Abstract
The human genome contains many thousands of long noncoding RNAs (lncRNAs). While several studies have demonstrated compelling biological and disease roles for individual examples, analytical and experimental approaches to investigate these genes have been hampered by the lack of comprehensive lncRNA annotation. Here, we present and analyze the most complete human lncRNA annotation to date, produced by the GENCODE consortium within the framework of the ENCODE project and comprising 9277 manually annotated genes producing 14,880 transcripts. Our analyses indicate that lncRNAs are generated through pathways similar to that of protein-coding genes, with similar histone-modification profiles, splicing signals, and exon/intron lengths. In contrast to protein-coding genes, however, lncRNAs display a striking bias toward two-exon transcripts, they are predominantly localized in the chromatin and nucleus, and a fraction appear to be preferentially processed into small RNAs. They are under stronger selective pressure than neutrally evolving sequences—particularly in their promoter regions, which display levels of selection comparable to protein-coding genes. Importantly, about one-third seem to have arisen within the primate lineage. Comprehensive analysis of their expression in multiple human organs and brain regions shows that lncRNAs are generally lower expressed than protein-coding genes, and display more tissue-specific expression patterns, with a large fraction of tissue-specific lncRNAs expressed in the brain. Expression correlation analysis indicates that lncRNAs show particularly striking positive correlation with the expression of antisense coding genes. This GENCODE annotation represents a valuable resource for future studies of lncRNAs.
The cellular economy is transacted by both proteins and nonprotein-coding RNAs. Historically, proteins (and the messenger RNAs that encode them) have tended to dominate our view of the cell and its genome due to their abundance and to the relative ease with which protein-coding genes, and their gene products, can be identified and studied. However, in recent years this paradigm has been undermined as new technologies have provided accelerating depths of RNA sequencing. We now appreciate the pervasive transcription of numerous long and small RNA species in mammalian genomes, forcing us to radically reinterpret our understanding of the genome (Carninci et al. 2005; ENCODE Project Consortium 2007). In particular, attention is now shifting toward one of the most poorly understood, yet most common RNA species: long noncoding RNAs (lncRNAs).
The discovery and study of lncRNAs is of major relevance to human biology and disease since they represent an extensive, largely unexplored, and functional component of the genome (Mattick 2009; Ponting et al. 2009). While enough lncRNAs have been implicated in human disease to justify major investment in genome-wide screens for new lncRNA candidates, such studies are hampered by the current lack of lncRNA annotation. Thus, there is a need for the curation of high-quality catalogs of lncRNAs and information on the tissues in which they are expressed. Similar information for protein-coding genes has long been available. The FANTOM consortium pioneered the genome-wide discovery of lncRNAs in mouse in the early 2000s, publishing a set of 34,030 lncRNAs based on cDNA sequencing (Maeda et al. 2006). Only recently was a catalog of 5446 human lncRNAs created, based on a computational pipeline of sequenced cDNAs by Jia et al. (2010). Meanwhile the large intergenic noncoding RNAs (“lincRNAs”) (Guttman et al. 2009; Khalil et al. 2009), discovered through epigenetic annotation of human and mouse genomes, represent a useful set of RNAs, but omit the many lncRNAs that reside within or overlap protein-coding loci, and do not explicitly provide lncRNA gene structures. Most recently, Cabili et al. (2011) created a catalog of 4662 human intergenic lncRNAs by combining partially complete GENCODE annotations and computational predictions based on RNA-seq data. Similarly for lncRNA expression data, efforts are underway to create databases and microarray expression platforms for mouse lncRNAs (Dinger et al. 2009), but no equivalent data has been available in human.
In fact, the GENCODE consortium within the ENCODE project has for several years been manually annotating a comprehensive set of human lncRNAs. Early releases of the GENCODE annotation have already been used to investigate the potential function of these transcripts (Ørom et al. 2010). Version 7 (v7) of GENCODE (Harrow et al. 2012), the reference annotation used for the ENCODE analysis (The ENCODE Project Consortium 2012) has introduced a number of biotypes to specifically define different classes of lncRNAs, consolidating a GENCODE lncRNA set. Here, we report on the annotation of this set, which includes 14,880 manually curated and evidence-based transcripts. We integrate these lncRNAs with other transcriptome and epigenome data sets produced within the ENCODE project and elsewhere. We show that lncRNAs have canonical gene structures and histone modifications. They tend to be under weaker evolutionary constraint than coding genes, and to be expressed at lower levels. As a class, lncRNAs are preferentially enriched in the chromatin and nucleus of the cell. We present expression maps of these transcripts throughout the human body and brain. Overall, the GENCODE lncRNA catalog represents a valuable resource for future studies on the role of lncRNAs in human biology.
Results
Identification and initial categorization of lncRNAs in the GENCODE gene annotation
The lncRNA catalog described in this study represents a subset of the manually annotated GENCODE human gene annotation catalog (Harrow et al. 2012; www.gencodegenes.org/) that consists of 15,512 transcripts grouped in 9640 gene loci. Thus, the GENCODE lncRNA annotation constitutes the largest manually curated catalog of human lncRNAs. These lncRNAs can been further reclassified into the following locus biotypes based on their location with respect to protein-coding genes:
Antisense RNAs, which have transcripts that intersect any exon of a protein-coding locus on the opposite strand, or published evidence of antisense regulation of a coding gene.
LincRNA are transcripts that are intergenic noncoding RNA loci with a length >200 bp.
Sense overlapping transcripts contain a coding gene within an intron on the same strand.
Sense intronic transcripts reside within introns of a coding gene, but do not intersect any exons.
Processed transcripts do not contain an open reading frame (ORF) and cannot be placed in any of the other categories.
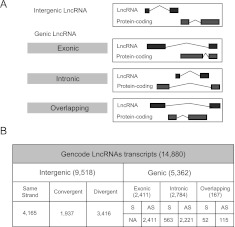
We have applied this categorization automatically to the GENCODE 7 lncRNA data set, resulting in the following distribution: antisense (3233), lincRNA (5094), sense intronic (378), and processed transcript (935). Since the exact boundaries of protein-coding loci are difficult to predict due to unannotated alternatively spliced transcripts, we have defined the boundaries of protein-coding genes to be 5 kb upstream of the start codon and 30 kb downstream from the stop codon (Fig. 1A).
Figure 1.
Manual annotation of lncRNAs in the human genome. (A) How lncRNAs were subclassified based on intersection with protein-coding genes. Priority was assigned to protein-coding exonic intersect over intronic or overlapping. Then, in cases where multiple protein-coding transcripts could be chosen, the protein-coding transcript having the longest intersect with the lncRNA was considered the best partner over the others (see Methods). (B) Number of lncRNA transcripts per subcategory. (S) Same sense; (AS) antisense.
For the analysis presented in this study, we removed transcripts shorter than 200 bp (n = 198) and also those containing at least one exon intersecting a protein-coding exon on the same strand (n = 594). This results in a final set of 14,880 transcripts originating from 9277 loci. We then subclassified this set of GENCODE lncRNAs biotypes according to their localization with respect to the nearest known protein transcripts (exonic, intronic, overlapping) (Fig. 1B). The majority of lncRNA transcripts (9518) do not intersect with any protein-coding genes and were therefore considered as intergenic (Fig. 1B) and analogous to the “lincRNAs” that were defined by chromatin signatures (Khalil et al. 2009). The remaining 5362 lncRNAs that intersect protein-coding transcripts in some way were further categorized by those covering protein-coding exons (2411), introns (2784), or overlapping protein-coding (i.e., where the protein-coding transcript lies within an intron of the lncRNA) (167) (Fig. 1B). Note that transcripts intersecting protein-coding exons on the same strand were previously omitted from this analysis.
