Background: The hallmark of Parkinson disease is α-synuclein aggregation, whose cytotoxic effects are reproducible in yeast.
Results: α-Synuclein aggregate clearance is impaired by vacuolar protease inhibitor PMSF but not by proteasome inhibitor MG132.
Conclusion: Yeast recovers from α-synuclein-induced toxicity by clearing aggregates via autophagy and vacuolar pathways.
Significance: The data provide insight into the mechanisms used by yeast cells to clear toxic aggregated proteins.
Keywords: α-Synuclein, Autophagy, Parkinson Disease, Proteasome, Yeast, Aggregate Clearance, Vacuole
Abstract
Parkinson disease is the second most common neurodegenerative disease. The molecular hallmark is the accumulation of proteinaceous inclusions termed Lewy bodies containing misfolded and aggregated α-synuclein. The molecular mechanism of clearance of α-synuclein aggregates was addressed using the bakers' yeast Saccharomyces cerevisiae as the model. Overexpression of wild type α-synuclein or the genetic variant A53T integrated into one genomic locus resulted in a gene copy-dependent manner in cytoplasmic proteinaceous inclusions reminiscent of the pathogenesis of the disease. In contrast, overexpression of the genetic variant A30P resulted only in transient aggregation, whereas the designer mutant A30P/A36P/A76P neither caused aggregation nor impaired yeast growth. The α-synuclein accumulation can be cleared after promoter shut-off by a combination of autophagy and vacuolar protein degradation. Whereas the proteasomal inhibitor MG-132 did not significantly inhibit aggregate clearance, treatment with phenylmethylsulfonyl fluoride, an inhibitor of vacuolar proteases, resulted in significant reduction in clearance. Consistently, a cim3-1 yeast mutant restricted in the 19 S proteasome regulatory subunit was unaffected in clearance, whereas an Δatg1 yeast mutant deficient in autophagy showed a delayed aggregate clearance response. A cim3-1Δatg1 double mutant was still able to clear aggregates, suggesting additional cellular mechanisms for α-synuclein clearance. Our data provide insight into the mechanisms yeast cells use for clearing different species of α-synuclein and demonstrate a higher contribution of the autophagy/vacuole than the proteasome system. This contributes to the understanding of how cells can cope with toxic and/or aggregated proteins and may ultimately enable the development of novel strategies for therapeutic intervention.
Introduction
Parkinson disease (PD),2 the second most common neurodegenerative disease after Alzheimer disease, is characterized by the progressive death of the dopaminergic neurons in the substantia nigra pars compacta of the brain. This associates with a gradual development of symptoms including bradykinesia, muscular rigidity, postural instability, and resting tremor (1). The molecular hallmark of the disease is the accumulation of proteinaceous inclusions termed Lewy bodies, which contain proteins such as α-synuclein, ubiquitin, synphilin-1, and cytoskeletal proteins (2). The major constituent of Lewy bodies is misfolded and aggregated α-synuclein (3), a small neuronal protein of 140 amino acids (4) encoded by the SNCA gene. Allelic duplication or triplication of the wild-type SNCA gene encoding α-synuclein was found to be linked to familial forms of PD and corroborates that α-synuclein is important for the disease (5, 6). Additionally, two missense mutations, A30P and A53T, are associated with autosomal dominant early-onset forms of PD (7, 8). Besides PD, α-synuclein inclusions have also been reported in other neurodegenerative diseases collectively referred to as α-synucleinopathies.
It is assumed that the aggregation pathway of α-synuclein in neurons starts with the formation of soluble unstable oligomeric species and is promoted by the initial binding to lipid membranes. The generation of Lewy bodies is a consequence of oligomerization/fibrillation followed by attachment of ubiquitin (9–12). Several studies have focused on the aggregation behavior and toxicity of wild-type and mutant forms of α-synuclein, including those specifically designed according to structural predictions. The designer mutant A30P/A53T/A76P (TP) α-synuclein shows enhanced oligomer and impaired amyloid fibril formation in vitro and does not form insoluble aggregates in animal models (13, 14) despite increased neurotoxicity in primary neurons, worms, and flies (13, 15). This further supported the idea that soluble prefibrillar α-synuclein oligomers and not the insoluble aggregates are associated with the detrimental effects found in PD (13, 16).
Yeasts, flies, worms, and mice have been used as model systems to understand the molecular basis of α-synuclein-mediated toxicity (17–21). In particular, the budding yeast Saccharomyces cerevisiae is a powerful model organism for PD due to the high conservation with higher eukaryotes, its rapid growth, and the existence of comprehensive genetic tools. Although yeast does not endogenously express α-synuclein homologues, α-synuclein-related effects can be efficiently mimicked in yeast, such as proteasome impairment, increased reactive oxygen species, lipid droplet accumulation, and vesicle trafficking dysfunction (20, 22). At the same time, the type of α-synuclein construct used in yeast as well as quantification of α-synuclein in cells is critical.
α-Synuclein-related neurotoxicity is generally attributed to a gain of toxic function of misfolded and aggregated α-synuclein. Therefore, a central question pertains to the mechanism of clearance of α-synuclein aggregates. Several studies support that soluble α-synuclein can be a target for the 26 S proteasome (23, 24). In contrast, α-synuclein oligomeric forms cannot be subject to proteasomal degradation (25) but rather inhibit the system (26). α-Synuclein was also reported to be degraded by autophagy (27, 28) and might inhibit macroautophagy (29). Given that α-synuclein complexes can impair the proteasome and that both PD patients and animal models display an elevated number of autophagic vesicles (30), autophagy, in particular macroautophagy, might provide a means for the cell to cope with aggregates.
Here, we studied the ability of cells to recover from α-synuclein exposure and to clear aggregates by a comparison of wild-type, A30P, A53T, and TP α-synuclein in the yeast model. We compared the contribution of the proteasome and of autophagy/vacuolar pathways in α-synuclein aggregate clearance and, additionally, show that α-synuclein interferes with the activation of autophagy. Our data will now enable further studies in higher model systems and may open novel possibilities for the design of therapeutic strategies.
EXPERIMENTAL PROCEDURES
Plasmids, Yeast Strains, and Growth Conditions
Plasmids and S. cerevisiae strains are listed in Tables 1 and 2. Wild-type α-synuclein encoding cDNA sequence (hereafter referred to as SNCA) and the corresponding mutant A30P and A53T sequences were cloned into pRS426 yeast high expression vector or into the integrative pRS306 vector (31) preceded by the GAL1 promoter and followed by the CYC1 terminator. A p416 vector containing GFP-Atg8 under the Copper promoter CUP1 was used for autophagy assays (32). Plasmids are listed in Table 1. For microscopic studies, all α-synuclein forms were tagged with GFP via linker, myeGFP with or without linker, mCherry with linker, and eGFP with linker. The human α-synuclein-encoding cDNA fused via the KLID linker to GFP was amplified by polymerase chain reaction on genomic DNA from the yeast strain HiTox (20). Wild-type α-synuclein was tagged either at the amino or the C terminus. N-terminal fusions were connected by SAAAG, and C-terminal fusions were connected by either AAAG or KLID as linkers. The GAL1-SNCA-GFP with the corresponding linkers was integrated into the triple-mutated ura3-52 locus of S. cerevisiae W303-1A using an intact URA3 gene on the corresponding integrative plasmid for selection. All constructs were verified by DNA sequencing. Yeast strains listed in Table 2 were grown in non-selective medium (YPD) at 30 °C and transformed as described (33). Selective synthetic complete (SC) media (34) contained 2% raffinose or 2% galactose and lacked the nutrient corresponding to the marker. α-Synuclein expression was induced by shifting yeast cultivated overnight in raffinose to galactose medium (A600 = 0.1).
TABLE 1.
