Background: NIK is a central component in the non-canonical NF-κB pathway, and its activity is associated with various diseases.
Results: An N-terminal extension is required for activity and stabilizes the kinase in an active conformation.
Conclusion: The NIK kinase domain adopts a constitutively active conformation.
Significance: This work presents the first NIK structure and provides a molecular basis for NIK regulation.
Keywords: NF-κB, Protein Kinases, Signal Transduction, Structural Biology, X-ray Crystallography, NIK, Nuclear Factor κB-inducing Kinase
Abstract
NF-κB-inducing kinase (NIK) is a central component in the non-canonical NF-κB signaling pathway. Excessive NIK activity is implicated in various disorders, such as autoimmune conditions and cancers. Here, we report the first crystal structure of truncated human NIK in complex with adenosine 5′-O-(thiotriphosphate) at a resolution of 2.5 Å. This truncated protein is a catalytically active construct, including an N-terminal extension of 60 residues prior to the kinase domain, the kinase domain, and 20 residues afterward. The structure reveals that the NIK kinase domain assumes an active conformation in the absence of any phosphorylation. Analysis of the structure uncovers a unique role for the N-terminal extension sequence, which stabilizes helix αC in the active orientation and keeps the kinase domain in the catalytically competent conformation. Our findings shed light on the long-standing debate over whether NIK is a constitutively active kinase. They also provide a molecular basis for the recent observation of gain-of-function activity for an N-terminal deletion mutant (ΔN324) of NIK, leading to constitutive non-canonical NF-κB signaling with enhanced B-cell adhesion and apoptosis resistance.
Introduction
The NF-κB family of transcription factors plays a crucial role in the regulation of diverse physiological processes, such as immune responses, inflammation, and cell survival and death (1, 2). Although many different stimuli activate NF-κB proteins to induce their nuclear translocation, NF-κB signaling is controlled through two major pathways: canonical (or classical) and non-canonical (or alternative) (1–5). The canonical NF-κB pathway involves activation of the IκB (inhibitor of κB) kinase (IKK)4 complex, which then phosphorylates IκBs, leading to the degradation of IκBs and subsequent nuclear translocation of NF-κB. The activation of the non-canonical NF-κB pathway is dependent upon the stabilization and activation of the NF-κB-inducing kinase (NIK), which activates IKKα, which in turn phosphorylates the NF-κB2 precursor p100, leading to the processing of p100 into its active form, p52 (6).
NIK was originally identified as an integral component in the canonical pathway downstream of the proinflammatory cytokines TNF-α and IL-1β (7). However, the essential role proposed for NIK in this pathway was soon open for debate when NIK-deficient cells showed normal NF-κB activation in response to TNF, IL-1β, and lymphotoxin-β stimulation (8). Intriguingly, both alymphoplasia (aly/aly) mice, which possess a naturally occurring NIK-defective mutation, and NIK knock-out mice demonstrated defects in lymphoid organogenesis (8, 9). Accumulating evidence suggests that NIK functions in the canonical NF-κB pathway only in a cell type- and receptor-specific manner (10–12). In contrast, NIK is essential for the phosphorylation and activation of IKKα and subsequent activation of NF-κB2 (6, 13–16). Thus, it plays a central role in the non-canonical NF-κB pathway.
As a direct upstream kinase for IKKα, the kinase function of NIK is required for NF-κB2 signaling, which is activated by a subset of TNF-β (lymphotoxin-α)-related cytokines, including BAFF (B-cell activation factor belonging to the TNF family), CD40 ligand, lymphotoxin-β, TWEAK (TNF-like weak inducer of apoptosis), and RANKL (receptor activator for NF-κB ligand) (10, 14). Excessive NF-κB2 activity is implicated in a wide range of disorders, including inflammation, osteoporosis, and many other autoimmune conditions (17). NIK mutations that lead to hyperactive NIK have emerged as one of the major causes of NF-κB deregulation in multiple myeloma (18–20). NIK thus presents a promising therapeutic target for the treatment of autoimmune disorders and cancers.
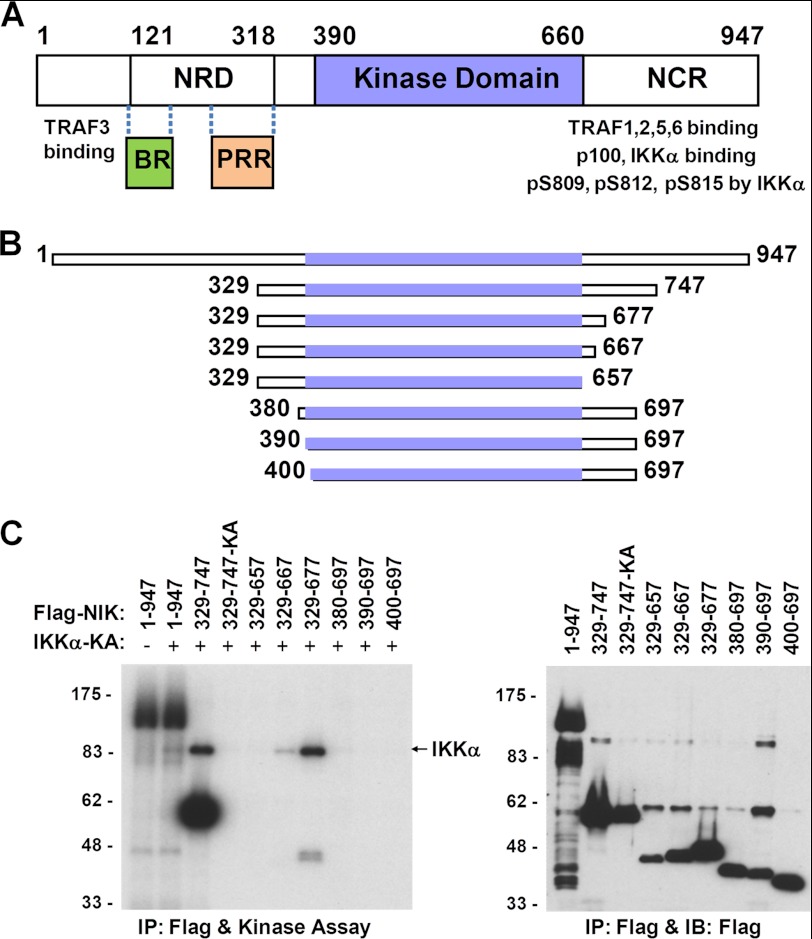
NIK, also known as MAP3K14 (MAPK kinase kinase 14), is a serine/threonine kinase in the MAP3K family (7). There are 947 amino acids in the human NIK sequence, which contains at least four identified domains, including an N-terminal TRAF3-binding domain (approximately residues 30–120), a negative regulatory domain (approximately residues 121–318), a central serine/threonine kinase domain (approximately residues 390–660), and a C-terminal non-catalytic region that is required for signaling (see Fig. 1A) (16, 21). The negative regulatory domain is composed of two structural motifs: a basic region with leucine zipper motifs (approximately residues 127–146) and a proline-rich repeat (approximately residues 250–317) (21). Deletion of the basic and proline-rich repeat regions in the negative regulatory domain enhanced its catalytic activity compared with the full-length protein. The C-terminal non-catalytic region has been reported to be responsible for interacting with various important components in the signaling cascade, including substrates IKKα and p100, and regulators such as TRAF1, TRAF2, TRAF5, and TRAF6 (7, 21, 22). In addition, a recent report also identified three phosphorylation sites in the non-catalytic region, i.e. Ser-809, Ser-812, and Ser-815, through IKKα-mediated phosphorylation as negative feedback in the non-canonical pathway (23).