The GENCODE lncRNA data set is larger than other available lncRNA data sets, and it shows limited intersect with them. A total of 42% (44 out of 96) of the long noncoding RNA database lncRNAdb (Amaral et al. 2011) are represented in GENCODE lncRNAs. We checked the same-strand overlap against recent lncRNA catalogs: GENCODE v7 lncRNAs contain 30% of Jia et al. (2010) lncRNAs, 33% of Cabili et al. (2011) (stringent) lincRNAs, 39% of Cabili et al. (2011) (all) lincRNAs, and 12% of vlincs (Supplemental Fig. S1; Kapranov et al. 2010). We also examined the overlap with the Khalil et al. (2009) set of human lincRNAs; however, we could not perform an accurate intersection analysis due to their lack of strand information. Manual curation of the remaining genes in lncRNAdb reveals many that intersect with protein-coding genes and/or small RNAs. A full characterization of the GENCODE v7 lncRNA set can be downloaded as Supplemental Table S1 from: http://big.crg.cat/bioinformatics_and_genomics/lncrna_data.
LncRNAs do not show evidence of protein-coding potential
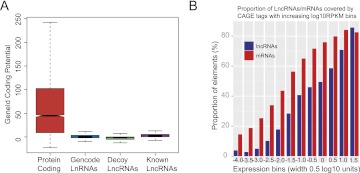
There is some evidence that transcripts thought to be purely noncoding lncRNAs may in fact encode proteins, in small or otherwise unrecognized ORFs (Chooniedass-Kothari et al. 2004; Kondo et al. 2010; Dinger et al. 2011). To assess the protein-coding status of the present lncRNAs catalog, we first used the program GeneID (Blanco et al. 2007) to (1) measure the protein-coding potential and (2) find the longest possible ORF in each lncRNA sequence. We compared the results in the set of GENCODE lncRNAs with (1) known protein-coding transcripts, (2) experimentally validated lncRNAs (such as XIST, H19), and (3) a set of “decoy” lncRNAs, obtained by mapping lncRNA gene structures randomly onto the genome (see Methods). Figure 2A shows that lncRNAs have a similar coding potential and contain ORFs similar in length to known lncRNAs and decoy lncRNA (Supplemental Fig. S2A), but very different from protein-coding genes (Wilcoxon-Mann-Whitney: P < 2.2 × 10−16). Thus, at least at a sequence level, the lncRNA catalog does not appear to have ORFs of higher quality than expected of random sequences.
Figure 2.
GENCODE lncRNAs are independent, noncoding transcripts. (A) Protein-coding potential of transcripts computed in four data sets: protein-coding (red), GENCODE v7 lncRNAs (blue), decoy lncRNAs (green), and known lncRNAs (XIST, H19…) (purple). (B) Proportion of GENCODE lncRNAs and mRNAs transcripts with CAGE clusters mapped around their transcription start sites (TSSs) (see Methods) in bins of increasing expression levels (log10 RPKM).
On the other hand, investigation of mass spectrometry (MS) conducted within the ENCODE project on nine compartment-specific proteome samples from GM12878 and K562 cell lines identified 350 peptides that matched the GENCODE lncRNA set (out of a total of 79,333 peptides). These were found in only 111 lncRNA transcripts from 69 distinct loci. Among those 69 loci, only 12 have multiple in-frame peptides, providing particularly strong evidence of translation. Overall, these results, reported in detail (Bánfai et al. 2012), support the conclusion that most GENCODE lncRNAs lack any protein-coding potential.
The majority of GENCODE lncRNAs are independent transcriptional units
Annotation of lncRNAs is made challenging by the low expression levels of these transcripts, which may lead to fragmentary annotation and poor definition of the transcript boundaries. Concerns have also been raised as to whether lncRNAs are independent transcripts, or whether they are simply unrecognized extensions of neighboring protein-coding transcripts (van Bakel et al. 2010). To test whether this is the case for the GENCODE lncRNAs, we used various high-throughput sequencing data produced in the context of the ENCODE project to search for evidence of physical linkage of lncRNA transcripts and neighboring protein-coding genes. We first used CAGE tags obtained in 12 experiments to search for experimental validation of the annotated start sites of lncRNAs (Djebali et al. 2012), and found support (at least one tag within ±100 bp from the annotated 5′ end in at least one experiment) for 15% of lncRNA transcripts, compared with 55% of protein-coding transcripts (Supplemental Table S3). This could, in principle, suggest that lncRNA promoters are poorly annotated. However, this analysis is confounded by the lower expression level of lncRNAs compared with protein-coding genes. To control for gene expression, we computed the fraction of genes that have CAGE tags by binning the genes according to expression levels. When controlling for gene expression, we find that for each expression bin, protein-coding genes have ∼15% greater CAGE tag coverage compared with lncRNAs (Fig. 2B). We then sought to further delineate the boundaries of lncRNA transcripts using RNA paired-end ditags (PETs), a method in which the extreme 5′ and 3′ regions of RNA molecules are sequenced (Ng et al. 2005). Using PETs obtained in 16 experiments (Djebali et al. 2012), we found support simultaneously at the 3′ and 5′ end for 10% of the lncRNAs, compared with 39% of protein-coding genes (Supplemental Table S3). When binning for gene expression levels, we found a similar behavior as for CAGE, with ∼15% greater PET coverage for protein-coding transcripts in each expression bin (Supplemental Fig. S2B). We also tested for the presence of known poly(A) motifs in the 3′ end (100 bp surrounding the annotated transcription termination site) of GENCODE lncRNAs in comparison with protein-coding transcripts (see Methods). Overall, 39% of LncRNAs transcripts contain at least one of the six most common poly(A) motifs, compared with 51% observed for coding transcripts. This difference may be explained by the higher proportion of non-poly(A) lncRNAs compared with protein-coding transcripts (Supplemental Table S3; Supplemental Fig. S14B). In contrast, the proportion of decoy lncRNAs containing poly(A) motifs at the 3′end was 26%.
Finally, we used RNA-seq paired-end reads (PE reads) from three ENCODE cell lines and three compartments (Djebali et al. 2012) to assess potential “bridging” between lncRNAs and protein-coding transcripts, i.e., cases where the lncRNA and the coding transcript appear to originate from a single RNA molecule. We reasoned that if lncRNA transcripts are indeed independent, then we should observe no more PE reads connecting them to neighboring protein-coding genes than between protein-coding genes themselves. We found that 9% of lncRNAs are connected by at least one PE read in at least one experiment (see Methods). In principle, while this could suggest that many lncRNAs are unannotated UTRs from protein-coding genes, the proportion of protein-coding genes that are connected by PE reads to neighboring protein-coding genes is actually larger at 17% (Supplemental Table S3). Binning for expression level, we found that this increased connectivity between protein-coding genes is not an artifact of their greater expression (Supplemental Fig. S2C). We also investigated whether all of the lncRNA classes had equally strong experimental support for their 5′ and 3′ annotations (Supplemental Table S8). This showed that sense intronic transcripts had generally weaker support for their annotated start and end sites, suggesting that the quality of their annotation is weaker in general than the other lncRNA subclasses. In summary, these data suggest that the majority of lncRNAs are unlikely to represent unannotated extensions of neighboring protein-coding genes.
