNA methylation in germline cells or early developmental stages might be more similar to H1HESC than other cell lines. We also examined the effect of using CpG content for classifying expressed and nonexpressed promoters. As shown in Figure 3H, this method of classification achieves its highest accuracy (AUC = 0.82; see Methods for details) in H1HESC, with much lower accuracy in HEPG2 cells.
Regulation of alternative TSS by TFs
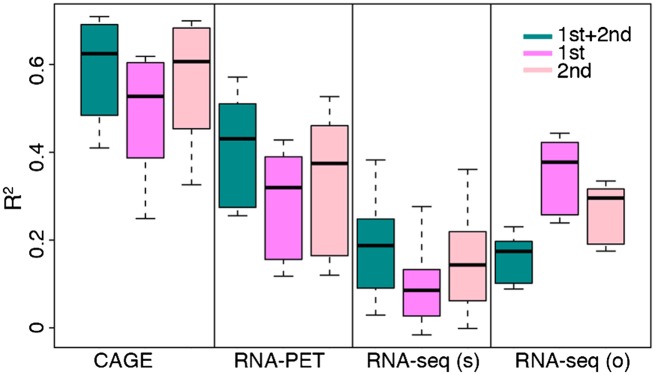
Many genes have multiple transcriptional start sites. Specifically, ∼35% of genes annotated by GENCODE possess more than one TSS (Harrow et al. 2012). To investigate whether there are systematic differences in the regulation of different classes of TSS, we selected all genes with alternative TSSs and collected the first and the second TSS of these genes to form two TSS sets (the average distance between the first and the second TSS is 236 bp). Then we constructed separate models for the first TSS and the second TSS sets. Using expression data from the CAGE and RNA–PET experiments, we achieved higher predictive accuracy for the second TSS set (Fig. 4). The same trend was observed in RNA-seq data only when the short RNA extraction protocol was adopted. Higher prediction accuracy was achieved for the first TSS set for RNA-seq data using other RNA extraction protocols. It is unlikely that these results are caused by the CpG content issue, because the two TSS sets are similar in their CpG contents (56.2% and 55.2% of TSSs in the first and second set, respectively, are HCPs). Moreover, there is no significant difference in the expression levels between the two TSS sets. Our results imply that expression levels of the downstream TSS might rely more on TF regulation, while other chromatin features might have more influence on the transcription of the first TSS. In addition, the relative importance of TFs is different between the two models (Supplemental Fig. S2). For instance, MXI1 is the second most predictive TF in the model for the first TSS set, but it shows only a low relative importance in the model for the second TSS set. Thus, there might exist distinct regulatory mechanisms between the first and the other TSSs as suggested in Davuluri et al. (2008) and Wray et al. (2003).
Figure 4.
Comparison of accuracies of the TF model for predicting the expression level of the first and second TSS of genes. The binding signals of 40 TFSSs are used as the predictors, and only promoters from genes with at least two TSSs are included in the models. The calculation is based on expression data from K562. RNA-seq (s) and RNA-seq (o) represent RNA-seq data using small-RNA extraction protocol and other protocols, respectively.
Correlation of differential gene expression with differential TF binding
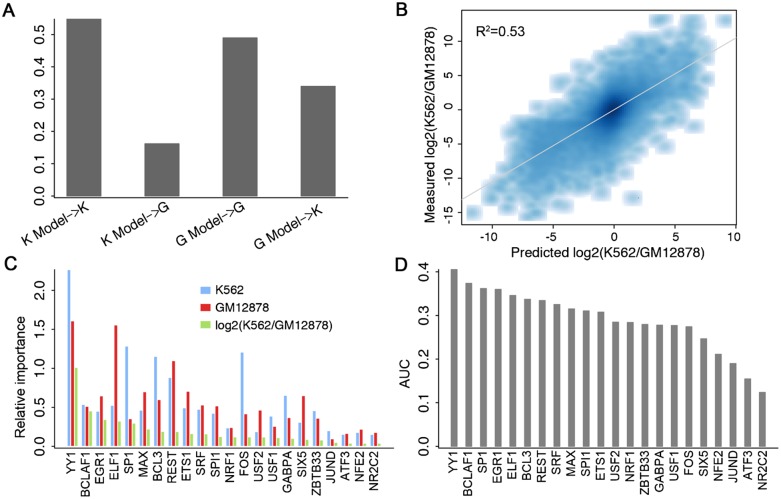
TF binding is regulated in a cell-type–specific manner, so we expect that in two different cell lines, differential TF binding should be correlated with differential TSS expression. We investigated this hypothesis using the data in K562 and GM12878, which were derived from erythroleukemia cells and normal lymphoblastoid cells, respectively. We selected promoters with more than fourfold expression difference between the two cell lines and constructed a K562-specific model (K-model) and a GM12878-specific model (G-model) (used 22 shared TFs in both models). When applied to whole-cell Poly A+ RNA expression data, the K-model explains 55% of the variance in the expression level of promoters in K562, but only 16% of the variance in GM12878 (Fig. 5A). Similarly, the G-model accounts for much more variance of expression in GM12878 (49%) than in K562 (34%). Moreover, TFs exhibit different relative importance in the two cell lines. For example, SP1 shows a relatively stronger effect on gene expression in GM12878, whereas MAX and ETS1 have a stronger effect in K562 (Fig. 5C).
Figure 5.
Cell line specificity of the TF model. (A) Models trained and tested on data from the same cell line result in higher predictive accuracies. K Model and G Model represent models trained with data from K562 and GM12878, respectively. (B) Consistency of predicted log2 fold changes with the experimentally measured differences between K562 and GM12878. Differential binding of 22 TFs are used as the predictors in a predictive model of differential expression. (C) The relative importance of TFs in K562- and GM12878-specific models as well as the predictive model for differential expression. (D) The power of each individual TF for classifying K562- and GM12878-specific promoters (log2 fold change >2). CAGE expression data in Poly A+ RNA extracted from K562 and GM12878 whole cells were used in the calculation.
We next examined the effectiveness of predicting differential expression based on differential binding of TFs in promoter regions. The binding differences (log2) in K562 versus GM12878 were calculated for 22 TFs for which the ChIP-seq data were available in both cell lines. A model using those differences as predictors explains 53% of the variance in expression differences (log2 ratios) of TSSs between K562 and GM12878 (whole cell Poly A+ RNA extraction) (Fig. 5B). We also explored the relative importance of TFs in the differential expression model. Interestingly, we find that the TFs important for differential expression (e.g., YY1) are in general those that are important in both the K-model and the G-model. TFs with higher RI scores in only one cell line (e.g., SP1, MAX, and ETS1) show quite limited contributions to predicting differential expression of promoters (Fig. 5C).
In addition to the regression models, we also constructed classification models. Specifically, we selected 4493 K562-specific (log2(K562/GM12878)>2) and 8183 GM12878-specific (log2(GM12878/K562)>2) TSSs, and examined the capability of each individual TF for discriminating these two TSS categories (using the TF as the single classifier). As shown in Figure 5D, all of these TFs can classify the two TSS categories, with YY1 achieving the highest classification accuracy (AUC = 0.86). Similar results were achieved when different thresholds were used to select K562 and GM12878 specific TSSs.
Relationship between histone modifications and TF-binding signals
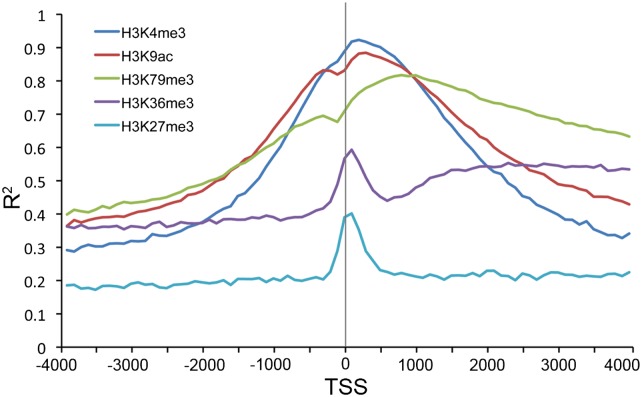
We have previously shown that both TF binding and histone modification are predictive of expression levels of genes (Cheng and Gerstein 2011; Cheng et al. 2011b). In fact, at promoter regions, TF-binding signals and histone modification signals are highly correlated. Active genes are generally bound by transcriptional activators in their promoters and associated with strong signals of active histone marks in their promoters and gene bodies. We thus quantified the relationship between histone modifications and TF-binding signals using the predictive models. We find that histone modification can be predicted accurately by the binding signals of TFs at the TSS regions. As shown in Figure 6, the TF-binding signal at the TSS of genes can predict H3K4me3 signals around the TSS with very high accuracy (R2 = 0.85). It is also highly predictive of the signals of other histone marks, such as H3K9ac and H3K79me3 (see Supplemental Fig. S3). More interestingly, the TF-binding signals can predict the patterns of histone marks, i.e., the positions where they are located. For example, the best prediction accuracy was achieved right at the TSS for H3K4me3, which is known to be a mark for active promoters (Koch et al. 2007). In contrast, high predictive accuracy was obtained at the TSS and in the transcribed region of genes for H3K36me3, which is a histone mark for the gene body (Kolasinska-Zwierz et al. 2009). The relative importance of TFs is different for predicting different histone modification types, but MAX, YY1, ETS1, and E2F6 are generally the most informative ones (see Supplemental Fig. S4; Supplemental Table S3).
Figure 6.
The effectiveness of TF-binding signals for predicting histone-modification patterns around the TSS of promoters. The binding signals of 40 TFSSs are used as the predictors. Both the TF-binding and the histone-modification data are from K562.
Interplay between TF binding and other chromatin features for regulating gene expression
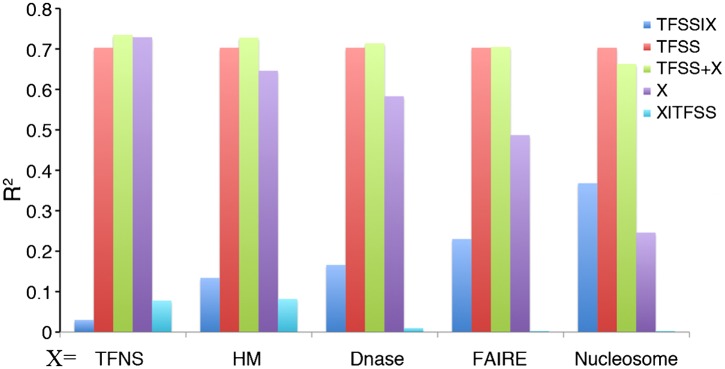
The expression levels of promoters are strongly correlated with the local chromatin structure around the promoter regions. On one hand, chromatin structure is largely determined by nucleosome density (Lee et al. 2007) and histone modifications (Kouzarides 2007), which are in turn influenced by TFs (Narlikar et al. 2002). On the other hand, chromatin structure influences accessibility of the underlying DNA to TFs (Li et al. 2007). The chromatin structure of DNA can be captured by two technologies: DNase hypersensitivity (Follows et al. 2006; Sabo et al. 2006) and Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) experiments (Giresi et al. 2007). We thus applied models to investigate the relationships between gene expression and TF binding (including both TFSSs and TFNSs), histone modifications, DNase, and FAIRE data generated by ENCODE. Given the TFSS-binding data and another chromatin feature X (where X can be histone modification, general TF binding, DNase, FAIRE, or nucleosome occupancy data), we constructed five models to calculate the fractions of variance of promoter expression levels (R2) explained by TFSS-binding data alone (TFSS model), X data alone (X model), a combination of TFSS binding and X data (TFSS + X model), the additional variance explained by TFSS-binding data after considering the X data (TFSS|X model), and the additional variances explained by X data after considering the TFSS binding data (X|TFSS model) (Fig. 7; Supplemental Table S4).
Figure 7.
The relationship of the TFSS-binding data with five types of chromatin features for predicting promoter expression. For each type of chromatin feature, we constructed five models to calculate the fraction of variance of promoter expression levels explained by the TFSS alone (TFSS), by each feature alone (X), by a combination of TFSS and feature X (TFSS+X), as well as the additional variance explained by TFSS after taking feature X into account (TFSS|X) and vice versa (X|TFSS). Feature X represents general transcription factors (TFNS), histone modifications (HM), DNase signal, FAIRE signal, or nucleosome occupancy. CAGE expression data in Poly A+ RNA extracted from K562 whole cells were used in the calculation.
The binding data of sequence-specific TFs and general TFs (Pol II, TATA-binding proteins, etc.) account for at least 74% of the variance in gene expression levels (the TFSS + TFNS model). The remaining variance of gene expression levels (26%) is mainly determined by post-transcriptional regulation. General TFs alone account for 73% of the variance (the TFNS model), and explain the 8% additional variance after considering the sequence-specific TF-binding data (the TFNS|TFSS model). This 8% additional variance is basically what is regulated at the transcriptional level but not captured by the binding data of those 40 TFSSs in the TFSS model, e.g., distal regulation by enhancers and regulation contributed by other factors. After taking into account general TF binding, the additional variance contributed by TFSS binding (the TFSS|TFNS model) is very limited (3%).
After considering the histone modification data, binding of TFSS accounts for a further 13% of additional variance in gene expression levels (the TFSS|HM model), and 8% vice versa (the HM|TFSS model). This suggests that the contributions of TFSS binding and histone modification to aggregate expression of TSS are highly but not completely redundant. Each provides extra information that is not accounted for by the other. We note that here we only use histone modification signals at the TSS regions (100 bp). Since histone modifications affect a broad region around genes, the actual variance that can be explained by the HM model should be even larger (Cheng et al. 2011b; Dong et al. 2012).
The additional variance explained by TFSS-binding data after considering the data of DNase (the TFSS|Dnase model), FAIRE (the TFSS|FAIRE model), and nucleosome occupancy (the TFSS|Nucleosome model) are 16%, 23%, and 37%, respectively. In contrast, after taking into account the TFSS-binding data, the additional variance further explained by these other chromatin features are negligible (<1%), and including them in a model cannot further improve the prediction accuracy for TSS expression. In fact, a combined model including all of these five categories of features leads to an accuracy of R2 = 0.74.
Discussion
TFs and histone modifications are two critical factors that coordinately regulate gene transcription. The regulatory mechanisms of these and other factors are summarized in Figure 8. First, TFs and histone modifications can regulate the initiation of transcription by interacting with RNA polymerase and other general TFs and recruiting them to the TSS (see points 5, 6, 7, and 8 in Fig. 8), or by changing the accessibility of promoters to them via modulating chromatin structure (see points 3 and 4 in Fig. 8) (Mitchell and Tjian 1989; Li et al. 2007). This regulation is achieved with the assistance of chromatin modifiers and other chromatin-associated proteins, e.g., proteins that specifically recognize and bind modified histones (Kouzarides 2007). For these reasons, TF-binding data, histone modification data, and the data that capture local chromatin structure (e.g., DNase and FAIRE) are all predictive of the expression levels of genes (Fig. 7). Second, these factors are inter-related and coordinately participate in transcriptional regulation. For example, TFs such as YY1 can influence histone modifications by recruiting histone modifiers to a DNA region (Yang et al. 1997); and conversely, histone modifications can affect TF binding by directly recruiting them or indirectly by changing the accessibility of DNA regions to them (Li et al. 2007). As a consequence, TF-binding and histone-modification signals are often highly correlated in TSS proximal regions. Due to this high coordination, they share a similar amount of information for “predicting” gene expression levels (Cheng and Gerstein 2011); i.e., they are redundant. Third, the transcription status of genes can in turn affect the TF-binding and histone modifications by interacting with TFs and histone modifiers (Okitsu et al. 2010). A recent study shows that TAF3, the TBP-associated core promoter factor, interacts with CTCF to form DNA loops that connect core promoters with promoter-distal sites, implying that general TFs might regulate chromatin structure of distal regions (Liu et al. 2011). This feedback complicates the cause and effect relationship between TF binding, histone modifications, and gene expression. Taken together with previous studies, our analysis reveals a highly coordinated system for regulation of gene expression that consists of TFs, histone modifications, RNA polymerase, and other chromatin-related proteins.
Figure 8.
Regulatory mechanism of TF binding, histone modification, and other chromatin features on gene expression.
In previous studies, it has been shown that TF binding and histone modifications are predictive of expression levels of mRNA transcripts measured by RNA-seq or microarrays (Ouyang et al. 2009; Cheng and Gerstein 2011). These studies also showed that expression levels from RNA-seq could be more accurately predicted than those from microarrays, indicating the higher precision of the former. In eukaryotes, many genes have multiple transcripts, which might start from different TSS. Technically, it is often difficult to quantify precisely the expression level of each transcript by RNA-seq or microarray. We overcame this problem in this study by focusing on TSS regions, relating TF-binding signal around TSS with expression levels of TSS. CAGE is by nature the technology to quantify expression levels of TSS. For RNA–PET and RNA-seq data, we also calculate the TSS expression levels by focusing on TSS proximal regions. Overall, at the TSS level we obtained higher predictive accuracy compared with those models for predicting expression of transcripts. Our results also suggest that CAGE can best capture the expression levels of TSS. In addition, the accuracy of TSS expression quantification is also dependent on the RNA-extraction protocol being used, with highest performance achieved in Poly A+ RNA. For RNA-seq data the expression levels for TSS and transcript are both available, and we find that the TF models can predict transcript expression with a slightly higher accuracy than TSS expression (Supplemental Table S5). This indicates that RNA-seq, unlike CAGE, more accurately quantifies the expression levels for transcripts than for TSSs.
TF-binding signals used in the TF models capture regulatory information at the transcriptional level. Gene expression levels, however, are also determined by post-transcriptional factors like mRNA degradation. It is therefore more difficult for the TF model to predict the expression levels of genes that are regulated strongly at the post-transcriptional level. We performed gene ontology (GO) analysis on poorly predicted genes (i.e., genes with the largest residuals in the TF model). We find significant enrichment for some GO categories, e.g., involvement in cell cycle control (Supplemental Table S6). In addition, TSSs whose expression levels are underestimated by the TF model ( ) tend to have higher expression variance across different cell lines.
) tend to have higher expression variance across different cell lines.
We have previously shown that the histone-modification model for gene expression prediction is tissue specific (Cheng and Gerstein 2011). In this work, we show that the TF model is also tissue specific, or more precisely, cell line specific (Fig. 5A). The best prediction accuracy is achieved when the TF-binding data and TSS-expression data from the same cell line are used. Note that to predict the expression in a cell line, we always use the TF-binding data from the same cell line, although the model might be trained from the other cell line. Thus, the higher performance of the model in the matched cell line is not caused by differential TF binding; instead, it reflects the different regulatory mechanisms between K562 and GM12878. In addition, TFs show different relative importance in different cell lines. A TF might be active and exhibit significant influence on gene expression in K562, but inactive with little effect on gene expression in GM12878. For example, SP1 shows a relatively stronger effect on gene expression in GM12878 than in K562, while MAX and ETS1 show the opposite trend. Conventionally, TF binding is often regarded as an on/off event. However, Figure 5B shows that the differential expression of TSS can be precisely reflected by the difference of TF-binding signals between two cell lines. This suggests that a quantitative way of TF binding should be used for studying the TF–gene regulatory relationship (Biggin 2011; Cheng et al. 2011a).
Based on normalized CpG content, TSSs can be categorized into HCPs and LCPs. TSSs in the former class tend to have higher expression levels. Our results show that the expression levels of HCPs can be more accurately predicted than those of LCPs by TF-binding signals. We also find that the relative importance of some TFs is different between HCPs and LCPs. Methylation of CpG sites around TSS can represent another mechanism of gene expression regulation. In fact, it has been shown that binding of E2F factors was affected by the methylation status of their binding sites (Campanero et al. 2000; Landolin et al. 2010). Thus, the difference between the HCP model and the LCP model might reflect cooperation between TF binding and DNA methylation for transcriptional regulation.
The first TSS of a gene might be recognized in a different way from the other TSSs by the transcriptional machinery. For instance, it might recognize different TSSs independently, or alternatively, it tends to recognize the most upstream TSS but skip it in a certain frequency to initiate transcription at a downstream TSS. A recent study of the glucocorticoid (GR) and estrogen (ER) nuclear receptors (Voss et al. 2011) found that GR is a “driver” TF, while ER is a “passenger” TF that benefits from “assisted loading” from GR. It was posited that driver TFs bind to closed but breathing chromatin and recruit chromatin remodeling factors to open the chromatin fully. Passenger TFs only bind to chromatin that has been opened already by driver TFs or some other chromatin remodeling mechanism, so they benefit from assisted loading. In our study of the first and second TSS of genes, we were better able to model the second TSS from TF-binding data. We also found that YY1 best predicts expression of the set of the first TSS. It is known that YY1 can recruit chromatin remodeling factors as expected from a driver TF (Yang et al. 1997). These facts lead us to postulate that, for most genes, driver TFs bind to the first TSS and recruit chromatin remodelers, which then open the chromatin around the second TSS. This hypothesis can explain the relative predictive power of our models: When a passenger TF binds near the second TSS, its power to predict second TSS expression is boosted by the fact that chromatin remodeling has already occurred near the first TSS. Likewise, some of the predictive power of TF binding at the first TSS goes to predict transcription of the second TSS.
We show here that TF binding is highly predictive of gene expression levels using human ENCODE data, and we have previously shown the same using mouse data (Cheng and Gerstein 2011). In yeast, several studies have been performed to relate gene expression with motif existence, TF–DNA-binding data, or histone modification data (Kurdistani et al. 2004; Yuan et al. 2006). For example, Yuan et al. (2006) constructed a linear regression model to predict transcription rates of yeast genes. They showed that three types of histone acetylations alone accounted for 18% of the variance (R2 = 0.18) of transcription rates, and the R2 increased to 33% if TF-binding motif and nucleosome occupancy data were also included in the model. Furthermore, Li et al. (2010) showed in another study that TF binding was predictive of intrinsic expression noise of yeast genes, indicating that TF binding impacts not only the levels but also the fluctuation of gene expression. In addition, many other studies focused on identifying regulatory motifs or TFs underlying a biological process via combining expression data with TF-binding data or sequence motif analysis (Conlon et al. 2003; Yu et al. 2003; Tsai et al. 2005; Li and Zhan 2008). In the future, with more data available it would be more practical to perform similar analysis in higher organisms.
Methods
Data processing
All of the data used in this work were generated by the ENCODE project. The expression data of GENCODE TSSs were produced using three different technologies (CAGE, RNA–PET, and RNA-seq). The data include a total of 267 expression profiles, representing expression profiles for RNA samples in 12 different cell lines extracted from six different cellular components (whole-cell, cytosolic, nuclear, and nuclear subcompartments, namely chromatin, nucleoplasm, and nucleolus) using four different protocols (Poly A+, Poly A-, total, and short RNA). Note that the samples are not evenly collected from different cell lines; a large fraction of them are from K562 and GM12878. To facilitate the comparison of data from different technologies, the RNA-sequencing data were processed to obtain expression levels of the TSSs (T Lassmann, P Carninci, in prep.). The RNA–PET expression of a TSS is defined as the total number of 5′ tags within a 101-bp window centered on the TSS. For RNA-seq experiments, the expression level of a TSS is calculated as the sum of expression levels of all transcripts initiated from it. TSS expression levels are normalized and represented as RPM (reads per million) for CAGE, RNA–PET, and short RNA-seq data, or RPKM (reads per kilobase per million) for long RNA-seq (Poly A+, Poly A- and total RNA) data. The expression levels of transcripts (based on GENCODE v7 annotation) were measured as RPKM and calculated using the software FLUX CAPACITOR.
The genome-wide TF-binding data were obtained from ChIP-seq experiments. The data include >400 binding profiles, representing the binding of >120 TFs and chromatin factors in many different cell lines. Again, the most complete data were available from K562 and GM12878. We calculated the binding strengths of each TF at all of the GENCODE TSSs. Specifically, we calculated and averaged the number of reads covering a 100-bp DNA region centering on each TSS, resulting in the binding signal for this TSS. We choose the 100-bp region for two reasons: (1) We have previously shown that TF binding signals in a narrowed DNA region around TSS achieves the highest prediction accuracy; (2) for genes with multiple TSSs the average distance between the first and the second TSSs is ∼200 bp. In fact, when we increased the window size from 100 to 300, 500 until 1500 bp, we observed a gradual decrease of predictive accuracy by the TF model (Supplemental Fig. S5).
The other data sets, including histone modification, DNase I hypersensitivity, FAIRE, and nucleosome occupation, were also generated by the ENCODE project using high-throughput sequencing technologies. The data were processed in the same way as for the TF-binding data. The human promoters/TSSs were annotated by the GENCODE project, version 7 (Harrow et al. 2012). In this work, we focus our analysis on ∼130,000 high-confidence TSSs.
Categorization of DNA-binding proteins
In this work, we mainly focus on using sequence-specific TFs for predicting the expression levels of promoters. In some cases, however, the model was extended to general TFs and other DNA-binding proteins. Basically, we categorized the DNA-binding proteins with ChIP-seq data available in six categories: sequence-specific TFs (TFSS), general or nonspecific TFs (TFNS), chromatin structure factors (ChromStr), chromatin remodeling factors (ChromRem), histone methyltransferases (HISase), and Pol3-associated factors (Pol3F).
Models for predicting TSS expression levels
To understand the relationship between TF-binding signals and the expression levels of promoters, we constructed predictive models based on four different machine-learning methods: RF (random forest), MARS (multivariate adaptive regression splines), SVR (support vector regression), and MLR (multivariate linear regression). In these models, the binding signals (the average read coverage at each nucleotide) in a particular bin (e.g., the 100-bp bin at the TSS) for a set of TFs (e.g., sequence-specific TFs) were used as the predictors to predict the response variable Y (i.e., the expression levels of promoters). The promoter expression levels are distributed over an exponential range, so to stabilize variance we use log2-transformed values as the response variable with 0.03 as pseudo-count.
To evaluate the performance of the predictive models, we randomly selected 2000 promoters as the training data and the remaining as the test data. A model was trained on the training data and applied to predicting the expression levels of promoters in the test data (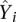 ). The predictive accuracy of the model can be measured by the correlation (R) between the predicted values (
). The predictive accuracy of the model can be measured by the correlation (R) between the predicted values (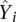 ) and the actual experimental expression levels (Yi). Predictive accuracy can also be measured by the coefficient of determination (R2), the fraction of variance of gene expression explained by the model, which is defined as follows:
) and the actual experimental expression levels (Yi). Predictive accuracy can also be measured by the coefficient of determination (R2), the fraction of variance of gene expression explained by the model, which is defined as follows:
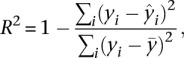 |
where 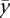 is the mean gene expression level.
is the mean gene expression level.
For each model, we generated 10 groups of training and test data, and averaged the resulting R or R2 as the predictive accuracy. The variation of R/R2 is low, indicating that the training data set with 2000 promoters is large enough to achieve stable predictions.
To estimate the predictive power of an individual TF, we predicted the expression levels using a SVR model with the binding signal of the TF as the single predictor. It is also informative to show the relative contribution of each predictor in a model with multiple predictors. We use the “%IncMSE” (increase of mean squared error) calculated from the Random Forest method to represent the relative importance (RI) of TFs. Specifically, the values of each TF of the test data were permuted and the prediction error (mean squared error of all genes) in the test data was recalculated using the original model. Compared with the unpermuted data, permutation of a TF will, in general, result in increase of prediction error. Such an increase (i.e., %IncMSE) is used as a measurement of relative importance of a TF in the model (Breiman 2001). A TF with higher IncMSE value relative to other TFs in the model has higher importance for predicting the gene expression level.
The R packages “randomForest”, “earth”, and “e1071” were utilized to implement these models (R Core Development Team 2011).
Models for predicting differential gene expression
In the differential gene-expression model, the response variable “Y” was calculated as the log2 ratio of the expression levels in K562 versus GM12878 (log2 K562/GM12878), and the predictors “Xs” were calculated as the log ratio of binding signals between the two cell lines. The predictors in this model are 22 TFs for which the binding data are available for both the K562 and GM12878 cell lines. The pseudo-count (0.03) was used during the calculation to avoid extreme values caused by small expression levels. The same approaches as described in the preceding section were used for evaluating model performance and calculating relative importance of TFs.
Classification of promoters specific to K562 and GM12878
In addition to the regression models, we also constructed classification models to examine the effectiveness of classifying individual TSS as either K562-specific or GM12878-specific TSSs based on the strength of TF-binding signals. We first identified K562-specific and GM12878-specific TSSs according to their expression in Poly A+ RNA extracted from whole cells. Promoters expressed with more than fourfold higher levels in one cell line versus the other were defined as cell-type–specific TSSs. We constructed models using RF and SVM (support vector machine) to classify the two types of TSSs. The classification accuracy was measured by the AUC (Area Under the ROC curve) in the cross-validation data, where the ROC curve (receiver operating characteristic) is a graphic plot of the sensitivity versus 1-specificity. The AUC takes a value within [0, 1], with a greater value indicating higher performance of a classification model.
Models for predicting histone modifications
We also constructed models to predict histone modification signals at different positions relative to the TSS by using the TF-binding signal in 100-bp bins around the TSS as the predictors. With these models, we examined the power of TF-binding signals for inferring histone-modification signals of 12 different types, including H3K4me1, H3K4me2, H3K4me3, H3K36me3, H3K9me1, H3K9me3, H3K27me3, H4K20me1, H3K79me2, H3K9ac, H3K27ac, and H2az. The DNA regions around TSS ([−4 kb, 4 kb]) were divided into 80 bins, each 100 bp in size. For each bin the histone modification signals associated with promoters were examined by the models. In these models the response variable Y (histone modification signal) was log2 transformed.
Models for understanding the relationships of different chromatin features
The expression levels of promoters are correlated with chromatin structure, which is influenced by histone modifications, nucleosome occupancy, and TF binding. Chromatin structure can also be captured by DNase I hypersensitivity and FAIRE data. Thus, all of these chromatin features are predictive of the expression levels of promoters. Using the ENCODE data, we investigated the relationship of five groups of chromatin features (general TF binding, histone modification, nucleosome occupancy, DNase I hypersensitity, and FAIRE signals with the TFSS-binding features in the context of predicting gene expression levels. For each group X, we constructed five different models. Three of the models use chromatin features in the group X (the X model), the binding signals of TFSS (the TFSS model), or a combination of them (the TFSS+X model) as the predictors, respectively. In the remaining two models, we examined the predictive power of features in X after considering the TFSS-binding signals (the X|TFSS model), and vice versa (the TFSS|X model). Specifically, for the X|TFSS model, we first predicted the expression levels of promoters (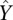 ) based on the binding signals, and then used the features in X to predict the residuals (
) based on the binding signals, and then used the features in X to predict the residuals ( ). We calculated the R2 for each of the five models. The R2 of the X|TFSS model indicates the additional variance explained by the chromatin features in group X after already taking into account the TFSS-binding signal.
). We calculated the R2 for each of the five models. The R2 of the X|TFSS model indicates the additional variance explained by the chromatin features in group X after already taking into account the TFSS-binding signal.
Calculation of normalized CpG content
We calculated the normalized CpG content of all GENCODE promoters in 2-kb DNA regions centered around their TSSs using the method described in Saxonov et al. (2006). Briefly, the normalized CpG content is calculated by dividing the observed number of CpG dinucleotides by the expected number in a promoter. Normalized CpG contents for promoters followed a bimodal distribution (Fig. 3A). Setting the cutoff value between low and high normalized CpG to 0.4 best separated the two peaks in the distribution. Promoters with a normalized CpG content above the cut-off value were classified as high CpG content promoters (HCP), and the remaining promoters were classified as low CpG content promoters (LCP). Approximately, the normalized CpG content reflects the existence of a CpG island nearby a TSS or not (e.g., many HCPs are located nearby a CpG island). It considers the CpG enrichment in the DNA regions centering directly on the TSS, and thereby is more practical than the CpG island-based method for classifying promoters.
Data access
All data are publicly available on the UCSC Genome Browser (http://genome.ucsc.edu/ENCODE/downloads.html).
Acknowledgments
We thank the ENCODE Consortium for the rich data and insightful discussions. We also thank Dr. Anshul Kundaje and Dr. Ben Brown for valuable comments and suggestions. We acknowledge support from the NIH and from the AL Williams Professorship funds.
Footnotes
[Supplemental material is available for this article.]
Article and supplemental material are at http://www.genome.org/cgi/doi/10.1101/gr.136838.111.
Freely available online through the Genome Research Open Access option.
References
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA 2004. Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 14: 283–291 [DOI] [PubMed] [Google Scholar]
- Biggin MD 2011. Animal transcription networks as highly connected, quantitative continua. Dev Cell 21: 611–626 [DOI] [PubMed] [Google Scholar]
- Breiman L 2001. Random Forests. Mach Learn 45: 5–32 [Google Scholar]
- Campanero MR, Armstrong MI, Flemington EK 2000. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci 97: 6481–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Gerstein M 2011. Modeling the relative relationship of transcription factor binding and histone modifications to gene expression levels in mouse embryonic stem cells. Nucleic Acids Res 40: 553–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Min R, Gerstein M 2011a. TIP: A probabilistic method for identifying transcription factor target genes from ChIP-seq binding profiles. Bioinformatics 27: 3221–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Yan KK, Yip KY, Rozowsky J, Alexander R, Shou C, Gerstein M 2011b. A statistical framework for modeling gene expression using chromatin features and application to modENCODE datasets. Genome Biol 12: R15 doi: 10.1186/gb-2011-12-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon EM, Liu XS, Lieb JD, Liu JS 2003. Integrating regulatory motif discovery and genome-wide expression analysis. Proc Natl Acad Sci 100: 3339–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH 2008. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet 24: 167–177 [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Greven MC, Kundaje A, Djebali S, Brown JB, Cheng C, Gingeras TR, Gerstein M, Guigó R, Birney E, et al. 2012. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follows GA, Dhami P, Gottgens B, Bruce AW, Campbell PJ, Dillon SC, Smith AM, Koch C, Donaldson IJ, Scott MA, et al. 2006. Identifying gene regulatory elements by genomic microarray mapping of DNaseI hypersensitive sites. Genome Res 16: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan K-K, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD 2007. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res 17: 877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res (this issue). doi: 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kadonaga JT 2004. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116: 247–257 [DOI] [PubMed] [Google Scholar]
- Koch CM, Andrews RM, Flicek P, Dillon SC, Karaoz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, et al. 2007. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 17: 691–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J 2009. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD 2007. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci 104: 12955–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M 2004. Mapping global histone acetylation patterns to gene expression. Cell 117: 721–733 [DOI] [PubMed] [Google Scholar]
- Landolin JM, Johnson DS, Trinklein ND, Aldred SF, Medina C, Shulha H, Weng Z, Myers RM 2010. Sequence features that drive human promoter function and tissue specificity. Genome Res 20: 890–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Young RA 2000. Transcription of eukaryotic protein-coding genes. Annu Rev Genet 34: 77–137 [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet 39: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Li H, Zhan M 2008. Unraveling transcriptional regulatory programs by integrative analysis of microarray and transcription factor binding data. Bioinformatics 24: 1874–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Li J, Min R, Vizeacoumar FJ, Jin K, Xin X, Zhang Z 2010. Exploiting the determinants of stochastic gene expression in Saccharomyces cerevisiae for genome-wide prediction of expression noise. Proc Natl Acad Sci 107: 10472–10477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD 2012. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484: 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Scannell DR, Eisen MB, Tjian R 2011. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PJ, Tjian R 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245: 371–378 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Okitsu CY, Hsieh JC, Hsieh CL 2010. Transcriptional activity affects the H3K4me3 level and distribution in the coding region. Mol Cell Biol 30: 2933–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Zhou Q, Wong WH 2009. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci 106: 21521–21526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y 2011. A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues. PLoS Genet 7: e1001316 doi: 10.1371/journal.pgen.1001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2011. http://www.R-project.org R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna.
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Ruan Y, Ooi HS, Choo SW, Chiu KP, Zhao XD, Srinivasan KG, Yao F, Choo CY, Liu J, Ariyaratne P, et al. 2007. Fusion transcripts and transcribed retrotransposed loci discovered through comprehensive transcriptome analysis using Paired-End diTags (PETs). Genome Res 17: 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. 2006. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods 3: 511–518 [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci 103: 1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470 [DOI] [PubMed] [Google Scholar]
- Schoenherr CJ, Anderson DJ 1995. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science 267: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, et al. 2003. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci 100: 15776–15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Tsai HK, Lu HH, Li WH 2005. Statistical methods for identifying yeast cell cycle transcription factors. Proc Natl Acad Sci 102: 13532–13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM 2009. A census of human transcription factors: Function, expression and evolution. Nat Rev Genet 10: 252–263 [DOI] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA 2003. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol 20: 1377–1419 [DOI] [PubMed] [Google Scholar]
- Yang WM, Yao YL, Sun JM, Davie JR, Seto E 1997. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem 272: 28001–28007 [DOI] [PubMed] [Google Scholar]
- Yu H, Luscombe NM, Qian J, Gerstein M 2003. Genomic analysis of gene expression relationships in transcriptional regulatory networks. Trends Genet 19: 422–427 [DOI] [PubMed] [Google Scholar]
- Yuan GC, Ma P, Zhong W, Liu JS 2006. Statistical assessment of the global regulatory role of histone acetylation in Saccharomyces cerevisiae. Genome Biol 7: R70 doi: 10.1186/gb-2006-7-8-r70 [DOI] [PMC free article] [PubMed] [Google Scholar]