FIGURE 1.
Identification of catalytically active NIK kinase domain constructs. A, NIK sequence motifs based on literature reports (16, 21). NRD, negative regulatory domain; NCR, non-catalytic region; BR, basic region; PRR, proline-rich repeat. B, schematic of a selected panel of truncated NIK constructs with the kinase domain shown as slate boxes. C, in vitro kinase phosphorylation assays. Left panel, NIK autophosphorylation and IKKα(K43A) phosphorylation by NIK. IKKα(K43A) and NIK construct 329–747(K429A) (329–747-KA) are inactive alanine mutants of the catalytic Lys-43 and Lys-429, respectively. Right panel, immunoblot (IB) analysis of total cell extracts. Positions of molecular mass standards (in kilodaltons) are shown on the left for both panels. IP, immunoprecipitate.
Significant progress toward elucidating the NIK mode of action has been made since NIK was first identified 14 years ago. However, a complete understanding of the regulation of NIK activity remains a challenge due to its complicated role in both the non-canonical and canonical NF-κB pathways (24). No structural information has been reported for NIK thus far. To gain molecular insight into the function of NIK, we have pursued biochemical and structural studies of NIK. Here, we first identified a truncated NIK construct around the kinase domain (residues 330–679) that retains catalytic activity. Subsequently, we solved the crystal structure of this truncated protein in complex with the non-hydrolyzable ATP analog ATPγS to 2.5 Å resolution. The structure reveals that the NIK kinase domain adopts an active conformation, in agreement with its catalytic activity. Unexpectedly, such a catalytically competent conformation is maintained by an N-terminal extension prior to the kinase domain rather than through a phosphorylation event.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
The NIK gene encoding human NIK residues 330–679 and harboring a surface mutation of S549D was subcloned into a pFastBac HT vector (Invitrogen) with a His6 tag at the N terminus and a tobacco etch virus protease cleavage site between the His6 tag and NIK. NIK protein was expressed in baculovirus High Five cells (Invitrogen) grown at 27 °C for 72 h in ESF 921 insect cell medium (Expression Systems). Cells were microfluidized, and the supernatant was loaded on an affinity column containing nickel-nitrilotriacetic acid beads for His-tagged protein (Qiagen) and eluted with buffer containing 50 mm Tris-HCl (pH 7.9), 250 mm NaCl, 10% glycerol, 5 mm imidazole, and 5 mm β-mercaptoethanol. The protein was cleaved with recombinant tobacco etch virus at 4 °C overnight and subsequently buffer-exchanged into 50 mm Tris-HCl (pH 7.9), followed by ion exchange on SOURCE Q30 (GE Healthcare). The resultant peak was pooled, and 200 mm NaCl, 10% glycerol, and 10 mm MgCl2 were added. The protein was then further purified with a Superdex 75 size exclusion column (GE Healthcare) and concentrated to 1 mg/ml. The static light scattering (SLS) measurements were carried out on a miniDAWN TREOS multiple-angle light scattering instrument (Wyatt Technology Corp., Santa Barbara, CA) in PBS.
For NIK crystallization, 1 mm ATPγS was added to the protein sample, incubated at 4 °C for 1 h, and concentrated to 5–8 mg/ml. Crystals were grown by sitting drop vapor diffusion, where protein was mixed with a reservoir solution of 12.5% polyethylene glycol 3350, 200 mm ammonium sulfate, and 0.1 m sodium citrate (pH 5.4) in a 1:1 ratio. Crystals grew to 0.3 × 0.2 × 0.1 mm after 1 week and were stepwise cryoprotected with 10, 15, and 20% ethylene glycol.
The co-crystals of wild-type construct 330–679 with ATPγS diffracted to about 3.5 Å. The S549D mutant showed improved crystal diffraction but with virtually no difference in terms of the phosphorylation state, biochemical activity, and three-dimensional structural features compared with the wild-type protein. The S549D mutation was initially introduced as a potential phosphomimetic, along with two other residue modifications, T552D/E and T559D/E, in the activation loop. Although of all these mutants showed similar activity compared with the wild-type protein, some displayed slightly better solubility and less batch-to-batch variation, such as S549D.
Data Collection and Structure Determination
X-ray diffraction data sets were collected on beamline 5.0.2 at the Advanced Light Source (Berkeley, CA). Data sets were processed and scaled with MOSFLM (25) and SCALA in the CCP4 Program Suite (26). A 2.5 Å structure of NIK in complex with ATPγS was solved via molecular replacement in CCP4 (26). An initial solution was obtained with the program Phaser (27) using a bundle of 20 kinase structures as an ensemble search model. The molecular replacement phases were further improved using the program DM (28). Model building was carried out using the programs O (29) and QUANTA (Accelrys, San Diego, CA). Subsequent model building and refinement were done using Coot (30) and REFMAC5 (31), respectively. The data collection and refinement statistics are shown in Table 1. All structural figures were prepared using PyMOL (Schrodinger).