LncRNAs have unusual exonic structure, but exhibit standard canonical splice site signals, and alternative splicing
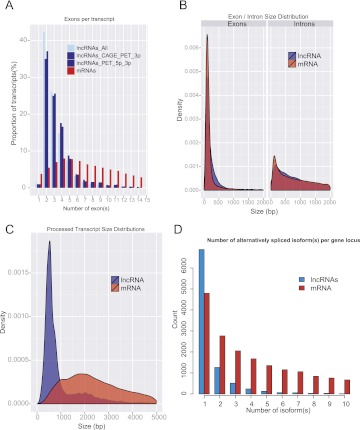
Most LncRNAs are spliced (98%), but they show a striking tendency to have only two exons (42% of lncRNA transcripts have only two exons compared with 6% of protein-coding genes) (Fig 3A). This does not seem to be an artifact of lncRNA's low expression, or poor annotation, since even subsets with experimental support for their 5′ and 3′ boundaries exhibit this effect (Fig. 3A). While lncRNA exons are slightly longer than those of protein-coding transcripts (medians 149 and 132 bp, respectively; t-test, P-value = 0.00014), introns from lncRNAs are longer that those from protein-coding genes (medians 2280 bp and 1602 bp, respectively: t-test, P-value <2.2 × 10−16) (Fig. 3B). Because they have less exons, overall lncRNA transcripts are shorter than protein-coding (median 592 bp compared with 2453bp for protein-coding transcript; t-test, P-value <2.2 × 10−16) (Fig. 3C). Interestingly, the longest lncRNA is NEAT1, a single-exon lincRNA of 22.7 kb, which was recently shown to be necessary for the formation of nuclear paraspeckles (Sunwoo et al. 2009). In addition, >25% of lncRNA genes show evidence of alternative splicing with at least two different transcript isoforms per gene locus (Fig. 3D). The most highly spliced lncRNA gene is PCBP1-AS1 with 40 annotated isoforms. This human lncRNA gene is situated at a complex locus with major gene structure differences between human and mouse orthologs (Supplemental Fig. S3).
Figure 3.
Features of lncRNA gene structure. (A) Number of exons per transcripts for all lncRNA transcripts (light blue), lncRNAs having CAGE or PET supports for either their 5′ or 3′ ends (blue), lncRNAs having PET tags mapping to both ends of the transcript (dark blue), and protein-coding transcripts (red). (B) Exon (left) and intron (right) size distributions for lncRNA and mRNAs. (C) Processed transcript size distributions of lncRNAs (blue) and protein-coding (red). (D) Distribution of the number of alternative spliced forms per lncRNA (blue) and protein-coding (red) gene locus.
The vast majority of lncRNA introns are flanked by canonical splices sites (GT/AG), and we find no differences in splicing signal usage compared with protein-coding genes (Supplemental Table S4; Supplemental Fig. S4). Finally, we have also identified 11 lncRNAs U12 introns (Alioto 2007) within the lncRNA catalog, of which eight belong to intergenic lncRNAs (lincRNAs) and three are in antisense of protein-coding introns.
Human lncRNAs are under weaker selective constraints than protein-coding genes, and many are primate specific
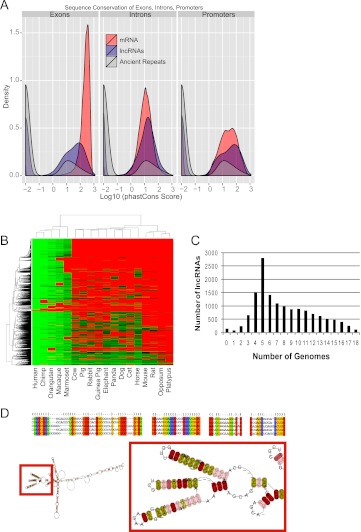
Purifying selection of genomic sequence represents powerful evidence for functionality and, thus, we sought to assess whether the GENCODE lncRNAs have experienced such selection. We used the precomputed, nucleotide-level calculations of evolutionary selection provided by the phastCons algorithm (Siepel et al. 2005). By this measure, lncRNA exons are significantly more conserved than neutrally evolving ancestral repeat (AR) sequences, albeit at lower levels than protein-coding genes (Fig. 4A). These findings are in agreement with studies of other lncRNA catalogs (Ponjavic et al. 2007; Guttman et al. 2009; Marques and Ponting 2009; Ørom et al. 2010). We also compared the sequence conservation of different regions of lncRNA genes: promoters, exons, and introns (Fig. 4A). In fact, lncRNA promoters are on average more conserved than their exons, and almost as conserved as protein-coding gene promoters, as observed in mouse lincRNAs (Guttman et al. 2009).
Figure 4.
Evolutionary conservation of lncRNAs. (A) Density plots of phastCons score distributions of protein genes (red curves), lncRNA genes (blue curves), and ancestral repeats (gray curve) for exons (left), introns (middle), and promoters (right). (B) Human lncRNA conservation in mammals: The heatmap summarizes the lncRNA orthologs discovered in 18 other mammalian genomes (see Methods). (Columns) Mammal species. (Rows) Query lncRNAs. The color scheme reflects the level of sequence similarity (percent identity) measured between query and target homologs. (Red) No reliable homolog was detected. (C) The number of orthologs discovered for each lncRNA. LnRNAs with zero orthologs are those that could not be reliably remapped to the human genome at the levels of stringency used in the analysis, due to high repeat content. (D) Example of a multiple sequence alignment of a five-member family. The position containing compensated mutations are labeled by orange columns (correlated) and red columns (correlated Watson-Crick). (Yellow columns) Perfect Watson-Crick matches; (green columns) neutral matches (including G-U pairs); (blue columns) incompatible matches. The putative 2D consensus structure shown is based on the full multiple sequence alignment (RNAaliFold minimum folding energy). (Red box) Details of the 2D structure, with the precise location of the groups of compensated mutations. The colors associated with the residues indicate mutational pattern with respect to the structure as reported by RNAalifold.
The relatively fast evolutionary change of lncRNAs reported here depends on phastCons-based analysis and, therefore, on the accuracy of an underlying multiple genome alignment (MGA). To avoid potential underestimation of conservation of noncoding sequence by this method, we completed this study with an MGA-independent assessment of the transcripts' conservation across mammalian genomes. We systematically BLASTed human lncRNAs against all available mammalian genomes, and subsequently used exonerate (Slater and Birney 2005) to reconstruct gene models on the genome sections yielding hits strong enough to support the presence of a homologous gene (Fig. 4B). With this method, ∼30% of lncRNA transcripts (n = 4546) appear to be primate specific. Altogether, this high primate conservation explains the large number of lncRNAs conserved in five species (2802) (Fig. 4C), and the derived clustering recapitulates well the most commonly accepted primate tree of life. A total of 0.7% (101) of transcripts appear to be specific to the human lineage. A similar number (134; 1.0%) is found reciprocally in all of the 18 species analyzed here. This figure increases to 3.4% (n = 506) if we ignore the two marsupials in the analysis, opossum and platypus. These widely conserved transcripts show an average intronic size about twice that of the full lncRNA set, whereas their exons are shorter.
We were interested in whether lncRNAs may belong to evolutionarily related families. We clustered all the transcripts by sequence similarity using BLASTClust (Altschul et al. 1990), identifying 194 families with between two and 96 members, and percent identity between 49% and 100% (Supplemental Table S1). It is worth noting that 138 out of 194 families contain only two members, and only three families contain more than 10 members. This may reflect a high turnover rate or the difficulty of effectively aligning fast-evolving sequences. A more stringent analysis also revealed that the majority of these families were defined by a degraded version of common repeats such as LINE, SINE, and LTRs within the lncRNA sequence (Supplemental Fig. S5). Interestingly, the multiple alignments revealed a high number of correlated positions (1100 in total; on average, five per kilobase of multiple sequence alignment) that may be interpreted as evidence of evolutionary conservation of RNA secondary structures. It is also worth pointing out that some of the families thus identified do not contain any identifiable repeat sequence, while others contain conserved structural elements. One such family is shown on Figure 4D along with its predicted fold and 10 identified compensatory mutations maintaining Watson-Crick base pairing, and clustered in the same predicted loop structure.