Yeast plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pME2795 | pRS426-GAL1-Promoter, CYC1-Terminator, URA3, 2 μm, pUC origin, AmpR | This study |
| pME3526 | pME2795 with GAL1::eGFP::SNCAWT (SAAAG linker) | This study |
| pME3527 | pME2795 with GAL1::eGFP::SNCAA30P (SAAAG linker) | This study |
| pME3528 | pME2795 with GAL1::eGFP::SNCAA53T (SAAAG linker) | This study |
| pME3759 | pME2795 with GFP | This study |
| pME3760 | pME2795 with GAL1::SNCAWT | This study |
| pME3761 | pME2795 with GAL1::SNCAA30P | This study |
| pME3762 | pME2795 with GAL1::SNCAA53T | This study |
| pME3941 | pME2795 with GAL1::SNCAA30P/A56P/A76P | This study |
| pME3763 | pME2795 with GAL1::SNCAWT::GFP (KLID linker) | This study |
| pME3764 | pME2795 with GAL1::SNCAA30P::GFP (KLID linker) | This study |
| pME3765 | pME2795 with GAL1::SNCAA53T::GFP (KLID linker) | This study |
| pME3942 | pME2795 with GAL1::SNCAA30P/A56P/A76P::GFP (KLID linker) | This study |
| pME3766 | pME2795 with GAL1::SNCAWT::myeGFP (AAAG linker) | This study |
| pME3943 | pME2795 with GAL1::SNCAWT::GFP (AAAG linker) | This study |
| pME3769 | pME2795 with GAL1::SNCAWT::myeGFP (no linker) | This study |
| pME3770 | pME2795 with GAL1::SNCAA30P::myeGFP (no linker) | This study |
| pME3771 | pME2795 with GAL1::SNCAA53T::myeGFP (no linker) | This study |
| pME3772 | pME2795 with GAL1::SNCAWT::mCherry (KLID linker) | This study |
| pME3773 | pME2795 with GAL1::SNCAWT::myeGFP (KLID linker) | This study |
| pME3774 | pRS306-GAL1-Promoter, CYC1-Terminator, URA3, integrative, pUC origin, AmpR | This study |
| pME3945 | pME3774 with SNCAWT::GFP (KLID linker) | This study |
| pME3946 | pME3774 with SNCAA30P::GFP (KLID linker) | This study |
| pME3947 | pME3774 with SNCAA53T::GFP (KLID linker) | This study |
| pME3948 | pCu416ATG8-GFP, Cu promoter | Ref. 32 |
TABLE 2.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MAT a; ura3-52; trp1D2; leu2-3_112; his3-11; ade2-1; can1-100 | EUROSCARF |
| Δpep4 | BY4741; Mat a; his3D1; leu2D0; met15D0; ura3D0; YPL154c::kanMX4 | EUROSCARF |
| Δerg6 | BY4741; Mat a; his3D1; leu2D0; met15D0; ura3D0; YML008c::kanMX4 | EUROSCARF |
| RH3465 | W303 containing GAL1::GFP in URA3 locus | This study |
| RH3466 | W303 containing 1 genomic copy GAL1::SNCAWT::GFP in URA3 locus (KLID linker) | This study |
| RH3467 | W303 containing 2 genomic copies GAL1::SNCAWT::GFP in URA3 locus (KLID linker) | This study |
| RH3468 | W303 containing 3 genomic copies GAL1::SNCAWT::GFP in URA3 locus (KLID linker) | This study |
| RH3469 | W303 containing 1 genomic copy GAL1::SNCAA30P::GFP in URA3 locus (KLID linker) | This study |
| RH3470 | W303 containing 2 genomic copies GAL1::SNCAA30P::GFP in URA3 locus (KLID linker) | This study |
| RH3471 | W303 containing 3 genomic copies GAL1::SNCAA30P::GFP in URA3 locus (KLID linker) | This study |
| RH3472 | W303 containing 1 genomic copy GAL1::SNCAA53T::GFP in URA3 locus (KLID linker) | This study |
| RH3473 | W303 containing 2 genomic copies GAL1::SNCAA53T::GFP in URA3 locus (KLID linker) | This study |
| RH3474 | W303 containing 3 genomic copies GAL1::SNCAA53T::GFP in URA3 locus (KLID linker) | This study |
| RH3475 | W303 with YGL180w::kanMX4 (Δatg1), GAL1::SNCAWT::GFP in HIS and TRP loci | This study |
| RH3477 | W303 with YGL180w::kanMX4, cim3–1, temperature sensitive (cim3–1Δatg1 double mutant), GAL1::SNCAWT::GFP in HIS and TRP loci | This study |
| RH3486 | W303 with cim3–1, temperature sensitive, GAL1::SNCAWT::GFP in HIS and TRP loci | This study |
Spotting Tests
To assess growth on solid media, cultures were grown to mid-log phase in minimal medium containing raffinose and lacking uracil or lacking both uracil and tryptophan. Cells were normalized to equal densities, serially diluted 10-fold starting with an A600 of 0.1, and spotted on SC−ura or on SC−ura-trp plates containing either 2% glucose or 2% galactose. After 2 days incubation at 30 °C the plates were photographed.
Immunoblotting
Overnight cultures of yeast strains harboring GAL1-SNCA-GFP integrations were grown in synthetic complete medium containing 2% raffinose and lacking uracil. For induction of the GAL1 promoter, cells were inoculated in a fresh 50-ml culture of SC−ura+galactose to an A600 = 0.1 and incubated for 6 h. Cell extracts were prepared, and the protein concentrations were determined with a Bradford assay. 10 μg of protein from each strain were run on a 12% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with a mouse anti α-synuclein monoclonal antibody (1:3000, AnaSpec) or with a rabbit anti eIF2α monoclonal antibody (1:1000, Invitrogen) for loading control. For the autophagy assays a mouse anti GFP monoclonal antibody (1:500, Santa Cruz Biotechnology) was used. Peroxidase-coupled goat anti-mouse or goat anti-rabbit immunoglobulins G was used as secondary antibody (Invitrogen, diluted 1:5000).
Southern Hybridization and Copy Number Determination
Several transformants were analyzed by Southern hybridization (35) for verification of the integration of α-synuclein into the mutated genomic ura3-52 locus. Isolation of genomic DNA from S. cerevisiae was performed according to standard procedures (36). 10 μg of genomic DNA were subjected to restriction digestion with HindIII. The restriction fragments were resolved on a 1% agarose gel, transferred to a nitrocellulose membrane, cross-linked by UV irradiation for 5 min and hybridized to a URA3 gene fragment probe. Copy numbers of the integrated vectors were estimated with the ImageJ software (37). One copy corresponded to 2.7 + 4.6 kb, two copies to 2.7 + 4.6 + 6.2 kb, and three copies to 2.7 + 4.6 + 6.2kb (higher intensity).
Fluorescence Microscopy, FM4-64 Stainings, and Quantifications
Yeast cell cultures were grown in synthetic complete medium with raffinose lacking uracil until mid-log phase and transferred to SC medium supplemented with galactose and lacking uracil. Cells were visualized using a Zeiss Axiovert S100 microscope with a GFP filter and a 100× magnification at different time points after induction. Stainings with the lipophilic dye FM4-64 were performed to observe yeast vacuoles as previously described (38). For quantification of aggregation at least 300 cells were counted per strain and per experiment. For each strain, the number of cells displaying cytoplasmic foci was reported to the total number of cells counted and displayed as percentage on a column chart. Cells exhibiting only bright peripheral halos (plasma membrane localization) or additional perivacuolar fluorescence and cells displaying cytoplasmic distribution were not counted as aggregation-displaying cells.
Immunofluorescence
Overnight cultures of W303 cells containing high copy plasmids with wild-type, A30P, or A53T α-synuclein were grown in synthetic complete medium containing 2% raffinose and lacking uracil. For induction of the GAL1 promoter, cells were inoculated into fresh SC medium containing 2% galactose and lacking uracil to an A600 = 0.1. After 6 h of incubation, the cells were collected, washed, and fixed with 37% formaldehyde for 30 min. Cell walls were digested with 20 mg/ml zymolyase, and the cells were permeabilized in 0.5% Triton X-100 phosphate buffer. After preblocking with 5% fetal bovine serum for 1 h, cells were incubated with a mouse anti-α-synuclein antibody (1:3000, AnaSpec). An anti-mouse Alexa Fluor 594(red)-conjugated secondary antibody (1:200, Invitrogen) was used. Cells were observed using an inverted Zeiss Axiovert S100 microscope and a dsRed filter.
Promoter Shut-off Studies and Drug Treatments
W303, BY4741, Δatg1, cim3-1, cim3-1Δatg1, and Δpep4 cells were pre-grown in SC medium containing raffinose and lacking uracil overnight then shifted to SC medium containing galactose and lacking uracil for 4 h to induce α-synuclein expression. After preincubation in galactose medium, cells were shifted to SC medium supplemented with glucose and lacking uracil to shut off the promoter, then observed with a fluorescent microscope at several time points. For experiments in which temperature-sensitive cim3-1 and cim3-1Δatg1 mutant strains were employed, the cells were grown at 25 °C. Induction was performed for 3 h at 25 °C and 1 hour additional at 37 °C. Incubation in glucose was done at 37 °C. For consistency, the parent strain W303 and the Δatg1 strain were grown under the same conditions. α-Synuclein was overexpressed from a 2-μm plasmid to achieve higher aggregation levels. The protein was also overexpressed in Δpep4 and in the BY4741 genetic background, which corresponds to this mutant. The -fold decrease in percentage of cells displaying cytoplasmic inclusions was recorded and plotted on a graph. Three drug treatments were separately applied to W303 cells concomitantly with the shift to glucose. Carbobenzoxyl-leucinyl-leucinyl-leucinal (MG132) dissolved in dimethyl sulfoxide (DMSO) was applied to the cell suspension in a concentration of 50 μm. In parallel, for control, 0.1% DMSO was added to W303 cells. Phenylmethanesulfonyl fluoride (PMSF) was dissolved in ethanol (EtOH) and added to the cell suspension to a final concentration of 1 mm. For control, 1% ethanol was added to the cells (39). Rapamycin dissolved in ethanol was added to cells incubated in galactose for 3 h to a final concentration of 100 nm. After 1 h of rapamycin exposure, cells were washed and shifted to glucose. An equal volume of ethanol was applied to cells as control.
Autophagy Monitoring Assays
W303 cells were co-transformed with pRS426 vectors containing different α-synuclein variants under the galactose-inducible GAL1 promoter and with a pGFP-Atg8(416) vector containing GFP-Atg8 under the copper promoter CUP1. Cells grown overnight at 30 °C in SC medium containing raffinose and lacking uracil and tryptophan were induced for 2 h with 50 μm CuSO4 to express GFP-Atg8. The cells were washed twice, diluted to an A600 = 0.5, and incubated in 50 ml of SC medium containing galactose and lacking uracil and tryptophan for α-synuclein induction. In a first experiment cells were induced for 8 h, and probes were taken at 2, 4, and 8 h. In a second experiment, cells were induced for 4 h in galactose, washed twice, and transferred to nitrogen starvation medium, SD(−N) (40) supplemented with galactose for another 4 h. Probes were taken at 2 and 4 h of starvation. From 10-ml cultures, collected cell extracts were prepared and subjected to Bradford analysis and immunoblotting as described.
Statistical Analysis
Statistical analysis for autophagy assays was carried out using one-way ANOVA test. A p value less than 0.05 was considered to indicate a significant difference.