TABLE 1.
Statistics of crystallographic data and refinement
| Data collection | |
| Space group | P41 |
| Unit cell dimensions (Å) | a = 85.06, b = 85.06, c = 115.36 |
| Wavelength (Å) | 1.000 |
| Resolution (Å) | 50–2.5 (2.64–2.50) |
| No. of total reflections | 137,194 (19,725) |
| No. of unique reflections | 28,420 (4101) |
| Wilson B-factor (Å2) | 51.82 |
| Average multiplicity | 4.8 (4.8) |
| Completeness (%) | 99.9 (100) |
| Intensity I/σ | 15.8 (3.4) |
| Rsyma | 0.067 (0.493) |
| Refinement (|F| > 0) | |
| Resolution (Å) | 30–2.5 (2.624–2.50) |
| Rwork/Rfreeb | 0.182/0.222 |
| No. of reflections | 26,957 |
| No. of atoms | 5322 |
| Protein | 5158 |
| Water | 70 |
| Ligand | 62 |
| Mg2+ | 2 |
| Other (ethylene glycol) | 30 |
| Ramachandran statistics (%) | |
| Residues in favored regions | 92 |
| Residues in allowed regions | 7.2 |
| Residues in disallowed regions | 0.7 |
| Average B-factors (Å2) | |
| Protein | 56.1 |
| Ligand | 38.1 |
| Water | 47.0 |
| r.m.s.d.c in bond length (Å) | 0.005 |
| r.m.s.d. in bond angles | 0.997° |
a Rsym = Σ|Iavg − Ii|/ΣIi, where Ii is the observed intensity for the ith measurement and Iavg is the average intensity of all measurements.
b Rfactor = Σ|Fo − Fc|/ΣFo, where Fo and Fc are the observed and calculated structure factors, respectively, Rfree was calculated from a randomly chosen 5% of reflections excluded from the refinement, and Rfactor was calculated from the remaining 95% of reflections.
c r.m.s.d. denotes the root mean square deviation from ideal geometry. Values in parentheses are for the highest resolution shell.
In Vitro Phosphorylation Assays
NIK Autophosphorylation
FLAG epitope-tagged NIK constructs were expressed for 24 h in HEK293 cells after transient transfection. Cell lysates were immunoprecipitated with anti-FLAG monoclonal antibody affinity resin (Eastman Kodak Co.), and bound proteins were eluted with 30 μl of cell lysis buffer containing FLAG peptide (300 μg/ml). In vitro autophosphorylations were performed with 1 μl of NIK-containing eluate in 15 μl of reaction buffer containing 20 mm Tris-HCl (pH 7.6), 20 mm magnesium chloride, 20 mm β-glycerophosphate, 20 mm p-nitrophenyl phosphate, 1 mm EDTA, 1 mm sodium orthovanadate, 0.4 mm PMSF, 1 mm ATP, 20 mm creatine phosphate, and 5μCi of [γ-32P]ATP at 37 °C for 30 min. Samples were analyzed by 10% SDS-PAGE and autoradiography.
In Vitro Phosphorylation of IKKα by NIK
HEK293 cells were transiently transfected with the indicated epitope-tagged expression vectors. Twenty-four hours after transfection, cell lysates were immunoprecipitated with anti-FLAG monoclonal antibody affinity resin, and bound proteins were eluted with 30 μl of cell lysis buffer containing FLAG peptide (300 μg/ml). Purified NIK constructs and the IKKα(K43A) (catalytically inactive) mutant protein (32) were used in in vitro kinase reactions with [γ-32P]ATP. Samples were analyzed by 10% SDS-PAGE and autoradiography.
Immunoblot Analysis of Total Cell Extracts
Aliquots of 10 μl of total cell extracts from the transfections described above were immunoblotted with anti-FLAG monoclonal antibody.
RESULTS
Search for a Catalytically Active and Soluble Kinase Domain Construct
When we expressed human full-length NIK protein in mammalian cells, the protein showed solid autophosphorylation activity but very weak activity for its substrate IKKα and limited solubility. Expression of the kinase domain-only construct (residues 390–660) resulted in very little soluble protein with no detectable catalytic activity. To obtain a soluble NIK protein with catalytic activity, we expressed a series of constructs with an N- or C-terminal deletion of every 50 residues in HEK293 cells and evaluated their autophosphorylation activity as well as their ability to phosphorylate the downstream substrate kinase IKKα in vitro. Both activities were retained for protein expression constructs with an N-terminal deletion of 328 residues (ΔN328) or a C-terminal deletion of 250 residues (ΔC250), but not for constructs with an N-terminal deletion of 380 residues (ΔN380) or a C-terminal deletion of 300 residues (ΔC300).
To further narrow down the boundaries that define the catalytic activity, a second set of constructs with finer deletion intervals from both the N- and C-terminal regions were generated, and their activities were evaluated again with in vitro phosphorylation assays. The results for a selected panel of these deletion constructs are shown in Fig. 1 (B and C). Constructs with an N terminus starting from the kinase domain or with a C terminus ending at the kinase domain failed to produce catalytically active proteins. The shortest soluble construct that still retained both catalytic activities contained residues 329–677 (ΔN328-ΔC270), composed of the kinase domain, an N-terminal extension of 60 residues to the kinase domain, and a C-terminal extension of 20 residues (Fig. 1C). The need for the 60-residue segment prior to the N terminus of the kinase domain is very clear for both catalytic activities, as exemplified by construct 380–697, which was completely inactive. The requirement for a C-terminal extension of ∼20 residues beyond the kinase domain might be rationalized by both the catalytic activity and protein expression yield. Taken together, the data indicate that the 60-residue N-terminal extension and the 20-residue C-terminal extension to the kinase domain are required for producing soluble and catalytically active NIK protein.
To generate sufficient amounts of soluble protein suitable for crystallographic studies, we expressed in insect cells various constructs designed around the region of the shortest catalytically active construct identified, with an N-terminal His6 tag followed by a tobacco etch virus protease cleavage site. The final construct used in our structural studies contained residues 330–679 (construct 330–679) harboring a surface mutation of S549D to improve protein solubility and crystal diffraction.