Expressed lncRNAs have typical histone modifications
Histone modifications are known to play a role in the regulation of gene expression (Barski et al. 2007) and have been successfully used as a proxy for the identification of novel lncRNAs (Guttman et al. 2009). We investigated the chromatin signatures of the GENCODE lncRNA genes and compared them with protein-coding genes based on ChIP-seq data from eight cell lines and eight chromatin marks (Ernst et al. 2011). We produced aggregate plots of ChIP-seq reads (see Methods) for histone methylation and acetylation patterns around TSSs (transcription start sites) of lncRNAs and coding transcripts (Supplemental Fig. S6). To eliminate the confounding influence of transcriptionally silent lncRNAs, we analyzed only those lncRNAs expressed in the same cell type where the histone modifications were measured. LncRNA TSS histone profiles are similar to those of protein-coding genes for several active histone marks (H3K4me2, H3K4me3, H3K9ac, H3K27ac), but have slightly excess levels of other marks associated with both silencing (H3K27me3) and activity (H3K36me3). Chromatin marks are more pronounced for the 2157 lncRNAs with 5′-support (see previous section) than for the total set of lncRNAs—probably due to the higher expression of the lncRNAs in this subset, and the greater precision of their 5′ annotation. In summary, expressed lncRNAs have histone modifications indicative of actively regulated gene promoters.
Some lncRNAs may be post-processed into smaller RNAs, particularly snoRNAs
Many lncRNAs may serve as precursors for functional small RNAs (sRNA), with or without having intrinsic functionality themselves (Askarian-Amiri et al. 2011). To evaluate this for the present lncRNA set, we compared their genomic position with small RNAs on the same strand, as annotated by GENCODE (Harrow et al. 2012). A total of 27% of all annotated small RNAs (tRNAs, miRNAs, snRNAs, and snoRNAs) map within the genic boundaries of 7% of all protein-coding genes, while 5% of small RNAs map within the boundaries of 4% of all lncRNAs. This does not necessarily rule out a propensity for lncRNAs to host small RNAs compared with protein-coding genes, because this analysis is biased by the greater number and length of protein-coding genes. To control for this, we computed the proportion of nucleotides in lncRNAs that overlap different classes of small RNAs, and compared it with similar data for protein-coding genes and intergenic background. This revealed that lncRNA exons are enriched for all classes of small RNAs, with the exception of snRNAs, compared with other genomic domains, including lncRNA introns. Particularly striking is the enrichment for snoRNAs, which are present in sixfold excess in lncRNA exons compared with other genomic domains (Supplemental Fig. S7). Nevertheless, it is important to note that, in absolute terms, more snoRNAs arise from lncRNA introns compared with exons, due to the far greater length of the former.
LncRNAs show lower and more tissue-specific expression than protein-coding genes
We investigated the expression patterns of lncRNAs in a wide range of human organs and cell lines using available RNA-seq data as well as a custom lncRNA microarray. We were particularly interested in understanding the magnitude of lncRNA expression, as well as its degree of tissue specificity.
Using RNA-seq
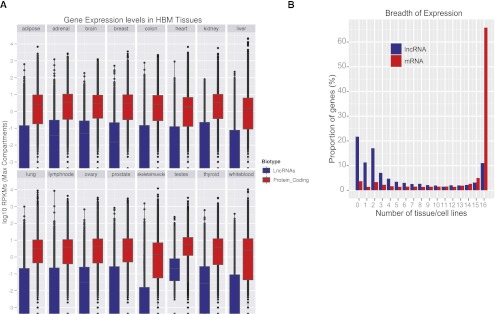
We used RNA-seq data obtained in various human tissues by the Illumina Human Body Map Project (HBM) (www.illumina.com; ArrayExpress ID: E-MTAB-513). HBM reads were mapped using the ENCODE RNA-seq pipeline (Djebali et al. 2012) and GENCODE lncRNA transcripts were quantified, as RPKM (read per kilobase of exon per million mapped reads) (Mortazavi et al. 2008), using the FluxCapacitor (Montgomery et al. 2010). We computed the distribution of expression of lncRNAs and protein-coding genes across the 16 tissues profiled in the HBM project (Fig. 5A). As shown previously (Ravasi et al. 2006; Ørom et al. 2010), lncRNAs show lower expression in all tissues compared with mRNAs, although lncRNAs show relatively high expression in testis. LncRNAs also show more tissue-specific patterns compared with protein-coding genes, although this may be a result of their lower expression levels and resultant false negative detection in some tissues when applying strict cutoff of expression (Supplemental Fig. S8). The majority (65%) of protein-coding genes were detected in all HBM tissues compared with 11% of lncRNAs (21% of lncRNAs were not be detected in any tissue, and 11% are only detected in a single tissue using an RPKM threshold greater than 0.1) (Fig. 5B). Consistent with this observation, we have also found that lncRNAs show higher expression variability, measured as the coefficient of variation across cell lines and tissues, than protein-coding genes (Supplemental Fig. S9).
Figure 5.
Characteristics of lncRNA expression in human tissues. (A) Distributions of lncRNA (blue) and protein-coding (red) transcripts' expression (log10 RPKM) in HBM tissues. (B) Distribution of the number of HBM tissues in which lncRNA and protein-coding transcripts' are detected (RPKM > 0.1).
Mapping lncRNA expression in the human body by custom microarray
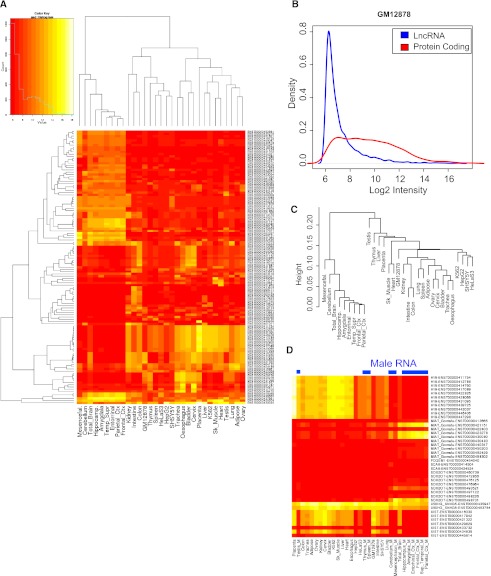
To get a deeper picture of lncRNA expression throughout the human body, we developed a custom microarray platform capable of quantifying the transcripts in the GENCODE lncRNA annotation. The array was printed with multiple nonredundant 60-mer oligonucleotides targeting 9747 lncRNA transcripts from the GENCODE version 3c annotation. We hybridized this array with human RNA from a range of sources: five common cell lines, of which four are used by ENCODE; nine brain regions; 17 other tissues from the adult body (Fig. 6A). The microarray results are available in the Supplemental Data online. Overall, we detected essentially all transcripts (99%) expressed in at least two cell types and 29% in all 31 cell types using standard microarray analysis (Supplemental Fig. S10). Using more stringent methods did not substantially alter the numbers of lncRNAs detected (95% and 28%, respectively). In accordance with previous microarray studies (Dinger et al. 2008) and RNA-seq (Fig. 5A), we found that lncRNAs are generally far lower expressed than protein-coding genes (Fig. 6B).
Figure 6.
