Abstract
The adult body plan of bilaterians is achieved by imposing regional specifications on pluripotential cells. The establishment of spatial domains is governed in part by regulating expression of transcription factors. The key to understanding bilaterian evolution is contingent on our understanding of how the regulation of these transcription factors influenced bilaterian stem-group evolution.
“Since the developmental process is inherited, the genome must contain the regulatory programs with which to control the pattern of genetic activity … We feel that to explain the magnitude of the functional and structural stage in evolution it is necessary to postulate changes in the regulatory apparatus … ” These words were written almost 30 years ago (1), but what was then inferred only as a construct of logic has now grown into a great repertoire of observed molecular mechanisms. These are the mechanisms by which the parts and pieces of the bilaterian body plan are generated during embryogenesis, as determined by the hardwired genetic regulatory programs that directly control development. Such programs define the morphology of each clade, and they underlie the cladistic organization of the Bilateria. It is now obvious that everything we learn experimentally about how developmental regulatory programs work and how they are encoded in the DNA enriches our understanding of animal evolution.
The changes in regulatory “wiring” that generate developmental control apparatus in evolution are of two sorts: those that can be considered conservation of prior assignments of regulatory interactions to given developmental jobs and those that can be considered cooptions, or reassignments of regulatory routines or subroutines to new jobs. But distinguishing between these categories is anything but easy. Ultimately it requires “regulatory cladistics”: exact knowledge, based on comparative observation, of whether a given set of regulatory interactions is shared among sister groups or is novel because the set is specific to a given lineage.
Thinking about the regulatory evolution of bilaterians in general is not the same as thinking about the regulatory origin of specific bilaterian structures, such as appendages or eyes. Suppose we understood how the parts of bilaterians are generated in development, how they formulate their anterior/posterior (A/P) axes, their segments, their guts, and their heads: this still begs the question of how they arose in evolution, because the required regulatory programs cannot have sprung forth full blown. However, detailed knowledge of developmental mechanisms at least specifies the end points of the evolutionary process that led to the bilaterians, a level of understanding that the field is now approaching at a pretty good clip.
What the bilaterians share is not only an A/P axis but also the ability to generate complex body parts by using a common type of regulatory process, whatever its morphological product. The A/P axis is indeed a character of all bilaterians, whereas only some generate appendages, segments, or eyes, but this does not mean that the mechanism for generating the A/P axis somehow contains the secret that explains how all of the other parts arise. Rather, all such morphogenetic programs are to be regarded as applications of a particular type of regulatory program architecture that bilaterians use to encode morphogenetic functions. As a shorthand designation, we refer to the bilaterian way of building morphological structures as pattern formation by regional specification. The conclusion is that to infer the evolutionary origins of the bilaterians, we need first to consider what are the general features of this kind of regulatory architecture and then to consider its likely antecedents.
Pattern Formation in the Morphogenesis of Adult Bilaterian Body Plans.
Regional specification has the following specific meaning, in respect to developmental mechanism. As discussed elsewhere (2, 3) and illustrated by many examples (4–9), the key initial stages of pattern formation consist of successive regional specification events, in each of which a particular regulatory state is imposed on a spatial domain of pluripotential cells. Regulatory states are established by the transcriptional expression of a given set of genes encoding transcription factors. The field of cells that is going to constitute a given structure is thus blocked out (e.g., a limb bud field), usually in response to signaling events that specify where the regulatory genes will and will not be expressed. Further boundaries are set within, by a succession of additional regulatory subdivisions, resulting in transcriptional definition of all of the future elements of the structure, and also of growth within these elements. Ultimately the batteries of genes encoding the properties of each part of the structure are called into play. It is important to realize that this is in essence an abstract sequential process: it sets up complex spatial organizations by a multistage mechanism, so that the pattern is laid out separately from the installation of terminal processes. It is also an extremely powerful process. Linked intimately with growth controls, it can be used to organize structures of any size; it is lineage independent until the very end; it operates de novo, requiring only spatial sources of signals; and it can be used at any stage of development and for any purpose, from setting up the major components of the body plan at the beginning of embryogenesis to determining the exact disposition of bristles or feathers at the end (10–12). As the enormous diversity of bilaterian morphologies shows, it can be used for almost anything.
Embryogenesis Without Regional Specification.
About a decade ago, a comparative survey of embryonic specification processes led to a surprising conclusion (13). We had all grown up with a series of classical embryological typologies, which sharply distinguished the embryos of different clades from one another according to properties such as “determinate” cleavage as opposed to “regulative” capacity. But viewed from a mechanistic point of view, most embryos are seen to operate in very similar ways, and there emerges the image of a basic, nearly panbilaterian mode of embryological development. Only clades that evolved after the initial appearance of most bilaterian groups, particularly highly derived forms, some of which equipped themselves to exploit terrestrial environments (for instance insects, vertebrates, and their immediate cousins), seem to have escaped this basic mechanism by inventing other ways of turning their eggs into embryos. The basic form of embryogenesis is referred to as the “Type 1” embryonic process (13, 14). The Type 1 mechanism of early development is so wide spread that it is evidently an ancient property of the bilaterians: the same rules can be stated for embryogenesis in molluscs and annelids, nematodes, echinoderms, ascidians, cephalochordates, lophophorates, and so forth, i.e., it is common to all three branches of the bilaterian cladogram (13). These rules are as follows: specification immediately produces cell types that express differentiation genes in these embryos; specification of cell lineages depends both on signaling between blastomeres during cleavage and on maternal cytoarchitectural coordinates; these embryos turn on their genes immediately, early in cleavage. What they do not do is pattern formation by regional specification, because their cells learn who they are to be on an individual basis, dependent on their lineage specification devices.
How far Type 1 embryonic processes alone can take the organism can be seen explicitly in what we call “maximal indirect development.” This type of development is basal to lophotrochozoans and to at least the echinoderm–hemichordate branch of the deuterostomes (15). Here the embryo gives rise to relatively simple feeding free-living larvae that bear no morphological relation to the adult form. Such larvae are known by the term “primary larvae” (16). The adult body plan is subsequently generated within the larva in a separate postembryonic developmental process. Indirectly developing larvae are philosophically fascinating little creatures. They are bilateral in organization; they consist of only a couple of thousand cells and 10 or 12 differentiated cell types, including a few neurons, a few muscle cells, a few mesenchymal cells, a single cell-thick gut, and ectodermal layers. There are no multilayered structures such as those that underlie the construction of all adult bilaterian body parts. Type 1 embryonic processes suffice to account for the final structure and organization of these larvae. Because the Hox gene complex is used to control specific aspects of the regional specification process, we predicted (2), contrary to dogma, that the Hox gene complex would not be used in the embryonic development of primary larvae, even though these are indeed free-living organisms of bilaterian grade. The prediction was based on the idea that Hox genes operate by controlling pattern formation processes and thus would seem to have no role in the Type 1 embryogenesis of such larvae. This prediction was confirmed for sea urchin embryos (17) and now, as described in this issue, for a polychaete embryo as well (18); but in both organisms, the Hox complex is used in the postembryonic developmental process by which the adult body plan forms.
A new theory for the origin of the bilaterians follows from the idea that Type 1 processes of embryogenesis were the original mode of development in stem- group bilaterians and that the regulators they used served as a platform for the evolution of regional specification mechanisms (2). This theory outlines the essential evolutionary changes in regulatory program that could have given rise to adult body plans of the complexity seen in modern and fossil Bilateria. The fundamental events leading to the bilaterians are proposed to be the coevolution, in remote Precambrian time, of genomic mechanisms for pattern formation by means of regional specification and of certain cell populations which remain unspecified throughout embryonic life. These we term “set-aside cells,” and it is in their progeny, which retain the capacity for growth, that pattern formation processes can generate useful new morphological structures. Set-aside cells are easily identified as the source of adult body parts in all maximally indirectly developing organisms that are alive today (15). The initial evolutionary experimentation with pattern formation mechanisms that operate by stepwise regional specification would at first have produced small and simple structures. These would have been superimposed on preexisting microfaunal forms developing by the more ancient Type 1 regulatory processes. Because regulatory apparatus consists essentially of genomic cis-regulatory systems and their linkages, the changes we envision are those that would have set up the control circuits linking genes that encode relevant transcription factors with those encoding signal system components and growth control components. It is striking how the same subcircuits linking certain of these components are still used throughout the bilaterians.
Bilaterian Origins.
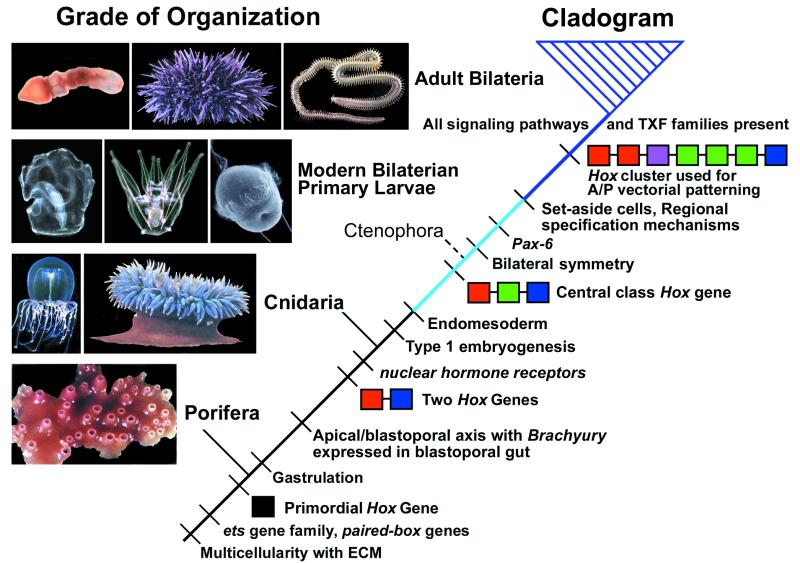
A phylogeny of basal metazoans and how they relate to bilaterians is shown in Fig. 1. At the base of the tree are the sponges and at the apex are the bilaterians (the crown group is shown by the hatched triangle). In between are the other two major nonbilaterian groups, cnidarians and ctenophores. The ctenophores are indicated by a dotted line to reflect the contradictory nature of evidence relating to their phylogenetic position (e.g., ref. 19), but because they share developmental (13, 20) and genetic (21) characters with bilaterians, they are shown as the bilaterian sister group. On the right are some of the important regulatory inventions or their developmental manifestations on the march toward the bilaterians. Also indicated are key innovations that result in new grades of animal organization. Thus, for example, gastrulation is among the developmental innovations that underlie the appearance of animals with true ectodermal and endodermal tissue layers. On the far left are pictures of animals that represent each particular grade of organization.
Figure 1.
A cladogram of basal metazoans and some of the important regulatory inventions leading to the crown group bilaterians (purple triangle). The dotted line leading to Ctenophora reflects the equivocal nature of evidence regarding their phylogenetic position. The change in grade of organization from a two-dimensional to a three-dimensional form required the evolution of endomesoderm. This stage is indicated by the light-blue line. With the evolution of set-aside cells and regional specification mechanisms, macroscopic bilaterian body plans are now evolvable, and this change is indicated by the purple line. By the time the crown group evolved, all signaling pathways and transcription factor (TXF) families had appeared. The single “primordial” Hox gene found in sponges is shown by the black box. Presumably this gene underwent tandem gene duplication resulting in two genes, an “anterior” gene related to Hox 1 and Hox 2 of bilaterians (shown in red) and a posterior gene related to Hox 9–13 (i.e., Abd-B relatives, shown in blue). A central class Hox gene has been found in ctenophores (Hox 4–8, shown in green). The latest common ancestor must have had at least seven Hox genes involving both gene duplications of previous classes (e.g., multiple anterior and middle genes) and new classes (Hox 3, violet box). This view of Hox cluster evolution devolves from studies of de Rosa et al. (33), Finnerty and Martindale (30), and others (see text). The adult enteropneust, the larval enteropneust, and the larval sea urchin photographs are from the authors' collections; the rest of the animal pictures are from ref. 40 [reproduced with permission from ref. 40 (Copyright 1980, Stanford University Press)].
At the bottom of the tree are some known aspects of the basic regulatory repertoire found in sponges. These include cell signaling systems such as integrins associated with the extracellular matrix (22), transcription factors, including representatives of the ets (23, 24), paired-box (25), and several different homeobox genes, including NK (26) and Hox (27) class genes. Despite concerted efforts, only one type of Hox-like gene has been found in sponges (black square) (28).
Examination of cnidarians has shown that not only are new transcription factor families present (e.g., nuclear hormone receptors, ref. 29), but there is now a “proto-Hox” cluster. This cluster consists of two types of genes (30), one whose sequence displays similarities with “anterior” Hox genes of bilaterian Hox clusters (red) and one that appears similar to “posterior” Hox genes (blue). The only evidence regarding expression of a Hox gene in cnidarians concerns a posterior type Hox gene, the expression of which is both stage and tissue specific (31). But there is so far no evidence that these two kinds of Hox gene are involved in any sort of vectorial patterning mechanism.
Endomesodermal cells provide the possibility of a third dimension in organismal morphology, and with their evolution a new grade of organization appears. This is represented among modern animals by ctenophores and also in the primary larvae of indirectly developing bilaterians (shown in Fig. 1 by the light-blue line). As we note above, the Type 1 embryonic processes needed to build free-living larval organisms such as those illustrated opposite the light-blue line in Fig. 1 are much simpler and more direct than are the pattern formation mechanisms required to build the adult body plans of crown group bilaterians. The ctenophores also may share with the bilaterians a central class Hox gene, shown here in green (21).
The evolution of set-aside cells and regional specification mechanisms allowed for the final transition in grade of organization, that giving rise to body plans of the complexity of modern Bilateria (purple line). In addition to the appearance of new transcription factors such as Pax-6 (32), the Hox complex is now a full-fledged cluster consisting of at least seven genes (33). At some point in the bilaterian stem lineage, the Hox cluster was recruited for A/P axis patterning (34). It is important to note that in modern bilaterians, the Hox cluster is not used for initiation of the A/P axis; this is accomplished by prior embryonic processes in all systems so far studied. The Hox gene cluster is a vectorial patterning device, used to establish different transcriptional states not only along the A/P axis of adult bilaterians but also, for example, along certain of the axial projections of vertebrate limbs and genital tracts (e.g., refs. 35 and 36).
This view of Hox gene evolution differs from the “zootype” hypothesis (37) and similar concepts, in which animals are essentially defined in terms of the Hox cluster and its function in A/P axis specification. As can be seen in Fig. 1, we think that many stages of animal evolution and many regulatory inventions preceded the advent of the complete Hox gene complex and its role in A/P patterning (see also ref. 38). In fact, this evolutionary event is the last entry in Fig. 1 to precede crown group bilaterian diversification. We note furthermore that taxa must be defined in terms of their genealogical or cladistic relationships (as in Fig. 1), not by the possession of particular characters or features, even if, like the Hox gene cluster, these are useful for recognizing to which taxon an organism belongs. Specifically, the key to bilaterian evolution lies in understanding the nature of bilaterian stem groups, and much bilaterian stem-group evolution must have preceded the appearance of the fully fledged Hox gene complex.
By the time the crown group of Bilateria appeared, all signaling pathways and transcription factor families were present, because they are common to all modern bilaterians. The important conclusion follows that evolution of phylum-specific body plans does not depend on invention of new developmental genes but rather on novel gene regulatory circuitry. The advent of this circuitry, that is, of regional specification mechanisms including the Hox gene complex, occurred by the latest Precambrian if not before. In our view, it is likely that this was preceded by a long prior history of bilaterian micrometazoans similar in grade of organization to the primary larvae of modern bilaterians. This paleontological prediction is strengthened now by the work of Chen et al. (39), who describe a series of remarkable microscopic fossils dating to mid-Neoproterozoic times. Not only are poriferan and cnidarian embryos present, but so are bilaterian embryos of the scale and form capable of producing the primary larval morphologies still to be seen in today's oceans (2). The coupling of micropaleontology and molecular biology is opening a new window into early animal evolution, a window on a sea filled with elegant micrometazoans whose descendants would come to dominate the macroscopic world of the Phanerozoic.
Acknowledgments
We thank Profs. Ellen Rothenberg (California Institute of Technology) and William McGinnis (University of California at San Diego) for helpful reviews. This work was supported by the Fundamental Biology Research Program of the Life Sciences Division of the National Aeronautic and Space Administration/Ames, Grant no. NAG 2–1368.
Abbreviation
- A/P
anterior/posterior
References
- 1.Britten R J, Davidson E H. Quant Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- 2.Davidson E H, Peterson K J, Cameron R A. Science. 1995;270:1319–1325. doi: 10.1126/science.270.5240.1319. [DOI] [PubMed] [Google Scholar]
- 3.Cameron R A, Peterson K J, Davidson E H. Am Zool. 1998;38:609–620. [Google Scholar]
- 4.Shubin N, Tabin C, Carroll S. Nature (London) 1997;388:639–648. doi: 10.1038/41710. [DOI] [PubMed] [Google Scholar]
- 5.Lumsden A, Krumlauf R. Science. 1996;274:1109–1115. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Skarmeta J L, Modolell J. Genes Dev. 1996;10:2935–2945. doi: 10.1101/gad.10.22.2935. [DOI] [PubMed] [Google Scholar]
- 7.Weatherbee S D, Halder G, Kim J, Hudson A, Carroll S. Genes Dev. 1998;12:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilder D, Graba Y, Scott M P. Development (Cambridge, UK) 1998;125:1781–1790. doi: 10.1242/dev.125.9.1781. [DOI] [PubMed] [Google Scholar]
- 9.Logan M, Tabin C J. Science. 1999;283:1736–1739. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- 10.Stern D L. Nature (London) 1998;396:463–466. doi: 10.1038/24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nellesen D T, Lai E C, Posakony J W. Dev Biol. 1999;213:33–53. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- 12.Chuong C-M. BioEssays. 1993;15:513–521. doi: 10.1002/bies.950150804. [DOI] [PubMed] [Google Scholar]
- 13.Davidson E H. Development (Cambridge, UK) 1991;113:1–26. doi: 10.1242/dev.113.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Davidson E H. Development (Cambridge, UK) 1990;108:365–389. doi: 10.1242/dev.108.3.365. [DOI] [PubMed] [Google Scholar]
- 15.Peterson K J, Cameron R A, Davidson E H. BioEssays. 1997;19:623–631. doi: 10.1002/bies.950190713. [DOI] [PubMed] [Google Scholar]
- 16.Jägersten G. Evolution of the Metazoan Life Cycle: A Comprehensive Theory. London: Academic; 1972. [Google Scholar]
- 17.Arenas-Mena C, Martinez P, Cameron R A, Davidson E H. Proc Natl Acad Sci USA. 1998;95:13062–13067. doi: 10.1073/pnas.95.22.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson K J, Irvine S Q, Cameron R A, Davidson E H. Proc Natl Acad Sci USA. 2000;97:4487–4492. doi: 10.1073/pnas.97.9.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins A G. Proc Natl Acad Sci USA. 1998;95:15458–15463. doi: 10.1073/pnas.95.26.15458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martindale M Q, Henry J Q. Am Zool. 1998;38:672–684. [Google Scholar]
- 21.Finnerty J R, Master V A, Irvine S, Kourakis M J, Warriner S, Martindale M Q. Mol Mar Biol Biotechnol. 1996;5:249–258. [PubMed] [Google Scholar]
- 22.Wimmer W, Perovic S, Kruse M, Schroder H C, Krasko A, Batel R, Müller W E G. Eur J Biochem. 1999;260:156–165. doi: 10.1046/j.1432-1327.1999.00146.x. [DOI] [PubMed] [Google Scholar]
- 23.Degnan B M, Degnan S M, Naganuma T, Morse D E. Nucleic Acids Res. 1993;15:3479–3484. doi: 10.1093/nar/21.15.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laudet V, Hanni C, Stehelin D, Duterque-Coquillaud M. Oncogene. 1999;18:1351–1359. doi: 10.1038/sj.onc.1202444. [DOI] [PubMed] [Google Scholar]
- 25.Hoshiyama D, Suga H, Iwabe N, Koyanagi M, Nikoh N, Kuma K, Matsuda F, Honjo H, Miyata T. J Mol Evol. 1998;47:640–648. doi: 10.1007/pl00006421. [DOI] [PubMed] [Google Scholar]
- 26.Master V A, Kourakis M J, Martindale M Q. Dev Dyn. 1996;207:404–419. doi: 10.1002/(SICI)1097-0177(199612)207:4<404::AID-AJA5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 27.Degnan B M, Degnan S M, Giusti A, Morse D E. Gene. 1995;155:175–177. doi: 10.1016/0378-1119(94)00908-b. [DOI] [PubMed] [Google Scholar]
- 28.Finnerty J R. Curr Topics Dev Biol. 1998;40:211–254. doi: 10.1016/s0070-2153(08)60368-3. [DOI] [PubMed] [Google Scholar]
- 29.Escriva H, Safi R, Hanni C, Langlois, Marie-C, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Proc Natl Acad Sci USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finnerty J R, Martindale M Q. Evo Dev. 1999;1:16–23. doi: 10.1046/j.1525-142x.1999.99010.x. [DOI] [PubMed] [Google Scholar]
- 31.Aerne B L, Baader C D, Schmid V. Dev Biol. 1995;169:547–556. doi: 10.1006/dbio.1995.1168. [DOI] [PubMed] [Google Scholar]
- 32.Callaerts P, Munoz-Marmos A M, Glardon S, Castillo E, Sun H, Li W-H, Gehring W J, Salo E. Proc Natl Acad Sci USA. 1999;96:558–563. doi: 10.1073/pnas.96.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Rosa R, Grenier J K, Andreeva T, Cook C E, Adoutte A, Akam M, Carroll S B, Balavoine G. Nature (London) 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- 34.Peterson K J, Cameron R A, Davidson E H. Dev Biol. 2000;219:1–17. doi: 10.1006/dbio.1999.9475. [DOI] [PubMed] [Google Scholar]
- 35.Nelson C E, Morgan B A, Burke A C, Laufer E, DiMambro E, Murtaugh L C, Gonzales E, Tessarollo L, Parada L F, Tabin C. Development (Cambridge, UK) 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- 36.Taylor H S, Vanden Heuvel G B, Igarashi P. Biol Reprod. 1997;57:1338–1345. doi: 10.1095/biolreprod57.6.1338. [DOI] [PubMed] [Google Scholar]
- 37.Slack J M W, Holland P W H, Graham C F. Nature (London) 1993;361:490–492. doi: 10.1038/361490a0. [DOI] [PubMed] [Google Scholar]
- 38.Schierwater B, Kuhn K. Mol Phylogenet Evol. 1998;9:375–381. doi: 10.1006/mpev.1998.0489. [DOI] [PubMed] [Google Scholar]
- 39.Chen J-Y, Oliveri P, Li C-W, Zhou G-Q, Gao F, Hagadorn J W, Peterson K J, Davidson E H. Proc Natl Acad Sci USA. 2000;97:4457–4462. doi: 10.1073/pnas.97.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris R H, Abbott D P, Haderlie E C. Intertidal Invertebrates of California. Stanford, CA: Stanford Univ. Press; 1980. [Google Scholar]