Overall Structure
The crystal structure of NIK construct 330–679 in complex with the non-hydrolyzable ATP analog ATPγS was determined at a resolution of 2.5Å. The structure was solved by molecular replacement using a bundle of 20 previously reported protein kinase structures as a search model. The final electron density is good throughout the molecule for residues 333–675, except for part of the activation loop (residues 543–555), which has no interpretable electron density.
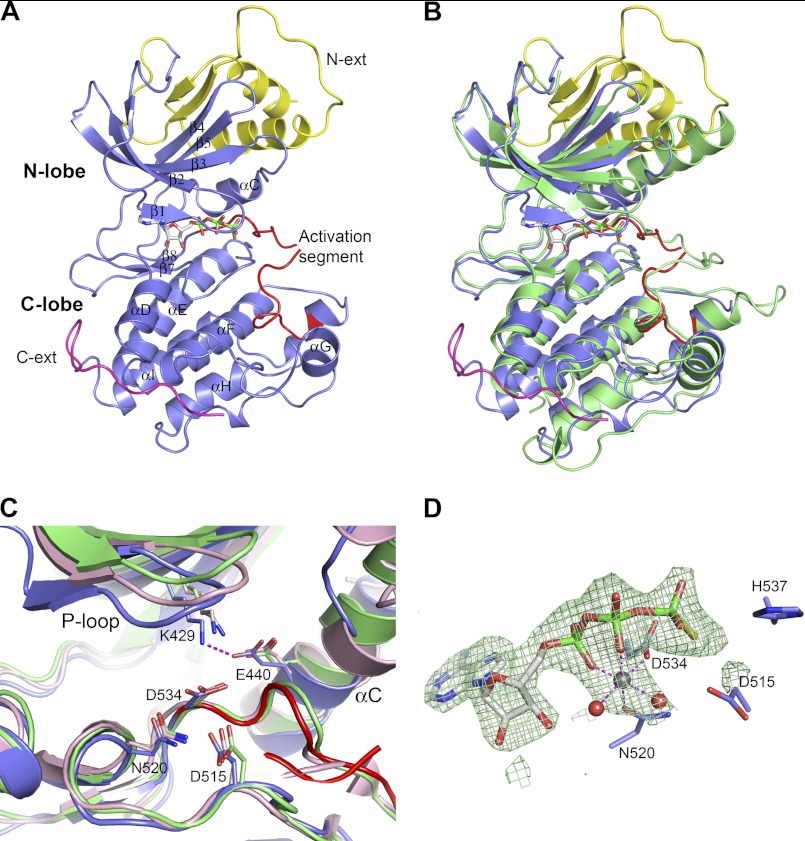
There are two NIK molecules per asymmetric unit. The two molecules can be superimposed with a root mean square deviation of 0.38 Å (main chain Cα) and can thus be considered structurally identical. The overall structure of the NIK kinase domain adopts a canonical two-lobe protein kinase fold, with the N-terminal lobe comprising mainly a five-stranded antiparallel β-sheet plus one helix and the C-terminal lobe containing an all-α-helical structure (Fig. 2A). Connecting the two lobes is the hinge region where ATPγS binds. In comparison with other kinase structures, the NIK kinase domain assumes a closed conformation, resembling other protein kinases in the activated form (Fig. 2B) (33).
FIGURE 2.
Crystal structure of NIK construct 330–679 in complex with ATPγS. A, overall NIK structure shown in ribbon diagram. The kinase domain is colored in slate with the activation segment highlighted in red. The N-terminal (N-ext) and C-terminal (C-ext) extensions to the kinase domain are colored in yellow and magenta, respectively. ATPγS is shown in stick representation and is atomic color-coded gray for carbon, red for oxygen, blue for nitrogen, yellow for sulfur, and green for phosphorus. N-lobe, N-terminal lobe; C-lobe, C-terminal lobe. B, overall structure comparison of NIK with a constitutively active PhK (Protein Data Bank code 2PHK). NIK is color-coded as described for A, and PhK is colored green. C, ribbon diagram of the NIK kinase domain active site compared with PhK and cAMP-dependent kinase (Protein Data Bank code 1ATP). Residues are shown in atomic color scheme with the carbon color scheme being slate for NIK, green for PhK, and pink for cAMP-dependent kinase. D, the bound nucleotide ATPγS in NIK with omit electron density map contoured at 2.5σ. A Mg2+ ion is shown as a gray sphere, and two water molecules are shown as red spheres.
Active Site and ATPγS Binding
Our structure reveals a partially disordered activation loop and no phosphorylation on any residues for this truncated NIK protein. The phosphorylation states of both the wild-type and S549D proteins were then further corroborated by a whole mass analysis, which revealed no phosphorylation modification. However, the active site of the NIK kinase domain bears a close structural resemblance to previously analyzed serine/threonine kinases in their active forms (33–35). The positions of several catalytically important residues, including the catalytic Lys-429, Glu-440 of helix αC as the salt bridge partner for Lys-429, the catalytic base Asp-515, the metal ion-chelating Asp-534 of the DFG motif, and Asn-520 (which orients the catalytic Asp-515), are in the correct constellation for catalysis. In fact, they are nearly identically positioned to those residues typically found in active protein kinase structures, such as phosphorylase kinase (PhK) (36) and cAMP-dependent kinase (Fig. 2C) (37).
The ATPγS binding mode in the NIK kinase domain resembles other nucleotides bound to protein kinases, with the adenine ring anchored to the hinge region through two hydrogen bonding interactions. Here, one Mg2+ is observed at the ATP site in NIK. The position of this metal ion is more similar to that of a secondary metal ion seen in other kinases, such as ATP in PhK and cAMP-dependent kinase and ATPγS in PknB (36–38). However, it seems to play a hybrid role of a primary and secondary metal ion by bridging the α- and β-phosphate oxygen atoms (Asn-520 Oδ1 and Asp-534 Oδ2), as well as being coordinated to two water molecules (Fig. 2D).