Microarray analysis of lncRNA expression in the human body. (A) The heatmap shows expression of the 121 most variably expressed lncRNAs (rows), defined as those with a coefficient of variation >0.2 across 31 cell/tissue types (columns). In the color scheme, yellow indicates higher expression, red indicates lower expression. (B) The intensity distribution of lncRNAs compared with protein-coding mRNAs. The data from GM12878 cells are shown, but similar results were observed in all samples. (C) A tree of expression correlation between samples; correlations were calculated using the expression of all lncRNAs in each sample. (D) The expression pattern of known lncRNAs. RNAs were manually curated from the literature. (Blue bars) Those RNA samples that do not contain any female component. Each row corresponds to a lncRNA transcript, and most lncRNA genes are represented on the array by several different transcript isoforms, resulting in multiple entries per lncRNA.
To gauge the reliability of the microarray platform, we performed extensive comparison to RNA-seq for the four ENCODE cell line samples that were analyzed using both methods. Comparison of the control protein-coding genes that were printed on the microarray to the RNA-seq data showed a high correlation of 0.6–0.7, consistent with other reports (Supplemental Fig. S11A; Fu et al. 2009). However, for lncRNA expression, the concordance between technology platforms was lower—with correlation coefficients from 0.24 to 0.31 (Supplemental Fig. S11B). This agrees with previous studies showing that the correlation between RNA-seq and microarrays is poor in genes that are either lowly (such as lncRNAs) or highly expressed (Wang et al. 2009). Although manual inspection of the correlation suggests that microarrays have lower accuracy in quantitating the absolute expression levels of lncRNAs compared with RNA-seq, it has to be noted that the microarray is more sensitive in detecting whether a lncRNA is expressed or not, compared with RNA-seq at the depth of sequencing in these experiments (Supplemental Figs. S8, S10).
Using the expression data from lncRNAs, we could cluster the 31 cell types and recover biologically meaningful relationships between them, particularly separating the brain from other tissues (Fig. 6C). Indeed, amongst the most differentially/expressed lncRNAs, a brain-specific cluster accounts for ∼40% (Fig. 6A). Finally, we examined the expression profiles of various known lncRNAs (Fig. 6D). As expected, the X chromosome-specific XIST transcript is only detected in RNA samples from females. MIAT (also known as Gomafu) and SOX2-OT have brain-specific expression, while H19 is highly expressed in the placenta.
Correlations of expression between lncRNAs/mRNAs genes reveal potential subclasses of interactions
The issue of whether lncRNA expression correlates with either neighboring (cis) or distal (trans) protein-coding genes has been a matter of debate in recent studies (Ørom et al. 2010; Cabili et al. 2011). We next analyzed whether any nonrandom coexpression patterns of lncRNA-mRNA exist in the ENCODE and HBM RNA-seq data.
Trans-acting correlation of expression
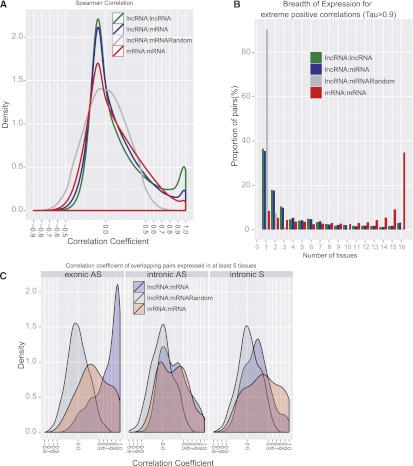
In order to highlight possible interactions between lncRNA and protein-coding genes, we computed all pairwise correlations between lncRNA and protein-coding genes (lncRNA-mRNA) using expression values (RPKM) from the 16 Human Body Map tissues (see Methods). The reliability of these correlations were estimated based on comparison with three different sets: (1) correlations of all-against-all protein-coding genes (mRNAs-mRNAs set), (2) lncRNA-mRNA correlation profiles where the expression of the coding genes were randomly shuffled (lncRNA-mRNA_Random) and lncRNAs-lncRNAs correlations (Supplemental Table S5). Within each of these data sets, every pairwise gene combination at a distance >1 Mb or involving interchromosomal elements were tested, therefore representing trans-correlations of expression (Fig. 7A). Overall, we observed that lncRNAs are more positively than negatively correlated with protein-coding genes (12.1% vs. 0.2% with Spearman coefficient rho (rs) >|0.5|, respectively, out of a total of about 100 million trans-correlations tested) (Supplemental Table S5). Yet, this tendency is also observed for correlations between protein-coding genes themselves (12.0% vs. 0.6%, respectively). However, lncRNA genes exhibit more extreme positive correlations (rs > 0.9) with protein-coding genes (2.6%) than protein-coding mRNAs with other mRNAs (1.3%) (Fig. 7A). This high proportion of positive correlations is mainly due to tissue-specific lncRNAs for which correlations of expression with lncRNAs or mRNAs only expressed in the same tissue (artificially) lead to high positive correlations. Two results support this hypothesis: (1) the higher frequency of lncRNAs-lncRNAs with extreme positive correlations (6.8%) compared with lncRNAs-mRNAs, and (2) the limited breadth of expression for pairwise correlations having a high coefficient of correlation (Fig. 7B). Indeed, when we removed tissue-specific genes for the calculation of coexpression, one could observe that the proportion of extreme positive correlations becomes significantly less pronounced (Supplemental Fig. S12A).
Figure 7.
Correlation of expression of lncRNAs and protein-coding transcripts. (A) Correlations of expression of all-against-all genes from different data sets involving trans-pairs of genes. (B) The breadth of expression for trans-pairs having a highly correlated profile of expression (rs > 0.9). (C) Correlations of expression of intersected genes for different categories: Intron AS (intronic antisense), intron S (intronic sense), and exonic AS (exonic antisense).
Cis-acting correlation of expression
We next focused our attention on pairwise correlations of expression involving neighboring genes (Supplemental Table S5). Several studies have demonstrated either positive (Kim et al. 2010; Ørom et al. 2010) or negative (Brockdorff et al. 1992; Nagano et al. 2008) regulation by intergenic lncRNAs (lincRNAs) of neighboring protein-coding genes. We found more positive (rs > 0.5) and more extreme positive (rs > 0.9) lncRNAs-mRNAs and mRNAs-mRNAs correlations in cis than in trans (Supplemental Table S5). But, in contrast to trans, we observed in cis more positive and more extreme positive lncRNA-mRNA correlations than mRNA-mRNA correlations (7.1% vs. 3.9%, of extreme positive correlations respectively, both significantly higher than random, 0.07%, Supplemental Table S5). Then, we asked whether the distance between neighboring pairs could have an impact on these coexpression patterns (Supplemental Fig. S12B,C). While 2.95% of lncRNAs have highly positive correlation with neighboring mRNAs within 20 kb, this proportion decreases to 0.44% when the neighbor mRNA is 80–100 kb away (Supplemental Table S6). For mRNAs-mRNAs cis-correlations, the proportion of proximal <20 kb extreme correlations is less important (1.5%), and thus, the fall of positive extreme correlation seems less distance dependent with 0.3% of highly correlated pairs being separated by 80–100 kb. To rule out that some of the correlations arise from lncRNAs being actually unannotated UTRs of the neighbor protein-coding genes, we focused on the set of intergenic lncRNAs that appear to be full-length as measured by PET data (see Section 3). A total of 31 such independent lncRNA units were highly correlated (rs > 0.7) with neighbor protein-coding genes at a median distance of 2.6 kb (Supplemental Table S7). Interestingly, using Gene Ontology (GO) analysis based on the DAVID web server (Huang da et al. 2009), we found that these lncRNAs are correlated with protein-coding genes enriched in “regulation of transcription” processes and nucleus compartment. These full-length lncRNAs, significantly coexpressed with nearby protein-coding genes, thus represent interesting candidates to be tested in further experimental studies.