RESULTS
Three Copies Wild-type (WT) and Two Copies A53T Are Thresholds for Cytotoxicity and Aggregation
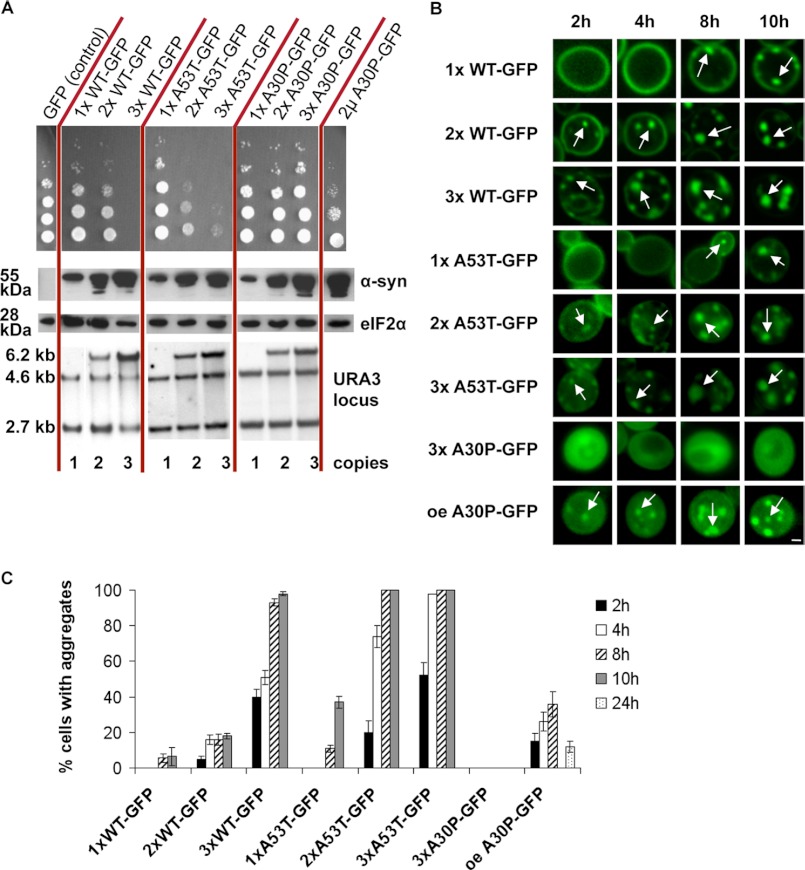
We aimed to monitor α-synuclein aggregate clearance with dependence on different cellular protein degradation pathways in yeast. First, we compared the growth impact of differently tagged α-synuclein variants with that of untagged α-synuclein (supplemental Fig. S1). Spotting assays revealed that expression of N-terminal α-synuclein GFP fusions or C-terminal fusions without a linker did not affect yeast growth in contrast to the corresponding untagged versions (supplemental Fig. S1, D and E). Overexpression of untagged A30P or A30P α-synuclein-GFP only partially affected cell growth (supplemental Fig. S1, A and B). Tagged or untagged triple proline TP designer α-synuclein behaved similarly to A30P. In several mammalian models the TP variant is unable to form aggregates and has a toxic effect (13). Tagged or untagged wild-type and A53T α-synucleins were equally toxic to the untagged counterparts only when GFP was fused C-terminally to α-synuclein by a linker. Several different fluorescent tags resulted in a similar intracellular morphology when provided in this C-terminal combination connected with a linker (supplemental Fig. S2). Previous studies reported that the α-synuclein-encoding cDNA integrated into one genomic locus had no effect on growth or aggregation. Upon a second integration, α-synuclein was toxic and aggregated (20). We constructed strains with single or multiple integrations at the single URA3 locus for a quantitative assay. We aimed to avoid genetic interference effects that might be caused by second-locus integrations. The number of integrated copies of α-synuclein was determined by Southern blot analysis (Fig. 1A, lower panel). Cells expressing one or two copies of wild-type α-synuclein at the same locus did not display growth inhibition (Fig. 1A, upper panel). However, expression of three copies impaired growth and, therefore, represents a threshold for multiple integrations of GAL1 promoter-driven α-synuclein at one locus. Cells expressing one copy of A53T α-synuclein showed no growth defect, in contrast to cells expressing two copies, which displayed a significant growth inhibition that was further enhanced when three copies were expressed. Therefore, the threshold of A53T is lower than three copies and lower than that of wild-type α-synuclein. Immunoblotting analysis was used to correlate the gene copy number with protein expression, further supporting these threshold values (Fig. 1A, middle panel).
FIGURE 1.
Threshold copy number for growth impairment by GAL1-driven WT and A53T α-synucleins integrated at the single yeast URA3 locus. A, shown is growth of yeast strains carrying increasing numbers of copies of GAL1-driven α-synuclein-KLID-GFP fusion alleles integrated into the triple-mutated ura3-52 locus. For comparison, yeasts expressing GAL1-driven A30P α-synuclein-KLID-GFP from a 2-μm plasmid are shown. Spotting analysis (top) reflects growth impairment with increasing copy numbers for WT or A53T but not A30P α-synuclein. Protein levels (middle) are visualized after induction (6 h) by Western analysis using anti-α-synuclein and as loading control anti-eiF2α antibodies. In addition to the integration strains, a spotting analysis of W303 cells expressing A30P α-synuclein from high copy plasmid and corresponding protein levels is shown. α-Synuclein gene copies (below) were determined by Southern hybridization using labeled URA3 as probe. Integrated α-synuclein genes correspond to 2.7 + 4.6 kb (1×), 2.7 + 4.6 + 6.2 kb (2×), 2.7 + 4.6 + 6.2 kb higher intensity (3×) as indicated. Copy numbers were determined with the ImageJ software. B, shown is copy number-dependent α-synuclein aggregate formation visualized by live-cell fluorescence microscopy at indicated time points. White arrows point at intracellular inclusions. Scale bar = 1 μm. oe, overexpression. C, aggregate quantification of yeast cells with indicated α-synuclein copies is shown.
Copy numbers of α-synuclein in yeast also correlated with aggregation kinetics as monitored in living cells (Fig. 1 B, and C). Non-toxic wild-type α-synuclein expressed from a single copy remained at the plasma membrane even 8 h after induction, when only a small percentage of cells started to form aggregates. Two gene copies of α-synuclein resulted in low aggregation. In contrast, three gene copies resulted in immediate wild-type α-synuclein aggregation after induction. Quantification of aggregation formation showed that three copies resulted in increased aggregation kinetics in comparison to two copies (Fig. 1C). One copy of A53T α-synuclein did not show aggregates until 8 h of induction, similarly to the wild-type version. However, A53T aggregate formation derived from two copies was significantly faster than two wild-type copies but slower than three A53T copies. These results illustrate how multiple integrations of various copies of GAL1-SNCA-linker-GFP fusions into a single locus correlate with impaired cell growth and the intracellular accumulation of fluorescent foci.
A30P α-Synuclein Can Transiently Form Aggregates in Yeast
Three integrated copies of the A30P α-synuclein variant at the URA3 locus did not impair yeast growth (Fig. 1A). Aggregate formation had not been described for this variant in yeast (20, 41) and could also not be observed for three integrated copies (Fig. 1B). However, under our conditions, the A30P fusion expressed from a high copy plasmid resulted in a reduced yeast growth rate (Fig. 1A). Therefore, we compared the intracellular distribution of α-synuclein in cells expressing high copies of C-terminal-tagged A30P α-synuclein with toxic or non-toxic variants in more detail.
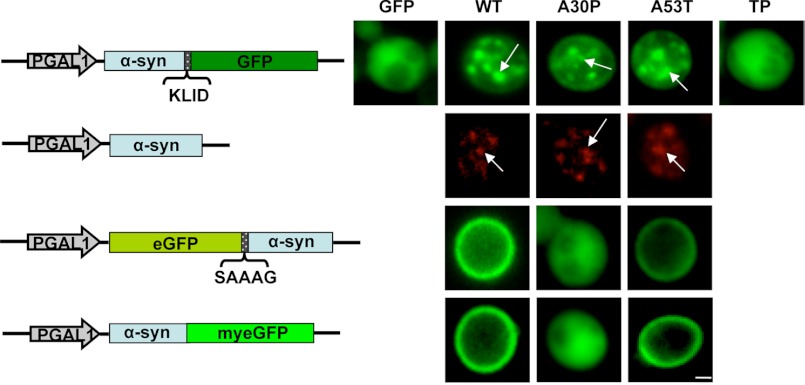
Overexpression of C-terminal-tagged A30P α-synuclein resulted in the formation of fluorescent foci, which are similar to the foci formed by the other variants (Fig. 1B and Fig. 2). Only the non-toxic TP mutant α-synuclein retained a homogeneous distribution throughout the cytoplasm and did not form aggregates. Immunofluorescence analysis with untagged α-synuclein revealed an identical distribution of the protein when compared with the C-terminal GFP-tagged α-synuclein. Non-toxic N-terminal-linked or non-toxic C-terminal A30P-GFP without linker displayed a similar cytoplasmic distribution as the non-toxic TP. In contrast, non-toxic GFP fusions of wild-type and A53T α-synuclein, which did not display aggregates, localized at the plasma membrane (Fig. 2).
FIGURE 2.
A30P α-synuclein aggregate formation upon high copy expression in yeast. Live-cell fluorescence microscopy of yeast cells expressing α-synuclein-KLID-GFP from high copy plasmids was compared with immunofluorescence of untagged α-synucleins. Yeast cells pre-grown to mid-log phase were induced in galactose-containing medium and examined for aggregates at 8 h of induction. GFP-expressing cells were used as control. Aggregate formation of untagged WT, A30P, and A53T α-synuclein was visualized by immunofluorescence. N-terminal eGFP-tagged α-synuclein via SAAAG linker and α-synuclein C-terminally fused to myeGFP was employed for further comparison. White arrows point at intracellular inclusions. Scale bar = 1 μm.
A30P requires much higher levels of protein to form fluorescent foci. Quantification of the A30P-KLID-GFP α-synuclein foci formation revealed a significantly reduced percentage of cells displaying aggregates in comparison to other variants (Fig. 1A, right panel, and C). The aggregation percentage was even further reduced from 35% after 8 h to 12% after 24 h of induction (Fig. 1C). This suggests that the aggregation of A30P α-synuclein is only transient and that the cells can then cope with the presence of this variant by activating protein clearance mechanisms that counteract aggregation.
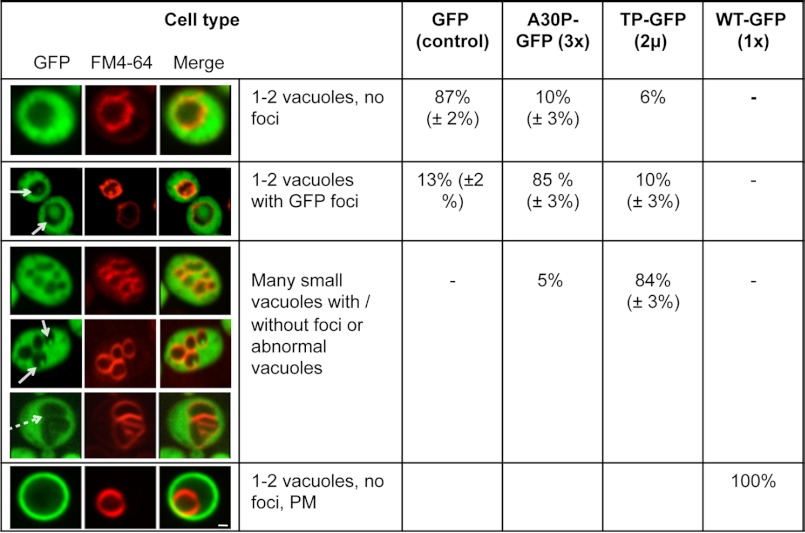
We further followed the localization of non-aggregated α-synuclein. We chose A30P-GFP (three copies integrated in the genome) and TP-GFP (expressed from a 2-μm vector). As additional controls we used GFP and WT α-synuclein-GFP expressed from a single copy. Cells expressing A30P-GFP and TP-GFP revealed diffuse cytoplasmic localization of the GFP signal, whereas the strain expressing WT-GFP α-synuclein revealed only plasma membrane localization. Next, we used the lipophilic membrane dye FM4-64 to determine whether α-synuclein could be targeted to the vacuole. A30P-GFP and TP-GFP formed fluorescent foci inside the vacuole. The foci were surrounded by a lipid membrane and moved inside the vacuole (supplemental Videos 1 and 2). These structures might result from budding of vesicles that are released into the vacuolar lumen and move freely until they are finally degraded. The GFP shift inside the vacuole was observed already after 15 min of α-synuclein induction. The effect was most pronounced after 24 h, when 85% of the A30P-GFP-expressing cells showed fluorescent foci of different sizes inside the vacuoles (Fig. 3). Fluorescent foci were observed also in 13% of the control cells (GFP alone).
FIGURE 3.
Vacuole morphology and GFP localization in yeast cells expressing non-aggregated α-synuclein-GFP variants. Live-cell fluorescence microscopy of yeast cells (W303) expressing GFP (control), A30P-GFP (3× integrated in the genome), TP-GFP (2-μm plasmid), and WT-GFP (1× integrated in the genome). GAL1-driven α-synuclein-GFP protein expression was induced in galactose-containing medium for 24 h. Yeast vacuoles were stained with FM4-64 and examined for morphology. White arrows point at GFP-foci inside the vacuole. The dashed arrow indicates tubular invaginations of the yeast vacuole. The quantification of different cell types represents an average of three independent experiments. PM, plasma membrane localization of GFP signal. Scale bar = 1 μm.
The TP-GFP mutant showed a special vacuole phenotype. Beside the strong shift into the vacuole, 85% of the population showed increased number of vacuoles (typically between 5 and 8 vacuoles/cell) or abnormal vacuoles with vacuolar invaginations. These results show that the accumulation of A30P-GFP and TP-GFP induces a strong shift of the fusion proteins into the vacuole and suggest the involvement of autophagy/vacuolar pathways in α-synuclein degradation.
Yeast Cells Recover from Transient α-Synuclein Exposure by Clearing Protein Aggregates
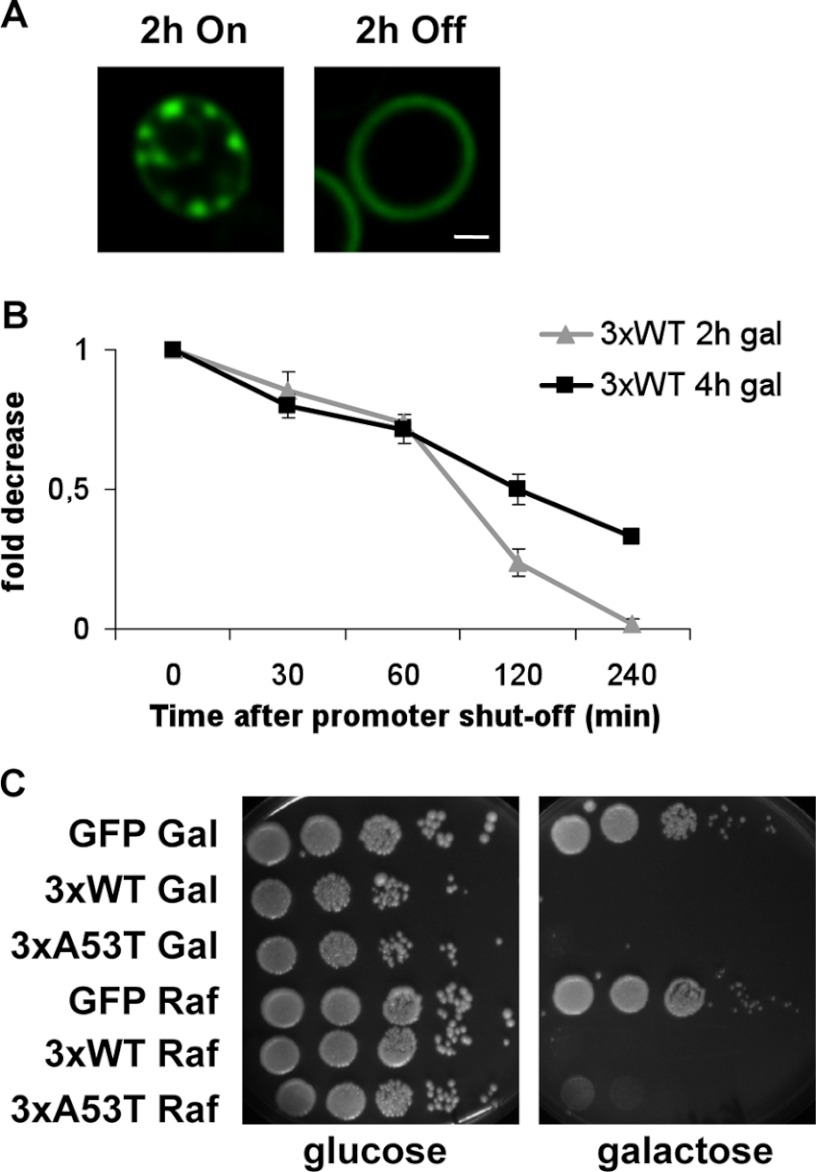
To investigate the clearance of wild-type α-synuclein, we performed GAL1 promoter shut-off experiments and determined the effect on cytoplasmic fluorescent foci. Promoter shut-off was achieved by transferring cells from galactose to glucose-containing medium, which represses the promoter (42). After 2 h of expression by preincubation in galactose, cells harboring three genomically integrated wild-type α-synuclein-GFP fusions displayed aggregates (Figs. 1B and 4A). Promoter shut-off for only 2 h resulted already in a distinct reduction of aggregates in these cells (Fig. 4, A and B). An increased preincubation time resulted in slower clearance kinetics, as it resulted from quantifying the percentage of cells displaying aggregates at several time points after transfer to glucose (Fig. 4B). Our data suggest that cells not only have an efficient clearing mechanism for A30P, which results in transient aggregation, but also that cells can efficiently clear wild-type α-synuclein foci in a dose-dependent manner.
FIGURE 4.
Clearing of α-synuclein aggregates and subsequent recovery of yeast cells from transient α-synuclein exposure. A, live-cell fluorescence microscopy of W303 yeast cells expressing WT α-synuclein-GFP three times-integrated in the genome (3×). Pictures were taken after 2 h of galactose preincubation (left) and 2 h after incubation in glucose (right). Scale bar = 1 μm. B, quantification of cells displaying wild-type α-synuclein-GFP aggregation after promoter shut-off at indicated time points is shown. Preincubation in galactose was done for 2–4 h. C, spotting analysis is shown. Yeast cells were induced in galactose-containing medium to express GFP-tagged WT and A53T α-synuclein from three genomically integrated copies for 8 h. GFP-expressing cells were used as control. The cells were then spotted on glucose and galactose. As the negative control, the same strains were grown in raffinose-containing medium.
Recovery of yeast cells carrying three copies of wild-type or A53T α-synuclein was examined to determine whether the detrimental effects of α-synuclein on growth could be reversed. The GAL1 promoter-driven α-synuclein variants were expressed for 8 h and subsequently spotted onto solid medium containing either glucose, to shut-off the promoter, or onto galactose, which further allows GAL1-driven α-synuclein expression. Cells plated on galactose were inviable, whereas cells plated on glucose regained the same ability to form colonies as controls, indicative of recovery from the transient α-synuclein exposure (Fig. 4C).
α-Synuclein Aggregate Clearance Requires Lysosomal/Vacuolar and Autophagy Pathways
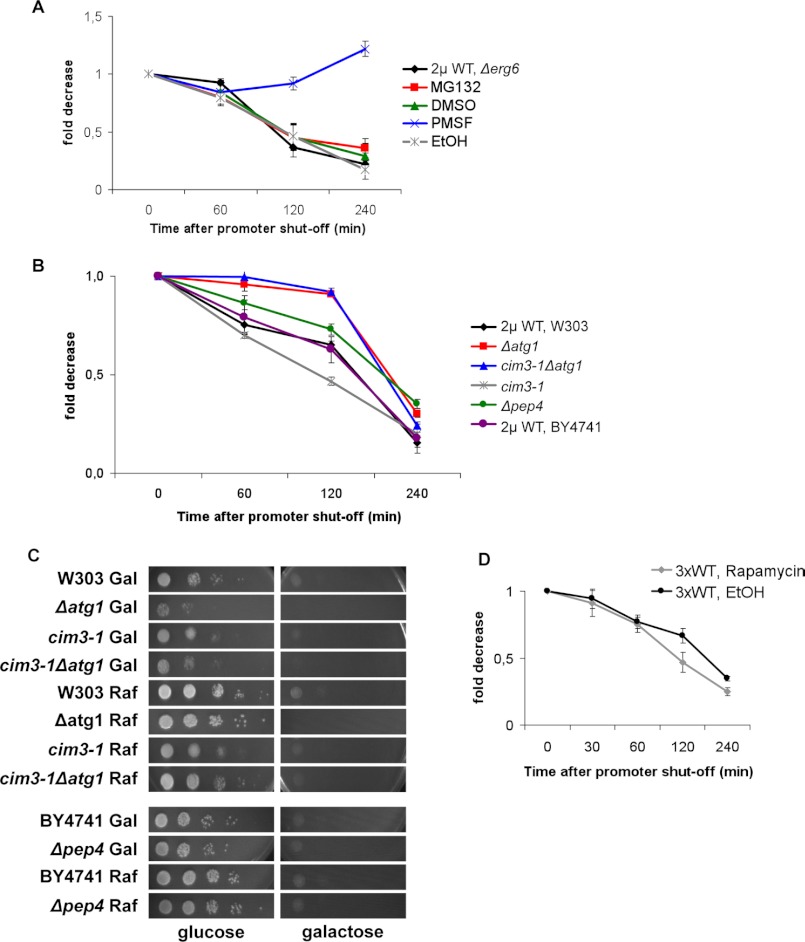
α-Synuclein can be degraded both by the proteasome and lysosomal/vacuolar pathways (28). Thus, we next analyzed the impact of blocking these systems, either pharmacologically or genetically, on the clearing of α-synuclein. Initially, we used MG132 as a proteasome inhibitor (43) in the cell wall-permeable Δerg6 mutant strain to facilitate drug uptake and increase the intracellular concentration of the drugs (39). To confirm that MG132 actually affected cells, immunoblotting analysis was performed and revealed a 3-fold increase in the amount of ubiquitinated proteins in treated cells (data not shown). Quantification of the reduction of aggregates formed in cells carrying high copy plasmids and induced for 4 h revealed that promoter shut-off resulted in equally efficient clearing of aggregates in MG132-treated cells when compared with vehicle-treated (DMSO) or untreated cells (Fig. 5A). These results suggested a minor contribution of the proteasome toward α-synuclein clearing in yeast.
FIGURE 5.
α-Synuclein aggregate clearance upon promoter shut-off in different degradation impairment conditions. A, drug treatments are shown. Aggregation quantification of Δerg6 cells expressing WT α-synuclein-GFP from a high copy plasmid upon promoter shut-off at indicated time points is shown. After 4 h of preincubation in galactose, cells were shifted to glucose medium supplemented with 50 μm MG132 in DMSO or only DMSO and to glucose medium supplemented with 1 mm PMSF dissolved in ethanol (EtOH) or only EtOH. Quantifications were the result of at least three independent experiments (S.D. were less than 10%). B, degradation-related mutants are shown. Aggregation quantification upon promoter shut-off. Wild-type (W303 and BY4741), Δatg1, cim3-1, and Δpep4 yeast cells were preincubated in galactose for 4 h to express WT α-synuclein, shifted to glucose, and counted at the indicated time points. C, spotting analyses are shown. W303, Δatg1, cim3-1, cim3-1Δatg1, BY4741, and Δpep4 yeast cells transformed with a high copy plasmid carrying WT α-synuclein-GFP were induced in galactose-containing medium for 8 h. The same strains were grown in raffinose-containing medium for control. Cells were spotted on glucose or galactose. D, clearance of α-synuclein aggregates after exposure to rapamycin is shown. Cells expressing GFP-tagged WT α-synuclein from three genomically integrated copies (3×) were induced for 4 h in galactose. At the 3-h time point of induction, the medium was supplemented with 100 nm rapamycin for 1 h followed by washing and shifting the cells to glucose. For control, cells were pretreated with the same volume of EtOH, the vehicle of rapamycin.
As a second line of evidence for a minor contribution of the proteasome for aggregate clearance, a genetic approach was performed with promoter shut-off studies of a temperature-sensitive cim3-1 mutant strain. This yeast mutant is deficient in one of the AAA ATPases of the 19 S proteasome regulatory subunit, which unfolds substrate proteins in an ATP-dependent manner (44). Compared with the parent strain, cim3-1 cultures presented lower percentages of cells with aggregates. The cells were able to clear aggregates in time, even with a faster kinetic than the parental strain (Fig. 5B), further corroborating a minor contribution of the proteasome for α-synuclein aggregate clearance.
The pharmacological approach to determine the contribution of the lysosomal/vacuolar pathways for aggregate clearing included PMSF dissolved in ethanol. PMSF blocks these pathways by inhibiting the activity of numerous vacuolar serine proteases (45) without affecting proteasome function (46). PMSF treatment of cell wall-permeable Δerg6 mutant strains prevented clearance of α-synuclein aggregates after promoter shut-off (Fig. 5A). This suggested that PMSF blocked pathways, which play an important role in aggregate clearance. PMSF affects autophagic body formation (40). Therefore, we questioned whether aggregate clearance could occur via autophagic pathways. We thus performed promoter shut-off studies with the mutant Δatg1, which renders cells unable to perform autophagy. ATG1 stands for autophagy-specific gene 1, and Atg1 is a serine/threonine kinase that acts in autophagy regulation and is essential for autophagy induction (47). α-Synuclein was expressed from a high copy plasmid, and shut-off studies were performed. The results indicated that Δatg1 cells were unable to clear aggregates for the first 120 min after promoter shut-off, as opposed to the wild-type parental strain (W303) (Fig. 5B). The double mutant cim3-1Δatg1 cleared aggregates upon promoter shut-off in a similar manner as Δatg1 cells. This suggests that autophagy contributes more to aggregate clearance than the proteasome.
A Δpep4 mutant was also included in the shut-off experiments to obtain additional insight into the extent to which vacuolar degradation is involved in α-synuclein aggregate clearance. The PEP4 gene of S. cerevisiae encodes the vacuolar protease proteinase A, which initiates the maturation and activation of several vacuolar hydrolases, such as carboxypeptidase Y, proteinase B, and aminopeptidase I (48, 49). The Δpep4 cells in the BY4741 background were able to clear α-synuclein aggregates less efficiently than control cells (Fig. 5B). Aggregate clearance in the BY4741 and W303 backgrounds were comparable. Furthermore, the BY4741 (supplemental Fig. S3) and W303 wild-type (supplemental Fig. S1A and Fig. 2) backgrounds show the same cytotoxicity and α-synuclein aggregation phenotypes. The Δpep4 cells displayed only an intermediate response, which further supports the involvement of vacuolar proteases in aggregate clearance. This intermediate response suggests that additional proteases might be involved in the process, which is not controlled by proteinase A.
The involvement of autophagy in α-synuclein clearance was supported by a spotting analysis. Δatg1 cells were unable to recover their normal growth after α-synuclein exposure. The cells had been induced to express the protein for 8 h then spotted on glucose or galactose media (Fig. 5C). Similarly, cim3-1Δatg1 cells presented an impaired ability to recover. The cim3-1 mutant showed growth recovery similar to the raffinose control. The Δpep4 mutant presented a slightly impaired ability to recover growth. Growth recovery thus correlated with the ability of cells to clear aggregates (Fig. 5B).
To investigate whether autophagy up-regulation would facilitate the process of aggregate clearance, cells expressing α-synuclein from three genetic copies were exposed to the autophagy-inducing drug rapamycin (50). Yeast cells were preincubated with the drug at a concentration of 100 nm for 1 h and washed before the shift to glucose to avoid the drug anti-fungal effects (51). Aggregate quantification after promoter shut-off (Fig. 5D) revealed that rapamycin accelerated the clearance kinetic, validating the importance of autophagy in this process. Taken together, our pharmacological and genetic approaches support a more pronounced role for autophagic and vacuolar pathways in the clearance of α-synuclein aggregates in comparison to proteasomal degradation.
Wild-type and A53T α-Synuclein Interfere with the Induction of Autophagy in Yeast
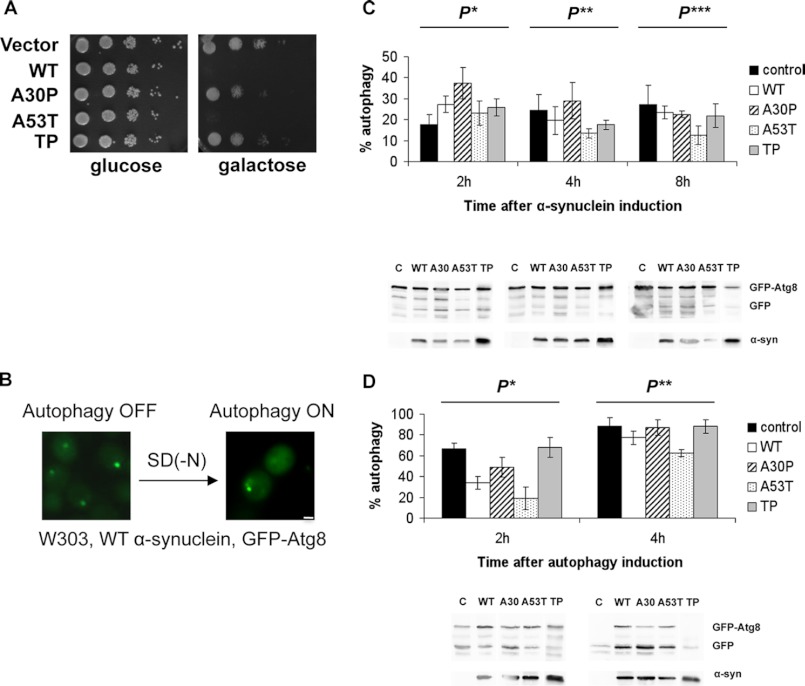
Next, we examined whether α-synuclein expression had an impact on the induction of autophagy in yeast. Nonspecific autophagy was monitored by a GFP-Atg8 assay (32). Co-transformation of high copy vectors expressing untagged α-synuclein and a pCu416 plasmid encoding GFP-Atg8 into wild-type cells did not interfere with the decreased growth caused by α-synuclein expression (Fig. 6A). Atg8 is an ubiquitin-like protein that is delivered to the vacuole by the cytoplasm-vacuole-targeting (Cvt) pathway and is essential for macroautophagy. Atg8 fused to GFP is degraded inside the vacuole, whereas GFP is released into the cellular cytoplasm. This could be visualized by a concentrated GFP signal before the induction of autophagy, which was distributed throughout the entire cytoplasm after induction of autophagy. α-Synuclein also did not interfere with GFP-Atg8 expression; hence, we observed the GFP signal before and after autophagy induction (Fig. 6B). Autophagy induction can be quantified by this assay by immunoblotting after the appearance of a band corresponding to GFP. The GFP percentage from the total GFP and GFP-Atg8 signals corresponds to the autophagy percentage in a specific set of cells (32). In an initial experiment we tested whether α-synuclein overexpression influenced autophagy throughout time. Autophagy ratios were monitored at different time points during induction, up to 8 h. The data showed that autophagy percentages varied among cells expressing different α-synuclein variants (Fig. 6C). Although differences between cells expressing wild-type α-synuclein, A53T, TP, and control were minor in the first 4 h of induction, the autophagy ratio of cells expressing A30P was substantially higher than that of control cells at the 2-h time point, suggesting that A30P can up-regulate autophagy. By 8 h of induction, this ratio became comparable with that of control, meaning that the effect of A30P was only transient. At 4 h of induction the autophagy ratio of cells expressing A53T was only slightly smaller than that of the control but became half of the control autophagy ratio at 8 h of induction, suggesting an increasing inhibitory effect of A53T on autophagy in time (Fig. 6C).
FIGURE 6.
α-Synuclein interference with the activation of autophagy. A, shown is spotting analysis of wild-type (W303) yeast cells co-transformed with high copy plasmids carrying different untagged α-synuclein versions and with a high copy plasmid carrying GFP-Atg8 under the control of a copper-inducible promoter. B, shown is live-cell microscopy of W303 cells during WT α-synuclein induction before shifting to SD(−N) starvation medium (autophagy off) and after (autophagy on). Scale bar = 1 μm. C, Western blots and quantifications present autophagy percents in cells expressing untagged α-synucleins during 8 h of induction in SC medium supplemented with galactose at the indicated time points. The values represent the average of four independent experiments. Significance of differences was tested using one-way ANOVA test (P* < 0.002; P**, P*** < 0.02; n = 4). D, Western blots and quantifications present autophagy percents of cells preinduced for 4 h to produce α-synuclein, then shifted to nitrogen starvation medium SD(−N) further supplemented with galactose. Probes were taken at 2 and 4 h after the shift. Previous to the shift, cells had been induced in galactose-containing SC medium for 4 h. The columns represent the average of 4 independent experiments. For control, W303 cells expressing the empty α-synuclein vector and the pCuGFP-Atg8 plasmid were used. Significance of differences was tested using one-way ANOVA test (P*, P** < 0.0002; n = 4).
We then tested whether α-synuclein can affect the induction of autophagy. Autophagy induction has long been achieved by means of nitrogen starvation, and a typical nitrogen starvation medium for yeast is SD(−N) (40). Thus, cells expressing α-synuclein for 4 h were shifted to SD(−N) medium further supplemented with galactose. Quantifications showed that A30P and TP α-synuclein did not influence autophagy significantly throughout time, as the autophagy ratios in cells expressing these variants were similar to the control (Fig. 6D). The autophagy ratios of cells expressing wild-type and A53T α-synuclein were considerably lower than those of control in the first 2 h of starvation. At 4 h starvation the differences were less pronounced. These results suggest a delay in autophagy induction and strengthen our hypothesis that α-synuclein affects autophagy in yeast.
DISCUSSION
In this study we used the yeast S. cerevisiae as a model to evaluate clearance pathways of α-synuclein aggregates. We used C-terminal tags where α-synuclein is connected to the tag via a linker. The N-terminal region needs to remain tag-free for α-synuclein to be toxic, presumably because it is essential for membrane interactions and oligomerization (52). The C terminus is highly acidic and tends to inhibit protein aggregation due to other protein-protein interactions (53–55). Capping the C terminus with a GFP tag might prevent its tendency to decrease aggregation.
Three copies of C-terminal-linker-tagged wild-type or two copies of A53T α-synuclein driven by the yeast GAL1-promoter and integrated into a single genomic locus were shown to be sufficient to cause growth inhibition and fluorescent foci formation/aggregation in yeast. Importantly, these data recapitulate the effect of allele multiplication of the wild-type α-synuclein gene in familial forms of PD (5, 6).
The A30P α-synuclein mutant forms aggregates when strongly overexpressed from a high copy vector. The aggregation was transient, suggesting a better clearance mechanism in comparison to that of the wild-type or A53T α-synuclein. A30P was previously reported not to aggregate in yeast (20, 41), in contrast to its behavior in mammalian systems and in vitro (3, 9, 53). Despite this, aggregate formation by A30P had only a minor effect on yeast cell growth. In comparison, triple-proline α-synuclein TP overexpression had no effect on yeast growth and was unable to form aggregates. The fact that no TP aggregates were detected in yeast is in line with previous results in vitro and in vivo (13, 14). In contrast to worms, flies, or mammalian neurons, however, TP was not toxic to yeast cell growth (13). In yeast, the TP mutant was found in the vacuole in most of the cells, similar to the low copy-expressed A30P α-synuclein. The mutant showed a special vacuolar phenotype characterized by increased number of vacuoles and formation of tube-like structures. Similar structures were described as autophagic tubes and are characteristic of yeast microautophagy (56). The number of autophagic tubes increases upon autophagy induction by starvation or stress conditions. This facilitates vesicle budding from the tip of the invaginations (56). These invaginations were observed for both A30P and TP α-synuclein. The two variants are presumably degraded by microautophagy in yeast.
Testing different constructs and comparing α-synucleins in different copy numbers elucidated the relationship between growth toxicity and aggregation. A lack of toxicity correlates with a lack of aggregation. Examples are the N-terminal GFP tagged constructs or the constructs carrying direct fusions α-synuclein-GFP. α-Synuclein can also form aggregates that can still be tolerated, with no impact on growth (e.g. 1× and 2× copies of wild-type or 1× A53T α-synuclein). This suggests that moderate amounts of aggregates are below a threshold for toxicity in yeast. It is still unknown whether some types of aggregates may even have a cytoprotective role in yeast, as it has been postulated for mammalian systems and in the human brain. Especially in the late stages of the disease, surviving neurons in PD brains are often found to contain Lewy bodies (2). Furthermore, evidence emerged that oligomeric species rather than aggregates of α-synuclein are responsible for toxicity (13, 57) and that in several PD models the rate of fibrillization and aggregation does not correlate with neurotoxicity (20, 58, 59). For example, the TP mutant is unable to aggregate, yet it causes neurotoxicity in several organisms (13). The fact that TP was not toxic to yeast growth suggested that yeast might be more resistant to oligomeric states of α-synuclein than higher eukaryotes.
Once the production of α-synuclein is stopped, aggregates can be cleared and cells can recover from α-synuclein exposure. This further supports that mild aggregate formation is likely a dynamic process that is non-toxic within a certain range. Aggregate clearance was previously investigated only in cell culture models (60–62) but not in yeast. Findings demonstrating that autophagy inhibition slows down aggregate clearance were previously reported in mammalian cell culture (63). Consistently, an increased number of autophagic structures can be found in PD patients as well as in animal PD models (30). Neuronal models focusing on α-synuclein degradation propose an important role of macroautophagy even though there degradation can additionally occur via chaperone-mediated autophagy (27). Here, we studied possible pathways responsible for α-synuclein aggregate clearance. Pharmacological treatment with the proteasome inhibitor MG132 or with vacuolar/lysosomal inhibitor PMSF demonstrated a more significant contribution of the vacuolar pathways to clearance. MG132 treatments did not inhibit aggregate clearance. Similarly, in neuroblastoma and oligodendroglial cells, inhibition of the proteasome by MG132 did not induce α-synuclein aggregation (62, 64), and in yeast, A30P α-synuclein was still degraded despite proteasomal inhibition with the same drug (65). Moreover, PD models propose that most α-synuclein in the cell is degraded not by the proteasome but by lysosomal enzymes (66). A possible reason for diminished involvement of the proteasome in α-synuclein degradation could be that α-synuclein itself alters the composition of the proteasome and impairs proteasomal degradation (67). Although α-synuclein may be degraded by the proteasome (23, 24), the protein could also inhibit this system (26), thus enhancing its accumulation and aggregation (68–70). Genetic analysis further supported this finding. Δpep4 cells with vacuolar defects or Δatg1 strains defective in autophagy were less efficient in clearing α-synuclein aggregates when compared with wild-type cells. The involvement of autophagy in α-synuclein aggregate clearance was supported by an inability of Δatg1 cells to recover growth defects after transient α-synuclein exposure. Additionally, wild-type cells accomplish an accelerated aggregate clearance kinetic when exposed to the autophagy-inducing drug rapamycin.
Although it is tempting to postulate that large aggregates of α-synuclein are preferentially degraded by (macro)autophagy, non-oligomeric species of α-synuclein may be degraded by the proteasome as well. Several reports suggest that once aggregate-prone substrates cannot efficiently be cleared by the proteasome, autophagy becomes the default degradation pathway (70, 71). Interestingly, the cim3-1Δatg1 yeast double mutant strain, impaired for both autophagy and proteasomal degradation, did not have a stronger impact than the autophagy deficiency alone. The double mutant was still able to clear aggregates, suggesting that other mechanisms might be involved in α-synuclein clearance. Recent evidence also proposes α-synuclein degradation by the endosomal-lysosomal pathway (72). Here, we showed that non-aggregated α-synuclein is presumably degraded by microautophagy.
α-Synuclein has long been hypothesized to affect autophagy. Our data reveal a transitory up-regulation of autophagy by A30P α-synuclein in the first 2 h of induction. This might correlate with low aggregation levels and the non-toxic phenotype of the A30P mutant, suggesting that autophagy could be part of a detoxification mechanism. Apart from that, the autophagy rates of cells expressing α-synuclein were comparable with the control, suggesting that WT and A53T α-synuclein overexpression does not activate autophagy. Conversely, 8 h of A53T α-synuclein induction inhibited autophagy. When autophagy was externally induced by means of nitrogen starvation, both wild-type and A53T α-synuclein delayed its activation. The more pronounced effect of A53T over that of wild-type α-synuclein on autophagy inhibition might correlate with the greater propensity of A53T to aggregate (11). The fact that WT and A53T α-synucleins perturbed autophagy correlates with previously reported vesicular trafficking defects (73, 74). Because autophagic vesicles fuse with endosomes to form an autophagosome that finally further fuses with the vacuole, impairment of endocytic traffic by the two α-synucleins would also affect autophagy. In contrast, A30P α-synuclein did not disturb autophagy, which correlates with the ability of A30P-containing vesicles to traffic to and merge with the vacuole (65).
Our autophagy-monitoring experiments revealed that α-synuclein exerts an inhibitory effect on autophagy, yet the molecular mechanism of inhibition has to be further elucidated. Additional observations that α-synuclein inhibits autophagic pathways in neuronal cells (29, 75) may suggest that autophagic inhibition by α-synuclein is conserved from yeast to higher organisms. Overall, our study opens new possibilities for the identification of novel genes involved in these processes and for the understanding of how cells can cope with toxic and/or aggregated proteins. This may ultimately enable the development of novel strategies for therapeutic intervention.
Supplementary Material
Acknowledgments
We thank Maria Mayer for excellent technical assistance. We thank Susan Lindquist (Whitehead Institute for Biomedical Research) for the IntTox strain and Dan Klionsky (Michigan University) for the GFP-Atg8 plasmid. We thank Nadine Smolinski for RH3475 strain and Mark Dumkow for pME3526, pME3527, and pME3528 plasmids.
This work was supported by the Deutsche Forschungsgemeinschaft, the Center for Molecular Physiology of the Brain, Bundesministerium für Bildung und Forschung (NGFN-Plus 01GS08190), European Union (NEURASYNC PITNGA-2009-238316), and the Volkswagen-Stiftung, and the Fonds der Chemischen Industrie.

This article contains supplemental Figs. S1–S3 and Videos 1 and 2.
- PD
- Parkinson disease
- TP
- A30P/A53T/A76P
- SC
- synthetic complete.
REFERENCES
- 1. Galvin J. E., Lee V. M., Trojanowski J. Q. (2001) Synucleinopathies. clinical and pathological implications. Arch. Neurol. 58, 186–190 [DOI] [PubMed] [Google Scholar]
- 2. Forno L. S., DeLanney L. E., Irwin I., Langston J. W. (1996) Electron microscopy of Lewy bodies in the amygdala-parahippocampal region. Comparison with inclusion bodies in the MPTP-treated squirrel monkey. Adv. Neurol. 69, 217–228 [PubMed] [Google Scholar]
- 3. Spillantini M. G., Schmidt M. L., Lee V. M., Trojanowski J. Q., Jakes R., Goedert M. (1997) α-Synuclein in Lewy bodies. Nature 388, 839–840 [DOI] [PubMed] [Google Scholar]
- 4. Maroteaux L., Campanelli J. T., Scheller R. H. (1988) Synuclein. A neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J. Neurosci. 8, 2804–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K. (2003) α-Synuclein locus triplication causes Parkinson disease. Science 302, 841. [DOI] [PubMed] [Google Scholar]
- 6. Hardy J., Cai H., Cookson M. R., Gwinn-Hardy K., Singleton A. (2006) Genetics of Parkinson disease and parkinsonism. Ann. Neurol. 60, 389–398 [DOI] [PubMed] [Google Scholar]
- 7. Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., Nussbaum R. L. (1997) Mutation in the α-synuclein gene identified in families with Parkinson disease. Science 276, 2045–2047 [DOI] [PubMed] [Google Scholar]
- 8. Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., Przuntek H., Epplen J. T., Schöls L., Riess O. (1998) A30P mutation in the gene encoding α-synuclein in Parkinson disease. Nat. Genet. 18, 106–108 [DOI] [PubMed] [Google Scholar]
- 9. Giasson B. I., Uryu K., Trojanowski J. Q., Lee V. M. (1999) Mutant and wild type human α-synucleins assemble into elongated filaments with distinct morphologies in vitro. J. Biol. Chem. 274, 7619–7622 [DOI] [PubMed] [Google Scholar]
- 10. Lashuel H. A., Petre B. M., Wall J., Simon M., Nowak R. J., Walz T., Lansbury P. T., Jr. (2002) α-Synuclein, especially the Parkinson disease-associated mutants, forms pore-like annular and tubular protofibrils. J. Mol. Biol. 322, 1089–1102 [DOI] [PubMed] [Google Scholar]
- 11. Conway K. A., Rochet J. C., Bieganski R. M., Lansbury P. T., Jr. (2001) Kinetic stabilization of the α-synuclein protofibril by a dopamine-α-synuclein adduct. Science 294, 1346–1349 [DOI] [PubMed] [Google Scholar]
- 12. Volles M. J., Lansbury P. T., Jr. (2002) Vesicle permeabilization by protofibrillar α-synuclein is sensitive to Parkinson disease-linked mutations and occurs by a pore-like mechanism. Biochemistry 41, 4595–4602 [DOI] [PubMed] [Google Scholar]
- 13. Karpinar D. P., Balija M. B., Kügler S., Opazo F., Rezaei-Ghaleh N., Wender N., Kim H. Y., Taschenberger G., Falkenburger B. H., Heise H., Kumar A., Riedel D., Fichtner L., Voigt A., Braus G. H., Giller K., Becker S., Herzig A., Baldus M., Jäckle H., Eimer S., Schulz J. B., Griesinger C., Zweckstetter M. (2009) Pre-fibrillar α-synuclein variants with impaired β-structure increase neurotoxicity in Parkinson disease models. EMBO J. 28, 3256–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taschenberger G., Garrido M., Tereshchenko Y., Bähr M., Zweckstetter M., Kügler S. (2012) Aggregation of α-synuclein promotes progressive in vivo neurotoxicity in adult rat dopaminergic neurons. Acta Neuropathol. 123, 671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gajula Balija M. B., Griesinger C., Herzig A., Zweckstetter M., Jäckle H. (2011) Pre-fibrillar α-synuclein mutants cause Parkinson disease-like non-motor symptoms in Drosophila. PLoS One 6, e24701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winner B., Jappelli R., Maji S. K., Desplats P. A., Boyer L., Aigner S., Hetzer C., Loher T., Vilar M., Campioni S., Tzitzilonis C., Soragni A., Jessberger S., Mira H., Consiglio A., Pham E., Masliah E., Gage F. H., Riek R. (2011) In vivo demonstration that α-synuclein oligomers are toxic. Proc. Natl. Acad. Sci. U.S.A. 108, 4194–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kahle P. J., Neumann M., Ozmen L., Haass C. (2000) Physiology and pathophysiology of α-synuclein. Cell culture and transgenic animal models based on a Parkinson disease-associated protein. Ann. N.Y. Acad. Sci. 920, 33–41 [DOI] [PubMed] [Google Scholar]
- 18. Matsuoka Y., Vila M., Lincoln S., McCormack A., Picciano M., LaFrancois J., Yu X., Dickson D., Langston W. J., McGowan E., Farrer M., Hardy J., Duff K., Przedborski S., Di Monte D. A. (2001) Lack of nigral pathology in transgenic mice expressing human α-synuclein driven by the tyrosine hydroxylase promoter. Neurobiol. Dis. 8, 535–539 [DOI] [PubMed] [Google Scholar]
- 19. Giasson B. I., Duda J. E., Quinn S. M., Zhang B., Trojanowski J. Q., Lee V. M. (2002) Neuronal α-synucleinopathy with severe movement disorder in mice expressing A53T human α-synuclein. Neuron 34, 521–533 [DOI] [PubMed] [Google Scholar]
- 20. Outeiro T. F., Lindquist S. (2003) Yeast cells provide insight into α-synuclein biology and pathobiology. Science 302, 1772–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lakso M., Vartiainen S., Moilanen A. M., Sirviö J., Thomas J. H., Nass R., Blakely R. D., Wong G. (2003) Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human α-synuclein. J. Neurochem. 86, 165–172 [DOI] [PubMed] [Google Scholar]
- 22. Cooper A. A., Gitler A. D., Cashikar A., Haynes C. M., Hill K. J., Bhullar B., Liu K., Xu K., Strathearn K. E., Liu F., Cao S., Caldwell K. A., Caldwell G. A., Marsischky G., Kolodner R. D., Labaer J., Rochet J. C., Bonini N. M., Lindquist S. (2006) α-Synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson models. Science 313, 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bennett M. C., Bishop J. F., Leng Y., Chock P. B., Chase T. N., Mouradian M. M. (1999) Degradation of α-synuclein by proteasome. J. Biol. Chem. 274, 33855–33858 [DOI] [PubMed] [Google Scholar]
- 24. Tofaris G. K., Layfield R., Spillantini M. G. (2001) α-Synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 509, 22–26 [DOI] [PubMed] [Google Scholar]
- 25. Ancolio K., Alves da Costa C., Uéda K., Checler F. (2000) α-Synuclein and the Parkinson disease-related mutant A53T-α-synuclein do not undergo proteasomal degradation in HEK293 and neuronal cells. Neurosci. Lett. 285, 79–82 [DOI] [PubMed] [Google Scholar]
- 26. Lindersson E., Beedholm R., Højrup P., Moos T., Gai W., Hendil K. B., Jensen P. H. (2004) Proteasomal inhibition by α-synuclein filaments and oligomers. J. Biol. Chem. 279, 12924–12934 [DOI] [PubMed] [Google Scholar]
- 27. Vogiatzi T., Xilouri M., Vekrellis K., Stefanis L. (2008) Wild type α-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J. Biol. Chem. 283, 23542–23556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Webb J. L., Ravikumar B., Atkins J., Skepper J. N., Rubinsztein D. C. (2003) α-Synuclein is degraded by both autophagy and the proteasome. J. Biol. Chem. 278, 25009–25013 [DOI] [PubMed] [Google Scholar]
- 29. Winslow A. R., Chen C. W., Corrochano S., Acevedo-Arozena A., Gordon D. E., Peden A. A., Lichtenberg M., Menzies F. M., Ravikumar B., Imarisio S., Brown S., O'Kane C. J., Rubinsztein D. C. (2010) α-Synuclein impairs macroautophagy. Implications for Parkinson disease. J. Cell Biol. 190, 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anglade P., Vyas S., Hirsch E. C., Agid Y. (1997) Apoptosis in dopaminergic neurons of the human substantia nigra during normal aging. Histol. Histopathol. 12, 603–610 [PubMed] [Google Scholar]
- 31. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C. W., Klionsky D. J. (2005) Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell 16, 3438–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gietz D., St Jean A., Woods R. A., Schiestl R. H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sherman F. (1991) Getting started with yeast. Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 35. Southern E. M. (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98, 503–517 [DOI] [PubMed] [Google Scholar]
- 36. Hoffman C. S., Winston F. (1987) A 10-min DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57, 267–272 [DOI] [PubMed] [Google Scholar]
- 37. Abramoff M., Magelhaes P., Ram S. (2004) Image processing with Image J. Biophotonics Int. 11, 36–42 [Google Scholar]
- 38. Vida T. A., Emr S. D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee D. H., Goldberg A. L. (1996) Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem. 271, 27280–27284 [DOI] [PubMed] [Google Scholar]
- 40. Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dixon C., Mathias N., Zweig R. M., Davis D. A., Gross D. S. (2005) α-Synuclein targets the plasma membrane via the secretory pathway and induces toxicity in yeast. Genetics 170, 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lamphier M. S., Ptashne M. (1992) Multiple mechanisms mediate glucose repression of the yeast GAL1 gene. Proc. Natl. Acad. Sci. U.S.A. 89, 5922–5926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee D. H., Goldberg A. L. (1998) Proteasome inhibitors. Valuable new tools for cell biologists. Trends Cell Biol. 8, 397–403 [DOI] [PubMed] [Google Scholar]
- 44. Ghislain M., Udvardy A., Mann C. (1993) S. cerevisiae 26 S protease mutants arrest cell division in G2/metaphase. Nature 366, 358–362 [DOI] [PubMed] [Google Scholar]
- 45. Jones E. W. (2002) Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 351, 127–150 [DOI] [PubMed] [Google Scholar]
- 46. Dubiel W., Ferrell K., Pratt G., Rechsteiner M. (1992) Subunit 4 of the 26 S protease is a member of a novel eukaryotic ATPase family. J. Biol. Chem. 267, 22699–22702 [PubMed] [Google Scholar]
- 47. Matsuura A., Tsukada M., Wada Y., Ohsumi Y. (1997) Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192, 245–250 [DOI] [PubMed] [Google Scholar]
- 48. Parr C. L., Keates R. A., Bryksa B. C., Ogawa M., Yada R. Y. (2007) The structure and function of Saccharomyces cerevisiae proteinase A. Yeast 24, 467–480 [DOI] [PubMed] [Google Scholar]
- 49. Woolford C. A., Daniels L. B., Park F. J., Jones E. W., Van Arsdell J. N., Innis M. A. (1986) The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol. Cell. Biol. 6, 2500–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kamada Y., Sekito T., Ohsumi Y. (2004) Autophagy in yeast. A TOR-mediated response to nutrient starvation. Curr. Top Microbiol. Immunol. 279, 73–84 [DOI] [PubMed] [Google Scholar]
- 51. Wong G. K., Griffith S., Kojima I., Demain A. L. (1998) Antifungal activities of rapamycin and its derivatives, prolylrapamycin, 32-desmethylrapamycin, and 32-desmethoxyrapamycin. J. Antibiot. 51, 487–491 [DOI] [PubMed] [Google Scholar]
- 52. Karube H., Sakamoto M., Arawaka S., Hara S., Sato H., Ren C. H., Goto S., Koyama S., Wada M., Kawanami T., Kurita K., Kato T. (2008) N-terminal region of α-synuclein is essential for the fatty acid-induced oligomerization of the molecules. FEBS Lett. 582, 3693–3700 [DOI] [PubMed] [Google Scholar]
- 53. Cookson M. R. (2005) The biochemistry of Parkinson disease. Annu. Rev. Biochem. 74, 29–52 [DOI] [PubMed] [Google Scholar]
- 54. Murray I. V., Giasson B. I., Quinn S. M., Koppaka V., Axelsen P. H., Ischiropoulos H., Trojanowski J. Q., Lee V. M. (2003) Role of α-synuclein carboxyl terminus on fibril formation in vitro. Biochemistry 42, 8530–8540 [DOI] [PubMed] [Google Scholar]
- 55. Tsigelny I. F., Bar-On P., Sharikov Y., Crews L., Hashimoto M., Miller M. A., Keller S. H., Platoshyn O., Yuan J. X., Masliah E. (2007) Dynamics of α-synuclein aggregation and inhibition of pore-like oligomer development by β-synuclein. FEBS J. 274, 1862–1877 [DOI] [PubMed] [Google Scholar]
- 56. Müller O., Sattler T., Flötenmeyer M., Schwarz H., Plattner H., Mayer A. (2000) Autophagic tubes. Vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J. Cell Biol. 151, 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Outeiro T. F., Putcha P., Tetzlaff J. E., Spoelgen R., Koker M., Carvalho F., Hyman B. T., McLean P. J. (2008) Formation of toxic oligomeric α-synuclein species in living cells. PLoS One 3, e1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen L., Feany M. B. (2005) α-Synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat. Neurosci. 8, 657–663 [DOI] [PubMed] [Google Scholar]
- 59. Volles M. J., Lansbury P. T., Jr. (2007) Relationships between the sequence of α-synuclein and its membrane affinity, fibrillization propensity, and yeast toxicity. J. Mol. Biol. 366, 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim Y. S., Laurine E., Woods W., Lee S. J. (2006) A novel mechanism of interaction between α-synuclein and biological membranes. J. Mol. Biol. 360, 386–397 [DOI] [PubMed] [Google Scholar]
- 61. Opazo F., Krenz A., Heermann S., Schulz J. B., Falkenburger B. H. (2008) Accumulation and clearance of α-synuclein aggregates demonstrated by time-lapse imaging. J. Neurochem. 106, 529–540 [DOI] [PubMed] [Google Scholar]
- 62. Riedel M., Goldbaum O., Schwarz L., Schmitt S., Richter-Landsberg C. (2010) 17-AAG induces cytoplasmic α-synuclein aggregate clearance by induction of autophagy. PLoS One 5, e8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fortun J., Dunn W. A., Jr., Joy S., Li J., Notterpek L. (2003) Emerging role for autophagy in the removal of aggresomes in Schwann cells. J. Neurosci. 23, 10672–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dyllick-Brenzinger M., D'Souza C. A., Dahlmann B., Kloetzel P. M., Tandon A. (2010) Reciprocal effects of α-synuclein overexpression and proteasome inhibition in neuronal cells and tissue. Neurotox Res. 17, 215–227 [DOI] [PubMed] [Google Scholar]
- 65. Flower T. R., Clark-Dixon C., Metoyer C., Yang H., Shi R., Zhang Z., Witt S. N. (2007) YGR198w (YPP1) targets A30P α-synuclein to the vacuole for degradation. J. Cell Biol. 177, 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Paxinou E., Chen Q., Weisse M., Giasson B. I., Norris E. H., Rueter S. M., Trojanowski J. Q., Lee V. M., Ischiropoulos H. (2001) Induction of α-synuclein aggregation by intracellular nitrative insult. J. Neurosci. 21, 8053–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen Q., Thorpe J., Keller J. N. (2005) α-synuclein alters proteasome function, protein synthesis, and stationary phase viability. J. Biol. Chem. 280, 30009–30017 [DOI] [PubMed] [Google Scholar]
- 68. Bedford L., Hay D., Devoy A., Paine S., Powe D. G., Seth R., Gray T., Topham I., Fone K., Rezvani N., Mee M., Soane T., Layfield R., Sheppard P. W., Ebendal T., Usoskin D., Lowe J., Mayer R. J. (2008) Depletion of 26 S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J. Neurosci. 28, 8189–8198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. McNaught K. S., Jenner P. (2001) Proteasomal function is impaired in substantia nigra in Parkinson disease. Neurosci. Lett. 297, 191–194 [DOI] [PubMed] [Google Scholar]
- 70. Rideout H. J., Lang-Rollin I., Stefanis L. (2004) Involvement of macroautophagy in the dissolution of neuronal inclusions. Int. J. Biochem. Cell Biol. 36, 2551–2562 [DOI] [PubMed] [Google Scholar]
- 71. Olanow C. W., McNaught K. S. (2006) Ubiquitin-proteasome system and Parkinson disease. Mov. Disord. 21, 1806–1823 [DOI] [PubMed] [Google Scholar]
- 72. Tofaris G. K., Kim H. T., Hourez R., Jung J. W., Kim K. P., Goldberg A. L. (2011) Ubiquitin ligase Nedd4 promotes α-synuclein degradation by the endosomal-lysosomal pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 17004–17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Soper J. H., Kehm V., Burd C. G., Bankaitis V. A., Lee V. M. (2011) Aggregation of α-synuclein in S. cerevisiae is associated with defects in endosomal trafficking and phospholipid biosynthesis. J. Mol. Neurosci. 43, 391–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gitler A. D., Bevis B. J., Shorter J., Strathearn K. E., Hamamichi S., Su L. J., Caldwell K. A., Caldwell G. A., Rochet J. C., McCaffery J. M., Barlowe C., Lindquist S. (2008) The Parkinson disease protein α-synuclein disrupts cellular Rab homeostasis. Proc. Natl. Acad. Sci. U.S.A. 105, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xilouri M., Vogiatzi T., Vekrellis K., Park D., Stefanis L. (2009) Abberant α-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS One 4, e5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.