Abstract
Prokaryotic cyanobacteria express robust circadian (daily) rhythms under the control of a clock system that appears to be similar to those of eukaryotes in many ways. On the other hand, the KaiABC-based core cyanobacterial clockwork is clearly different from the transcription-translation feedback loop model of eukaryotic clocks in that the cyanobacterial clock system regulates gene expression patterns globally, and specific clock gene promoters are not essential in mediating the circadian feedback loop. A novel model, the oscilloid model, proposes that the KaiABC oscillator ultimately mediates rhythmic changes in the status of the cyanobacterial chromosome, and these topological changes underlie the global rhythms of transcription. The authors suggest that this model represents one of several possible modes of regulating gene expression by circadian clocks, even those of eukaryotes.
Keywords: circadian, biological clock, kai, prokaryote, global gene expression, supercoiling
Animals, plants, fungi, and cyanobacteria all display daily or circadian rhythms in their biochemistry, physiology, and/or behavior that are controlled by biological clocks (Dunlap et al., 2004). A variety of biological processes in various organisms are controlled by biological clocks such as gene expression, photosynthesis, sleeping/waking, and development, and this regulation is thought to help organisms adapt to the daily changes in light, temperature, and other factors in their environment. Circadian regulation of gene expression (i.e., the levels of specific proteins in cells) can be accomplished by clock control of transcription, mRNA stability, translation, and protein degradation. A particularly fascinating example of translational control by a circadian clock is that in the dinoflagellate alga, Gonyaulax (Rossini et al., 2003). However, we focus our discussion specifically on clock control of cyanobacterial gene expression at the level of transcription.
In general, studies of circadian gene expression in eukaryotes have often used microarray analyses to show that a relatively small proportion of genes (~5%–15%) in eukaryotic genomes display rhythms in mRNA abundance. While microarray technology is well advanced, it should be noted that microarrays are not very sensitive to small changes of mRNA abundance, and moreover, they are not a good measure of rhythmic transcriptional activity for mRNAs that are either very unstable or very stable. Therefore, microarray results might not be reflective of transcriptional control in detail.
For example, in the model plant Arabidopsis thaliana, microarray studies have demonstrated that approximately 10% of genes exhibited circadian rhythms of mRNA abundance, identifying circadian control of genes involved in photosynthesis/carbon metabolism, stress and hormone responses, transcription, and other pathways (Harmer et al., 2000; Schaffer et al., 2001). However, a promoter trap experiment showed that ~35% of Arabidopsis promoters were rhythmically controlled (Michael and McClung, 2003). This discrepancy implies that the promoter activity of many genes is controlled by the biological clock but that posttranscriptional control mechanisms counterbalance the rhythmic transcription of some genes such that the mRNA abundance remains more or less constant.
In the filamentous fungus, Neurospora crassa, microarray analyses showed that approximately 5% of genes from starving cultures and 20% of genes from rapidly growing cultures were clock regulated (Nowrousian et al., 2003; Correa et al., 2003). Microarrays revealed that about 1% of genes in Drosophila melanogaster displayed circadian rhythms in mRNA abundance (McDonald and Rosbash, 2001), and in mammals, microarray analyses of SCN and peripheral tissues showed that the mRNA abundance of approximately 8% to 10% of all genes is regulated by the circadian clock (Akhtar et al., 2002; Panda et al., 2002; Storch et al., 2002; Ueda et al., 2002). Based on the observations in Arabidosis (Michael and McClung, 2003) and the limitations of microarrays, it is quite likely that the degree to which circadian clocks control promoter activity is underestimated.
THE CIRCADIAN CLOCK SYSTEM IN CYANOBACTERIA
Unlike the cases of the eukaryotic systems, the clock system in prokaryotic cyanobacteria appears to globally regulate transcriptional activity. The cyanobacterial system was the first to use the promoter trap technique (Liu et al., 1995), and the application of this methodology may explain why pervasive clock control over gene expression was found. This profound control of transcriptional activity in the cyanobacterium Synechococcus elongatus PCC 7942 is regulated by a molecular clockwork that is encoded by 3 genes—kaiA, kaiB, and kaiC—that form a cluster on the chromosome; inactivation of any of the kai genes abolishes clock function (Ishiura et al., 1998). The proteins encoded by these genes interact with one another (Iwasaki et al., 1999; Taniguchi et al., 2001; Xu et al., 1999) to form large complexes in vivo with KaiC as the core (Kageyama et al., 2003). KaiC exists in both phosphorylated and nonphosphorylated forms in vivo, and its phosphorylation status is correlated with clock speed (Nishiwaki et al., 2000; Iwasaki et al., 2002; Xu et al., 2003; Johnson, 2004). In one of the more remarkable discoveries in the field in recent years, it was shown that a circadian oscillation of KaiC phosphorylation could be reconstituted in vitro with the purified KaiA, KaiB, and KaiC proteins (Nakajima et al., 2005). KaiA enhances KaiC autophosphorylation, while KaiB antagonizes the effects of KaiA (Iwasaki et al., 2002; Williams et al., 2002; Kitayama et al., 2003; Xu et al., 2003).
The enzymatic activity of the KaiABC system in regulating circadian output rhythms in vivo is not yet known, but evidence suggests that KaiC-containing protein complexes interact with DNA, either directly or indirectly. Neither kaiA nor kaiB show obvious similarities to any other known genes; however, kaiC appears to be an internally duplicated version of a bacterial recA/dnaB-like gene (Iwasaki et al., 1999; Leipe et al., 2000). In bacteria, RecA functions as a DNA recombinase, and DnaB is a DNA helicase; thus, the similarity implies that KaiC might also interact directly with DNA. Indeed, KaiC binds DNA substrates in vitro with low affinity (Mori et al., 2002). KaiC also interacts with a 2-component histidine protein kinase, SasA (Iwasaki et al., 2002), and stimulates the autophosphorylation of SasA (Smith and Williams, in press), but SasA does not alter the phosphorylation status of KaiC. Recently, a candidate protein that may function as the cognate response regulator with SasA has been identified (Takai et al., 2006); this response regulator, called RpaA, interacts with SasA and contains a leucine zipper DNA-binding motif. Therefore, it is likely that KaiC interacts with DNA either directly (Mori et al., 2002) or indirectly through SasA/RpaA to regulate circadian gene expression (Takai et al., 2006).
RHYTHMIC CLOCK OUTPUTS IN CYANOBACTERIA
In addition to regulating gene expression, a number of other metabolic and cellular processes in cyanobacteria are under the control of the biological clock. This observation suggests that circadian cycles allow cellular processes to occur in a temporal sequence that is most beneficial to the organism. For example, some species of cyanobacteria (e.g., the genera Trichodesmium, Cyanothece, and Synechococcus) show a daily rhythm of photosynthesis that is in reverse phase to the daily cycle of nitrogenase expression, a key enzyme for nitrogen fixation (Chen et al., 1999; Schneegurt et al., 1994; Huang et al., 1999). The peak of nitrogenase expression is during the dark phase of the cycle, allowing the temporal separation of expression of the oxygen-sensitive nitrogenase enzyme during periods when oxygen-evolving photosynthesis is not occurring. In Synechococcus RF-1, circadian changes in the uptake of amino acids has been observed (Chen et al., 1991), and more recently, infection of S. elongatus PCC 7942 by the cyanophage AS-1 has been shown to be influenced by light (Kao et al., 2005). In particular, adsorption of AS-1 appeared to be more efficient in the light phase than in the dark phase. For years, it was thought that a circadian clock would be of no benefit to prokaryotic organisms since cell division often occurs more than once daily in many prokaryotic species. However, S. elongatus cells, dividing on average more than once per day, display robust daily rhythms of gene expression (Kondo et al., 1997) and have the timing of their cell division “gated” to occur only during the light phase of the circadian cycle (Mori et al., 1996). These rhythms of gene expression persist when cell division, but not DNA replication, is inhibited (Mori and Johnson, 2001). Apparently, cell division is another output that is gated by the circadian clock, and control of the circadian timing mechanism is independent of the cell division cycle (Mori and Johnson, 2001; Kondo et al., 1997).
Using bacterial luciferase genes (luxAB) of Vibrio harveyi as a reporter for promoter activity, circadian rhythms of gene expression have been detected in a number of cyanobacterial species. By fusing the psbA1 promoter, which drives expression of a component of photosystem II, to a promoterless luxAB gene cassette, transcription from this promoter was shown to be under circadian control in S. elongatus PCC 7942 (Kondo et al., 1993). A similar approach was used to show that expression from the dnaK promoter was rhythmic in Synechocystis sp. strain PCC 6803 (Aoki et al., 1995). Rhythmic transcription from the psbA1 promoter in the thermophilic cyanobacterium, Thermosynechococcus elongatus BP-1, has also been demonstrated (Onai et al., 2004). A survey of 40 cyanobacterial species, including representatives of all 5 sections, indicated the presence of a kaiC gene. This discovery suggested that a kaiC-regulated circadian system is universal to cyanobacteria (Lorne et al., 2000). Therefore, regulation of promoter activities by the circadian clock appears to be a general phenomenon in cyanobacteria.
Despite the fact that the expression of genes for a number of photosynthetic proteins (e.g., psbAI) is regulated by the circadian clock in S. elongatus PCC 7942, photosynthesis itself (as monitored by oxygen evolution) is not rhythmic in constant light (Yen et al., 2004). Therefore, the significance of the rhythmic expression of photosynthesis genes in S. elongatus is not known. It is possible that this observation means that the transcription of some photosynthesis genes in S. elongatus is regulated by the circadian clock but that mRNA abundance or protein stability is regulated by a different mechanism as a design feature to supply more or less constant levels of photosynthetic proteins over the daily cycle (many proteins involved in photosynthesis are known to be damaged during the day and therefore must be replaced so that photosynthesis can proceed). On the other hand, in other closely related cyanobacterial species (Synechocystis sp. PCC 6803, Synechococcus RF-1), there are rhythms of oxygen evolution in constant light (Yen et al., 2004). It is not yet clear what these differences in gene expression patterns versus photosynthetic capabilities portend.
EVIDENCE FOR GLOBAL REGULATION OF TRANSCRIPTION BY THE CIRCADIAN CLOCK IN SYNECHOCOCCUS ELONGATUS
As mentioned above, in the eukaryotic organisms examined to date, genes from a relatively minor fraction of the genome show circadian rhythms of mRNA abundance; thus, control of transcription by the circadian clock does not appear to be pervasive. However, since mRNA abundance is due to both transcription and mRNA stability, this measure could be misleading. In S. elongatus, it is not a fraction of the total number of genes that is regulated by the biological clock; rather, promoter activities throughout the genome are controlled by the clock (Liu et al., 1995). In fact, we have not yet found even a single promoter whose activity is reproducibly constitutive. These conclusions are based on a sensitive and comprehensive promoter trap experiment in which a promoterless luciferase gene set (luxAB) was randomly inserted throughout the genome. Whenever the luxAB gene set inserted into a locus that was correctly positioned and oriented to a cyanobacterial promoter, luciferase was expressed. In our 1995 paper (Liu et al., 1995), the luxAB expression patterns of more than 800 independent colonies were analyzed; all of the colonies that expressed luciferase displayed circadian rhythms with the same period.
The pattern of the rhythmic luxAB expression differed among the promoters isolated, both in terms of phasing and waveform (Figure 1A,B). Most of the promoters (class 1 promoters) were activated during the day, consistent with an interpretation that their corresponding proteins are produced to replace proteins that become progressively inactivated during the daylight hours (Figure 1A). In contrast, a smaller number of promoters were activated only during the night (class 2 promoters). Even the promoter activity for a gene that would be expected to be constitutively expressed (e.g., a “housekeeping gene” such as a ribosomal RNA gene [rrnA]) was shown to be rhythmic (Liu et al., 1995). Furthermore, the activity of heterologous promoters, such as the Escherichia coli promoters, conIIp and trcp, is rhythmic when inserted into the cyanobacterial chromosome (Katayama et al., 1999; Xu et al., 2003). The data from the conIIp and trcp promoters demonstrate that even promoters that have not evolved within the cyanobacterial system are recognized and expressed rhythmically in S. elongatus. Together, these observations indicate that the cyanobacterial clock controls gene expression in a global manner by regulating the activity of all promoters.
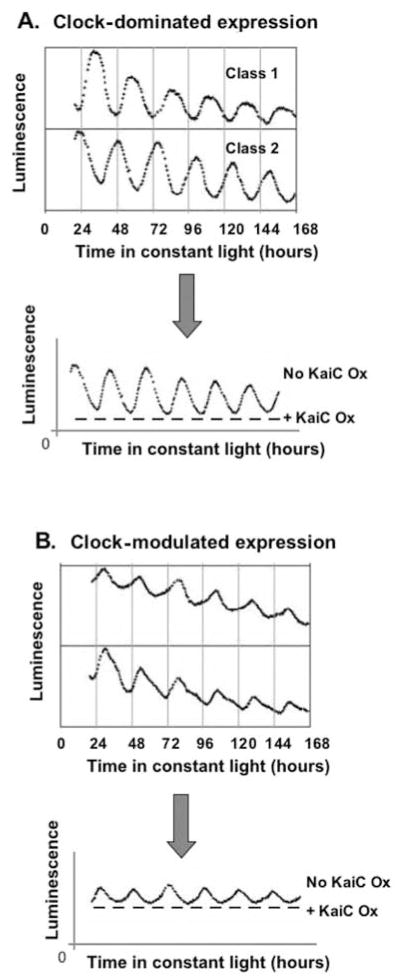
Figure 1.
Global circadian regulation of transcriptional activities in cyanobacteria. Representations of rhythmic waveforms from a promoter trap experiment (Liu et al., 1995) and the effect of KaiC overexpression on promoter activities (Nakahira et al., 2004) are shown in (A) and (B). Promoter activity was measured as luminescence from a bacterial luciferase (luxAB) reporter. (A) “Clock-dominated” genes. Waveforms representing both class 1 and class 2 promoter activities are shown. Promoters of clock-dominated genes display a high peak-to-trough amplitude with a relatively low basal level of activity. Overexpression of KaiC (lower panel, ± KaiC Ox) causes the high-amplitude waveform of these promoters to be abolished, and only low basal activity is observed (dashed line). (B) “Clock-modulated” genes. Promoters of clock-modulated genes display waveforms of luxAB expression with a low peak-to-trough amplitude but with a relatively high basal activity. Overexpression of KaiC abolishes the rhythmic expression, but a relatively high, nonrhythmic basal promoter activity remains (dashed line).
A microarray analysis of mRNA abundances in S. elongatus has recently been performed, and about 30% of the genome was found to exhibit circadian rhythms of mRNA abundance (Dr. Hideo Iwasaki, personal communication, 2006). Therefore, it appears that in S. elongatus, some of the genes whose promoter activities are rhythmic exhibit effectively non-rhythmic patterns of mRNA abundance (see below). In other cyanobacterial species, a similar pattern was found. For example, a promoter trap experiment using a promoterless luxAB reporter in Synechocystis sp. strain PCC 6803 showed that 80% of the 72 bioluminescent colonies isolated showed circadian rhythms of luxAB expression (Aoki et al., 1995), but in a genomewide screen using microarrays, the mRNA abundances of only ~9% of genes displayed circadian rhythms (Kucho et al., 2005). In the thermophilic cyanobacterium, T. elongatus BP-1, microarrays revealed that the mRNA abundance of ~6% of genes was rhythmic (Kucho et al., 2004).
One possible explanation for the discrepancies between the results obtained with microarrays and those of promoter trap assays is the different sampling intervals that can lead to differences in sensitivity to changes of gene expression (Kucho et al., 2005). For example, in the promoter trap analyses, luminescence (as a gauge of promoter activity) was monitored every 30 min to 1 h, but in the microarray experiments, the time between measurements of mRNA abundances was 4 h (in Synechocystis) and 12 h (in Thermosynechococcus). It is difficult to assess the significance of cyclic changes in genes that display low-amplitude rhythms when the sampling is infrequent (as in most microarray experiments). In addition, microarray analyses detect changes in mRNA abundance, while the promoter trap approach measures changes in transcriptional activity; therefore, genes that produce mRNAs that have either a very short or a very long half-life in the cell would show, at best, a small peak-to-trough amplitude in mRNA abundance within a high background, even though the promoter driving transcription may be under circadian control. On the other hand, turnover issues (i.e., protein half-life) can also complicate the interpretation of reporter assays as gauges of transcriptional activity. For example, if the half-life of a reporter protein such as luciferase is either (a) very short or (b) very long, the reporter can (1) underestimate the magnitude of an oscillation or (2) totally miss the oscillation, respectively (Hastings and Johnson, 2003).
If the current interpretation is correct that all promoters in cyanobacteria are rhythmically controlled but that some mRNA abundances are regulated by a different mechanism, this conclusion could have important implications for the experiments in eukaryotes in which the data are derived from measurements of mRNA abundance. Perhaps there are more pervasive rhythms in transcriptional activity in eukaryotes than originally thought, but many mRNAs are regulated posttranscriptionally such that their abundance appears to be more or less constant over the day.
POSSIBLE GENE REGULATORY PATHWAYS IN S. ELONGATUS
It is not yet known how temporal information is transmitted from the circadian clock globally to the promoters that control the expression of the ~2600 genes in S. elongatus. We consider herein 3 potential (and not mutually exclusive) mechanisms to explain this global regulation of transcription. The first mechanism we consider is that the circadian clock directly controls the basic transcriptional machinery of the cyanobacterial cell. In bacteria, promoter recognition by the transcription machinery is mediated by the binding of protein subunits, called sigma factors, to the RNA polymerase holoenzyme. Most bacteria contain several families or groups of sigma factors that modify the RNA polymerase holoenzyme, allowing recognition of different families of promoters. Thus, by temporally regulating the “pool” of available sigma factors, the circadian clock could control the promoter activity in a global manner.
In S. elongatus, an essential gene, rpoD1, encodes a sigma factor that has been characterized; a number of closely related genes encoding group 2 sigma factors (rpoD2, rpoD3, rpoD4, and sigC) that are not essential for growth have also been detected (Tanaka et al., 1992). Inactivation of genes that encode these alternative group 2 sigma factors, either singly or pairwise, results in altered circadian expression from the psbAI promoter; circadian regulation of psbAI expression was not dramatically disrupted, but modest changes in amplitude, phase angle, waveform, or period were observed (Tsinoremas et al., 1996; Nair et al., 2002). Inactivation of rpoD2 and pairwise inactivation of rpoD2rpoD3 or rpoD3rpoD4 resulted in altered period lengths of gene expression from the kaiBC promoter (Nair et al., 2002). These results indicate that these alternative group 2 sigma factors modulate the output pathway of temporal information from the circadian clock to at least a subset of cyanobacterial genes. However, the observation that rhythmic gene expression, although altered, was neither completely abolished nor drastically disrupted by inactivation of the genes for these alternate sigma factors implies that control of the pool of available sigma factors by the circadian clock does not solely account for global regulation of promoter activities in cyanobacteria.
A second mechanism that could explain how temporal information is transmitted from the KaiABC protein oscillator to globally affect circadian gene expression is by directly controlling the signal transduction pathway that in turn regulates the transcriptional machinery. A protein that could potentially play this role is the aforementioned 2-component histidine kinase, SasA (Iwasaki et al., 2000), and its cognate response regulator protein, RpaA (Takai et al., 2006). Two-component or histidyl-apartyl phosphorelay systems are the predominant mode of signal transduction in bacteria (Stock et al., 2000). Following autophosphorylation at a conserved histidine residue in response to some environmental stimulus, the kinase acts as a phospho-donor to the response regulator. When phosphorylated, the response regulator undergoes a conformational change to affect changes in cellular processes such as gene expression or locomotion. In S. elongatus, the core clock protein KaiC interacts with SasA (Iwasaki et al., 2000), and even though KaiC does not have an identifiable kinase domain, it stimulates the autophosphorylation of SasA (Smith and Williams, 2006; Takai et al., 2006). SasA, however, does not alter the phosphorylation status of KaiC, implying that information from the clock is being transmitted to SasA and not vice versa (Smith and Williams, 2006). It is possible that SasA transduces information from the KaiABC oscillator by interacting with its putative response regulator to globally control transcription of cyanobacterial genes or by controlling the expression of a subset of genes, the products of which cascade to regulate the expression of downstream genes in a circadian manner (Tomita et al., 2005; Takai et al., 2006).
Regulating sigma factor pools and/or a master signal transduction pathway implies the interaction of the transcriptional regulatory machinery with specific DNA sequences in clock gene promoters. Although both of these mechanisms could be involved in global clock control of transcription in cyanobacteria, several recent observations are difficult to explain by those mechanisms. It has been known for some time that continuous overexpression of KaiC represses transcription from the kaiBC promoter, suggesting negative feedback of KaiC on its own promoter reminiscent of eukaryotic clock systems (Ishiura et al., 1998). However, we now know that the kaiBC promoter is not the only one repressed by overexpression of KaiC; rhythms of transcription from all cyanobacterial promoters are repressed by KaiC overexpression (Nakahira et al., 2004; see Figure 1).
It was also unexpected to discover that the promoters that mediate (1) rhythmic transcription from the kai genes or (2) KaiC repression are not specifically required; heterologous E. coli promoters can be used to drive rhythmic expression of either kaiA or kaiBC (Xu et al., 2003; Nakahira et al., 2004). The transcriptional machinery of cyanobacteria recognizes the E. coli promoter, trcp, even though this promoter is obviously not one that evolved in conjunction with the cyanobacterial clockwork. Functional replacement of both the kaiA and kaiBC promoters with the inducible trcp promoter has been reported, and expression of the Kai proteins at a permissive level permits rhythmic transcription (Xu et al., 2003; Nakahira et al., 2004). Remarkably, temperature compensation, a defining characteristic of circadian clocks, was preserved when trcp replaces the kaiBC promoter (Nakahira et al., 2004). Taken together, these results indicate that the circadian clockwork in cyanobacteria does not require negative feedback of clock proteins on specific clock promoters; all that is required is the expression of appropriate levels of the Kai proteins. On one level, this result is explainable now that we know that the 3 Kai proteins together are sufficient to generate an oscillation in the phosphorylation status of KaiC in vitro (Nakajima et al., 2005); autoregulatory feedback on clock gene transcription is not required for this oscillation (at least not in constant darkness; see below). Nevertheless, the mechanism(s) that link the core KaiABC oscillator to global gene expression patterns is unknown.
The fact that the cyanobacterial clockwork does not require specific clock gene promoters, as well as the pervasiveness of KaiC regulation over rhythmic transcriptional activity, suggests a more general and broad mechanism for global clock control of promoter activities. Therefore, a third mechanism to explain pervasive circadian gene expression is the proposal that KaiC mediates both its own negative feedback regulation and global regulation of transcription throughout the genome by orchestrating oscillations in the condensation and/or supercoiling status of the entire cyanobacterial chromosome (Mori and Johnson, 2001). The chromosomes of most bacteria are arranged in a highly organized architecture based on condensation and coiling of DNA, called the nucleoid (Trun and Marko, 1998), and it is also well recognized that changes in the local supercoiling status of DNA can affect the transcriptional rate of bacterial genes (Pruss and Drlica, 1989; Straney et al., 1994; Schneider et al., 2000; Unniraman and Nagaraja, 2001). Furthermore, it has been shown recently that treatment of E. coli cells with a gyrase inhibitor causes a dramatic alteration of chromosomal supercoiling status that triggers genomewide changes in transcription (Peter et al., 2004).
THE OSCILLOID MODEL FOR CIRCADIAN CONTROL OF GENE EXPRESSION IN CYANOBACTERIA
We call this third mechanism for global promoter regulation by the circadian clockwork in cyanobacteria the oscilloid model (Mori and Johnson, 2001). This model posits that the condensation and/or supercoiling status of the cyanobacterial chromosome changes rhythmically such that it becomes an oscillating nucleoid, or “oscilloid.” These oscillations in chromosome topology promote rhythmic modulation of the transcriptional rates of all genes, accounting at least in part for global regulation of gene expression. The model proposes that the gene-specific cis-elements that mediate rhythmic gene expression might be (at least partially) responsive to chromosomal status rather than exclusively to specific trans-acting factors. Heterologous promoters from other species of bacteria that are integrated into the cyanobacterial chromosome are rhythmically active because they are also subject to the oscillating chromosomal status (Xu et al., 2003; Nakahira et al., 2004). Moreover, a KaiC-containing protein complex regulates, directly or indirectly, these changes of chromosomal status (Nakahira et al., 2004), and the phosphorylation status of KaiC is important in regulating the activity of this protein complex (Iwasaki et al., 2002; Xu et al., 2003).
A number of observations support the feasibility of this hypothesis. In the eukaryotic alga Chlamydomonas reinhardtii, there is evidence for daily rhythms of topological changes in the eubacterial-like chloroplast chromosome (Salvador et al., 1998). When integrated into the cyanobacterial chromosome, the promoter of the E. coli gene, fis, whose activity is known to be topology dependent, displays rhythmic activity suggesting that the cyanobacterial chromosome may undergo rhythmic changes in topology (Min et al., 2004). The observation that circadian clock function in cyanobacteria does not require negative feedback of clock proteins on DNA sequences in specific clock promoters and that overexpression of KaiC represses the activity of all promoters (Xu et al., 2003; Nakahira et al., 2004) is also consistent with this hypothesis. Furthermore, Smith and Williams (2006) have recently provided direct evidence to support this hypothesis. DAPI staining of chromosomes in S. elongatus cells has shown that the cyanobacterial circadian clock regulates a daily rhythm of chromosome compaction (Smith and Williams, 2006); this rhythm of chromosome compaction is kai dependent but not dependent on SasA.
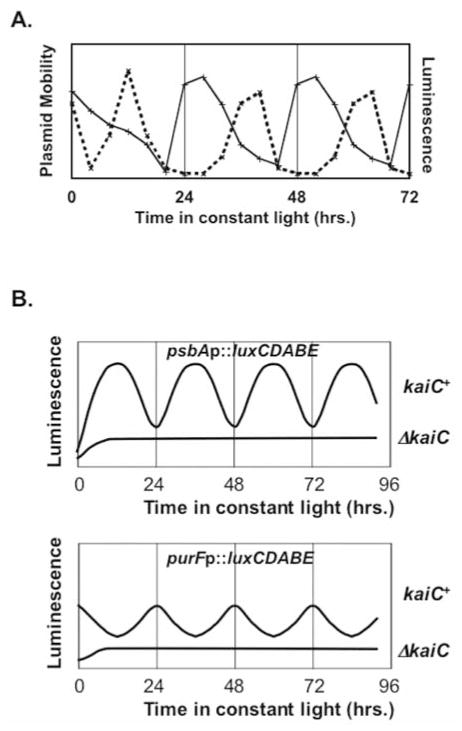
Unpublished data from our laboratory also support the “oscilloid” model of clock control of transcription. Some strains of S. elongatus harbor a nonessential endogenous plasmid called pANS. Using chloroquine agarose gel electrophoresis (a technique that separates plasmid topoisomers based on superhelicity), we have shown that the relative super-helicity of the pANS plasmid changes in a circadian manner and that these changes in superhelicity correlate with changes in the transcriptional activity of a chromosomal PsbAIp::luxAB reporter (Figure 2A; Woelfle et al., manuscript in preparation). Furthermore, plasmids containing either psbAIp (class 1) or purFp (class 2) driving luxCDABE expression in cyanobacteria display rhythms of transcription with the same period and phase as when these same reporters are incorporated into the chromosome (Figure 2B; Woelfle et al., manuscript in preparation). Moreover, rhythmic expression of luxCDABE from the plasmid is abolished in cyanobacterial cells in which kaiC has been deleted (Figure 2B). Assuming that plasmids in S. elongatus accurately reflect a phenomenon that also occurs in the chromosome, these results suggest that changes in DNA topology are regulated by the circadian clock of cyanobacteria and that these changes in DNA topology affect promoter activity. Interestingly, the kai-dependent rhythms of chromosomal compaction (Smith and Williams, 2006) are in more or less the same phase as the rhythms of plasmid supercoiling that we have observed. In general, as the cyanobacterial chromosome compacts during the course of the day, the endogenous plasmid displays increased supercoiling, and as the chromosome decompacts during the night, supercoiling in the endogenous plasmid is reduced. This observation suggests that the plasmid reporter system is an accurate reporter of chromosomal changes.
Figure 2.
Circadian rhythms of plasmid supercoiling and cyanobacterial promoter activity. (A) Plot of plasmid DNA mobility versus time in constant light. Wild-type cyanobacterial cells were given a 12-h dark pulse prior to release into constant light, and plasmid DNA from cells was isolated; luminescence from a chromosomal psbaIp::luxAB reporter was measured in the same culture at various times after cells had been released into constant light. The mobility of an endogenous plasmid (pANS) was determined by chloroquine agarose gel electrophoresis (solid curve), and luminescence from the chromosomal reporter (dashed curve) is plotted versus time in constant light (Woelfle et al., manuscript in preparation). (B) A schematic representation of cyanobacterial promoter activity when the promoters driving a luxCDABE reporter are contained on a plasmid. Luminescence versus time in constant light is shown for kaiC+ and ΔkaiC cyanobacterial strains carrying the plasmid reporters. In kaiC+ cells, the psbAI promoter (a class 1 promoter) drives rhythmic transcription of luxCDABE in the same phase as when the promoter is located in the chromosome (upper panel), but promoter activity is arhythmic from the plasmid in cells in which kaiC is deleted (ΔkaiC). The purF promoter (class 2) drives transcription of luxCDABE from a plasmid displaying a rhythm that is in antiphase to that of psbAIp in kaiC+ cells (lower panel); this rhythmic promoter activity is abolished in ΔkaiC cells (Woelfle et al., manuscript in preparation).
Nakahira and coworkers have shown that rhythms of transcription from all cyanobacterial promoters are repressed by KaiC overexpression (Nakahira et al., 2004) and that cyanobacterial promoters could be divided into 2 classes based on their transcriptional response to KaiC repression. The first type of response, exhibited by 5% to 10% of the promoters, including the kaiBC promoter, displays a high-amplitude oscillation that is eliminated by KaiC overexpression (Figure 1A). This response is reflective of promoters whose activity is “clock dominated”; the promoter shows almost no basal activity at trough phases of the circadian cycle or during KaiC overexpression (Nakahira et al., 2004; Johnson, 2004). In addition to the kaiBC promoter, other promoters that display “clock-dominated” activity are those controlling expression of sigC, chlB, rpoBC1C2, and clpC genes. The second type of response to KaiC overexpression is a lower amplitude oscillation in promoter activity in which the rhythmic component is abolished, but a significant nonrhythmic basal level of promoter activity remains (Figure 1B). This “clock-modulated” response is exhibited by 90% to 95% of promoters, including the psbA1 promoter (Nakahira et al., 2004). These results indicate that KaiC or, more likely, a KaiC-containing complex coordinates global control of transcription. For the majority of genes, the kaiABC oscillator controls a minor rhythmic component in the expression of genes that have a large constitutive basal transcriptional activity (clock modulated); in the smaller subset of clock-dominated genes, the kaiABC oscillator dictates the entire profile of transcriptional activity. The clock-dominated class of promoters might drive the transcription of genes that encode proteins intrinsically involved in the circadian clock system that, in turn, rhythmically alters the activity of the clock-modulated promoters.
As discussed earlier, the core protein of the KaiABC complex, KaiC, exists in both phosphorylated and nonphosphorylated forms in vivo, and its phosphorylation status is correlated with clock speed (Nishiwaki et al., 2000; Iwasaki et al., 2002; Xu et al., 2003; Johnson, 2004). In LL and LD cycles, a circadian oscillation of both KaiC phosphorylation and plasmid supercoiling is observed in vivo (Iwasaki et al., 2002; Xu et al., 2003; Woelfle et al., manuscript in preparation). Since KaiC appears to have a low-affinity binding to DNA (Mori et al., 2002), and based on KaiC’s similarity to helicases and recombinases (Leipe et al., 2000), it is possible that a KaiC-containing protein complex directly modulates DNA topology. However, when rhythmically growing cyanobacterial cells are released into constant darkness, the circadian oscillation of plasmid supercoiling is abolished (Woelfle, unpublished data), even though an oscillation of KaiC phosphorylation remains (Tomita et al., 2005). Since S. elongatus is an obligate photoautotroph, and transcription and translation are inhibited in constant darkness (Tomita et al., 2005), one can conclude that (1) a minimal core circadian oscillator is possible in cyanobacteria without any transcription-translation feedback loop (TTFL) properties, and (2) the KaiC phosphorylation rhythm alone is not sufficient to modulate changes in chromosome topology. Interestingly, the circadian oscillation of KaiC phosphorylation status that can be reconstituted in vitro with purified KaiA, KaiB, and KaiC proteins (Nakajima et al., 2005) appears to be a good model for the in vivo rhythm in constant darkness.
When cyanobacterial cells are in LL or LD, however, rhythms of global transcriptional activity and chromosomal topology/supercoiling are activated, and concomitantly, KaiB and KaiC are rhythmically transcribed, translated, and degraded such that there are rhythms of KaiB and KaiC protein abundance (Xu et al., 2000; Tomita et al., 2005). Therefore, in LL and LD, a TTFL loop becomes relevant. It is as though there are 2 states of the cyanobacterial clock: (1) a core KaiABC clockwork that is entirely posttranslational and keeps track of time under conditions of metabolic repression and (2) an extended clock system that involves a TTFL and regulates global transcriptional activity when metabolism is churning. The “switch” between these 2 states is conditional: Metabolic repression (DD) supports the minimal posttranslational timer, whereas metabolic activity (LL and LD) activates the global system that robustly turns over its molecular components. The switch between these states might be light dependent or energy charge dependent (e.g., adenosine triphosphate/ adenosine diphosphate [ATP:ADP] ratio). This switch is like a condition-dependent “clutch” (Figure 3). In LL/LD, the clutch is engaged, and therefore the clockwork drives global transcription; in DD, the clutch is disengaged, and therefore the KaiABC oscillator ticks away without driving the transcriptional outputs. The constant thread between these 2 states of the clockwork is presumably the rhythm of KaiC phosphorylation that is characteristic of both conditional states. However, it is curious that the KaiC phosphorylation rhythm is maintained in the LL/LD conditions since KaiC is nearly totally degraded at the end of night under these conditions (Imai et al., 2004). If KaiC is degraded, how can it keep track of its phosphorylation status? And how does the switch from metabolic repression to metabolic activity activate clock control over transcription?
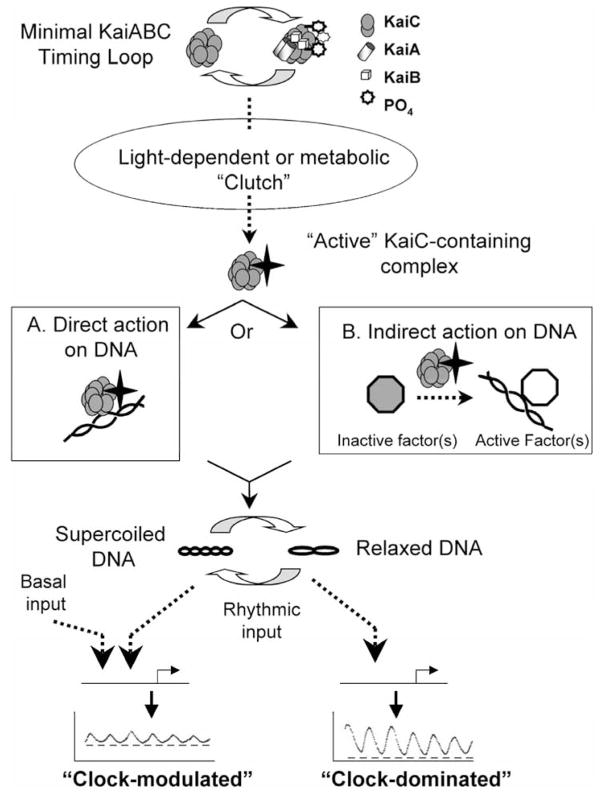
Figure 3.
The oscilloid model for the circadian system of cyanobacteria. KaiA, KaiB, and KaiC are transcribed and translated from the kaiABC gene cluster. KaiA (white cylinders) and KaiB (white cubes) interact with hexamers of KaiC (gray) to form KaiC-containing protein complexes forming the minimal KaiABC timing loop, which is capable of displaying rhythms of KaiC phosphorylation both in vivo and in vitro. The level of KaiC phosphorylation (indicated by stars on KaiC hexamers) in these complexes increases during the subjective day, reaching its peak around dusk; the level of phosphorylated KaiC decreases during the subjective night, reaching its lowest level around dawn. This minimal KaiABC-based timing loop is inactive in terms of regulating promoter activities and is subject to actions of light-dependent or metabolic “clutch,” presumably due to the interaction of an unknown protein(s). This clutch activates KaiC-containing protein complexes for either direct (inset A) or indirect (inset B) interaction with the cyanobacterial chromosome. The exact protein composition of “active” KaiC-containing protein complexes is not known. Inset A: Direct action of KaiC-containing complexes on DNA. Active KaiC-containing protein binds directly to chromosomal DNA to modulate levels of supercoiling. Inset B: Indirect action of KaiC-containing complexes on DNA. KaiC-containing complexes activate an intermediate factor or factors (e.g., second component pathways, DNA gyrases, topoisomerases, or other nucleoid-associated proteins) that in turn modulate levels of chromosomal supercoiling. Regardless of whether interaction of KaiC-containing protein complexes is direct or indirect, changes in chromosomal supercoiling affect the activity of both “clock-dominated” and “clock-modulated” promoters. Clock-dominated promoter activity is turned on and off by changes in chromosomal supercoiling. For clock-modulated promoters, the peak-to-trough amplitude, but not the basal level of activity (determined by the “basal input”), is affected by changes in chromosomal supercoiling. Gene products from the clock-dominated genes or other protein factors may influence the activity of clock-modulated genes.
The latter question leads us back to the 3 models for clock orchestration of transcription in cyanobacteria: (1) transcriptional factors such as sigma subunits, (2) 2-component signal transduction pathways such as SasA/RpaA, and (3) the oscilloid model. Again, these 3 models are not mutually exclusive; indeed, we may ultimately discover that all 3 mechanisms are involved. Figure 3 presents an updated version of the oscilloid model. There are several possibilities of how the KaiABC oscillator modulates changes in chromosomal topology. The first possibility is a direct modulation of chromosomal topology that in turn affects promoter activity (inset A of Figure 3). Transcriptional activity of “clock-dominated” promoters is totally dependent on these topology changes, possibly due to specific DNA sequences or to their chromosomal location. On the other hand, the rhythmic component (= peak-to-trough amplitude) but not the basal level of “clock-modulated” promoters is affected by changes in chromosomal topology.
An alternate possibility is that there is a multifactorial linkage between the core KaiABC oscillator and transcriptional activity that is mediated by an intermediate protein factor(s) (inset B of Figure 3). Possible candidates for the intermediate factor(s) that could link the KaiABC oscillator to changes in DNA topology are proteins such as bacterial DNA gyrases and/or topoisomerases or nucleoid proteins. In S. elongatus, there are obvious homologs of topA, gyrA, and gyrB that encode type I and type II topoisomerases, respectively, as well as genes that appear to encode nucleoid-associated proteins similar to HU and HNS. It is also possible that the intermediate factors that interact directly with the KaiABC oscillator are SasA/RpaA, which transmit rhythmic directions to proteins (e.g., gyrases and/or topoisomerases) that regulate chromosomal topology; however, the observation that SasA is not required for the rhythm of chromosomal compaction (Smith and Williams, in press) argues against this possibility (at least in dim light; Takai et al., 2006). Whether the simple alternative (inset A of Figure 3) or the multifactorial alternative (inset B of Figure 3) is correct, the bottom line is that the core clockwork may regulate the activities of the clock-dominated and clock-modulated promoters by changes in chromosome topology.
CONCLUDING REMARKS/IMPLICATIONS FOR OTHER CIRCADIAN SYSTEMS
While the oscilloid model can incorporate the wealth of new observations regarding global regulation of transcription by the cyanobacterial circadian clock, it is a difficult hypothesis to prove due to the fact that transcription itself introduces supercoils into DNA. Therefore, we have a “chicken and egg” problem: (1) is primary regulation of supercoiling globally modulating the transcriptional rate, or (2) is primary regulation of transcription causing a concomitant rhythm of supercoiling? Moreover, we cannot rule out the possibility that to some degree or another, the circadian clockwork regulates both transcription and chromosomal topology. Knocking out nonlethal sigma factors singly or in combination does not abolish the circadian patterns of transcriptional activity (Nair et al., 2002), but perhaps the relevant sigma factors are essential and cannot be deleted. It is also quite possible that global regulation of gene expression by the circadian clock incorporates a number of redundant mechanisms, as is the case for many important biological processes; inactivation of any one of these mechanisms may show only small effects on the circadian phenotype.
As mentioned earlier, there is a circadian rhythm of KaiC phosphorylation in vivo in S. elongatus (Iwasaki et al., 2002), and this rhythm is maintained when cells are transferred to constant darkness (Tomita et al., 2005), a condition in which transcription and translation are inhibited. This observation indicates that a circadian oscillator is possible in cyanobacteria without any TTFL properties. The circadian oscillation of KaiC phosphorylation status can also be reconstituted in vitro with purified KaiA, KaiB, and KaiC proteins (Nakajima et al., 2005). Thus, the in vitro KaiABC system could serve as an excellent model for the in vivo cyanobacterial clock in constant darkness. An issue of considerable interest now is how the system is changed in LL/LD to include molecular turnover (transcription, translation, and protein degradation) and whether a TTFL is necessary in these conditions.
At the present time, the circadian clock system in cyanobacteria differs in a number of ways from that of eukaryotes. However, it now appears that the cyanobacterial clock system in LL/LD is possibly a TTFL as in eukaryotes, whereas the DD system is purely post-translational. Might it be possible that the eukaryotic clock systems include an underlying timing circuit that is also totally posttranslational? If so, perhaps there is a technique by which such an underlying posttranslational clock can be “teased out.” At the moment, there is good evidence of chromosomal topological alteration that links the cyanobacterial clock to its molecular outputs, whereas we tend to assume that eukaryotic clock-controlled genes are regulated by rhythmic activity of specific transcriptional factors. However, we might find in time that regulation of chromosomal remodeling is also very important in the regulation of transcription by eukaryotic clocks; indeed, recent evidence suggests that the mammalian clockwork does just that (Etchegaray et al., 2003; Ripperger and Schibler, 2006; Doi et al., 2006). In eukaryotes, activation and repression of transcription are often associated with modifications to histones (e.g., phosphorylation, methylation, or acetylation) that condense/decondense chromatin. In mammals, the activation of several CLOCK-BMAL target genes requires histone acetylation (Etchegaray et al., 2003; Ripperger and Schibler, 2006); histone acetylation activates transcription by decondensing chromatin. Interestingly, CLOCK has recently been shown to have histone acetyltransferase activity (HAT) and that this activity is necessary to rescue circadian rhythmicity in Clock mutant cells (Doi et al., 2006). The HAT activity of CLOCK and its apparent requirement for rhythmicity implies that regulation of chromatin structure by the circadian clock is an important mechanism for transcriptional regulation. Furthermore, the similarity of mammalian CLOCK to that of Drosophila dCLOCK suggests that this control mechanism may be widespread among eukaryotic circadian clocks.
Indeed, the recent report of kai-dependent rhythms of chromosomal compaction (Smith and Williams, 2006) and our observation of rhythmic changes in supercoiling status in S. elongatus reveal similarities between the cyanobacterial mechanism of rhythmic gene expression and that of eukaryotic clock-controlled genes whose regulatory elements undergo cyclic changes in chromatin structuring. Perhaps interactions between clockworks and output pathways that maintain chromosomal structure are widespread. Thus, although the molecular details may differ, investigations of the molecular mechanisms of the cyanobacterial clock system, especially into the apparently conditional linkage between the phosphorylation rhythm of KaiC and rhythmic transcription, may lead to insights that are more broadly applicable to circadian clock systems of other organisms.
Acknowledgments
We would like to thank Drs. Takao Kondo, Susan Golden, Hideo Iwasaki, and Masahiro Ishiura (and their colleagues and students) for a productive and exciting partnership to understand cyanobacterial clocks. We particularly thank Dr. Hideo Iwasaki for sharing information regarding RpaA and microarray studies prior to publication. Finally, we are grateful for support from the National Science Foundation and the National Institutes of Health.
References
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Aoki S, Kondo T, Ishiura M. Circadian expression of the dnaK gene in the cyanobacterium Synechocystis sp. Strain PCC 6803. J Bacteriol. 1995;177:5606–5611. doi: 10.1128/jb.177.19.5606-5611.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TH, Chen TL, Huang LM, Huang TC. Circadian rhythm in amino acid uptake by Synechococcus RF-1. Plant Physiol. 1991;97:55–59. doi: 10.1104/pp.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YB, Dominic B, Zani S, Mellon MT, Zehr JP. Expression of photosynthesis genes in relation to nitrogen fixation in the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. IMS 101. Plant Mol Biol. 1999;41:89–104. doi: 10.1023/a:1006231805030. [DOI] [PubMed] [Google Scholar]
- Correa A, Lewis ZA, Greene AV, March IJ, Gomer RH, Bell-Pedersen D. Multiple oscillators regulate circadian gene expression in Neurospora. Proc Natl Acad Sci U S A. 2003;100:13597–13602. doi: 10.1073/pnas.2233734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P. Circadian Regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: Biological Timekeeping. Sunderland (MA): Sinauer; 2004. [Google Scholar]
- Etchegaray J-P, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Hastings JW, Johnson CH. Bioluminescence and chemiluminescence. Methods Enzymol. 2003;360:75–104. doi: 10.1016/s0076-6879(03)60107-2. [DOI] [PubMed] [Google Scholar]
- Huang TC, Lin RF, Chu MK, Chen HM. Organization and expression of nitrogen-fixation genes in the aerobic nitrogen-fixing unicellular cyanobacterium Synechococcus sp. Strain RF-1. Microbiology. 1999;145:743–753. doi: 10.1099/13500872-145-3-743. [DOI] [PubMed] [Google Scholar]
- Imai K, Nishiwaki T, Kondo T, Iwaski H. Circadian rhythms in the synthesis and degradation of a master clock protein KaiC in cyanobacteria. J Biol Chem. 2004;279:36534–36539. doi: 10.1074/jbc.M405861200. [DOI] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci U S A. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Taniguchi Y, Kondo T, Ishiura M. Physical interactions among circadian clock proteins, KaiA, KaiB and KaiC, in cyanobacteria. EMBO J. 1999;18:1137–1145. doi: 10.1093/emboj/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- Johnson CH. Global orchestration of gene expression by the biological clock of cyanobacteria. Genome Biol. 2004;5:217.1–217.4. doi: 10.1186/gb-2004-5-4-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama H, Kondo T, Iwasaki H. Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J Biol Chem. 2003;278:2388–2395. doi: 10.1074/jbc.M208899200. [DOI] [PubMed] [Google Scholar]
- Kao CC, Green S, Stein B, Golden SS. Diel infection of a cyanobacterium by a contractile bacteriophage. Appl Environ Microbiol. 2005;71:4276–4279. doi: 10.1128/AEM.71.8.4276-4279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama M, Tsinoremas NF, Kondo T, Golden SS. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J Bacteriol. 1999;181:3516–3524. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:1–8. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Mori T, Lebedeva NV, Aoki S, Ishiura M, Golden SS. Circadian rhythms in rapidly dividing cyanobacteria. Science. 1997;275:224–227. doi: 10.1126/science.275.5297.224. [DOI] [PubMed] [Google Scholar]
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: Luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci U S A. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Okamoto K, Tsuchiya Y, Nomura S, Nango M, Kanehisa M, Ishiura M. Global analysis of circadian expression in the cyanobacterium Synechocystis sp. Strain PCC 6803. J Bacteriol. 2005;187:2190–2199. doi: 10.1128/JB.187.6.2190-2199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Tauchiya Y, Okumoto Y, Harada M, Yamada M, Ishiura M. Construction of unmodified oligonucleotide-based microarrays in the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: Screening of the candidates for circadianly expressed genes. Genes Genet Syst. 2004;79:319–329. doi: 10.1266/ggs.79.319. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Aravind L, Grishin NV, Koonin EV. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res. 2000;10:5–16. [PubMed] [Google Scholar]
- Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- Lorne J, Scheffer J, Lee A, Painter M, Miao VPW. Genes controlling circadian rhythm are widely distributed in cyanobacteria. FEMS Microbiol Lett. 2000;189:129–133. doi: 10.1111/j.1574-6968.2000.tb09218.x. [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- Michael TP, McClung CR. Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 2003;132:629–639. doi: 10.1104/pp.021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min HY, Liu Y, Johnson CH, Golden SS. Phase determination of circadian gene expression in Synechococcus elongatus PCC 7942. J Biol Rhythms. 2004;19:103–112. doi: 10.1177/0748730403262056. [DOI] [PubMed] [Google Scholar]
- Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci U S A. 1996;93:10183–10188. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Johnson CH. Circadian programming in cyanobacteria. Semin Cell Dev Biol. 2001;12:271–278. doi: 10.1006/scdb.2001.0254. [DOI] [PubMed] [Google Scholar]
- Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, Johnson CH. Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci U S A. 2002;99:17203–17208. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Ditty JL, Min H, Golden SS. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. J Bacteriol. 2002;184:3530–3538. doi: 10.1128/JB.184.13.3530-3538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira Y, Katayama M, Miyashita H, Kutsuna S, Iwasaki H, Oyama T, Kondo T. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc Natl Acad Sci U S A. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci U S A. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian M, Duffield GE, Loros JJ, Dunlap JC. The frequency gene is required for temperature-dependent regulation of many clock-controlled genes in Neurospora crassa. Genetics. 2003;164:923–933. doi: 10.1093/genetics/164.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai K, Morishita M, Itoh S, Okamoto K, Ishiura M. Circadian rhythms in the thermophilic cyanobacterium Thermosynechococcus elongatus: Compensation of period length over a wide temperature range. J Bacteriol. 2004;186:4972–4977. doi: 10.1128/JB.186.15.4972-4977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Arsuaga J, Breier AM, Khodursky AB, Brown PO, Cozzarelli NR. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss GJ, Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989;56:521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Rossini C, Taylor W, Fagan T, Hastings JW. Lifetimes of mRNAs for clock-regulated proteins in a dinoflagellate. Chronobiol Int. 2003;20:963–976. doi: 10.1081/cbi-120025248. [DOI] [PubMed] [Google Scholar]
- Salvador ML, Klein U, Bogorad L. Endogenous fluctuations of DNA topology in the chloroplast of Chlamydomonas reinhardtii. Mol Cell Biol. 1998;18:7235–7242. doi: 10.1128/mcb.18.12.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneegurt MA, Sherman DM, Nayar S, Sherman LA. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J Bacteriol. 1994;176:1586–1597. doi: 10.1128/jb.176.6.1586-1597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Travers A, Muskhelishvilli G. The expression of the Escherichia coli fis gene is strongly dependent of the superhelical density of DNA. Mol Microbiol. 2000;38:167–175. doi: 10.1046/j.1365-2958.2000.02129.x. [DOI] [PubMed] [Google Scholar]
- Smith RA, Williams SB. Circadian rhythms of gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci U S A. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:8–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Straney R, Krah R, Menzel R. Mutations in the -10 TATAAT sequence of the gyrA promoter affect both promoter strength and sensitivity to DNA super-coiling. J Bacteriol. 1994;176:5999–6006. doi: 10.1128/jb.176.19.5999-6006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci U S A. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Masuda S, Takahasi H. Multiple rpoD-related genes of cyanobacteria. Biosci Biotechnol Biochem. 1992;56:1113–1117. doi: 10.1271/bbb.56.1113. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Yamaguchi A, Hijikata A, Iwasaki H, Kamagata K, Ishiura M, Go M, Kondo T. Two KaiA-binding domains of cyanobacterial circadian clock protein KaiC. FEBS Lett. 2001;496:86–90. doi: 10.1016/s0014-5793(01)02408-5. [DOI] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H. Circadian rhythm of KaiC phosphorylation without transcription-translation feedback. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- Trun NJ, Marko JF. Architecture of a bacterial chromosome. ASM News. 1998;64:276–283. [Google Scholar]
- Tsinoremas NF, Ishiura M, Kondo T, Andersson K, Tanaka K, Takahasi H, Johnson CH, Golden SS. A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO J. 1996;19:2488–2495. [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- Unniraman S, Nagaraja V. Axial distortion as a sensor of supercoil changes: A molecular model for the homeostatic regulation of DNA gyrase. J Genet. 2001;80:119–124. doi: 10.1007/BF02717907. [DOI] [PubMed] [Google Scholar]
- Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc Natl Acad Sci U S A. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Johnson CH. Circadian clock-protein expression in cyanobacteria: Rhythms and phase-setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: Roles of KaiA, KaiB, and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22:2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Piston D, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: Application to interacting circadian clock proteins. Proc Natl Acad Sci U S A. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen UC, Huang TC, Yen TC. Observation of the circadian photosynthetic rhythm in cyanobacteria with a dissolved-oxygen meter. Plant Sci. 2004;166:949–952. [Google Scholar]