A T559A mutation was reported to dominantly interfere with TNF-α-induced NF-κB signaling (22). It should be noted that Thr-559 is a conserved threonine residue in the P+1 substrate-binding area, which is believed to contribute to the specificity of Ser/Thr kinases versus Tyr kinases (39). Here, Thr-559 engages in hydrogen bonding interactions with the serine/threonine kinase conserved Lys-517 in the catalytic loop and the catalytic base Asp-515, which also hydrogen bonds with Asn-520, signifying a common characteristic of the active state of serine/threonine kinases (34). Mutation of T559A is likely to disrupt such a concerted hydrogen bonding network in the active state, thus impeding catalysis.
The elucidated active structural attribute for construct 330–679 is consistent with its catalytic activity in the activity assays. The crystallized protein is not phosphorylated, but it permits autophosphorylation during ATP incubation owing to its intrinsic kinase activity. Note that the longer construct 329–747 shows a much stronger autophosphorylation activity in comparison with construct 329–677. These data suggest that sequence 680–747 contains the autophosphorylation sites, in which one or more residues among the four threonine and six serine residues are likely the participants.
N-terminal Extension Prior to the Kinase Domain
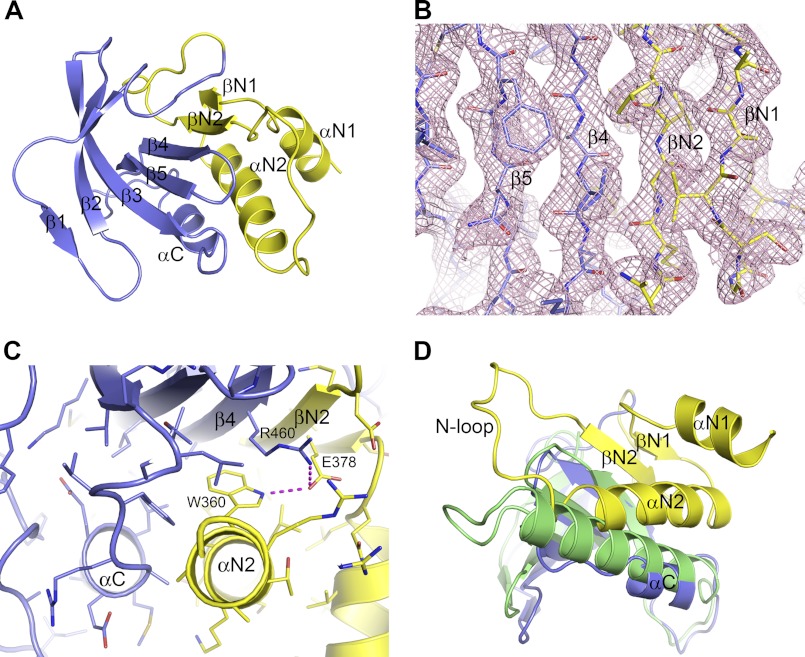
A surprising feature we observed in this structure is the packing of the N-terminal extension against the N-terminal lobe of the kinase domain. As mentioned previously, this 60-residue segment prior to the kinase domain was necessary to yield a catalytically active protein. Our structure reveals that the N-terminal extension becomes an integral part of the N-terminal lobe of the kinase domain by forming an additional two α-helices, two β-strands, and two loops before the first strand β1 in the kinase domain (Fig. 3A). The short strand βN1 of Val-345–Ser-347 runs parallel to strand βN2 (Asn-377–Leu-381), which is antiparallel to strand β4 in the kinase domain (Fig. 3B). These two additional β-strands from the N-terminal extension align very well with the edge of the five-stranded antiparallel β-sheet in the kinase domain, resulting in an extended curved seven-stranded β-sheet in the N-terminal lobe. The second helix αN2 (Ala-350–Arg-363) packs against the β-sheet and is roughly antiparallel to helix αC (Fig. 3C). The first helix αN1 of Val-333–Leu-341 forms an overhang outside of the kinase domain, packing loosely against the second helix αN2. The loop between αN2 and βN2 consisting of Ser-365–Asp-376 (N-loop) protrudes out of the kinase domain, forming virtually no interactions with the N-terminal lobe.
FIGURE 3.
N-terminal extension to the NIK kinase domain. A, extended N-terminal lobe of the NIK kinase domain. The typical N-terminal lobe of the kinase domain is shown in slate, and the N-terminal extension is shown in yellow. B, packing of strand βN2 of the N-terminal extension with strand β4 in the kinase N-terminal lobe. The pink mesh represents the 2Fo − Fc electron density map. C, packing interactions between the N-terminal extension and the kinase N-terminal lobe. D, position of helix αC in NIK in comparison with that in PhK, shown in green.
Although helix αN1 and strand βN1 have no direct interactions with the N-terminal lobe of the kinase domain, both helix αN2 and strand βN2 engage in close interactions with the kinase domain, especially helix αN2, which interacts with helix αC of the kinase domain. The interactions among the helices and the β-sheet include one salt bridge between Arg-460 of β4 and Glu-378 of βN2, one hydrogen bond between Glu-378 of βN2 and Trp-360 of αN2, and numerous hydrophobic and van der Waals contacts (Fig. 3C). One notable difference in the N-terminal lobe of the NIK kinase domain is that the classic helix αC is shorter by at least one helical turn compared with other protein kinases. In fact, the C terminus of helix αN2 occupies a similar position comparable to where longer αC helices from other protein kinases sit, such as PhK (Fig. 3D), compensating for the short helix αC of NIK. Therefore, the packing of helix αN2 of the N-terminal extension plays a critical role in stabilizing helix αC of NIK kinase domain in the active orientation.
Our findings provide a molecular basis for the earlier biochemical observations reported by Xiao and Sun (21); they demonstrated that residues 348–377 are critical to NIK activity. Here, our structure reveals that sequence 348–377 folds into helix αN2, the N-loop, and strand βN2 of the N-terminal extension, with both αN2 and βN2 being intimately involved in stabilizing helix αC of the kinase domain in the active conformation.