Correlation of expression between overlapping lncRNAs and mRNAs
Almost 40% (3934 lncRNA genes, 5361 transcripts) of GENCODE lncRNAs intersect protein-coding gene loci, and it is possible that such lncRNAs somehow contribute to the regulation of the latter (Fig. 1B; Gingeras 2007; Pasmant et al. 2011). We therefore computed correlations of expression between these lncRNA genes and their “host” mRNAs and again found that lncRNAs have more positive correlations with intersecting mRNAs elements than expected by chance (Supplemental Table S5). Strikingly, compared with that observed in a control set of mRNAs or the random set, lncRNAs intersecting protein-coding exons in antisense orientation (“exonic antisense”) appear to be specifically enriched in positive correlations with the mRNA host (Fig. 7C), as observed previously. Much weaker correlations were observed between mRNAs and their intronic lncRNAs, regardless of whether the lncRNA is on the same or opposite strand. In addition, contrary to our observations above, this finding still holds true when we included tissue-specific lncRNAs (Supplemental Fig. S12D). Altogether, we identified 186 lncRNA genes intersecting in antisense mRNAs exons and exhibiting a strong pattern of coexpression (Spearman rs > 0.95) with their mRNAs (Supplemental Fig. S13D illustrates two examples).
LncRNAs are enriched in the nucleus
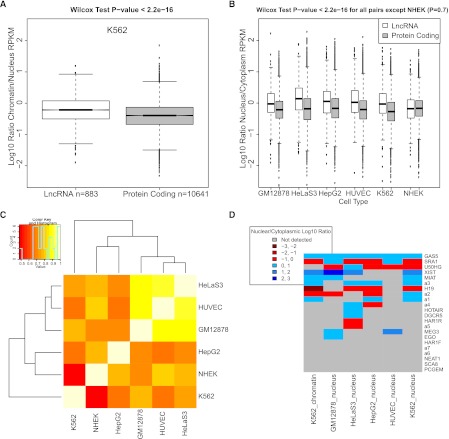
There is mounting evidence that many lncRNAs are recruited to chromatin and epigenetically regulate gene expression (Mondal et al. 2010). Thus, we would expect that lncRNAs should be preferentially localized in the chromatin and nuclear RNA fractions, in contrast to protein-coding mRNAs that are trafficked to the cytoplasm for translation. Analysis of the ENCODE RNA-seq data for nucleus and cytoplasm from six different cell lines indicates that this is indeed the case (Djebali et al. 2012). In Figure 8B, we display the ratio of transcript abundances in the nucleus over the cytoplasm. In all cell lines, except for NHEK, we observed a robust and highly statistically significant enrichment of lncRNAs in the nucleus, compared with protein-coding mRNAs. Furthermore, the lncRNAs nuclear/cytoplasmic enrichment is consistent between cell types (Fig. 8C). We also tested whether particular classes of lncRNA—intergenic, intronic, antisense—had distinct nuclear localization propensities, but we could find no significant difference, suggesting that nuclear enrichment is a general property of long non-protein-coding transcripts (data not shown).
Figure 8.
LncRNAs are enriched in the cell nucleus and chromatin. (A) Shown are the chromatin/cytoplasm expression ratios of lncRNAs and protein-coding transcripts in K562 cells. Data are represented as log10-transformed ratios of RPKM values (log10[chromatin RPKM/cytoplasm RPKM]). The data correspond to the 310 lncRNA and 10,287 protein-coding transcripts that fall below a 0.1 IDR threshold in both nuclear and chromatin data. (B) The boxplot, similar to that in A, shows the nucleus/cytoplasm expression ratios for the six ENCODE cell lines where data is available. Between 290 and 758 lncRNAs passed IDR cutoff and are shown, compared with between 16,561 and 20,666 protein-coding transcripts. (C) Nuclear enrichment of lncRNAs is correlated between cell types. The heatmap shows pairwise Pearson correlation values for the set of 98 lncRNA transcripts that passed IDR cutoff in all six ENCODE cell lines. Correlation was calculated for the nuclear/cytoplasmic enrichment value for this set of transcripts between each pair of cells. (D) Subcellular localization of known lncRNAs. The set of known lncRNAs was manually curated from the literature and lncRNAdb database (Amaral et al. 2011). (Not detected) The RPKM values did not meet the IDR 0.1 threshold.
We further investigated whether lncRNAs are enriched in particular compartments within the nucleus. We asked whether lncRNAs were enriched in chromatin by calculating the ratio of chromatin/nuclear RPKM for both lncRNAs and mRNAs (Fig. 8A). LncRNAs are significantly more enriched in chromatin than mRNAs: The median chromatin/nucleus expression ratio for lncRNAs is more than twice that of mRNAs (0.55 vs. 0.26, respectively). This lends further support to the idea that lncRNAs are specifically recruited to chromatin, where they play a regulatory role.
We examined the subcellular location of a number of well-known lncRNAs (Fig. 8D). Unsurprisingly, the X-chromosome inactivating transcript XIST was extremely highly enriched in the nucleus for all cells we examined (with a maximum enrichment of 273-fold in the nucleus of GM12878 cells) (Fig. 8D). Other regulatory lncRNAs such as GAS5 (Kino et al. 2010), LINC00568 (also known as ncRNA-a1), CYP4A22-AS1 (also known as ncRNA-a3) (Ørom et al. 2010), MIAT (Ishii et al. 2006), and MEG3 (Zhou et al. 2007) were nuclear enriched in at least two different cell types, consistent with their reported roles in gene regulation. Other transcripts, including the bifunctional transcript SRA1, which acts as both a regulatory RNA and a protein-coding sequence, have more variable subcellular location depending on cell type. As reported previously, the H19 transcript is consistently enriched in the cytoplasm, especially when comparing with the chromatin fraction (cytoplasmic/chromatin enrichment 167-fold) (Brannan et al. 1990).
We next used a more sophisticated method based on a negative binomial regression to identify individual lncRNAs and mRNAs with statistically significant enrichment in either the nuclear or the cytoplasmic compartments (see Methods). Only the subset of 1339 lncRNAs and 13,933 mRNAs with expression of RPKM > 1 were analyzed. At a statistical cutoff of P < 0.01 and FDR = 0.1, we found 228 lncRNAs enriched in the nucleus (17% of those tested) and 53 (4%) in the cytoplasm. These proportions are significantly different (Pearson's χ2 test <2.2 × 10−16) than those for protein-coding genes with 2064 (15%) in the nucleus and 3611 (26%) in the cytoplasm. As before, the two most nuclear-enriched lncRNAs are XIST and MEG3, a highly spliced lncRNA involved activation of TP53 (P-value 2.2 × 10−214 and 3.9 × 10−73, respectively) (Zhou et al. 2007). Multidimensional scaling (MDS) representation of this data (Supplemental Fig. S14A) reveals the presence of two distinct clusters corresponding to nuclear and cytoplasmic compartments, with a predominance for lncRNAs to be localized in the latter (Supplemental Fig. S14A, right cluster).
Finally, we examined whether lncRNAs preferentially exist as polyadenylated or nonpolyadenylated transcripts (Kapranov et al. 2010). Using a similar analysis as above, we calculated the simple ratio of poly(A)+ to poly(A)- nuclear RNA-seq reads mapping to lncRNAs, and compared this with protein-coding mRNAs (Supplemental Fig. S14B). In all cell types that we examined in this way, we found that lncRNAs are significantly enriched in poly(A)-, compared with mRNAs.
Discussion
Until very recently, RNA's main cellular role was assumed to be that of merely a messenger, mediating the transfer of information from DNA to proteins—the true effectors of biological function. This view has been altered, however, as a plethora of novel RNA species has been discovered (see, for instance, Carninci et al. 2005; Denoeud et al. 2007; Kapranov et al. 2007; Findeiss et al. 2010). Among these, long noncoding RNAs are emerging as central players in cell biology. Likely outnumbering protein-coding transcripts, a small but rapidly growing number of them have been shown to participate in the epigenetic regulation of gene expression. The precise biological role of the vast majority, however, is still unknown. Here, we report on the generation and characterization of the largest catalog of lncRNAs to date. This catalog has been created through manual curation of available cDNA and EST data by the GENCODE team within the framework on the ENCODE project. Using computational and experimental analysis, we have annotated GENCODE lncRNAs with a wide range of biologically relevant features. Since lncRNAs may contribute to explaining the phenotypic output encoded in the genome not currently explained by protein-coding genes, this repository and the associated annotation are likely to become central resources for research in molecular and cell biology.
Our analyses have shown that most GENCODE lncRNAs are likely to exist as independent transcriptional units, although it cannot be ruled out that a fraction of them can also eventually act, in a regulated fashion, as UTRs of neighbor protein-coding genes. While it has been recently suggested that lncRNAs have nonrandomly long ORFs (Dinger et al. 2011), we have found that with a small number of exceptions they exhibit minimal protein-coding capacity. The genomic structure of lncRNAs is very similar to that of protein-coding genes, although a surprisingly high proportion of the former are two-exon transcripts. Overall, we observe that while lncRNAs have exons of similar length to protein-coding mRNAs, they have fewer of them—in agreement with the recent findings of Cabili et al. (2011). One advantage of manual annotation of lncRNAs is that it provides accurate transcription start site, termination site, and exon–intron boundary information—although some of the lncRNAs we observed might be processed from longer precursor transcripts, meaning that the true start and end sites are beyond our annotations. Nevertheless, our analysis of promoter-specific histone modifications, and poly(A) signal frequencies, would suggest that in the majority of cases the transcript boundaries we annotate are accurate.
While lncRNAs exons are much less conserved than protein-coding genes, they are significantly more conserved than ancient repeat sequences, used here as a proxy of neutral evolution (Ponjavic et al. 2007). The evolutionary constraint on their sequence, even if weak, is thus an indication of functionality. The high proportion of primate-specific RNAs suggest that lncRNA may have a higher turnover rate than proteins. Nonetheless, many of the transcripts reported here (44%) appear to be evolutionarily conserved across the majority of placental mammals, and owing to the difficulty of comparing divergent nucleic acids sequences, one cannot rule out the possibility that lncRNA may be relatively conserved as gene units, but too rapidly evolving for conventional sequence analysis. Yet, this ancient evolutionary history suggests that these genes should have properties similar to those of proteins with families of homologs, or domains shared across otherwise unrelated sequences. We looked for such families and found them to exist, although the common domains appear to be mostly degraded versions of common repeat elements (LINE, SINE, LTR). An analysis based on multiple sequence conservation suggests the secondary structures associated with these modules to be actively maintained through the course of evolution—a finding incompatible with decaying repeats. It may therefore be that repeats have been exapted as functional RNA sequence modules with lncRNAs, as has recently been observed (Gong and Maquat 2011).
We have found lncRNAs to be particularly enriched in the nucleus relative to the cytoplasm when compared to protein-coding genes. Within the nucleus, they are particularly enriched in the chromatin fraction. While lncRNAs have previously been detected in the chromatin fraction (Mondal et al. 2010), the analysis of the rich diversity of RNA populations carried out within the ENCODE project (Djebali et al. 2012) represents the first demonstration that lncRNAs as a class are preferentially located in the chromatin and nucleus of the cell. While this is consistent with the major role so far proposed for lncRNAs as epigenetic regulators of gene expression, it could also reflect a higher rate of degradation (i.e., reduced stability) of lncRNAs compared with protein-coding genes. Because lncRNAs lack ORFs, and therefore have premature stop codons, degradation could occur through mechanisms such as translation-linked nonsense mediated decay (NMD) pathway, or translation-independent degradation pathways. On the other hand, the striking preponderance of single intron transcripts could also be a mechanism to escape NMD-like pathways. In any case, there is still a population of lncRNAs that are consistently enriched in the cytoplasm. Therefore, we must stress that although lncRNAs as a class are enriched in the nucleus compared with mRNAs, many may nevertheless function in other compartments. This is supported by various examples of lncRNAs that appear to operate in the cytoplasm, including MALAT1 (Wilusz et al. 2008), NRON (Willingham et al. 2005), GAS5 (Kino et al. 2010), or Thy-ncR1 (Aoki et al. 2010).
The GENCODE annotation is an ongoing process. As additional sequencing data from a variety of sources becomes available, GENCODE annotators will refine and reevaluate gene models. Given the low expression level and tissue specificity of lncRNAs, increased availability of deep RNA-seq data is particularly relevant to refine and expand the set of lncRNAs. Indeed, while the number of protein-coding genes has remained relatively stable, even slightly decreasing, across GENCODE versions (22,500 in version 3c, November 2009; 20,700 in version 7, April 2011), the number of lncRNA genes has substantially increased from 6000 to more than 10,000 during this period. It is not unlikely that lncRNA genes outnumber protein-coding genes in the human genome. The present annotation, version 7, covers almost 85% of the human genome; therefore, we expect at least 2000 more lncRNA genes remain to be discovered with present annotation methods (Supplemental Fig. S15). Indeed, it is important to note that the transcripts originating from lncRNA gene loci represent only a fraction of the lncRNA world: ∼50% of the transcripts arising from protein-coding loci (i.e., sense-exonic transcripts) are also noncoding and have not been analyzed in the present study. While the function of the vast majority of lncRNAs is unknown, the full importance of their contribution to the cell's phenotype is presently impossible to gauge. However, given the demonstrated biological significance of lncRNA, combined with their large number and the diversity of their mechanisms of action, it is likely that they constitute a crucial layer of gene regulation that has evolved in complex organisms.
Methods
LncRNA identification and classification
The GENCODE lncRNAs set version 7 (Harrow et al. 2012) was downloaded from the official GENCODE ftp repository: ftp://ftp.sanger.ac.uk/pub/GENCODE/. The protein-coding set used to define the lncRNAs category was extracted from the whole GENCODE 7 annotation (GENCODE.v7.annotation.gtf.gz on the ftp) and corresponds to transcripts having both gene and transcript biotypes annotated as “protein_coding” with the “known” status. This results in a protein-coding set of 20,646 genes, 76,006 transcripts, and 743,827 exons. Then, lncRNA and protein-coding genomic coordinates were intersected at the exons, introns, and gene levels using both the Bedtools suite (Quinlan and Hall 2010) and custom scripts. In an initial filtering step, we removed all lncRNAs that were shorter than 200 nt or overlapped a protein-coding exon on the same strand. The resulting set was divided into categories “intergenic” and “genic.” An lncRNA not intersecting any protein-coding loci was defined as intergenic and then subclassified according to its transcription orientation with the closest protein-coding gene (same sense, convergent, or divergent). The genic lncRNA set was classified as exonic if at least one of its exons intersects a protein-coding exon by at least 1 bp. lncRNAs intersecting a protein-coding exon on the same strand were discarded from all analyses. Otherwise, lncRNAs were classified as “intronic,” i.e., completely contained within protein-coding introns (sense or antisense) or overlapping (sense or antisense), i.e., when the protein-coding transcript was located within the intron of the lncRNA. For each category, a best mRNA partner is defined according to (1) its closer proximity to the lncRNA (intergenic category) or (2) a higher number of nucleotides intersecting with the candidate lncRNA (genic category). Finally, for the comparative analysis with the lncRNAs sets, we defined a stringent set of mRNAs which corresponds to transcripts having both gene and transcripts annotated as “protein_coding” with status “known,” a “ccdsid” tag and no match with “{start/stop}_NF” (Not Found). This results in a stringent protein coding set of 17,998 genes, 30,046 transcripts, and 319,048 exons. A full breakdown of subclassifications of lncRNA can be found in Supplemental Table S1. Furthermore, the number of genes used in every analysis is detailed in Supplemental Table S9.
Computational data analysis
We have developed and implemented a number of programs and scripts for bioinformatic and statistical analysis of the lncRNA data, including (1) assessment of the completeness of the annotated lncRNA sequences, (2) cross-species conservation of lncRNAs, (3) chromatin marks, (4) microarray processing, (5) expression correlation, (6) cellular compartmentalization. These methods, in addition to experimental details, are described in the Supplemental Methods file.
Data access
Raw RNA-seq reads can be accessed from the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE30567 (Long RNAs) and GSE24565 (Short RNAs). Additional detailed methods for RNA sequencing can be obtained in the Production Documents under “CSHL Long RNA-seq” and under “CSHL Sm RNA-seq” at: http://genome.ucsc.edu/ENCODE/downloads.html. Microarray data have been deposited under accession number GSE34894. Human Body Map (HBM) RNA-seq data can be downloaded from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-513. The lncRNA annotations can be found on the Guigo group website (http://big.crg.cat/bioinformatics_and_genomics/lncrna). Finally, the GENCODE annotation is freely available at http://www.gencodegenes.org.
Acknowledgments
We thank the CRG Microarray Facility for microarray hybridizations. Isidre Ferrer and colleagues (Hospital Bellvitge, Barcelona) kindly provided postmortem human brain samples. We are also very grateful to Laurens Wilming (from the HAVANA group) for the design of the figures and the HAVANA team for producing the annotation of the human genome. We thank Ignacio Gonzalez for his help using the mixOmics package. This work has been carried out under grants RD07/0067/0012, BIO2006-03380, and CSD2007-00050 from the Spanish Ministry of Science, grant SGR-1430 from the Catalan Government, grants 1U54HG004557-01 and 1U54HG004555-01 from the National Institutes of Health, and INB-ISCIII from Instituto de Salud Carlos III and FEDER. This research project has been cofinanced by the European Commission, within the 7th Framework Programme, Grant Agreement KBBE-2A-222664 (“Quantomics”). C.N. is financed by the Plan Nacional BFU2008-00419. G.B. is supported by the La Caixa Ph.D. program.
Footnotes
[Supplemental material is available for this article.]
Article and supplemental material are at http://www.genome.org/cgi/doi/10.1101/gr.132159.111.
Freely available online through the Genome Research Open Access option.
References
- Alioto TS 2007. U12DB: A database of orthologous U12-type spliceosomal introns. Nucleic Acids Res 35: D110–D115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS 2011. lncRNAdb: A reference database for long noncoding RNAs. Nucleic Acids Res 39: D146–D151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Harashima A, Sano M, Yokoi T, Nakamura S, Kibata M, Hirose T 2010. A thymus-specific noncoding RNA, Thy-ncR1, is a cytoplasmic riboregulator of MFAP4 mRNA in immature T-cell lines. BMC Mol Biol 11: 99 doi: 10.118/1471-2199-11-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani SR, et al. 2011. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 17: 878–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling W, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. 2012. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res (this issue). doi: 10.1101/gr.134767.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Blanco E, Parra G, Guigó R 2007. Using geneid to identify genes. Curr Protoc Bioinformatics 18: 4.3.1–4.3.28 [DOI] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM 1990. The product of the H19 gene may function as an RNA. Mol Cell Biol 10: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526 [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Chooniedass-Kothari S, Emberley E, Hamedani MK, Troup S, Wang X, Czosnek A, Hube F, Mutawe M, Watson PH, Leygue E 2004. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett 566: 43–47 [DOI] [PubMed] [Google Scholar]
- Denoeud F, Kapranov P, Ucla C, Frankish A, Castelo R, Drenkow J, Lagarde J, Alioto T, Manzano C, Chrast J, et al. 2007. Prominent use of distal 5′ transcription start sites and discovery of a large number of additional exons in ENCODE regions. Genome Res 17: 746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS 2008. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput Biol 4: e1000176 doi: 10.1371/journal.cbi.1000176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Crowe ML, Grimmond SM, Mattick JS 2009. NRED: A database of long noncoding RNA expression. Nucleic Acids Res 37: D122–D126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Gascoigne DK, Mattick JS 2011. The evolution of RNAs with multiple functions. Biochimie 93: 2013–2018 [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi AM, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeiss S, Schmidtke C, Stadler PF, Bonas U 2010. A novel family of plasmid-transferred anti-sense ncRNAs. RNA Biol 7: 120–124 [DOI] [PubMed] [Google Scholar]
- Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, Menzel C, Chen W, Li Y, Zeng R, et al. 2009. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics 10: 161 doi: 10.1186/1471-2164-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras TR 2007. Origin of phenotypes: Genes and transcripts. Genome Res 17: 682–690 [DOI] [PubMed] [Google Scholar]
- Gong C, Maquat LE 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res (this issue). doi: 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. 2006. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51: 1087–1099 [DOI] [PubMed] [Google Scholar]
- Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L 2010. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA 16: 1478–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kapranov P, St Laurent G, Raz T, Ozsolak F, Reynolds CP, Sorensen PH, Reaman G, Milos P, Arceci RJ, Thompson JF, et al. 2010. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol 8: 149 doi: 10.1186/1741-7007-8-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP 2010. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8 doi: 10.1126/scisignal.2000568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y 2010. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329: 336–339 [DOI] [PubMed] [Google Scholar]
- Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engström PG, Lenhard B, Aturaliya RN, Batalov S, Beisel KW, et al. 2006. Transcript annotation in FANTOM3: Mouse gene catalog based on physical cDNAs. PLoS Genet 2: e62 doi: 10.1371/journal.pgen.0020062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques AC, Ponting CP 2009. Catalogs of mammalian long noncoding RNAs: Modest conservation and incompleteness. Genome Biol 10: R124 doi: 10.1186/gb-2009-20-22-r124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS 2009. The genetic signatures of noncoding RNAs. PLoS Genet 5: e1000459 doi: 10.1371/journal.pgen.1000459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C 2010. Characterization of the RNA content of chromatin. Genome Res 20: 899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, Guigo R, Dermitzakis ET 2010. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464: 773–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Ng P, Wei CL, Sung WK, Chiu KP, Lipovich L, Ang CC, Gupta S, Shahab A, Ridwan A, Wong CH, et al. 2005. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat Methods 2: 105–111 [DOI] [PubMed] [Google Scholar]
- Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E, Sabbagh A, Vidaud M, Bièche I 2011. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J 25: 444–448 [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G 2007. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 17: 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and functions of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. 2006. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31 doi: 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL 2009. MEN ɛ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR 2010. Most “dark matter” transcripts are associated with known genes. PLoS Biol 8: e1000371 doi: 10.1371/journal.pbio.1000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M 2009. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG 2005. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A 2007. Activation of p53 by MEG3 non-coding RNA. J Biol Chem 282: 24731–24742 [DOI] [PubMed] [Google Scholar]