Abstract
Context:
A broad spectrum of GnRH-deficient phenotypes has been identified in individuals with both mono- and biallelic GNRHR mutations.
Objective:
The objective of the study was to determine the correlation between the severity of the reproductive phenotype(s) and the number and functional severity of rare sequence variants in GNRHR.
Subjects:
Eight hundred sixty-three probands with different forms of GnRH deficiency, 46 family members and 422 controls were screened for GNRHR mutations. The 70 subjects (32 patients and 38 family members) harboring mutations were divided into four groups (G1-G4) based on the functional severity of the mutations (complete or partial loss of function) and the number of affected alleles (monoallelic or biallelic) with mutations, and these classes were mapped on their clinical phenotypes.
Results:
The prevalence of heterozygous rare sequence variants in GNRHR was significantly higher in probands vs. controls (P < 0.01). Among the G1-G3 groups (homozygous subjects with successively decreasing severity and number of mutations), the hypogonadotropic phenotype related to their genetic load. In contrast, subjects in G4, with only monoallelic mutations, demonstrated a greater diversity of clinical phenotypes.
Conclusions:
In patients with GnRH deficiency and biallelic mutations in GNRHR, genetic burden defined by severity and dose is associated with clinical phenotype. In contrast, for patients with monoallelic GNRHR mutations this correlation does not hold. Taken together, these data indicate that as-yet-unidentified genetic and/or environmental factors may combine with singly mutated GNRHR alleles to produce reproductive phenotypes.
Isolated GnRH deficiency (IGD) in humans is a clinically and genetically heterogenous condition (1–16). Increasingly, reports of the range and number of nucleotide variants identified in patients with IGD across a variety of genes raise new questions about the precision of genotype-phenotype correlations (17, 18). While some patients harbor homozygous or compound heterozygous mutations, increasing numbers of patients with monoallelic heterozygous changes are being reported (13, 19). There is an expanding body of data that indicates that heterozygote mutations may lead to reductions in total gene product, whether through gene deletion, degradation of an unstable mRNA, or the creation of a poorly functioning protein. Some proteins, such as transcription factors, are quite sensitive to such reductions in gene dosage, whereas others, i.e. G protein-coupled receptors have traditionally been associated with recessive traits wherein heterozygous are typically asymptomatic. However increasing numbers of heterozygote mutations in patients with reproductive phenotypes are being discovered in genes that encode G protein-coupled receptors including GNRHR (GnRH receptor), PROKR2 (prokineticin 2 receptor), KISS1R (kisspeptin receptor), and TACR3 (neurokinin B receptor) (11–13, 15, 20).
Functional determinations of total mutation burden can improve the predictive power of genotypic information in certain rare diseases such as the ciliopathies (21). Applying this concept, this study focuses on the role of gene dosage and severity of loss of function in a large cohort of individuals bearing either monoallelic or biallelic mutations in GNRHR, one of the first genes to be identified as a cause of GnRH deficiency (6, 22). In addition to examining gene dosage effects, this study also integrates the homo/heterozygosity of each variant with in vitro and in silico information regarding its functional severity and thus expands the genotypic profiling of GnRH deficiency.
The following hypotheses are specifically examined: 1) both homozygous and heterozygous variants in GNRHR occur more commonly in patients with GnRH deficient states than in healthy controls, 2) the reproductive phenotype of patients harboring biallelic mutations is correlated with the functional severity of the GNRHR variants, and 3) in contrast, the reproductive phenotype of subjects harboring monoallelic mutations does not correlate with mutation severity.
In the course of examining these hypotheses, unique reproductive phenotypes were uncovered, suggesting that GNRHR mutation may also exert direct effects on gonadal function.
Patients and Methods
All activities were approved by the Massachusetts General Hospital Institutional Review Board. All subjects provided written informed consent.
Patient cohorts
GNRHR (accession no. NM_010323) was screened in 863 probands with IGD [n = 375, 280 males, 95 females; 23 of which had adult-onset idiopathic hypogonadotropic hypogonadism (AOIHH), Kallmann syndrome (KS) (n = 360, 272 males, 88 females), hypothalamic amenorrhea (HA) (n = 77 females), and constitutional delay of puberty (CDP) (n = 51, 29 males, 22 females)].
Controls
GNRHR was screened in volunteers with normal reproductive function by history and physical examination (252 Caucasians, 50 African-Americans). Sequence data from 120 subjects from the 1000 Genomes Project (http://www.1000genomes.org) was also used to expand this data set.
GnRH deficiency
The diagnosis of GnRH deficiency was based on previously published criteria (23). Olfactory testing was performed using a smell identification test (University of Pennsylvania Smell Identification Test) (24). A score of the fifth percentile or greater based on sex/age was deemed normal [normosmic (n = 130); anosmic (n = 292)]. Patients with reduced olfaction were diagnosed with KS. For the remaining patients, the assignment of diagnosis was informed by self-reported sense of smell.
Hypothalamic amenorrhea
Women had HA if they were between 18 and 40 yr old and had secondary amenorrhea for 6 months or longer with low/normal gonadotropins and low estradiol levels in the presence of weight loss (>15% of body weight), more than 15 h/wk exercise (25), or an eating disorder (26). The Eating Attitudes Test was administered to exclude clinical eating disorders (27).
Adult-onset idiopathic hypogonadotropic hypogonadism
Participants were diagnosed with AOHH based on previously published criteria (28).
Constitutional delay of puberty
The diagnosis of CDP was based on initiation of puberty at an age greater than 2 sd later than the general population without apparent pathology followed by eventual completion of pubertal development [females: no thelarche by 13 yr and/or no menarche by 15 yr; males: testes <4 cc and/or no growth spurt by age 14 yr (29, 30)].
Ethnicity/inheritance
The 863 probands were Caucasian (n = 646), Asian (n = 47), Black/African-American (n = 19), Native Hawaiian/Pacific Islanders (n = 2), American Indian or Alaska Natives (n = 2), and mixed ancestry (n = 11), with the remainder of unknown race. Five hundred eighty-eight probands were male and 275 were female. One hundred eighty-eight probands had one or more family member with some form of GnRH deficiency or associated phenotype, 267 had no family history of GnRH deficiency, and the remaining were unknown as to their mode of inheritance. Of the familial cases, 148 appeared to be autosomal dominant; 40 autosomal recessive; 18 X-linked recessive; and 23 had affected brothers in a single sibling pair. Subjects were designated as monoallelic (one or more changes on the same allele) or biallelic (homozygous or compound heterozygous) on the basis of the pedigree.
Neuroendocrine studies
Two hundred fifty-two of 863 probands (170 males, 82 females) were admitted to the Clinical Research Center of Massachusetts General Hospital for blood sampling every 10 min for 12–24 h to assess LH secretion (31).
Assays
Samples were assayed for LH by RIA (32) or an automated microparticle enzyme immunoassay (AxSYM System; Abbott Laboratories, Abbott Park, IL). The second assay was calibrated using the same reference preparations as the RIA to make results comparable across data sets. Data regarding LH pulses are based on the limits of detection of each system and integrated by virtue of a common standard in both assays that permitted interconversion.
Mutation analysis
Exon segments of genomic DNA were sequenced and all sequence variants were confirmed (33) in all probands and control subjects. Nucleotide changes were assessed in dbSNP (Database for Single Nucleotide Polymorphisms), dbEST (Database for Expressed Sequence Tags), and among control alleles. Data for heterozygous patients were analyzed for single nucleotide polymorphism (SNP) heterozygosity using 13 different SNP to ensure that intragenic deletions were not missed.
The 863 probands were also screened for mutations in other genes involved in idiopathic hypogonadotropic hypogonadism (IHH), including FGFR1 (n = 818), KISS1R (n = 769), NELF (n = 743), TAC3 (n = 456), TACR3 (n = 459), FGF8 (n = 818), GNRH1 (n = 475), KAL1 (n = 807), PROK2 (n = 840), PROKR2 (n = 839), and CHD7 (n = 188).
Functional studies
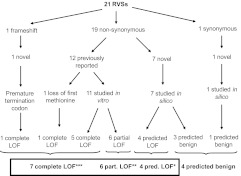
The severity of GNRHR variants was determined by in vitro studies (6, 34–36) or prediction programs [PolyPhen (http://genetics.bwh.harvard.edu/pph/) (37), Mutation Taster (http://www.mutationtaster.org), and Panther (http://www.pantherdb.org/tools/csnpScoreForm.jsp) (38) for nonsynonymous changes; and Mutation Taster and NNSPLICE 0.9 version (http://www.fruitfly.org/seq_tools/splice.html) for synonymous changes]. Changes leading to a frame shift, a loss of the first methionine, or in vitro studies showing the abolition of receptor function were considered to be a complete loss of function (cLOF) mutations (n = 7). Six variants were categorized as a partial loss of function (pLOF) mutations. Variants that were not studied in vitro but were predicted to impair GnRH receptor function in silico across all programs (n = 4) were categorized as presumed pLOF mutations. Three variants were either cLOF (A171T) or pLOF (N10K, Q11K) in vitro but were classified as benign by at least one prediction program and were included among cLOF/pLOF mutations.
Probands and family members (the latter including both affected and unaffected subjects) carrying GNRHR mutations were divided into four groups: G1, subjects with both alleles carrying cLOF mutations (cLOF/cLOF); G2, subjects with one allele harboring a cLOF mutation and the other having a pLOF mutation (cLOF/pLOF); G3, subjects with both alleles having pLOF mutations (pLOF/pLOF); and G4, subjects with only one allele harboring a mutation (cLOF/NL or pLOF/NL) [Figs. 1 and 2and Supplemental Tables 1–3, published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org, showing that family members carrying rare sequence variants (RSV) were given the same identification number as the proband followed by a unique letter designation].
Fig. 1.
Functional assessment of RSV.
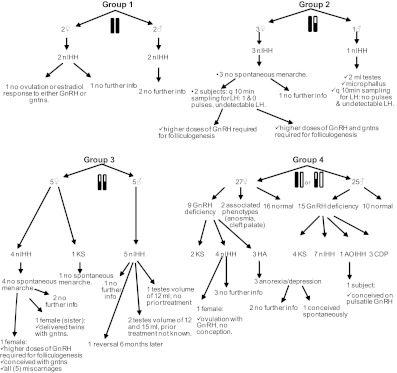
Fig. 2.
Phenotype of individuals included in the four groups. White bar, normal allele; black bar, complete loss of function mutation(s); white and black bar, partial loss of function mutation(s).
LH secretion patterns were compared between groups. When a subject had more than one sampling study, the mean number of LH pulses and mean LH levels were used in analyses. Values were compared by one-way ANOVA.
Results
Identification of variants and analysis of pathogenicity
Twenty-one RSV in GNRHR (i.e. ≤1% of control allele) were identified in 35 of 863 probands (4.0%) (Tables 1 and 2 and Supplemental Fig. 1), none of which have been reported in dbSNP or double-banded expressed sequence tag. Figure 1 outlines these RSV, their amino acid changes, in vitro studies, and predictions of pathogenicity. Seven of the 21 variants (33%) were cLOF (M1T, Q11fsX23, R139H, A171T, L266R, C279Y, P320L), six of 21 (29%) were pLOF (N10K, Q11K, T32I, Q106R, S217R, R262Q), four of 21 (19%) were predicted to be deleterious [and considered pLOF for the purpose of this study (P96S, L117P, P146S, L166P)], and four of 21 (19%) were predicted to be benign (A50V, L83V, S168A, F216F) (Table 1). The three subjects with only presumed benign RSV were not placed into any of the aforementioned groups (Supplemental Tables 1 and 2).
Table 1.
Studied and predicted functional consequences of rare variants in GNRHR
| Change (AA) | Functional studies | PolyPhen | Mutation Taster | Panther (score) | NNSPLICE9.0 | Classification for this study | In controls? | Reference |
|---|---|---|---|---|---|---|---|---|
| M1Ta | Complete LOF | LOF | LOF | — | — | Complete LOF | No | (55) |
| N10Kb | Partial LOF | Benign | LOF | −7.1 | — | Partial LOF | No | (34) |
| Q11Kb | Partial LOF | Benign | LOF | −4.8 | — | Partial LOF | No | (34) |
| Q11fsX23a | Complete LOF | — | — | — | — | Complete LOF | No | |
| T32Ib | Partial LOF | LOF | LOF | −5.1 | — | Partial LOF | No | (35) |
| A50V | Not published | Benign | Benign | −1.4 | — | Benign | No | |
| L83V | Not published | Benign | LOF | −7.7 | — | Benign | No | |
| P96Sc | Not published | LOF | LOF | −8.0 | — | Predicted LOF | No | |
| Q106Rb | Partial LOF | LOF | LOF | −4.9 | — | Partial LOF | 2d | (6) |
| L117Pc | Not published | LOF | LOF | −7.2 | — | Predicted LOF | No | |
| R139Ha | Complete LOF | LOF | LOF | −9.6 | — | Complete LOF | No | (56) |
| P146Sc | Not published | LOF | LOF | −6.9 | — | Predicted LOF | No | |
| L166Pc | Not published | LOF | LOF | −7.7 | — | Predicted LOF | No | |
| S168A | Not published | Benign | LOF | −5.1 | — | Benign | No | |
| A171Ta | Complete LOF | Benign | LOF | −5.3 | — | Complete LOF | No | (35) |
| F216F | Not published | — | Benign | — | Benign | Benign | No | |
| S217Rb | Partial LOF | LOF | LOF | −7.8 | — | Partial LOF | No | (36) |
| R262Qb | Partial LOF | LOF | LOF | −6.1 | — | Partial LOF | No | (6) |
| L266Ra | Complete LOF | LOF | LOF | −6.9 | — | Complete LOF | No | (35) |
| C279Ya | Complete LOF | LOF | LOF | −9.9 | — | Complete LOF | No | (35) |
| P320 litera | Complete LOF | LOF | LOF | −9.5 | — | Complete LOF | No | (34) |
Panther uses the score 0, neutral to −10, deleterious. LOF, Loss of function; —, not assessable.
Complete loss of function.
Partial loss of function.
Predicted to be pathologic; no asterisk: predicted to be benign.
Found in two controls in a heterozygous state.
Table 2.
Probands harboring rare variants in GNRHR
| Proband (and reference) | Gender | Diagnosis | Smell test (centile) | Ethnicity | Change (bp) | Change (AA) | Inheritance | Coding mutations in other genes |
|---|---|---|---|---|---|---|---|---|
| Complete loss of GNRHR, group 1, cLOF/cLOF | ||||||||
| 1 | F | nIHH | — | A | [c.32delA] + [c.32delA] | Q11fsX23 (homo) | NA | Nonea |
| 2 | M | nIHH | — | A | [c.836 G>A] + [c.836 G>A] | C279Y | AR | Noneb |
| Severe loss of GNRHR, group 2, cLOF/pLOF | ||||||||
| 3 | F | nIHH | 82 | C | [c.2 T>C] + [ = ]; [c.317 A>G] + [ = ]; [c.416 G>A] + [ = ] | M1T, Q106R, R139H | AR | Nonec |
| 4 (35) | F | nIHH | 60 | C | [c.30 T>A] + [ = ]; [c.31 C>A] + [ = ]; [ = ] + [c.959 C>T] | N10K, Q11K, P320L | S | Nonec |
| 5 | M | nIHH | 50 | C | [c.95 C>T] + [ = ]; [c.416 G>A] + [ = ] | T32I, R139H | AD | Noned |
| 6 | F | nIHH | — | AA | [c.317 A>G] + [ = ]; [c.797 T>G] + [ = ] | Q106R, L266R | S | Nonec |
| Severe loss of GNRHR, group 3, pLOF/pLOF | ||||||||
| 7 | M | Fertile eunuch | 41 | C | [c.286 C>T] + [ = ]; [c.317 A>G] + [ = ] | P96S, Q106R | S | Nonec |
| 8 (42) | M | nIHH reversal | — | C | [c.317 A>G] + [c.317 A>G] | Q106R (homo) | S | Nonec |
| 9 | F | KS | <5 | C | [c.317 A>G] + [c.317 A>G] | Q106R (homo) | S | Noned |
| 10 | M | nIHH/fertile eunuch | — | C | [c.317 A>G] + [c.317 A>G]; [c.651 C>A] + [ = ] | Q106R, S217R | AD | Nonec |
| 11 (41) | F | nIHH | 12 | C | [c. 317 A>G] + [ = ]; [c.785 G>A] + [ = ] | Q106R, R262Q | AD | FGFR1: R470Lc |
| 12 | M | nIHH/fertile eunuch | — | A | [c.497 T>C] + [ = ]; [c.785 G>A] + [ = ] | L166P, R262Q | AD | Nonec |
| 13 | F | nIHH | — | C | [c.785 G>A] + [c.785 G>A] | R262Q (homo) | NA | Nonec |
| Moderate loss of GNRHR, group 4, cLOF/NL or pLOF/NL | ||||||||
| 14 | F | nIHH | — | UK (Hispanic) | [c.247 C>G] + [ = ]; [c.317 A>G] + [ = ] | L83V, Q106R | AR | FGFR1: N117Se |
| 15 | M | nIHH | — | UK | [c.317 A>G] + [ = ] | Q106R | NA | Nonef |
| 16 | M | nIHH | 25 | A | [c.317 A>G] + [ = ] | Q106R | NA | PROKR2: V331Mc |
| 17 | M | nIHH | — | UK | [c.317 A>G] + [ = ] | Q106R | NA (Brother/Brother) | Noned |
| 18 | M | AOIHH | 14 | C | [c.317 A>G] + [ = ] | Q106R | S | Nonec |
| 19 | M | KS | — | C | [c.317 A>G] + [ = ] | Q106R | NA | Noned |
| 20 | F | KS | — | C | [c.317 A>G] + [ = ] | Q106R | AR | Noned |
| 21 | M | KS | <5 | C | [c.317 A>G] + [ = ] | Q106R | AD | Noned |
| 22 | M | nIHH | 30 | NA | [c.317 A>G] + [ = ] | Q106R | NA | Noneg |
| 23 | M | nIHH | 50 | C | [c.317 A>G] + [ = ] | Q106R | S | Noneg |
| 24 | F | nIHH | — | C | [c.317 A>G] + [ = ]; [c.742-132 A>G] + [ = ] | Q106R | AR | Nonec |
| 25 | F | HA | 32 | C | [c.317 A>G] + [ = ]; [c.504 T>A] + [ = ] | Q106R, S168A | S | Noned |
| 26 | M | CDP | 59 | C | [c.350 T>C] + [ = ] | L117P | S | Noneh |
| 27 | F | nIHH | — | C | [c.436 C>T] + [ = ] | P146S | NA | Nonec |
| 28 | M | KS | <5 | C | [c.436 C>T] + [ = ] | P146S | AD | Nonei |
| 29 | M | KS | <5 | C | [c.511 G>A] + [ = ] | A171T | NA | Noned |
| 30 | M | nIHH | — | C | [c.785 G>A] + [ = ] | R262Q | S | FGFR1: K618Nj |
| 31 | F | KS | — | C | [c.785 G>A] + [ = ] | R262Q | NA | Nonec |
| 32 | F | HA | — | UK (Hispanic) | [c.785 G>A] + [ = ] | R262Q | NA | Noned |
| Normal GNRHR (not included in the four groups) | ||||||||
| 33 | F | CDP | <5 | C | [c.149 C>T] + [ = ] | A50V | NA | PROKR2: N325Ke |
| 34 | M | nIHH | 25 | UK | [c.648 C>T] + [ = ] | F216F | NA | Nonec |
| 35 | M | nIHH | — | C | [c.649 C>T] + [ = ] | F216F | NA | Nonec |
C, Caucasian; A, Asian; AA, African-American; UK, unknown. AR, autosomal recessive; AD, autosomal dominant; S, sporadic; NA, not assessable; M, male; F, female.
Screened for FGF8, FGFR1, KAL1, NELF, PROK2, PROKR2, TAC3, and TACR3.
Screened for FGF8, FGFR1, KAL1, PROK2, and PROKR2.
Screened for FGF8, FGFR1, GNRH1, KAL1, KISS1, KISS1R, NELF, PROK2, PROKR2, TAC3, and TACR3.
Screened for FGF8, FGFR1, KAL1, KISS1, KISS1R, NELF, PROK2, and PROKR2.
Screened for FGF8, FGFR1, GNRH1, KAL1, KISS1R, KISS1, PROK2, PROKR2, TAC3, and TACR3.
Screened for GF8, FGFR1, GNRH1, KISS1, and KISS1R.
Screened for KISS1R, TAC3, TACR3, GnRH1, PROK2, and PROKR2.
Screened for FGF8, FGFR1, GNRH1, KAL1, KISS1R, NELF, PROK2, PROKR2, TAC3, and TACR3.
Screened for FGF8, FGFR1, KAL1, KISS1, PROK2, and PROKR2.
Screened for FGF8, FGFR1, KAL1, KISS1R, KISS1, NELF, PROK2, PROKR2, TAC3, and TACR3.
Although 20 of 21 RSV were not observed in controls, one variant (Q106R) was heterozygous in two controls (Table 1). The frequency of heterozygous GNRHR RSVs in probands (22 of 863 individuals, 2.5%) was significantly higher than that of controls (2 of 422 individuals, 0.5%) (χ2, P < 0.01).
Inheritance and segregation
None of the identified variants arose de novo (Supplemental Fig. 2). Of the 35 probands bearing variants, 12 came from pedigrees in which the disease phenotype appeared to be familial [dominant (n = 5), recessive (n = 6), or brother-brother (n = 1)]. Nine were sporadic and 14 had an unknown mode of inheritance (Table 2). Of the 46 family members who provided DNA, 38 carried GNRHR RSV. Ten of 38 had IGD, two had isolated anosmia or cleft lip/palate, and the remaining 26 had no known reproductive or other abnormalities (Supplemental Tables 1–3). Although variants were frequently identified in reproductively normal family members, all the family members with a GnRH-deficient phenotype carried the same GNRHR variant(s) as the proband.
In G1 (cLOF/cLOF, n = 4, two males, two females), all individuals had normosmic GnRH deficiency [three homozygous C279Y (no. 2, no. 2b, no. 2c), and one homozygous Q11fsX23 (no. 1)] (Table 2 and Supplemental Tables 1 and 2). Baseline LH pulsatility was not ascertained in any of these individuals. Proband no. 1, after receiving estrogen/progestin supplementation, was treated with pulsatile GnRH (one cycle, 250 ng/kg) and exogenous FSH (dose escalations of 75–225 IU/d over three cycles) from ages 34 to 35 yr. However, her estradiol levels remained low and she did not ovulate in response to either therapy (Supplemental Tables 1 and 2 and Fig. 2).
In G2 (cLOF/pLOF, n = 4, one male, three females), all individuals had normosmic IGD [compound heterozygous for M1T/Q106R/R139H (no. 3); N10K/Q11K/P320L (no. 4); T32I/R139H (no. 5), and Q106R/L266R (no. 6)] (Table 2 and Supplemental Tables 1 and 2). None of the females had spontaneous menarche. Two females (no. 3 and no. 4) demonstrated no and one low-amplitude LH pulse(s) in 12 h, respectively, with the mean LH levels below the limit of detection of the assay. Both patients received estrogen/progestin supplementation before initiating fertility treatments. These patients developed follicles with increasing doses of pulsatile GnRH, but their cycles had abnormal dynamics. Subject no. 4 (N10K/Q11K/P320L) required GnRH dose escalations from the usually effective dose of 75 ng/kg, iv, to 500 ng/kg to achieve folliculogenesis with appropriate estradiol secretion. Despite estradiol levels of 126–256 pg/ml, she failed to mount an induced LH surge. She was subsequently treated with gonadotropins to which she was normally responsive, eventually delivering twins on her fourth treatment cycle (34).
Subject no. 3 (M1T/Q106R/R139H) also required higher-than-normal doses of GnRH (100–250 ng/kg) to achieve folliculogenesis. She ovulated spontaneously on 250 ng/kg GnRH, iv, with a transiently positive human chorionic gonadotropin (hCG), but folliculogenesis was disordered in two further cycles with early luteinization and persistent progesterone secretion. This cycle was followed by two gonadotropin cycles in which she achieved poor folliculogenesis with higher-than-usual doses. At age 35 yr, she underwent in vitro fertilization and conceived using a protocol of high-dose recombinant FSH in addition to a small daily dose of hCG. Although she demonstrated a good follicular response by ultrasound, estradiol secretion remained low, requiring follicular phase supplementation with estradiol, and fertilization rates were poor. However, she delivered a healthy son (no. 3d) without cryptorchidism or micropenis (Supplemental Table 2 and Fig. 2).
The one male subject in group 2 (no. 5, T32I/R139H) presented with a severe GnRH deficiency [2 ml testes and microphallus (stretched penile length <10.5 cm [39])] and no LH pulses over 12 h (mean LH <1.6 IU/liter). He declined treatment with pulsatile GnRH (Supplemental Table 1 and Fig. 2).
G3 contained 10 individuals (five males, five females) classified as pLOF/pLOF [two homozygous Q106R (no. 8, no. 9), one homozygous R262Q (no. 13), and seven compound heterozygous (P96S/Q106R (no. 7), Q106R/S217R (no. 10, no. 10c), Q106R/R262Q (no. 11, no. 11b), L166P/R262Q (no. 12, no. 12b)]. All had normosmic GnRH deficiency except for one female (no. 9) with KS. None of the females had spontaneous menarche. Although the size of the groups is too small to allow statistical comparisons, in contrast to G1 and G2, individuals in G3 presented a greater evidence for endogenous GnRH induced LH pulsatility (n = 3, mean number of LH pulses = 3.5 ± 1.3 over 12 h, mean LH concentration = 3.6 ± 1.6 IU/liter) (Supplemental Table 2 and Fig. 2). One female (no. 11, Q106R/R262Q) was initially treated with pulsatile GnRH (75 ng/kg, one cycle) but had no follicular development. During 100 ng/kg treatments (three cycles), she had an endogenous LH surge but inadequate progesterone levels. Finally, at 250 ng/kg (eight cycles), monofolliculogenesis and normal neuroendocrine dynamics were documented. This patient conceived three times on pulsatile GnRH (250 ng/kg) and then twice with exogenous gonadotropins, but all five conceptions resulted in miscarriages (40). However, her sister (no. 11b, same mutations) bore three children after exogenous gonadotropins. Both sisters were also heterozygous for a deleterious FGFR1 mutation [R470L (17)] (Supplemental Table 2 and Fig. 2). In summary, female patients in G3 were able to achieve follicular development with increasing doses of either GnRH or gonadotropins, but in one case multiple miscarriages occurred.
G3 (pLOF/pLOF) also contained two males with attenuated forms of GnRH deficiency (Table 2, Supplemental Table 1, Fig. 2). Patient no. 8 (homozygous Q106R) presented with microphallus but after fathering a baby after 4 months of hCG monotherapy (1000 IU sc every other day) (41), he demonstrated spontaneous recovery (five LH pulses over 12 h, testosterone 271 ng/dl, testes 17 ml bilaterally, and sperm count 42 million/ml 6 months after discontinuation of hCG) (23). Subject no. 7 (P96S/Q106R) presented with the fertile eunuch syndrome, with 12 ml testes before treatment initiation. Two other males (no. 10, Q106R/S217R; no. 12, L166P/R262Q) in this group also had normosmic IHH (nIHH) with normal-sized testes (12 and 15 ml), but no information is available regarding prior treatment (Supplemental Table 1 and Fig. 2).
G4 [cLOF/normal (NL) or pLOF/NL] contained 52 individuals with only one impaired allele, including 26 patients (15 males, 11 females) (no. 5b, 10b, 10d, 12c, 14, 14b, 15, 16, 17, 18, 19, 20, 21, 21b, 22, 23, 24, 24b, 25, 26, 27, 28, 29, 30, 31, 32) and 26 normal family members (10 males, 16 females) (no. 3b, 3c, 3d, 4b, 4c, 4d, 6b, 6c, 8b, 8c, 8d, 8e, 11c, 11d, 11e, 11f, 11 g, 12d, 12e, 14c, 14d, 18b, 24c, 29b, 30b, 32b) (Supplemental Tables 1–3). Of 36 individuals in whom genomic sequence data were available, 33 of 36 (91.7%) were heterozygous by SNP analysis (Supplemental Table 4). Six had KS (no. 19, 20, 21, 28, 29, and 31) and 11 had nIHH (no. 14, 14b, 15, 16, 17, 22, 23, 24, 24b, 27, 30). Others had milder forms of GnRH deficiency: three females had HA (no. 25, 10d, and 32); one male had AOHH (no. 18); and three males had CDP (no. 10b, 12c, 26). Two females presented with isolated anosmia or cleft lip/palate (no. 5b and 21b, respectively) (Supplemental Tables 1 and 2 and Fig. 2). Five individuals underwent blood sampling every 10 min and, similar to group 3 (pLOF/pLOF), had discernible pulses: mean of 3.4 ± 2.3 LH pulses over 12 h and a mean LH concentration of 2.8 ± 2.0 IU/liter. Patient no. 30 (heterozygous R262Q) responded well to increasing doses of GnRH (ED50 = 13 and 23 ng/kg in two different studies), and patient no. 18 (heterozygous Q106R) fathered a baby on pulsatile GnRH treatment (5–25 ng/kg dose escalation over 1 yr).
The number of LH pulses and mean LH levels were not statistically different among G2, G3, and G4.
Probands were screened for mutations in genes implicated in the pathogenesis of GnRH deficiency. RSV were identified in only five of 33 probands [heterozygous FGFR1 in three (no. 14, 11, 30) and heterozygous PROKR2 in two (no. 33, no. 16)] and one of 36 family members (no. 11b, heterozygous FGFR1).
Discussion
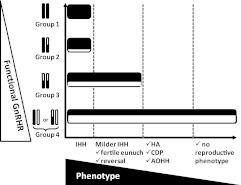
The central aim of this study was to examine the spectrum of reproductive phenotypes in a large cohort of individuals bearing mutations in GNRHR and to relate this to the apparent genotypic burden. Although the number of individuals in groups 1 and 2 are modest, the phenotypes of the patients are consistent with severe GnRH deficiency. More attenuated forms of GnRH deficiency, such as the fertile eunuch syndrome and reversible hypogonadotropism, were identified in group 3 (pLOF/pLOF) but not in groups 1 or 2.
In contrast, patients in G4 (pLOF/NL or cLOF/NL) do not follow the trend established by patients in G1-G3. Patients in G4 demonstrated a broad phenotypic spectrum, ranging from severe GnRH-deficient states such as KS (12%) and IHH (21%), to attenuated GnRH deficiency (13% with HA, AOIHH, or CDP), to seemingly normal GnRH neuronal function (normal puberty and sexual function (50%) (Fig. 3). The possibility that patients in G4 had a gene deletion on their nonmutated allele was excluded as the vast majority of patients were clearly heterozygous for common polymorphisms. In addition, the prevalence of heterozygous RSV in GNRHR was significantly higher in probands compared with controls (P < 0.01), supporting the concept that these monoallelic changes do contribute to abnormal reproductive function.
Fig. 3.
Relationship between the phenotype of patients harboring loss-of-function mutations and the number/severity of GnRH mutations.
Several possibilities exist to explain how monoallelic changes in GNRHR contribute to the abnormal reproductive phenotypes. Dominant-negative effects of GNRHR mutations have been described in vitro (35, 42). However, if that was the mechanism of all heterozygote mutations, a greater proportion of individuals in G4 would be expected to have GnRH deficiency. Instead, 50% of the individuals carrying heterozygote GNRHR mutations had seemingly normal reproductive function.
Given that GnRH deficiency has been established to be an oligogenic disease in a subset of patients (17, 18), a second possibility is that patients with GNRHR mutations may have additional genetic defects yet to be discovered. Indeed, the presence in G4 of seven individuals with abnormal olfaction as well as the subject who presented with only cleft lip/palate strongly suggests the presence of additional genetic defects in pathways that affect olfactory bulb and facial development/function. As recently shown by Sarfati et al. (19), the overwhelming majority of GnRH-deficient patients with mutations in PROKR2 have monoallelic mutations (11, 13) without dominant-negative effects (43), suggesting that these individuals carry mutations in other genes for GnRH deficiency (18, 19). Although only six of 70 patients actually had a second genetic abnormality identified in the current study, it is likely that increasingly availability of sequencing will accelerate the pace of gene discovery and the occurrence of demonstrable second mutations.
Finally, it is possible that heterozygote GNRHR mutations, through modest reductions in gene dosage, may confer susceptibility to environmental, behavioral, or psychosocial constraints on GnRH secretion (44). This hypothesis may be exemplified in two families in this study, each with biallelic siblings with nIHH (families of no. 10 and no. 12). Both sets of fathers (no. 10b and no. 12c) harbored monoallelic changes and presented with CDP (Supplemental Fig. 2 and Supplemental Table 1). One of the mothers (no. 10d) had a normal puberty but subsequently developed HA triggered by anorexia (Supplemental Table 2); the other mother (no. 12e) had normal puberty and sexual function, but it is not known whether she ever experienced excessive exercise or food deprivation (20) (Supplemental Table 3). In this study, the frequency of heterozygous changes in GNRHR was significantly more common in probands than in controls, suggesting that GNRHR variants are not merely innocent bystanders but may contribute to reproductive pathogenicity. Understanding how environmental inputs interact with genetic variation to modify phenotypic expression remains a great challenge in contemporary genetics.
Although the association of GNRHR mutations with decreased responsiveness to exogenous GnRH is expected, the impaired response to gonadotropin treatments reported in one female in G1 (no. 1) and another in G2 (no. 3) suggests additional gonadal defects. Both GNRH and GNRHR are expressed in the ovary of several species (45) including humans (46). GNRHR mRNA expression in granulosa-luteal cells increases during follicular development (47), and GNRHR binding has been demonstrated in granulosa cells of preovulatory follicles and corpora lutea, although it is lacking in primordial and early antral follicles (48, 49). This stage-specific expression and function raises the hypothesis that GNRHR mutations may have a role in folliculogenesis and contribute to abnormal gonadal responses to fertility treatments.
In G3, a female with a compound heterozygous GNRHR mutation and a FGFR1 mutation conceived five times on pulsatile GnRH but had five miscarriages (40). Both GNRHR and FGFR1 are expressed in the placenta and have been proposed as regulators of placental function through hCG secretion and angiogenesis, respectively (50–54). Although fetal gnrhr is not thought to be essential for maintenance of early pregnancy in mice (53), the potential significance of GnRH and its receptor on the maternal side of the placenta is still being investigated.
In conclusion, mutations in GNRHR are relatively common causes of hypogonadotropic hypogonadism, occurring in approximately 4% of a large population of patients with GnRH deficiency (5.6% of normosmic patients and 1.9% of KS). Receptor function in patients harboring biallelic mutations in GNRHR appears to correlate with the phenotypic spectrum of GnRH deficiency. However, patients harboring monoallelic mutations in GNRHR demonstrate a wider spectrum of GnRH-deficient states, suggesting the presence of yet-to-be-identified genetic and/or nongenetic factors that work in combination with the mutated GNRHR allele to produce reproductive phenotypes.
Supplementary Material
Acknowledgments
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institutes of Health through Cooperative Agreement U54 5U54HD028138 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research, and Grants R01-HD-15788, R01-HD-19938, R01-HD-42708, and R01-HD-043341.
Disclosure Summary: E.G., J.E.H., M.G.A., U.B.K., R.Q., J.A.S., D.L.M., N.P., V.M., P.M.M., L.L.L., L.I., M.L.-M, V.Y.F., R.G.D., M.L.C., W.F.C., L.P., and S.B.S. have nothing to declare.
Footnotes
- AOIHH
- Adult-onset idiopathic hypogonadotropic hypogonadism
- CDP
- constitutional delay of puberty
- cLOF
- complete loss of function
- dbEST
- Database for Expressed Sequence Tags
- dbSNP
- Database for Single Nucleotide Polymorphisms
- HA
- hypothalamic amenorrhea
- hCG
- human chorionic gonadotropin
- IGD
- isolated GnRH deficiency
- IHH
- idiopathic hypogonadotropic hypogonadism
- KS
- Kallmann syndrome
- pLOF
- partial loss of function
- nIHH
- normosmic IHH
- NL
- normal
- RSV
- rare sequence variant
- SNP
- single-nucleotide polymorphism.
References
- 1. Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, Carrozzo R, Maestrini E, Pieretti M, Taillon-Miller P, Brown CJ, Willard HF, Lawrence C, Graziella Persico M, Camerino G, Ballabio A. 1991. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature 353:529–536 [DOI] [PubMed] [Google Scholar]
- 2. Dodeé C, Levilliers J, Dupont JM, De Paepe A, Le Dû N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, Pêcheux C, Le Tessier D, Cruaud C, Delpech M, Speleman F, Vermeulen S, Amalfitano A, Bachelot Y, Bouchard P, Cabrol S, Carel JC, Delemarre-van de Waal H, Goulet-Salmon B, Kottler ML, Richard O, Sanchez-Franco F, Saura R, Young J, Petit C, Hardelin JP. 2003. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet 33:463–465 [DOI] [PubMed] [Google Scholar]
- 3. Falardeau J, Chung WC, Beenken A, Raivio T, Plummer L, Sidis Y, Jacobson-Dickman EE, Eliseenkova AV, Ma J, Dwyer A, Quinton R, Na S, Hall JE, Huot C, Alois N, Pearce SH, Cole LW, Hughes V, Mohammadi M, Tsai P, Pitteloud N. 2008. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest 118:2822–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouligand J, Ghervan C, Tello JA, Brailly-Tabard S, Salenave S, Chanson P, Lombès M, Millar RP, Guiochon-Mantel A, Young J. 2009. Isolated familial hypogonadotropic hypogonadism and a GNRH1 mutation. N Engl J Med 360:2742–2748 [DOI] [PubMed] [Google Scholar]
- 5. Chan YM, de Guillebon A, Lang-Muritano M, Plummer L, Cerrato F, Tsiaras S, Gaspert A, Lavoie HB, Wu CH, Crowley WF, Jr, Amory JK, Pitteloud N, Seminara SB. 2009. GNRH1 mutations in patients with idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA 106:11703–11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Roux N, Young J, Misrahi M, Genet R, Chanson P, Schaison G, Milgrom E. 1997. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med 337:1597–1602 [DOI] [PubMed] [Google Scholar]
- 7. de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. 2003. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA 100:10972–10976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O'Rahilly S, Carlton MB, Crowley WF, Jr, Aparicio SA, Colledge WH. 2003. The GPR54 gene as a regulator of puberty. N Engl J Med 349:1614–1627 [DOI] [PubMed] [Google Scholar]
- 9. Miura K, Acierno JS, Jr, Seminara SB. 2004. Characterization of the human nasal embryonic LHRH factor gene, NELF, and a mutation screening among 65 patients with idiopathic hypogonadotropic hypogonadism (IHH). J Hum Genet 49:265–268 [DOI] [PubMed] [Google Scholar]
- 10. Matsumoto S, Yamazaki C, Masumoto KH, Nagano M, Naito M, Soga T, Hiyama H, Matsumoto M, Takasaki J, Kamohara M, Matsuo A, Ishii H, Kobori M, Katoh M, Matsushime H, Furuichi K, Shigeyoshi Y. 2006. Abnormal development of the olfactory bulb and reproductive system in mice lacking prokineticin receptor PKR2. Proc Natl Acad Sci USA 103:4140–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dodé C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, Morgan G, Murat A, Toublanc JE, Wolczynski S, Delpech M, Petit C, Young J, Hardelin JP. 2006. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet 2:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pitteloud N, Zhang C, Pignatelli D, Li JD, Raivio T, Cole LW, Plummer L, Jacobson-Dickman EE, Mellon PL, Zhou QY, Crowley WF., Jr 2007. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA 104:17447–17452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cole LW, Sidis Y, Zhang C, Quinton R, Plummer L, Pignatelli D, Hughes VA, Dwyer AA, Raivio T, Hayes FJ, Seminara SB, Huot C, Alos N, Speiser P, Takeshita A, Van Vliet G, Pearce S, Crowley WF, Jr, Zhou QY, Pitteloud N. 2008. Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: molecular genetics and clinical spectrum. J Clin Endocrinol Metab 93:3551–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Topaloglu AK, Reimann F, Guclu M, Yalin AS, Kotan LD, Porter KM, Serin A, Mungan NO, Cook JR, Ozbek MN, Imamoglu S, Akalin NS, Yuksel B, O'Rahilly S, Semple RK. 2009. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for neurokinin B in the central control of reproduction. Nat Genet 41:354–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gianetti E, Tusset C, Noel SD, Au MG, Dwyer AA, Hughes VA, Abreu AP, Carroll J, Trarbach E, Silveira LF, Costa EM, de Mendonça BB, de Castro M, Lofrano A, Hall JE, Bolu E, Ozata M, Quinton R, Amory JK, Stewart SE, Arlt W, Cole TR, Crowley WF, Kaiser UB, Latronico AC, Seminara SB. 2010. TAC3/TACR3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin B in neonatal life followed by reversal in adulthood. J Clin Endocrinol Metab 95:2857–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim HG, Kurth I, Lan F, Meliciani I, Wenzel W, Eom SH, Kang GB, Rosenberger G, Tekin M, Ozata M, Bick DP, Sherins RJ, Walker SL, Shi Y, Gusella JF, Layman LC. 2008. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet 83:511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, Li WP, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi OA, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley W. 2007. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest 117:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sykiotis GP, Plummer L, Hughes VA, Au M, Durrani S, Nayak-Young S, Dwyer AA, Quinton R, Hall JE, Gusella JF, Seminara SB, Crowley WF, Jr, Pitteloud N. 2010. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc Natl Acad Sci USA 107:15140–15144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sarfati J, Guiochon-Mantel A, Rondard P, Arnulf I, Garcia-Piñero A, Wolczynski S, Brailly-Tabard S, Bidet M, Ramos-Arroyo M, Mathieu M, Lienhardt-Roussie A, Morgan G, Turki Z, Bremont C, Lespinasse J, Du Boullay H, Chabbert-Buffet N, Jacquemont S, Reach G, De Talence N, Tonella P, Conrad B, Despert F, Delobel B, Brue T, Bouvattier C, Cabrol S, Pugeat M, Murat A, Bouchard P, Hardelin JP, Dodé C, Young J. 2010. A comparative phenotypic study of Kallmann syndrome patients carrying monoallelic and biallelic mutations in the prokineticin 2 or prokineticin receptor 2 genes. J Clin Endocrinol Metab 95:659–669 [DOI] [PubMed] [Google Scholar]
- 20. Caronia LM, Martin C, Welt CK, Sykiotis GP, Quinton R, Thambundit A, Avbelj M, Dhruvakumar S, Plummer L, Hughes VA, Seminara SB, Boepple PA, Sidis Y, Crowley WF, Jr, Martin KA, Hall JE, Pitteloud N. 2011. A genetic basis for functional hypothalamic amenorrhea. N Engl J Med 364:215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, Stoetzel C, Szymanska K, Ramaswami G, Logan CV, Muzny DM, Young AC, Wheeler DA, Cruz P, Morgan M, Lewis LR, Cherukuri P, Maskeri B, Hansen NF, Mullikin JC, Blakesley RW, Bouffard GG, Gyapay G, Rieger S, Tönshoff B, Kern I, Soliman NA, Neuhaus TJ, Swoboda KJ, Kayserili H, Gallagher TE, Lewis RA, Bergmann C, Otto EA, Saunier S, Scambler PJ, Beales PL, Gleeson JG, Maher ER, Attié-Bitach T, Dollfus H, Johnson CA, Green ED, Gibbs RA, Hildebrandt F, Pierce EA, Katsanis N. 2011. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet 43:189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Layman LC, Cohen DP, Jin M, Xie J, Li Z, Reindollar RH, Bolbolan S, Bick DP, Sherins RR, Duck LW, Musgrove LC, Sellers JC, Neill JD. 1998. Mutations in gonadotropin-releasing hormone receptor gene cause hypogonadotropic hypogonadism. Nat Genet 18:14–15 [DOI] [PubMed] [Google Scholar]
- 23. Raivio T, Falardeau J, Dwyer A, Quinton R, Hayes FJ, Hughes VA, Cole LW, Pearce SH, Lee H, Boepple P, Crowley WF, Jr, Pitteloud N. 2007. Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med 357:863–873 [DOI] [PubMed] [Google Scholar]
- 24. Doty RL, Shaman P, Kimmelman CP, Dann MS. 1984. University of Pennsylvania Smell Identification Test: a rapid quantitative olfactory function test for the clinic. Laryngoscope 94:176–178 [DOI] [PubMed] [Google Scholar]
- 25. Thome JL, Espelage DL. 2007. Obligatory exercise and eating pathology in college females: replication and development of a structural model. Eat Behav 8:334–349 [DOI] [PubMed] [Google Scholar]
- 26. Santoro N, Filicori M, Crowley WF., Jr 1986. Hypogonadotropic disorders in men and women: diagnosis and therapy with pulsatile gonadotropin-releasing hormone. Endocr Rev 7:11–23 [DOI] [PubMed] [Google Scholar]
- 27. Garner DM, Olmsted MP, Bohr Y, Garfinkel PE. 1982. The eating attitudes test: psychometric features and clinical correlates. Psychol Med 12:871–878 [DOI] [PubMed] [Google Scholar]
- 28. Nachtigall LB, Boepple PA, Pralong FP, Crowley WF., Jr 1997. Adult-onset idiopathic hypogonadotropic hypogonadism–a treatable form of male infertility. N Engl J Med 336:410–415 [DOI] [PubMed] [Google Scholar]
- 29. Marshall WA, Tanner JM. 1969. Variations in pattern of pubertal changes in girls. Arch Dis Child 44:291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marshall WA, Tanner JM. 1970. Variations in the pattern of pubertal changes in boys. Arch Dis Child 45:13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hayes FJ, McNicholl DJ, Schoenfeld D, Marsh EE, Hall JE. 1999. Free α-subunit is superior to luteinizing hormone as a marker of gonadotropin-releasing hormone despite desensitization at fast pulse frequencies. J Clin Endocrinol Metab 84:1028–1036 [DOI] [PubMed] [Google Scholar]
- 32. Filicori M, Butler JP, Crowley WF., Jr 1984. Neuroendocrine regulation of the corpus luteum in the human. Evidence for pulsatile progesterone secretion. J Clin Invest 73:1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beranova M, Oliveira LM, Bédécarrats GY, Schipani E, Vallejo M, Ammini AC, Quintos JB, Hall JE, Martin KA, Hayes FJ, Pitteloud N, Kaiser UB, Crowley WF, Jr, Seminara SB. 2001. Prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab 86:1580–1588 [DOI] [PubMed] [Google Scholar]
- 34. Meysing AU, Kanasaki H, Bedecarrats GY, Acierno JS, Jr, Conn PM, Martin KA, Seminara SB, Hall JE, Crowley WF, Jr, Kaiser UB. 2004. GNRHR mutations in a woman with idiopathic hypogonadotropic hypogonadism highlight the differential sensitivity of luteinizing hormone and follicle-stimulating hormone to gonadotropin-releasing hormone. J Clin Endocrinol Metab 89:3189–3198 [DOI] [PubMed] [Google Scholar]
- 35. Leaños-Miranda A, Ulloa-Aguirre A, Janovick JA, Conn PM. 2005. In vitro coexpression and pharmacological rescue of mutant gonadotropin-releasing hormone receptors causing hypogonadotropic hypogonadism in humans expressing compound heterozygous alleles. J Clin Endocrinol Metab 90:3001–3008 [DOI] [PubMed] [Google Scholar]
- 36. Costa EM, Bedecarrats GY, Mendonca BB, Arnhold IJ, Kaiser UB, Latronico AC. 2001. Two novel mutations in the gonadotropin-releasing hormone receptor gene in Brazilian patients with hypogonadotropic hypogonadism and normal olfaction. J Clin Endocrinol Metab 86:2680–2686 [DOI] [PubMed] [Google Scholar]
- 37. Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. 2001. Prediction of deleterious human alleles. Hum Mol Genet 10:591–597 [DOI] [PubMed] [Google Scholar]
- 38. Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O. 2003. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res 31:334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gabrich PN, Vasconcelos JS, Damião R, Silva EA. 2007. Penile anthropometry in Brazilian children and adolescents. J Pediatr (Rio J) 83:441–446 [DOI] [PubMed] [Google Scholar]
- 40. Seminara SB, Beranova M, Oliveira LM, Martin KA, Crowley WF, Jr, Hall JE. 2000. Successful use of pulsatile gonadotropin-releasing hormone (GnRH) for ovulation induction and pregnancy in a patient with GnRH receptor mutations. J Clin Endocrinol Metab 85:556–562 [DOI] [PubMed] [Google Scholar]
- 41. Pitteloud N, Boepple PA, DeCruz S, Valkenburgh SB, Crowley WF, Jr, Hayes FJ. 2001. The fertile eunuch variant of idiopathic hypogonadotropic hypogonadism: spontaneous reversal associated with a homozygous mutation in the gonadotropin-releasing hormone receptor. J Clin Endocrinol Metab 86:2470–2475 [DOI] [PubMed] [Google Scholar]
- 42. Knollman PE, Janovick JA, Brothers SP, Conn PM. 2005. Parallel regulation of membrane trafficking and dominant-negative effects by misrouted gonadotropin-releasing hormone receptor mutants. J Biol Chem 280:24506–24514 [DOI] [PubMed] [Google Scholar]
- 43. Monnier C, Dodé C, Fabre L, Teixeira L, Labesse G, Pin JP, Hardelin JP, Rondard P. 2009. PROKR2 missense mutations associated with Kallmann syndrome impair receptor signalling activity. Hum Mol Genet 18:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marcus MD, Loucks TL, Berga SL. 2001. Psychological correlates of functional hypothalamic amenorrhea. Fertil Steril 76:310–316 [DOI] [PubMed] [Google Scholar]
- 45. Ramakrishnappa N, Rajamahendran R, Lin YM, Leung PC. 2005. GnRH in non-hypothalamic reproductive tissues. Anim Reprod Sci 88:95–113 [DOI] [PubMed] [Google Scholar]
- 46. Cheung LW, Wong AS. 2008. Gonadotropin-releasing hormone: GnRH receptor signaling in extrapituitary tissues. FEBS J 275:5479–5495 [DOI] [PubMed] [Google Scholar]
- 47. Peng C, Fan NC, Ligier M, Väänänen J, Leung PC. 1994. Expression and regulation of gonadotropin-releasing hormone (GnRH) and GnRH receptor messenger ribonucleic acids in human granulosa-luteal cells. Endocrinology 135:1740–1746 [DOI] [PubMed] [Google Scholar]
- 48. Brus L, Lambalk CB, de Koning J, Helder MN, Janssens RM, Schoemaker J. 1997. Specific gonadotrophin-releasing hormone analogue binding predominantly in human luteinized follicular aspirates and not in human pre-ovulatory follicles. Hum Reprod 12:769–773 [DOI] [PubMed] [Google Scholar]
- 49. Choi JH, Gilks CB, Auersperg N, Leung PC. 2006. Immunolocalization of gonadotropin-releasing hormone (GnRH)-I, GnRH-II, and type I GnRH receptor during follicular development in the human ovary. J Clin Endocrinol Metab 91:4562–4570 [DOI] [PubMed] [Google Scholar]
- 50. Cheng KW, Nathwani PS, Leung PC. 2000. Regulation of human gonadotropin-releasing hormone receptor gene expression in placental cells. Endocrinology 141:2340–2349 [DOI] [PubMed] [Google Scholar]
- 51. Anteby EY, Natanson-Yaron S, Hamani Y, Sciaki Y, Goldman-Wohl D, Greenfield C, Ariel I, Yagel S. 2005. Fibroblast growth factor-10 and fibroblast growth factor receptors 1–4: expression and peptide localization in human decidua and placenta. Eur J Obstet Gynecol Reprod Biol 119:27–35 [DOI] [PubMed] [Google Scholar]
- 52. Arany E, Hill DJ. 1998. Fibroblast growth factor-2 and fibroblast growth factor receptor-1 mRNA expression and peptide localization in placentae from normal and diabetic pregnancies. Placenta 19:133–142 [DOI] [PubMed] [Google Scholar]
- 53. Wu S, Wilson MD, Busby ER, Isaac ER, Sherwood NM. 2010. Disruption of the single copy gonadotropin-releasing hormone receptor in mice by gene trap: severe reduction of reproductive organs and functions in developing and adult mice. Endocrinology 151:1142–1152 [DOI] [PubMed] [Google Scholar]
- 54. Siler-Khodr TM, Khodr GS, Valenzuela G, Rhode J. 1986. Gonadotropin-releasing hormone effects on placental hormones during gestation: I. α-Human chorionic gonadotropin, human chorionic gonadotropin and human chorionic somatomammotropin. Biol Reprod 34:245–254 [DOI] [PubMed] [Google Scholar]
- 55. Wolczynski S, Laudanski P, Jarzabek K, Mittre H, Lagarde JP, Kottler ML. 2003. A case of complete hypogonadotropic hypogonadism with a mutation in the gonadotropin-releasing hormone receptor gene. Fertil Steril 79:442–444 [DOI] [PubMed] [Google Scholar]
- 56. Arora KK, Cheng Z, Catt KJ. 1997. Mutations of the conserved DRS motif in the second intracellular loop of the gonadotropin-releasing hormone receptor affect expression, activation, and internalization. Mol Endocrinol 11:1203–1212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.