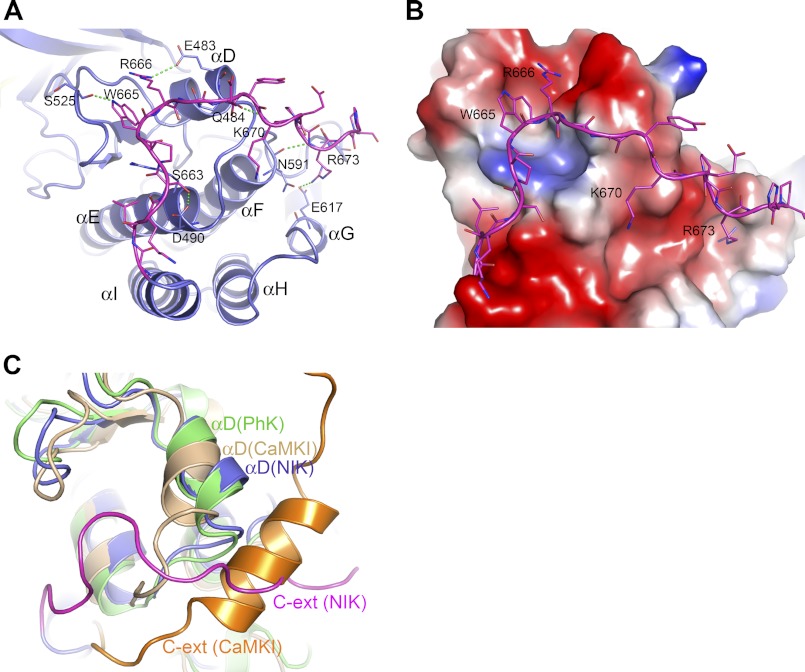
C-terminal Extension to the Kinase Domain
The kinase domain of NIK ends around Gly-660 at the end of helix I. Most of the residues in the C-terminal extension (residues 661–675) of the truncated construct pack tightly with the termini of helices αD, αE, αF, and αG in the C-terminal lobe of the kinase domain. This interface is dominated by numerous electrostatic interactions, hydrogen bonding interactions, and van der Waals contacts, as shown in Fig. 4A. Of particular note are Trp-665 and Arg-666, which project into the highly negatively charged surface depression formed by helix αD and loop β7-β8, with Arg-666 forming a salt bridge with Glu-483 of αD and Trp-665 donating a hydrogen bond to the backbone carbonyl of Ser-525 of loop β7-β8 (Fig. 4B). Here, the C-terminal extension in fact becomes an integral part of the C-terminal lobe of the kinase domain, likely providing stabilization to the kinase C-terminal lobe.
FIGURE 4.
C-terminal extension to the NIK kinase domain. A, extensive interactions from the C-terminal extension with the kinase C-terminal lobe. The color scheme is the same as described in the legend to Fig. 2A. B, interactions of the C-terminal extension with the kinase C-terminal lobe, which is shown in electrostatic molecular surface representation. C, superposition of the structures of active PhK and autoinhibited calcium/calmodulin-dependent protein kinase I (CaMKI; Protein Data Bank code 1A06) with NIK in the C-terminal extension (C-ext) area. The position of helix αD in NIK resembles that in the active PhK.
The C-terminal extension to the kinase domain has been shown to have a direct effect on the kinase activity in other protein kinases. In calcium/calmodulin-dependent protein kinases as well as twitchin kinase (40–42), the C-terminal extension in fact caused a rotation in helix αD that contributed to the inactivation of the kinases. Superposition of NIK with the inactive calcium/calmodulin-dependent protein kinase I and active PhK structures reveals that the position of helix αD in NIK resembles that in the active form of PhK (Fig. 4C), which is consistent with the active state of this NIK protein. Thus, the C-terminal extension might also contribute to the active conformation of the NIK kinase domain by stabilizing helix αD of the C-terminal lobe in the active orientation.
Dimer Versus Monomer
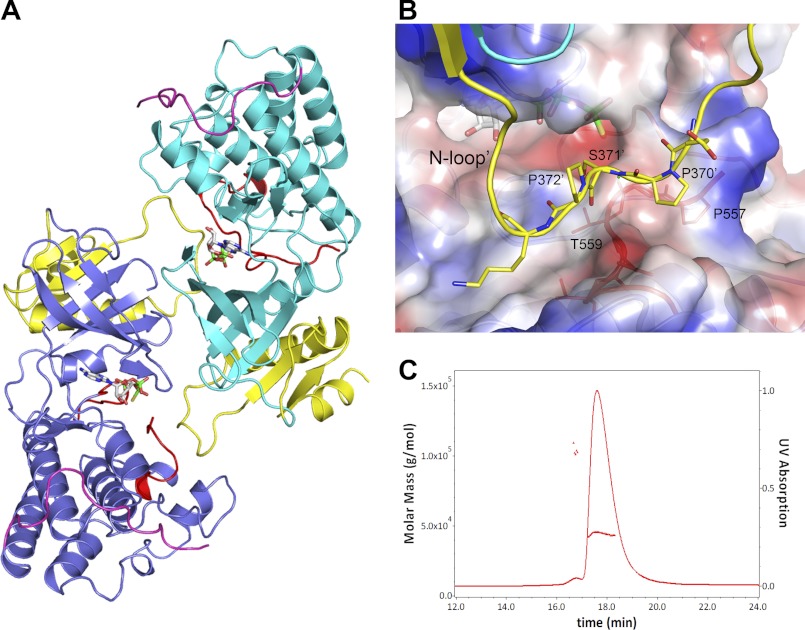
The two independent NIK molecules in the asymmetric unit form a head-to-tail dimer by non-crystallographic 2-fold symmetry as shown in Fig. 5A. Calculation from the PISA server (43) indicates that 2900 Å2 of total solvent-accessible surface area is buried in the NIK dimer interface; this exceeds that usually seen for a nonspecific crystal contact but is within the range for a homodimer interface (44). Notably, the N-loop docks at the typical P−1 to P+1 substrate-binding area of the dimer partner (Fig. 5B), reminiscent of substrate peptide binding in other kinases. Hence, such a dimer arrangement as seen in the crystal might interfere with substrate peptide binding.
FIGURE 5.
Dimer versus monomer. A, head-to-tail dimer of two independent NIK molecules in the asymmetric unit. The color scheme is the same as described in the legend to Fig. 2, except that the kinase domain of one monomer is colored cyan. B, N-loop binding in the dimer. The N-loop of one monomer is shown in ribbon and stick representation, binding to a region of the P+1 substrate-binding pocket of the dimer partner shown in electrostatic molecular surface representation. C, solution characterization of NIK construct 330–679. SEC-SLS of NIK showed a major peak and a minor peak with measured molecular masses of ∼44 and ∼100 kDa, respectively.
To investigate whether the observed dimer in the crystal structure is a crystallographic artifact, we performed size exclusion chromatography (SEC)-SLS analysis to determine the absolute molar mass for the SEC peak. Fig. 5C shows a SEC-SLS chromatogram of the NIK protein in solution, demonstrating the presence of a minor peak in addition to the major one. Both peaks were confirmed to be NIK by SDS-PAGE and mass spectrum analysis. SLS showed measured molecular masses of ∼44 and ∼100 kDa for the main and minor peaks, which correspond to the monomeric and dimeric forms of the construct, respectively. Thus, the truncated NIK protein presents both monomeric and dimeric forms in solution. However, the majority of the protein is in a monomeric form in solution, which is consistent with its observed catalytic activity. Homodimers or oligomers were reported for mammalian expressed full-length NIK protein (22). It remains to be determined whether the observed dimer arrangement in the crystal structure here might represent the actual dimer interface for the full-length protein.
DISCUSSION
NIK is a critical kinase in the non-canonical NF-κB pathway. Excessive activity of NIK leading to enhanced non-canonical signaling has been implicated in various pathological conditions of autoimmune disorders and cancers. Thus, understanding the molecular mechanism of NIK regulation becomes an imperative task. Even though NIK is characterized as a MAP3K serine/threonine kinase, it has a sequence of HGD in its catalytic loop, thus classifying NIK in the non-RD kinase family (33, 34), in which the catalytic aspartate is not preceded by the arginine residue that serves a critical role in neutralizing phosphoamino acids. The catalytic activity of non-RD kinases is often regulated by mechanisms different from the common phosphorylation event(s) occurring on the activation loop. Consistent with this concept, the NIK kinase domain assumes an active conformation in the absence of any phosphorylation, and its active state is maintained largely by a sequence segment outside of the kinase domain. The N-terminal 60 residues prior to the kinase domain are integrated into the N-terminal lobe of the kinase domain and stabilize its active conformation. The earlier evidence of the accumulation of NIK being sufficient to trigger the processing of p100 has raised the question of whether NIK is a constitutively active kinase (24, 45). Our finding that the NIK kinase domain is held in the intrinsically active conformation without a phosphorylation activation event provides a framework to resolve this long-standing query at the molecular level.
A very interesting recent study reported by Rosebeck et al. (46) showed that the proteolytic cleavage of NIK at Arg-325 by the API2-MALT1 (mucosa-associated lymphoid tissue) fusion oncoprotein preserves the kinase activity and resistance to proteasomal degradation, leading to constitutive non-canonical NF-κB signaling with enhanced B-cell adhesion and apoptosis resistance. Our structural elucidation of NIK construct 330–679 being constitutively active provides a molecular basis for such a gain-of-function activity of this N-terminal deletion mutant. Together with the identification of sequence 121–318 in NIK as a negative regulatory domain in an earlier biochemical report (21), these studies suggest a possible mechanism for NIK regulation in cells. Under normal conditions, the full-length NIK protein may be present in an autoinhibited form in which its constitutively active kinase domain is shielded by the N-terminal inhibitory element. Upon cytokine induction, NIK may undergo a conformational change in which the N-terminal domain releases the kinase domain or facilitates a proteolytic process that removes the N-terminal domain, as shown in the study of Rosebeck et al., unlocking its constitutive kinase activity. Structural elucidation of full-length NIK or the NIK kinase domain in complex with the inhibitory element may shed more light on the intricate regulation of NIK activity. Our mapping of the minimal domain required for NIK catalytic activity and our elucidation of the NIK kinase structure have not only paved the way for these advanced studies but have also laid a solid foundation for inhibitor development targeting NIK activity.
Acknowledgments
We thank David Goeddel for valuable guidance and advice. The Advanced Light Source at the Lawrence Berkeley National Laboratory is supported by United States Department of Energy Contract DE-AC03-76SF00098.
The atomic coordinates and structure factors (code 4DN5) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IKK
- IκB kinase
- NIK
- NF-κB-inducing kinase
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- SLS
- static light scattering
- PhK
- phosphorylase kinase
- SEC
- size exclusion chromatography.
REFERENCES
- 1. Vallabhapurapu S., Karin M. (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 [DOI] [PubMed] [Google Scholar]
- 2. Hayden M. S., Ghosh S. (2008) Shared principles in NF-κB signaling. Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 3. Bonizzi G., Karin M. (2004) The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25, 280–288 [DOI] [PubMed] [Google Scholar]
- 4. Senftleben U., Cao Y., Xiao G., Greten F. R., Krähn G., Bonizzi G., Chen Y., Hu Y., Fong A., Sun S. C., Karin M. (2001) Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science 293, 1495–1499 [DOI] [PubMed] [Google Scholar]
- 5. Pomerantz J. L., Baltimore D. (2002) Two pathways to NF-κB. Mol. Cell 10, 693–695 [DOI] [PubMed] [Google Scholar]
- 6. Xiao G., Harhaj E. W., Sun S. C. (2001) NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell 7, 401–409 [DOI] [PubMed] [Google Scholar]
- 7. Malinin N. L., Boldin M. P., Kovalenko A. V., Wallach D. (1997) MAP3K-related kinase involved in NF-κB induction by TNF, CD95, and IL-1. Nature 385, 540–544 [DOI] [PubMed] [Google Scholar]
- 8. Yin L., Wu L., Wesche H., Arthur C. D., White J. M., Goeddel D. V., Schreiber R. D. (2001) Defective lymphotoxin-β receptor-induced NF-κB transcriptional activity in NIK-deficient mice. Science 291, 2162–2165 [DOI] [PubMed] [Google Scholar]
- 9. Shinkura R., Kitada K., Matsuda F., Tashiro K., Ikuta K., Suzuki M., Kogishi K., Serikawa T., Honjo T. (1999) Alymphoplasia is caused by a point mutation in the mouse gene encoding NF-κB-inducing kinase. Nat. Genet. 22, 74–77 [DOI] [PubMed] [Google Scholar]
- 10. Ramakrishnan P., Wang W., Wallach D. (2004) Receptor-specific signaling for both the alternative and the canonical NF-κB activation pathways by NF-κB-inducing kinase. Immunity 21, 477–489 [DOI] [PubMed] [Google Scholar]
- 11. Smith C., Andreakos E., Crawley J. B., Brennan F. M., Feldmann M., Foxwell B. M. (2001) NF-κB-inducing kinase is dispensable for activation of NF-κB in inflammatory settings but essential for lymphotoxin-β receptor activation of NF-κB in primary human fibroblasts. J. Immunol. 167, 5895–5903 [DOI] [PubMed] [Google Scholar]
- 12. Garceau N., Kosaka Y., Masters S., Hambor J., Shinkura R., Honjo T., Noelle R. J. (2000) Lineage-restricted function of nuclear factor κB-inducing kinase (NIK) in transducing signals via CD40. J. Exp. Med. 191, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun S. C. (2011) Non-canonical NF-κB signaling pathway. Cell Res. 21, 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dejardin E. (2006) The alternative NF-κB pathway from biochemistry to biology: pitfalls and promises for future drug development. Biochem. Pharmacol. 72, 1161–1179 [DOI] [PubMed] [Google Scholar]
- 15. Aya K., Alhawagri M., Hagen-Stapleton A., Kitaura H., Kanagawa O., Novack D. V. (2005) NF-κB-inducing kinase controls lymphocyte and osteoclast activities in inflammatory arthritis. J. Clin. Invest. 115, 1848–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liao G., Zhang M., Harhaj E. W., Sun S. C. (2004) Regulation of the NF-κB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 279, 26243–26250 [DOI] [PubMed] [Google Scholar]
- 17. Saitoh Y., Yamamoto N., Dewan M. Z., Sugimoto H., Martinez Bruyn V. J., Iwasaki Y., Matsubara K., Qi X., Saitoh T., Imoto I., Inazawa J., Utsunomiya A., Watanabe T., Masuda T., Yamamoto N., Yamaoka S. (2008) Overexpressed NF-κB-inducing kinase contributes to the tumorigenesis of adult T-cell leukemia and Hodgkin Reed-Sternberg cells. Blood 111, 5118–5129 [DOI] [PubMed] [Google Scholar]
- 18. Keats J. J., Fonseca R., Chesi M., Schop R., Baker A., Chng W. J., Van Wier S., Tiedemann R., Shi C. X., Sebag M., Braggio E., Henry T., Zhu Y. X., Fogle H., Price-Troska T., Ahmann G., Mancini C., Brents L. A., Kumar S., Greipp P., Dispenzieri A., Bryant B., Mulligan G., Bruhn L., Barrett M., Valdez R., Trent J., Stewart A. K., Carpten J., Bergsagel P. L. (2007) Promiscuous mutations activate the non-canonical NF-κB pathway in multiple myeloma. Cancer Cell 12, 131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Annunziata C. M., Davis R. E., Demchenko Y., Bellamy W., Gabrea A., Zhan F., Lenz G., Hanamura I., Wright G., Xiao W., Dave S., Hurt E. M., Tan B., Zhao H., Stephens O., Santra M., Williams D. R., Dang L., Barlogie B., Shaughnessy J. D., Jr., Kuehl W. M., Staudt L. M. (2007) Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Demchenko Y. N., Glebov O. K., Zingone A., Keats J. J., Bergsagel P. L., Kuehl W. M. (2010) Classical and/or alternative NF-κB pathway activation in multiple myeloma. Blood 115, 3541–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xiao G., Sun S. C. (2000) Negative regulation of the nuclear factor κB-inducing kinase by a cis-acting domain. J. Biol. Chem. 275, 21081–21085 [DOI] [PubMed] [Google Scholar]
- 22. Lin X., Mu Y., Cunningham E. T., Jr., Marcu K. B., Geleziunas R., Greene W. C. (1998) Molecular determinants of NF-κB-inducing kinase action. Mol. Cell. Biol. 18, 5899–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Razani B., Zarnegar B., Ytterberg A. J., Shiba T., Dempsey P. W., Ware C. F., Loo J. A., Cheng G. (2010) Negative feedback in non-canonical NF-κB signaling modulates NIK stability through IKKα-mediated phosphorylation. Sci. Signal. 3, ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thu Y. M., Richmond A. (2010) NF-κB-inducing kinase: a key regulator in the immune system and in cancer. Cytokine Growth Factor Rev. 21, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leslie A. G. W. (1992) Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography, Vol. 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 26. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cowtan K. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography, Vol. 31, pp. 34–38, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 29. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 32. Woronicz J. D., Gao X., Cao Z., Rothe M., Goeddel D. V. (1997) IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science 278, 866–869 [DOI] [PubMed] [Google Scholar]
- 33. Johnson L. N., Noble M. E., Owen D. J. (1996) Active and inactive protein kinases: structural basis for regulation. Cell 85, 149–158 [DOI] [PubMed] [Google Scholar]
- 34. Nolen B., Taylor S., Ghosh G. (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 [DOI] [PubMed] [Google Scholar]
- 35. Huse M., Kuriyan J. (2002) The conformational plasticity of protein kinases. Cell 109, 275–282 [DOI] [PubMed] [Google Scholar]
- 36. Lowe E. D., Noble M. E., Skamnaki V. T., Oikonomakos N. G., Owen D. J., Johnson L. N. (1997) The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition. EMBO J. 16, 6646–6658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng J., Trafny E. A., Knighton D. R., Xuong N. H., Taylor S. S., Ten Eyck L. F., Sowadski J. M. (1993) 2.2 Å refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. D Biol. Crystallogr. 49, 362–365 [DOI] [PubMed] [Google Scholar]
- 38. Young T. A., Delagoutte B., Endrizzi J. A., Falick A. M., Alber T. (2003) Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat. Struct. Biol. 10, 168–174 [DOI] [PubMed] [Google Scholar]
- 39. Taylor S. S., Radzio-Andzelm E., