Abstract
The GENCODE Consortium aims to identify all gene features in the human genome using a combination of computational analysis, manual annotation, and experimental validation. Since the first public release of this annotation data set, few new protein-coding loci have been added, yet the number of alternative splicing transcripts annotated has steadily increased. The GENCODE 7 release contains 20,687 protein-coding and 9640 long noncoding RNA loci and has 33,977 coding transcripts not represented in UCSC genes and RefSeq. It also has the most comprehensive annotation of long noncoding RNA (lncRNA) loci publicly available with the predominant transcript form consisting of two exons. We have examined the completeness of the transcript annotation and found that 35% of transcriptional start sites are supported by CAGE clusters and 62% of protein-coding genes have annotated polyA sites. Over one-third of GENCODE protein-coding genes are supported by peptide hits derived from mass spectrometry spectra submitted to Peptide Atlas. New models derived from the Illumina Body Map 2.0 RNA-seq data identify 3689 new loci not currently in GENCODE, of which 3127 consist of two exon models indicating that they are possibly unannotated long noncoding loci. GENCODE 7 is publicly available from gencodegenes.org and via the Ensembl and UCSC Genome Browsers.
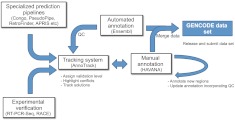
Launched in September 2003, the Encyclopedia of DNA Elements (The ENCODE Project Consortium 2011) project brought together an international group of scientists tasked with identifying all functional elements in the human genome sequence. Initially focusing on 1% of the genome (The ENCODE Project Consortium 2007), the pilot project was expanded to the whole genome in 2007. As part of the initiative, the GENCODE collaboration was established whose aim was to annotate all evidence-based gene features on the human genome at high accuracy, again initially focusing on the 1% (Harrow et al. 2006). The process to create this gene annotation involves manual curation, different computational analysis, and targeted experimental approaches. Eight groups in Europe and the United States directly contribute data to this project, with numerous additional sources of evidence also used for the annotation. Figure 1 shows how the different elements of the GENCODE Consortium interact together.
Figure 1.
The GENCODE pipeline. This schematic diagram shows the flow of data between the groups of the GENCODE Consortium. Manual annotation is central to the process but relies on specialized prediction pipelines to provide hints to first-pass annotation and quality control (QC) for completed annotation. Automated annotation supplements manual annotation, the two being merged to produce the GENCODE data set and also to apply QC to the completed annotation. A subset of annotated gene models is subject to experimental validation. The Annotrack tracking system contains data from all groups and is used to highlight differences, coordinate QC, and track outcomes.
The ability to sequence genomes has far exceeded the techniques for deciphering the information they encode. Selecting the correct reference gene annotation for a particular project is extremely important for any downstream analysis such as conservation, variation, and assessing functionality of a sequence. The type of gene annotation applied to a particular genome is dependent on its quality; therefore, next-generation sequencing assemblies (Metzker 2010) have had automatic gene annotation applied to them, whereas high-quality finished genomes such as the human (International Human Genome Sequencing Consortium 2004), mouse (Church et al. 2009), and zebrafish (Becker and Rinkwitz 2011) have manual annotation projects associated with them. Publicly available gene sets such as RefSeq (Pruitt et al. 2012), AceView (Thierry-Mieg and Thierry-Mieg 2006), and GENCODE are generated by a combination of manual and automatic annotation and have developed different methods to optimize their annotation criteria. For example, RefSeq annotates cDNAs rather than genomic sequence to optimize full-length gene annotation and is thus able to ignore sequencing errors in the genome. This publication will describe the generation of the GENCODE gene set and its strengths over other publicly available human reference annotation and the reasons it has been adopted by the ENCODE Consortium (The ENCODE Project Consortium 2011), The 1000 Genomes Project Consortium (2010), and The International Cancer Genome Consortium (2010) as their reference gene annotation.
Production of the GENCODE gene set: A merge of manual and automated annotation
The GENCODE reference gene set is a combination of manual gene annotation from the Human and Vertebrate Analysis and Annotation (HAVANA) group (http://www.sanger.ac.uk/research/projects/vertebrategenome/havana/) and automatic gene annotation from Ensembl (Flicek et al. 2011). It is updated with every Ensembl release (approximately every 3 mo). Since manual annotation of the whole human genome is estimated to take until the end of 2012, the GENCODE releases are a combination of manual annotation from HAVANA and automatic annotation from Ensembl to ensure whole-genome coverage.
Manual annotation process
The group's approach to manual gene annotation is to annotate transcripts aligned to the genome and take the genomic sequences as the reference rather than the cDNAs. Currently only three vertebrate genomes—human, mouse, and zebrafish—are being fully finished and sequenced to a quality that merits manual annotation. The finished genomic sequence is analyzed using a modified Ensembl pipeline (Searle et al. 2004), and BLAST results of cDNAs/ESTs and proteins, along with various ab initio predictions, can be analyzed manually in the annotation browser tool Otterlace (http://www.sanger.ac.uk/resources/software/otterlace/). The advantage of genomic annotation compared with cDNA annotation is that more alternative spliced variants can be predicted, as partial EST evidence and protein evidence can be used, whereas cDNA annotation is limited to availability of full-length transcripts. Moreover, genomic annotation produces a more comprehensive analysis of pseudogenes. One disadvantage, however, is that if a polymorphism occurs in the reference sequence a coding transcript cannot be annotated, whereas cDNA annotation, for example, performed by RefSeq (Pruitt et al. 2012), can select the major haplotypic form as it is not limited by a reference sequence.
Automatic annotation process
Protein-coding genes were annotated automatically using the Ensembl gene annotation pipeline (Flicek et al. 2012). Protein sequences from UniProt (Apweiler et al. 2012) (only “protein existence” levels 1 and 2) were included as input, along with RefSeq sequences. Untranslated regions (UTRs) were added using cDNA sequences from the EMBL Nucleotide Archive (ENA) (Cochrane et al. 2011). Long intergenic noncoding RNA (lincRNA) genes were annotated using a combination of cDNA sequences and regulatory data from the Ensembl project. Short noncoding RNAs were annotated using the Ensembl ncRNA pipelines, using data from mirBase (Griffiths-Jones 2010) and Rfam (Gardner et al. 2011) as input.
GENCODE gene merge process
This process of combining the HAVANA and Ensembl annotation is complex. During the merge process, all HAVANA and Ensembl transcript models are compared, first by clustering together transcripts on the same strand which have any overlapping coding exons, and then by pairwise comparisons of each exon in a cluster of transcripts. The merge process is summarized in the Supplemental Figures and Tables, including the rules involved in each step. Ensembl have developed a new module, HavanaAdder, to produce this GENCODE merged gene set. Prior to running the HavanaAdder code, the HAVANA gene models are passed through the Ensembl health-checking system, which aims to identify any inconsistencies within the manually annotated gene set. Annotation highlighted by this system is passed back to HAVANA for further inspection. In addition, the HAVANA transcript models are queried against external data sets such as the consensus coding sequence (CCDS) (Pruitt et al. 2009) gene set and Ensembl's cDNA alignments of all human cDNAs. If annotation described in these external data sets is missing from the manual set, then this is stored in the AnnoTrack system (see below) (Kokocinski et al. 2010) so that a record is kept for the annotators to inspect these loci.
The genes in the GENCODE reference gene set are classified into three levels according to their type of annotation. Level 1 highlights transcripts that have been manually annotated and experimentally validated by RT-PCR-seq (Howald et al. 2012), as well as pseudogenes that have been validated by three-way consensus, namely, that have been independently validated by three different strategies. Level 2 indicates transcripts that have been manually annotated. Some Level 2 transcripts have been merged with models produced by the Ensembl automatic pipeline, while other Level 2 transcripts are annotated by HAVANA only. Level 3 indicates transcripts and pseudogene predictions arising from Ensembl's automated annotation pipeline. GENCODE 7 consists of 9019 transcripts at Level 1, 118,657 transcripts at Level 2, and 33,699 transcripts at Level 3. Many of the protein-coding genes in Level 3 are contributed by Ensembl's genome-wide annotation in regions where HAVANA has not yet provided manual annotation.
Locus level classification
Manually annotated GENCODE gene features are subdivided into categories on the basis of their functional potential and the source of the evidence supporting their annotation. Annotated gene models are predominantly supported by transcriptional and/or protein evidence. Once the structure of a model has been established, it is classified into one of three broad locus level biotypes: protein-coding gene, long noncoding RNA (lncRNA) gene, or pseudogene. In addition, more detailed biotypes are associated with transcripts to attempt to assign a functionality, for example, protein-coding or subject to nonsense mediated decay (NMD) (see landscape Supplemental Tables).
To provide a more complete description of the gene model, a “status” is assigned at both the locus and transcript level. Loci can be assigned the status “known,” “novel,” or “putative” depending on their presence in other major databases and the evidence used to build their component transcripts. In brief, loci have the status “known” if they are represented in the HUGO Gene Nomenclature Committee (HGNC) database (Seal et al. 2011) and RefSeq (Pruitt et al. 2012); loci with the status “novel” are not currently represented in those databases but are well supported by either locus-specific transcript evidence or evidence from a paralogous or orthologous locus. Finally loci with status “putative” are supported by shorter, more sparse transcript evidence. A similar status categorization is employed at the transcript level (see Supplemental Figures and Tables).
In addition to the information captured by biotype and status, controlled vocabulary attributes are attached to both transcripts and loci. They are used to describe other features relevant to the structure or functional annotation of a transcript. Attributes may be subdivided into three main categories: those that explain features related to splicing, those related to the translation of the transcript, and those related to the transcriptional evidence used to build the transcript model. For a comprehensive list of all attributes along with the definitions used in the GENCODE annotation, see the landscape Supplemental Tables. Where further explanation of annotation is required, free text remarks are added. New controlled vocabulary is developed wherever possible so that annotation text strings can be searched computationally.
Analyzing long noncoding transcript annotation
Over the last decade, evidence from numerous high-throughput array experiments has indicated that evolution of the developmental processes regulating complex organisms can be attributed to the noncoding regions and not only to the protein-coding regions of the genome (Bertone et al. 2004; Mattick 2004; Kapranov et al. 2007; Clark et al. 2011). The GENCODE gene set has always attempted to catalog this noncoding transcription utilizing a combination of computational analysis, human and mammalian cDNAs/ESTs alignments, and extensive manual curation to validate their noncoding potential. GENCODE 7 contains 9640 lncRNA loci, representing 15,512 transcripts, which is the largest manually curated catalog of human lncRNAs currently publicly available. All the lncRNA loci in the catalog originate from the manual annotation pipeline and are initially classified as noncoding due to the lack of homology with any protein, no reasonable-sized open reading frame (ORF; not subject to NMD), and no high conservation, confirmed by PhyloCSF (see later section), through the majority of exons. The transcripts are not required to be polyadenylated but 16.8% are, and chromatin marks have been identified for 13.9% (Derrien et al. 2012). These lncRNAs can be further reclassified into the following locus biotypes based on their location with respect to protein-coding genes:
Antisense RNAs: Locus that has at least one transcript that intersects any exon of a protein-coding locus on the opposite strand, or published evidence of antisense regulation of a coding gene.
LincRNA: Locus is intergenic noncoding RNA.
Sense overlapping: Locus contains a coding gene within an intron on the same strand.
Sense intronic: Locus resides within intron of a coding gene but does not intersect any exons on the same strand.
Processed transcript: Locus where none of its transcripts contain an ORF and cannot be placed in any of the other categories because of complexity in their structure.
The GENCODE lncRNA data set is larger than other available lncRNA data sets, and it shows limited intersection with them. Forty-two percent (44 out of 96) of the lncRNA database lncRNAdb (Amaral et al. 2011) are represented in GENCODE lncRNAs. We checked the same strand overlap against recent lncRNA catalogs: GENCODE v7 lncRNAs contain 30% of Jia et al. (2010) lncRNAs, 39% of Cabili et al. lincRNAs (Cabili et al. 2011), and 12% of vlincs (Kapranov et al. 2007) (for more details, see Derrien et al. 2012). While this level of overlap between data sets shows how lncRNA annotation is improving, it also shows that substantial additional work is still required. There are likely to be a number of reasons for the limited overlap between the published lincRNAs and GENCODE, not least that a substantial fraction of transcript annotations are currently incomplete (see below). Another reason is that some of the published transcripts are single exons, which up to now have not been annotated in GENCODE unless there is additional support, for example, polyA features, conservation, submitted sequence, or publications. We are addressing this weakness and re-examining single exons lincRNAs based on annotation from Jia et al. (2010) in collaboration with the Lipovich group, and the data will be incorporated into GENCODE 10.
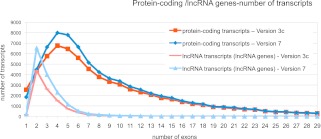
Although the current definition of lncRNAs requires the transcript to be >200 bp (Wang and Chang 2011), the GENCODE ncRNA set contains 136 spliced transcripts <200 bp (all of them single transcript loci) to highlight that there is evidence of expression at that position in the genome. We currently group the transcripts into loci, which is different compared with other lncRNA analysis groups, for example, the Fantom Consortium (Katayama et al. 2005). Multiple lncRNA transcripts appear to start from the same transcription start site (TSS), for example, the DLX6-AS1 locus shown in Supplemental Figure 2. To estimate the completeness of the lncRNA transcripts, we took advantage of CAGE tags from 12 different cell lines and manually annotated polyA features to assess the TSS and 3′ end of transcripts (Djebali et al. 2012). The beginning and end of 15% and 16.8% of lncRNA are supported, respectively, indicating that the majority of transcripts are incomplete. Interestingly lncRNA transcripts have an unusual exon structure compared with protein-coding transcripts, with their distribution peaking at two and five exons, respectively (see Fig. 2). This lower number does not appear to be an artifact or the product of incomplete annotation but most probably is a bona fide characteristic of the lncRNAs, as it is also observed in potential lncRNA models identified using the Illumina Human Body Map 2.0 (HBM) RNA-seq data generated in 2010 on HiSeq 2000 instruments (described below), which are not built from partial ESTs.
Figure 2.
Analysis of exon number of protein-coding and noncoding RNA transcripts. The numbers of exons for each individual transcript annotated at protein-coding and lncRNA loci are plotted for GENCODE 3c (red lines) and GENCODE 7 (blue lines). For each release, darker lines indicate protein-coding transcripts, and lighter lines indicate lncRNA transcripts. The 5′ and 3′ UTR exons of protein-coding transcripts are included.
Small noncoding RNAs are automatically annotated from the Ensembl pipeline and included within the GENCODE gene set. The number has remained relatively stable at 8801 since release 4. Protein-coding and noncoding transcripts that contain a small ncRNA within at least one intron or exon will be annotated with the attribute ncrna_host. Thirty-three percent of small ncRNAs map within the boundaries of a GENCODE gene, the majority of which reside in introns. The GENCODE 7 release contains 1679 protein coding and 301 lncRNA genes with ncrna_host attributes, and there is a sixfold enrichment of small nuclear RNAs (snoRNAs) within exons of lncRNAs (Derrien et al. 2012).
In summary, the lncRNAs data set in GENCODE 7 consists of 5058 lincRNA loci, 3214 antisense loci, 378 sense intronic loci, and 930 processed transcripts loci. Manually evaluating the RNA-seq models generated from HBM data and ENCODE data could potentially double this number in later releases of GENCODE and produce a uniform data set.
Integration of pseudogenes into GENCODE
Within most gene catalogs, pseudogenes have been annotated as a byproduct of protein-coding gene annotation, since a transcript has been identified with a frameshift or deletion, rather than an important entity in its own right. However recent analysis of retrotransposed pseudogenes such at PTENP1 (Poliseno et al. 2010) and DHFRL1 (McEntee et al. 2011) have found some retransposed pseudogenes to be expressed and functional and to have major impacts on human biology. The GENCODE catalog is unique in its annotation of the comprehensive pseudogene landscape of the human genome using a combination of automated, manual, and, more recently, experimental methods.
The assignment process for pseudogenes is described in detail by Pei et al. (2012). Briefly, in silico identification of pseudogenes is obtained from routine implementation of Yale's Pseudopipe (Zhang et al. 2006) with every new release of Ensembl. Pseudopipe identified 18,046 pseudogenes based on the human genome release in Ensembl 61. These pseudogenes were compared to a recent run of UCSC's RetroFinder, which included 13,644 pseudogenes, and HAVANA's latest annotations of 11,224 pseudogenes based on GENCODE 7, level 2. A three-way Yale, UCSC, and HAVANA pseudogene consensus set was obtained by using an overlap criteria of 50 bp and was developed for the annotation of 1% ENCODE Regions (Zheng et al. 2007). This resulted in a consensus set of 7183 pseudogenes, which are tagged level 1. The functional paralog of a pseudogene is often referred to as the “parent” gene. Currently, we have successfully identified parents for 9369 pseudogenes of the manually annotated pseudogenes, whereas the parents for the remaining 1847 pseudogenes are still ambiguous and may require further investigation. It is important to note, however, that it is not always possible to identify the true parent of a pseudogene with certainty, for example, when a pseudogene is highly degraded and is derived from a parent gene with highly similar paralogs or when the parent contains a common functional domain. We have added this information to the pseudogene annotation if known (based on protein alignments), and it is also available from the pseuodgene decorated resource (psiDR described in Pei et al. 2012) http://www.pseudogene.org/psidr/psiDR.v0.txt.
A pseudogene ontology was created to associate a variety of biological properties—such as sequence features, evolution, and potential biological functions—to pseudogenes and is incorporated into the GENCODE annotation file. The hierarchy of these properties is shown in Supplemental Figure 3. The ontology allows not only comprehensive annotation of pseudogenes but also automatic queries against the pseudogene knowledge database (Holford et al. 2010). The breakdown of the different biotypes within the GENCODE data set can be seen in Supplemental Table 4. A schematic to describe the different manually annotated pseudogene biotypes is presented in Figure 3. For example, unitary pseudogenes (i.e., genes that are active in mouse but pseudogenic in the human lineage) were all manually checked for false positives due to genomic sequencing errors or incorrect automated gene predictions in the mouse (Zhang et al. 2010).
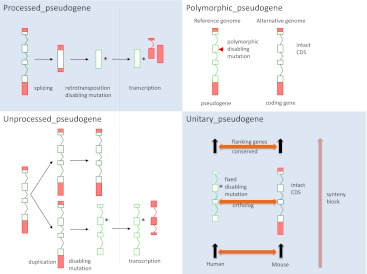
Figure 3.
A schematic showing the structural annotation of different pseudogene biotypes. The schematic diagram illustrates the categorization of GENCODE pseudogenes on the basis of their origin. Processed pseudogenes are derived by a retrotransposition event and unprocessed pseudogenes by a gene duplication event in both cases, followed by the gain of a disabling mutation. Both processed and unprocessed pseudogenes can retain or gain transcriptional activity, which is reflected in the transcribed_processed and transcribed_unprocessed_pseudogene classification. Polymorphic pseudogenes contain a disabling mutation in the reference genome but are known to be coding in other individuals, while unitary pseudogenes have functional protein-coding orthologs in other species (we have used mouse as a reference) but contain a fixed disabling mutation in human.
Computational approaches followed by experimental validation were implemented to examine how many GENCODE pseudogenes appeared to be transcribed (Pei et al. 2012). Briefly, transcribed pseudogenes were identified manually and tagged by the HAVANA team examining locus-specific transcription evidence (by aligning of mRNAs or ESTs). This identified 171 transcribed processed and 309 unprocessed pseudogenes. The locus-specific transcriptional evidence must indicate a best-in-genome alignment and clear differences compared with the parent locus. Interestingly, there was over one-third more unprocessed pseudogenes annotated as transcribed compared with processed pseudogenes, even though there are approximately four times as many processed pseudogenes present in the genome than unprocessed pseudogenes (see Supplemental Table 4). In addition, automated pipeline analysis of RNA-seq data from the total RNA of ENCODE cell line GM12878 and K562 plus HBM RNA-seq resource (Pei et al. 2012) generated an additional 110 and 344 transcribed processed and unprocessed pseudogenes, respectively. Specific primers could be designed for 162 potentially transcribed pseudogenes and have been subjected to experimental validation of transcription by the RT-PCR-seq pipeline within the GENCODE Consortium (Howald et al. 2012). After the validation experiments, 63 pseudogenes were found to be transcribed within at least one of eight tissues.
To summarize, we have manually annotated 11,224 pseudogene loci that are in GENCODE 7 level 2 category. These have been compared with automatic models to produce a consensus set of 7183 loci that have been elevated to level 1. Of these, a total of 480 pseudogenes have been manually tagged as “transcribed” based on publicly available EST or cDNA evidence, and an additional 454 pseudogenes have been identified as transcribed due to alignment of HBM data (Pei et al. 2012).
Evolution of the GENCODE gene set
The GENCODE gene set has developed substantially between releases 3c and 7 (see Fig. 4). Release 3c was the first complete merge set containing all the CCDS transcripts and used by the 1000 Genomes Consortium as its reference annotation. GENCODE release 7 is the reference for the analysis of ENCODE project data carried out in 2011. First-pass manual annotation has been done on 18 chromosomes (chr), and HAVANA still has chr14–19 to complete before the whole genome has been fully manually annotated. Supplemental Figure 4 demonstrates how the number of lncRNAs has increased dramatically with the full manual annotation of the chromosomes. The number of protein-coding loci has decreased significantly between GENCODE releases 3c and 7; however, this is almost entirely due to the removal of poorly supported automatic annotation models, particularly between releases 3c and 3d, where 1004 models were removed from the automatic annotation set. All GENCODE small noncoding RNAs are Level 3 and, as such, show a different pattern to other locus biotypes with their numbers dropping between 3d and 4 as incorrect automated gene models are removed and remaining stable thereafter.
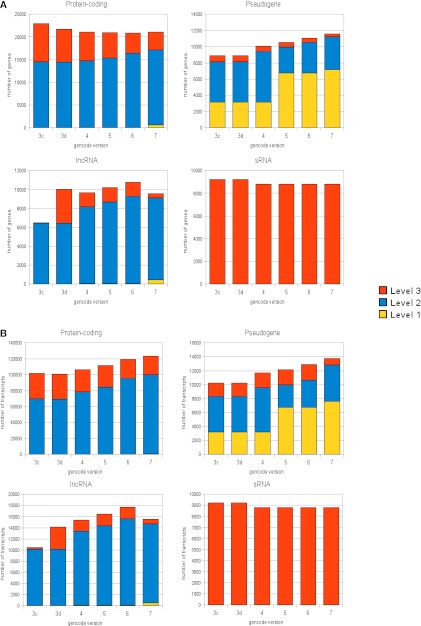
Figure 4.
Analysis of GENCODE annotation in 3c through 7. (A) The content of the GENCODE 3c to 7 at the locus level for four broad biotypes: protein-coding, pseudogene, long noncoding RNA (lncRNA), and small RNA (sRNA). The yellow section of each column indicates the proportion of loci classified as Level 1 (validated), the blue part as Level 2 (manually annotated), and the red part as Level 3 (automatically annotated). (B) The analysis of the content of the GENCODE 3c to 7 at the level of the individual transcript. Again, the yellow section of each column indicates the proportion of transcripts classified as Level 1, the blue part as Level 2, and the red part as Level 3.
The patterns of change in the GENCODE loci across releases 3c to 7 are reproduced at the transcript level. It is clear from the data that the vast majority (>75%) of transcripts are associated with protein-coding loci. While the total number of protein-coding loci is decreasing, the number of coding locus transcripts is increasing with each release. The lncRNA transcript numbers show less stability than protein-coding loci and pseudogenes because of the novel status of the whole area of lncRNAs and the method of identification has changed, for example, based on chromatin signatures or position relative to a protein coding gene. A key change in lncRNA transcripts between releases 3c and 7 is the introduction of a more refined set of biotypes for Level 1 and 2 transcripts (see Supplemental Table 4), specifically the number of transcripts with the biotype processed_transcript reduced significantly and the number of antisense, lincRNA, noncoding and sense_intronic biotypes correspondingly increases.
Assessing the completeness of transcript structures in the GENCODE 7 set
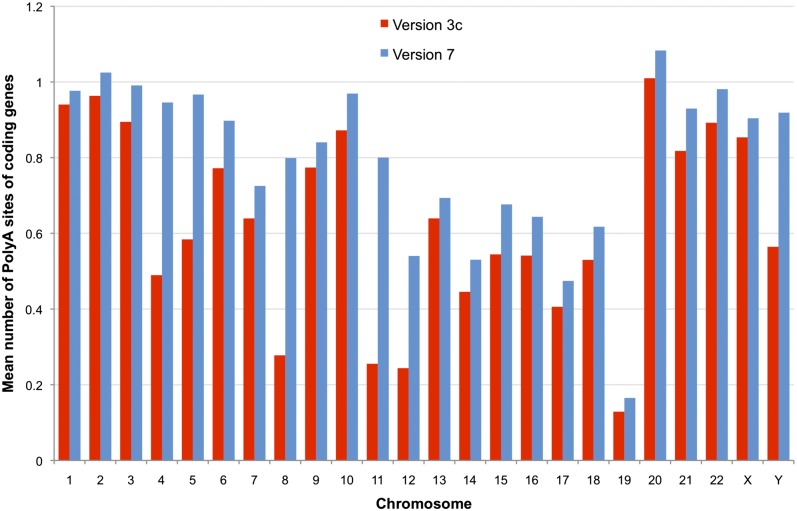
To assess whether a locus or transcript is full-length, it is necessary to identify the TSS and transcription termination site (TTS). TSSs may be tested by determining overlap with CAGE tags (Takahashi et al. 2012) and the TTS by the presence of polyadenylation feature (polyA signals and polyA sites) (Ara et al. 2006). The number of protein-coding genes with at least one polyadenylation feature annotated increased by ∼10% between 3c and 7, and in the later release, the majority (∼62%) of protein-coding loci have one or more polyA sites annotated. However, the mean number of individual transcripts with a polyA site decreased between versions 3c and 7 as the overall number of annotated transcripts increased (Supplemental Table 6). Transcripts at both coding and lncRNA loci show a reduction in the percentage with polyA site of ∼2.5%. Figure 5 shows the number of polyA features increases in every annotated chromosome between release 3c and 7 for protein-coding gene loci.
Figure 5.
Comparison of polyA features annotated across all chromosomes. The mean number of polyA features (sites plus signals) for all protein-coding loci are plotted for every chromosome for GENCODE 3c (red columns) and 7 (blue columns).
While around 62% of GENCODE coding loci have support for identification of the TTS of the locus, the picture is less clear at the TSS. Around 65% of GENCODE annotated TSSs do not overlap with CAGE tag clusters generated by the ENCODE transcript subgroup (Djebali et al. 2012). One likely reason for the disparity between the validation of the two ends of the gene is the limited tissue coverage of the TSS set, which is derived from 24 whole-cell line polyA+ CAGE experiments (Djebali et al. 2012).
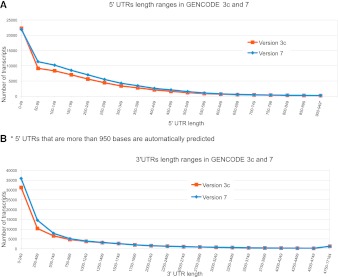
To assess the consistency in the structure of annotated gene models between releases, the number of exons per transcript was plotted for all splice variants at protein-coding and lncRNA loci in releases 3c and 7 (see Fig. 2). It is clear that, although their numbers have increased in release 7, the distribution of the numbers of exons per transcript suggests that the models themselves are very consistent in structure. Transcripts annotated at protein-coding loci demonstrate a peak at four exons per transcript, while lncRNAs show a very similar pattern given the large increase in their numbers between 3c and 7, with a distinct peak at two exons. This analysis confirms a high degree of homogeneity in the structure of transcripts annotated between releases 3c and 7. While the structure of annotated transcripts is invariant, there is a difference between the annotation of UTRs in those models in releases 3c and 7 (Fig. 6). Both the mean 5′ UTR and 3′ UTR length increase with each release between 3c and 7, with the mean 5′ UTR more than 41 bases longer in release 7 and the mean 3′ UTR 180 bases longer. Both increases are likely to be due to an increase in the amount of transcript data (ESTs and mRNAs) available to support the extension of transcripts.
Figure 6.
Examining the length of 5′ and 3′ UTRs between GENCODE 3c and 7. (A) The length of 5′ UTR sequence (in 50-bp bins) for each protein-coding transcript. 5′ UTR annotation from GENCODE 3c (red) and 7 (blue). (*) A cutoff was made at 949 bases; longer 5′ UTRs do exist. (B) The length of 3′ UTR sequence (in 250-bp bins) for each protein-coding transcript. 3′ UTR annotation from GENCODE 3c (red) and 7 (blue).
Comparing different publicly available data sets against the GENCODE 7 reference set
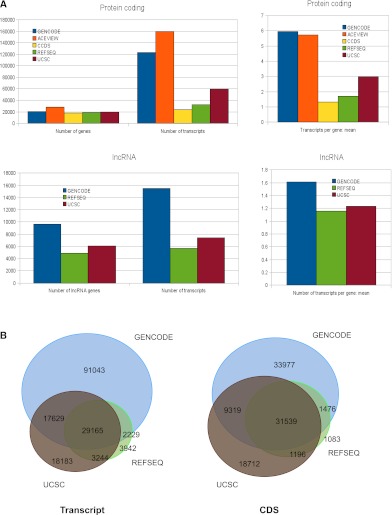
We compared the composition of annotation across the five major gene sets publicly available in UCSC, GENCODE, CCDS, RefSeq, and AceView. Both the number of protein-coding loci and transcripts at those loci were investigated. The CCDS set has the lowest number of protein-coding loci and alternatively spliced transcripts since it is a high-quality conservative gene set derived from RefSeq and Ensembl/HAVANA gene merge (Pruitt et al. 2009). In CCDS, every splice site of every transcript must agree in both the RefSeq and Ensembl/Havana gene set and all transcripts must be full-length. While the number of protein-coding loci in RefSeq, GENCODE, and UCSC is comparable, AceView has ∼20,000 more coding loci. One likely source of inflation is the predisposition for AceView to add a CDS to transcript model and hence create novel loci from lncRNAs and pseudogenes (e.g., PTENP1). AceView predicts 31,057 single exon loci compared with 1724 in GENCODE, 3234 in RefSeq, and 4731 in UCSC genes. Excluding single exon loci predicted by AceView from this analysis, the number of AceView gene loci is much closer to the number in other gene sets (Fig. 7A).
Figure 7.
(A) Comparing different publicly available gene sets. The protein-coding content of five major publicly available gene sets— GENCODE, AceView, consensus coding sequence (CCDS), RefSeq, and UCSC—were compared at the level of total gene number, total transcript number, and mean transcripts per locus. (Blue) GENCODE data; (orange) AceView; (yellow) CCDS; (green) RefSeq; (red) UCSC. The lncRNA content of three of these gene sets—GENCODE, RefSeq, and UCSC—were also compared at the level of total gene number, total transcript number, and mean transcripts per locus. Again, GENCODE data are shown in blue, RefSeq in green, and UCSC in red. (B) Overlap between GENCODE, RefSeq, and UCSC at the transcript and CDS levels. Both protein-coding and lncRNA transcripts of all data sets were compared at the transcript level. Two transcripts were considered to match if all their exon junction coordinates were identical in the case of multi-exonic transcripts, or if their transcript coordinates were the same for mono-exonic transcripts. Similarly, the CDSs of two protein-coding transcripts matched when the CDS boundaries and the encompassed exon junctions were identical. Numbers in the intersections involving GENCODE are specific to this data set, otherwise they correspond to any of the other data sets.
The GENCODE gene set contains 140,066 annotated alternative transcripts at coding loci compared with 66,612 in UCSC genes and 38,157 RefSeq. However it must be noted that not all GENCODE transcripts are full length, and if an annotated transcript is partial, it is tagged with start_not_found or end_not_found to highlight this to the user. The GENCODE gene set has 9640 lncRNA loci compared with 6056 in UCSC genes and 4888 in RefSeq. The three transcript data sets (UCSC, RefSeq, and GENCODE) were compared computationally to see how many transcripts were contained in all data sets and how many were unique to each data set (Fig. 7B). As expected, the majority (89%) of CDSs from RefSeq matched in all data sets exactly since Ensembl and UCSC genes use RefSeq cDNA in their automatic pipelines. However, GENCODE has 33,977 unique coding sequences outside RefSeq compared with 18,712 in UCSC genes. Of these unique transcripts, there are only 9319 exact matches in both these sets, indicating the different methods of annotation and the way they interpret EST data.
Analyzing the protein-coding complement of the GENCODE 7 reference set
We analyzed the annotated CDS in GENCODE 7 using the data in the APPRIS database (http://appris.bioinfo.cnio.es/). APPRIS defines the principal variant by combining protein structural, functional, and conservation information from related species in order to determine the proportion of transcripts that would generate functional isoforms with changes to their protein-coding features relative to the constitutional variant. Of the 84,408 transcripts annotated as translated in the GENCODE 7 release, 30,148 (35.7% of all transcripts or 47.3% of alternative transcripts) would generate protein isoforms either with fewer Pfam functional domains (Finn et al. 2010) or with damaged Pfam domains with respect to the constitutional variant for the same gene. Twenty-six thousand nine hundred fifty-five isoforms (31.9% of all isoforms or 42.3% of alternative isoforms) would have lost or damaged structural domains, based on alignments with Protein Data Bank (PDB) structures, and 16,540 isoforms (19.6% of all isoforms or 26% of alternative isoforms) would lose functionally important residues.
In total, 44.9% of the translated isoforms (59.9% of the alternative isoforms) would lose either functional or structural domains or functional residues relative to the constitutive isoform. For the “putative” and “novel” biotypes and for those isoforms predicted to be NMD targets, these figures were much higher: 71.5% of all putative transcripts, 62.7% of all novel transcripts, and 79.5% of all predicted NMD transcripts would give rise to isoforms with loss of protein functional or structural information.
The location of potential alternative isoforms would also be affected for some isoforms. A total of 1287 genes would generate isoforms with differing numbers of cellular or mitochondrial signal sequences, and 2697 genes would generate isoforms with differing numbers of trans-membrane helices. In addition, 8842 genes contained at least one transcript that appeared to be subject to non-neutral evolution. We were able to select a constitutive variant for 16,553 protein-coding genes (80%) based on the output of the methods in APPRIS (Tress et al. 2008).
Assessing quality of evidence and splice sites
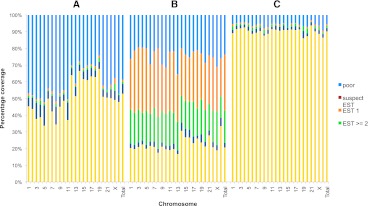
It is important that users understand how to assess transcript annotations that they see in GENCODE. Some transcript models have a high level of support through the entirety of their structure, while other transcripts are poorly supported with one or two ESTs. A new method is being developed to differentiate this level of support. This method relies on mRNA and EST alignments supplied by UCSC and Ensembl. The mRNA and EST alignments are compared to the GENCODE transcripts, and the transcripts are scored according to how well the alignment matches over its full length (for more details, see the Supplemental Methods). Figure 8 shows support level statistics on a set broken down by annotation method for GENCODE 7. The annotations are partitioned into those produced only by the automated process, and those only from the manual method and the merged annotations, where both processes result in the same annotation. For the analyzed subset of GENCODE, 38% of the transcripts have good full-length support and an additional 39% have a lower level. The different distribution of support levels between the annotations is consistent with the underlying approaches. The merged subset is heavily weighted toward well-supported transcripts. The higher fraction of manual-only annotations supported only by ESTs is due to Ensembl not directly using ESTs in their pipeline.
Figure 8.
Quality of evidence used to support automatic, manually, and merged annotated transcripts. The level of supporting evidence for automatic only (A), manual only (B), and merged (C) annotated transcripts is shown for each chromosome. (Yellow) The proportion of models with good support; (dark blue) those supported by suspect mRNAs from libraries with known problems with quality; (light green) those with multiple EST support; (orange) those with support from a single EST; (red) those supported by ESTs from suspect libraries; (pale blue) those lacking good support. The number of transcripts across all chromosomes represented in A is 23,855; B, 89,669; and C, 22,535.
We have focused on removing nonconsensus introns, those not matching the known splicing patterns of GT..AG, GC..AG, and AT..AC. We found ∼2200 such introns in GENCODE 7, which is a reduction from 3c, where there were ∼3300 nonconsensus introns. For version 7, 13% of these show good cDNA support. A small number of nonconsensus splice sites are believed to be other donor/acceptor combinations recognized by U12 spliceosomes. Others are suspected polymorphisms, some with SNP support. Only 2% could be converted to canonical splice sites by known SNPs from dbSNP version 132 in some members of the population, but this proportion has doubled since 3c, reflecting the recent increase in SNP discovery from genome-wide projects. There may be more of these splice sites that are polymorphic but the SNPs are as yet unknown. There are a number of nonconsensus splice sites that are only one base different from canonical, suggesting that they could have been formed due to mutation, but some may represent low frequency polymorphisms. As the 1000 Genomes Project (http://www.1000genomes.org) and other human sequencing projects progress, more SNPs will be discovered, and therefore, the number of known polymorphic splice sites should increase.
Experimental validation
The aim of the experimental validation group of GENCODE is to identify gene models that have limited or lower confidence transcribed evidence and to systematically experimentally validate them. Predicted exon–exon junctions were evaluated by RT-PCR amplification in eight different tissues followed by highly multiplexed sequencing readout, a method referred to as RT-PCR-seq (Howald et al. 2012). Eighty-two percent of all assessed junctions (n = 5871) are confirmed by this evaluation procedure, demonstrating the high quality of the annotation reached by the GENCODE gene set. RT-PCR-seq was also efficient at screening gene models predicted using the HBM RNA-seq data. We validated 73% of these predictions, thus confirming 1168 novel genes, mostly noncoding, which will further complement the GENCODE annotation (Howald et al. 2012). Our RT-PCR-seq–targeted approach can also be exploited to identify novel exons. We discovered unannotated exons in ∼10% of assessed introns. We thus estimate that at least 18% of loci contain as yet unannotated exons.
This novel experimental validation pipeline is significantly more effective than unbiased transcriptome profiling through RNA sequencing, which is becoming the norm. Exon–exon junctions specific to a single GENCODE annotated transcript are five times more likely to be corroborated with our targeted approach than with extensive large human transcriptome profiling such as the HBM and ENCODE RNA-seq (validation rates of 82% and 16%, respectively), as random sampling of RNA molecules leads to poor assessment of low expressed transcripts. It should be noted that the RNA-seq samples were deeply sequenced with the resulting data sets and contained a total of 4.99 and 5.55 billion sequence reads, respectively.
Our work demonstrates that the cataloging of all the genic elements encoded in the human genome will necessitate a coordinated effort between unbiased and targeted approaches, like RNA-seq and RT-PCR-seq, respectively (Howald et al. 2012).
Using next-generation sequencing to find novel protein-coding and lncRNA genes outside GENCODE
To identify novel coding and noncoding genes represented in RNA-seq data, we studied transcript models reconstructed using Exonerate (Howald et al. 2012) and Scripture (Guttman et al. 2010), based on the high-depth HBM transcriptomic data from 16 tissues made publicly available from Illumina (ArrayExpress accession: E-MTAB-513; ENA archive: ERP000546).
Assessing coding potential of RNA-seq models using PhyloCSF
We analyzed the resulting transcripts that did not overlap any GENCODE loci for coding potential using PhyloCSF (Lin et al. 2011), which examines evolutionary signatures within UCSC vertebrate alignments, including 33 placental mammals. There were 136 Ensembl HBM models with positive PhyloCSF scores out of a total of 3689 loci, although only five of these had sufficient support for manual reannotation as coding genes (see Supplemental Table 8). The remaining 131 transcripts showed varying quality and evidence; ∼50% overlap novel processed transcripts and could be a result of misalignment of reads or actual expressed pseudogenes. Two hundred Scripture transcript predictions that were outside GENCODE but had high PhyloCSF scores were also manually examined. Of these, 15 were added as novel loci, and only nine were annotated as coding genes (see Supplemental Table 9) and will be added to the next release of GENCODE). Considering the depth of reads of the HBM data (averaging over a billion read depth) from the 16 different tissues, we have not identified many missing coding genes based on PhyloCSF. Indeed, since 3127 HBM Ensembl genes consist of only two exons, it is highly likely these constitute new lncRNAs we have not yet annotated and will be merged into a later release of GENCODE.
Assessing coding potential of putative models using mass spectrometry data
A pipeline has been set up to verify the annotation of gene models with mass spectroscopy data from human proteomics experiments (M Tress, P Maietta, I Ezkurdia, A Valencia, J-J Wesselink, G Lopez, A Pietrelli, and JM Rodriguez, in prep.). The data from tandem mass spectrometry experiments are stored in two huge proteomics data repositories, the GPM (Craig et al. 2004) and Peptide Atlas (Desiere et al. 2006). Peptides are detected by mapping spectra from individual proteomics experiments to the gene products from the GENCODE annotation using the search engine X!Tandem (Craig and Beavis 2004). A single peptide may be detected in many different experiments, though only once per experiment. We generate P-values for all detected peptides by combining the X!Tandem P-values for each individual experimental peptide-spectrum match, and a target-decoy approach is used to determine false-discovery rates.
The pipeline was able to map peptides to almost 40% of the protein-coding genes in the GENCODE 7 release. The 83,054 tryptic peptides detected at a false-discovery rate of 1%, mapped unambiguously to 8098 of the 20,700 annotated protein-coding genes. We were able to detect the translation of multiple splice isoforms for 194 genes, and within this set of genes, we validated the expression of eight isoforms that were tagged as candidates for NMD degradation. We found peptide support for 33 transcripts annotated as “putative” and another 50 transcripts annotated as “novel.” With the mass spectroscopy data we generated for the GENCODE 3c release, we also detected the expression of peptides that mapped to a pseudogene (MST1P9) (Pei et al. 2012).
GENCODE error tracking using Annotrack
The AnnoTrack software system (Kokocinski et al. 2010) was developed as part of the GENCODE collaboration to facilitate the processing and tracking of the HAVANA annotation and the heterogeneous sources of information used in the genome annotation. It integrates data from distributed sources via DAS (Distributed Annotation System) servers set up by the GENCODE partners following a defined format (http://www.gencodegenes.org/gencodedasformats.html), a direct database connection to Ensembl, and flat file adaptors. The system highlights conflicts and facilitates the quick identification, prioritization, and resolution of problems during the process of genome annotation. Using controlled terms for the solutions chosen by the manual annotators when resolving conflicts allows a retrospective assessment in order to improve the analysis methods or external data sources. More than 4000 issues have been resolved this way in the last year. AnnoTrack also helps to track the progress of the overall HAVANA annotation and to inform external scientists about current issues about the genes they might be working with. The interface can be accessed at http://annotrack.sanger.ac.uk/human.
Accessing data via UCSC, Ensembl, and FTP
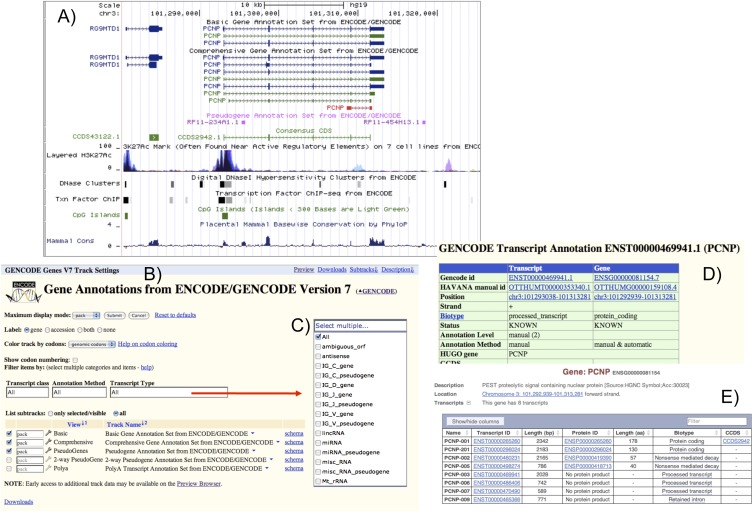
As the GENCODE gene set is now built partly through the Ensembl pipeline, the GENCODE data release cycle is coupled to the trimonthly Ensembl releases. Dates and release notes, as well as more details of the data sets and formats, are listed at http://www.gencodegenes.org. The GENCODE releases contain updated gene sets where either new data from the manual annotation has been integrated as described above or additionally the automated gene set was rebuilt or refined. Users can view GENCODE data in the UCSC browser (Fig. 9), and also it is the default gene set shown in Ensembl. All genes and pseudogenes within the release have stable Ensembl (ENS) identifications and the manual annotated genes have additional Vega (OTT) IDs (Wilming et al. 2008). All OTT identifications are also versioned so the user can identify when a transcript was last manually updated.
Figure 9.
Accessing the GENCODE gene set through UCSC and Ensembl. (A) The composite of screenshots from the UCSC browser shows GENCODE gene annotation displayed in the basic and comprehensive display mode, along with the GENCODE pseudogenes, CCDS models, and a subset of histone modification tracks, DNaseI hypersensitivity clusters, and transcription factor binding site tracks. (B) The configuration display where the user can filter on biotype, annotation method, and transcript type (C). (D) The transcript page in UCSC where the different identifications and version of the transcript can be seen, as well as the evidence used to build the transcript. From the page, the user can click on the Ensembl identification and immediately jump to the Ensembl gene view page (E) and see an overview of the different transcripts in the locus as well as which is a CCDS.
Figure 9 shows the UCSC browser with the “basic” view, which only displays full-length transcripts for every loci unless a partial transcript is the only representative of that locus. This is in demand from users that want a more compact view when browsing the genome. However, the “comprehensive” view shows everything annotated and is designed for the user that is interested in all annotated transcripts in a particular region of the genome. The data set can also be filtered by biotype and annotation method to display the transcript, and submitted nucleotide evidence that each particular transcript was built on is also available on the transcript page. GENCODE data are released and accessible in various formats, which are described in Supplemental Table 8. The gene sets and supporting data are submitted as GTF files to the ENCODE Data Coordination Center (DCC) for integration in the UCSC genome browser and for redistribution.
Discussion
As sequencing technology steadily improves and becomes cheaper a thousand dollar genome will soon become a reality. However, utilizing this genomic data efficiently is still dependent on which reference annotation is chosen to highlight its gene landscape and variation between other individuals. The GENCODE human reference set is a merge of automatic and manual annotation, thus producing the most comprehensive gene set publicly available. Its regular release cycle of every 3 mo ensures that models are continually refined and assessed based on new experimental data deposited in the public databases. In addition, users can send updates or queries concerning individual genes using the Annotrack system or via the Gencodegenes.org website to trigger investigation and updates of specific issues alongside those flagged by global quality-control (QC) checks.
Interestingly, there is still uncertainty about the number of protein-coding and long noncoding loci in the genome. Assessing how many protein-coding loci are missing from the catalog is difficult, but our analysis of coding potential using conservation indicates that the number is likely to be small, namely, around 100 protein-coding genes. A similar figure was suggest by Lindblad-Toh et al. (2011), who recently reported the sequencing and comparative analysis of 29 eutherian mammals. However the recent publication by Ingolia et al. (2011) suggests that there is a new class of small “polycistronic” ribosome-associated coding RNAs encoding small proteins that can now be detected using ribosome profiling. They highlight that the majority of predicted lncRNAs in the mouse from Guttman et al. (2009) actually show comparable translatability to that of protein-coding genes. In addition, Cabili et al. (2011) have found 2798 lincRNAs not in GENCODE 4 using a combination of HBM RNA-seq and additional RNA-seq from eight additional cell lines and tissues totaling 4 billion reads. This indicates that there are still many thousands of lncRNA loci to add to the GENCODE catalog, and completeness will be dependent on the depth and variety of tissues and cell lines sequenced.
The GENCODE catalog is highly specific in its subcategorization of protein-coding and noncoding transcripts highlighting transcripts subject to phenomena such as NMD and nonstop decay (Mazzoni and Falcone 2011). Interestingly mass spectrometry analysis within the project has identified four transcripts, annotated to be subject to NMD, that produce peptides. A recent publication by Bruno et al. (2011) has identified a brain-specific microRNA (miR-128) that represses the NMD pathway by binding to the RNA helicase UPF1. Thus suggesting certain miRNAs can induce cell-specific transcription/translation during development by inhibition of the NMD pathway.
Analysis of transcriptomic and proteomic data is also revealing pseudogenes that are potentially expressed. The GENCODE reference set aids such analysis since it is the only gene set to contain comprehensively manually annotated pseudogenes to the same level as protein and noncoding genes. We currently predict a total of around 10,000 pseudogenes within the human genome. Recent publications highlight the implications of pseudogenes as regulators of gene expression (Han et al. 2011) and specifically a role in tumor biology (Poliseno et al. 2010), and thus we will have to rethink the classification of pseudogenes as nonfunctional entities on the genome.
The GENCODE gene set gives the most comprehensive overview of alternative splicing than of any gene set available. However improved transcriptome sequencing is being facilitated by high-depth RNA sequencing (Mercer et al. 2011), which reveals that the full extent at which different transcripts are expressed in different cells and tissues and at different developmental stages is still to be fully characterized. The RT-PCR-seq methodology developed within the GENCODE project has identified novel exons within 10% of annotated introns being targeted (Howald et al. 2012). Therefore, even when the first pass of whole-genome manual annotation is finished in 2012, reannotation updates will be required on a large percentage of loci to correctly classify the new alternatively spliced variants being revealed by next-generation transcriptomics. However the major drawback with using next-generation short reads to assemble transcripts de novo is that the correct structure of the transcript is hard to predict, as was investigated in the RNA-seq Genome Annotation Assessment Project (RGASP) (J Harrow, T Steijger, F Kokocinski, JF Abril, C Howald, A Reymond, A Mortazavi, B Wold, T Gingeras, R Guigó, et al., in prep.). We therefore look forward to “third-generation” sequencing methods such as Pacific Biosciences (Schadt et al. 2010) that show promise of generating longer reads of 1–2k, which will enable improvements in transcriptome annotation, facilitating the investigation of expression of each transcript structure within a cell during its development.
Methods
Manual annotation
Manual annotation of protein-coding genes, lncRNA genes, and pseudogenes was performed according to the guidelines of the HAVANA, available at ftp://ftp.sanger.ac.uk/pub/annotation. In summary, the HAVANA group produces annotation largely based on the alignment of transcriptomic (ESTs and mRNAs) and proteomic data from GenBank and Uniprot. These data were aligned to the individual BAC clones that make up the reference genome sequence using BLAST (Altschul et al. 1997) with a subsequent realignment of transcript data by Est2Genome (Mott 1997). Transcript and protein data, along with other data useful in their interpretation, were viewed in the Zmap annotation interface. Gene models were manually extrapolated from the alignments by annotators using the otterlace annotation interface (Searle et al. 2004). Alignments were navigated using the Blixem alignment viewer (Sonnhammer and Wootton 2001). Visual inspection of the dot-plot output from the Dotter tool (Sonnhammer and Wootton 2001) was used to resolve any alignment with the genomic sequence that was unclear or absent from Blixem. Short alignments (less than 15 bases) that cannot be visualized using Dotter were detected using Zmap DNA Search (essentially a pattern matching tool; http://www.sanger.ac.uk/resources/software/zmap/). The construction of exon–intron boundaries required the presence of canonical splice sites, and any deviations from this rule were given clear explanatory tags. All nonredundant splicing transcripts at an individual locus were used to build transcript models, and all splice variants were assigned an individual biotype based on their putative functional potential. Once the correct transcript structure had been ascertained, the protein-coding potential of the transcript was determined on the basis of similarity to known protein sequences, the sequences of orthologous and paralogous proteins, the presence of Pfam functional domains (Finn et al. 2010), possible alternative ORFs, the presence of retained intronic sequence, and the likely susceptibility of the transcript to NMD (Lewis et al. 2003).
Amplification and sequencing
Double-stranded cDNA of eight human tissues (brain, heart, kidney, testis, liver, spleen, lung, and skeletal muscle) were generated with the Marathon cDNA amplification kit (Clontech). The cDNA concentration was normalized by quantitative PCR against AGPAT1 and EEF1A1 genes. The PCRs were performed in 386-well plates in a total volume of 12.5 μLl. One microliter of normalized cDNA was mixed with JumpStart REDTaq ReadyMix (Sigma) and primers (4 μM) with a Freedom evo robot (TECAN). The 10 first cycles of amplification were performed with a touchdown annealing temperature decreasing 1°C per cycle from 65°C to 55°C; annealing temperature of the next 30 cycles was carried out at 55°C. For each tissue, 2 μL of each RT-PCR reaction were pooled together and purified with the QIAquick PCR purification Kit (Qiagen) according to the manufacturer's recommendations. This purified DNA was directly used to generate a sequencing library with the “Genomic DNA sample prep kit” (Illumina) according to the manufacturer's recommendations with the exclusion of the fragmentation step. This library was subsequently sequenced on an Illumina Genome Analyzer 2 platform.
Mapping and validation of amplified exon–exon junction
Thirty-five- or 75-nucleotide (nt)-long reads were mapped both on to the reference human genome (hg19) and the predicted spliced amplicons with Bowtie 0.12.5 (Langmead et al. 2009). Only uniquely mapping reads with no mismatch were considered to validate a splice site (transcript). Splice junctions were validated if a minimum of 10 reads with the following characteristics spanned the predicted splice junctions. For 35- and 75-nt-long reads, we required at least 4 and 8 nt on each side of the breakpoints (i.e., on each targeted exon), respectively.
Comparison of RefSeq, UCSC, AceView, and GENCODE transcripts
Transcripts belonging to four different data sets (GENCODE, RefSeq, UCSC, and AceView) were compared to assess to which extent these data sets overlap. Releases compared were GENCODE 7, RefSeq and UCSC Genes freeze July 2011, and AceView 2010 release. First, the exon coordinates of all protein-coding and lncRNA transcripts, respectively, were compared among different data sets. If transcripts were multi-exonic, the transcript boundaries were ignored, thus allowing for some flexibility in the annotations of their 5′ and 3′ ends. Same exon coordinates implied that a transcript was shared between two data sets. Second, the CDS coordinates of protein-coding transcripts, including the intervening exon junctions, were also compared, and an exact match was required to consider that a CDS was shared between two data sets. The overlaps between different data set combinations were graphically represented as three-way Venn diagrams using the Vennerable R package (https://r-forge.r-project.org/projects/vennerable/) and edited manually.
PhyloCSF analysis
We used PhyloCSF (Lin et al. 2011) to identify potential novel coding genes in RNA-seq transcript models based on evolutionary signatures. For each transcript model generated from the Illumina HBM data using either Exonerate or Scripture, we generated a mammalian alignment by extracting the alignment of each exon from UCSC's vertebrate alignments (which includes 33 placental mammals) and “stitching” the exon alignments together. We then ran PhyloCSF on each transcript alignment using the settings “-f 6–orf StopStop3–bls,” which cause the program to evaluate all ORFs in six frames and report the best-scoring. The “–bls” setting causes the program to additionally report a branch length score (BLS), which measures the alignment coverage of the best-scoring region as the percentage of the neutral branch length of the 33 mammals actually present in the alignment (averaged across the individual nucleotide columns). We selected transcripts containing a region with a PhyloCSF score of at least 60 (corresponding to a 1,000,000:1 likelihood ratio in favor of PhyloCSF's coding model) and a BLS of at least 25% for manual examination by an annotator.
APPRIS (CNIO)
The APPRIS annotation pipeline deploys a range of computational methods to provide value to the annotations of the human genome. The server flags variants that code for proteins with altered structure, function or localization, and exons that are evolving in a non-neutral fashion. APPRIS also selects one of the CDS for each gene as the principal functional isoform. The pipeline is made up of separate modules that combine protein structure and function information and evolutionary evidence. Each module has been implemented as a separate web service.
firestar (Lopez et al. 2007, 2011) is a method that predicts functionally important residues in protein sequences.
Matador3D is locally installed and checks for structural homologs for each transcript in the PDB (Berman et al. 2000).
SPADE uses a locally installed version of the program Pfamscan (Finn et al. 2010) to identify the conservation of protein functional domains.
INERTIA detects exons with non-neutral evolutionary rates. Transcripts are aligned against related species using three different alignment methods, Kalign (Lassmann and Sonnhammer 2005), multiz (Blanchette et al. 2004), and PRANK (Loytynoja and Goldman 2005), and evolutionary rates of exons for each of the three alignments are contrasted using SLR (Massingham and Goldman 2005).
CRASH makes conservative predictions of signal peptides and mitochondrial signal sequences by using locally installed versions of the SignalP and TargetP programs (Emanuelsson et al. 2007) .
THUMP makes conservative predictions of trans-membrane helices by analyzing the output of three locally installed trans-membrane prediction methods, MemSat (Jones 2007), PRODIV (Viklund and Elofsson 2004), and PHOBIUS (Kall et al. 2004).
CExonic is a locally developed method that uses exonerate (Slater and Birney 2005) to align mouse and human transcripts and then looks for patterns of conservation in exonic structure.
CORSAIR is a locally installed method that checks for orthologs for each variant in a locally installed vertebrate protein sequence database.
Data access
Dates and release notes are listed on the website http://www.gencodegenes.org, as well as more details of the data access, which are listed in Supplemental Table 11.
Acknowledgments
We thank all of the HAVANA team that contributed to the manual annotation and the Ensembl gene builders, and the VEGA team that produced the release of GENCODE. In addition, we thank the ZMAP/ANACODE group for developing the annotation tools and running the analysis pipeline for HAVANA. We also thank Sarah Grubb for help with the figures and formatting of the manuscript. This work was supported by the National Institutes of Health (grant no. 5U54HG004555) and the Wellcome Trust (grant no. WT098051).
Footnotes
[Supplemental material is available for this article.]
Article and supplemental material are at http://www.genome.org/cgi/doi/10.1101/gr.135350.111.
Freely available online through the Genome Research Open Access option.
References
- The 1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS 2011. lncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res 39: D146–D151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Jesus Martin M, O'Donovan C, Magrane M 2012. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40: D71–D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ara T, Lopez F, Ritchie W, Benech P, Gautheret D 2006. Conservation of alternative polyadenylation patterns in mammalian genes. BMC Genomics 7: 189 doi: 10.1186/1471-2164-7-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker TS, Rinkwitz S 2011. Zebrafish as a genomics model for human neurological and polygenic disorders. Dev Neurobiol 72: 415–428 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE 2000. The Protein Data Bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14: 708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, Song HW, Corbett MA, Gifford WD, Gecz J, et al. 2011. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell 42: 500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, Bult CJ, Agarwala R, Cherry JL, DiCuccio M, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol 7: e1000112 doi: 10.1371/journal.pbio.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A et al. 2011. The reality of pervasive transcription. PLoS Biol 9: e1000625 doi: 10.1371/journal.pbio.1000625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane G, Karsch-Mizrachi L, Nakamura Y; International Nucleotide Sequence Database Collaboration. 2011. The International Nucleotide Sequence Database Collaboration. Nucleic Acid Res39: D15–D18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, Beavis RC 2004. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics 20: 1466–1467 [DOI] [PubMed] [Google Scholar]
- Craig R, Cortens JP, Beavis RC 2004. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res 3: 1234–1242 [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res (this issue). doi: 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiere F, Deutsch EW, King NL, Nesvizhskii AI, Mallick P, Eng J, Chen S, Eddes J, Loevenich SN, Aebersold R 2006. The PeptideAtlas project. Nucleic Acids Res 34: D655–D658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi AM, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046 doi: 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. 2010. The Pfam protein families database. Nucleic Acids Res 38: D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2011. Ensembl 2011. Nucleic Acids Res 39: D800–D806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. 2012. Ensembl 2012. Nucleic Acids Res 40: D84–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. 2011. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res 39: D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S 2010. miRBase: microRNA sequences and annotation. Curr Protoc Bioinformatics 29: 12.9.1–12.9.10 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. 2010. Ab initio reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28: 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YJ, Ma SF, Yourek G, Park YD, Garcia JG 2011. A transcribed pseudogene of MYLK promotes cell proliferation. FASEB J 25: 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D et al. 2006. GENCODE: Producing a reference annotation for ENCODE. Genome Biol 7: S4–S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holford ME, Khurana E, Cheung KH, Gerstein M 2010. Using semantic web rules to reason on an ontology of pseudogenes. Bioinformatics 26: i71–i78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howald C, Tanzer A, Chrast J, Kokocinski F, Derrien T, Walters N, Gonzalez JM, Frankish A, Aken BL, Hourlier T, et al. 2012. Combining RT-PCR-seq and RNA-seq to catalog all genic elements encoded in the human genome. Genome Res (this issue). doi: 10.1101/gr.134478.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Cancer Genome Consortium 2010. International network of cancer genome projects. Nature 464: 993–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L 2010. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 16: 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT 2007. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23: 538–544 [DOI] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Kokocinski F, Harrow J, Hubbard T 2010. AnnoTrack: A tracking system for genome annotation. BMC Genomics 11: 538 doi: 10.1186/1471-2164-11-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL 2005. Kalign: An accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6: 298 doi: 10.1186/1471-2105-6-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci 100: 189–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M 2011. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G, Valencia A, Tress ML 2007. firestar—prediction of functionally important residues using structural templates and alignment reliability. Nucleic Acids Res 35: W573–W577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G, Maietta P, Rodriguez JM, Valencia A, Tress ML 2011. firestar—advances in the prediction of functionally important residues. Nucleic Acids Res 39: W235–W241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A, Goldman N 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci 102: 10557–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massingham T, Goldman N 2005. Detecting amino acid sites under positive selection and purifying selection. Genetics 169: 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS 2004. RNA regulation: A new genetics? Natl Rev 5: 316–323 [DOI] [PubMed] [Google Scholar]
- Mazzoni C, Falcone C 2011. mRNA stability and control of cell proliferation. Biochem Soc Trans 39: 1461–1465 [DOI] [PubMed] [Google Scholar]
- McEntee G, Minguzzi S, O'Brien K, Ben Larbi N, Loscher C, O'Fagain C, Parle-McDermott A 2011. The former annotated human pseudogene dihydrofolate reductase-like 1 (DHFRL1) is expressed and functional. Proc Natl Acad Sci 108: 15157–15162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL 2011. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol 30: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML 2010. Sequencing technologies: The next generation. Natl Rev 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Mott R 1997. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl Biosci 13: 477–478 [DOI] [PubMed] [Google Scholar]
- Pei B, Sisu C, Frankish A, Howald C, Habegger L, Mu XJ, Harte R, Balasubramanian S, Tanzer A, Diekhans M, et al. 2012. The GENCODE pseudogene resource. Genome Biol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP 2010. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 465: 1033–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. 2009. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res 19: 1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Brown GR, Maglott DR 2012. NCBI Reference Sequences (RefSeq): Current status, new features and genome annotation policy. Nucleic Acids Res 40: D130–D135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Turner S, Kasarskis A 2010. A window into third-generation sequencing. Hum Mol Genet 19: R227–R240 [DOI] [PubMed] [Google Scholar]
- Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA 2011. genenames.org: The HGNC resources in 2011. Nucleic Acids Res 39: D514–D519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle SM, Gilbert J, Iyer V, Clamp M 2004. The otter annotation system. Genome Res 14: 963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31 doi: 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Wootton JC 2001. Integrated graphical analysis of protein sequence features predicted from sequence composition. Proteins 45: 262–273 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kato S, Murata M, Carninci P 2012. CAGE (cap analysis of gene expression): A protocol for the detection of promoter and transcriptional networks. Methods Mol Biol 786: 181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry-Mieg D, Thierry-Mieg J 2006. AceView: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol 7: S12–S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress ML, Wesselink JJ, Frankish A, Lopez G, Goldman N, Loytynoja A, Massingham T, Pardi F, Whelan S, Harrow J, et al. 2008. Determination and validation of principal gene products. Bioinformatics 24: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund H, Elofsson A 2004. Best α-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci 13: 1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY 2011. Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL 2008. The vertebrate genome annotation (Vega) database. Nucleic Acids Res 36: D753–D760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carriero N, Zheng D, Karro J, Harrison PM, Gerstein M 2006. PseudoPipe: An automated pseudogene identification pipeline. Bioinformatics 22: 1437–1439 [DOI] [PubMed] [Google Scholar]
- Zhang ZD, Frankish A, Hunt T, Harrow J, Gerstein M 2010. Identification and analysis of unitary pseudogenes: Historic and contemporary gene losses in humans and other primates. Genome Biol 11: R26 doi: 10.1186/gb-2010-11-3-r26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Frankish A, Baertsch R, Kapranov P, Reymond A, Choo SW, Lu Y, Denoeud F, Antonarakis SE, Snyder M, et al. 2007. Pseudogenes in the ENCODE regions: Consensus annotation, analysis of transcription, and evolution. Genome Res 17: 839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]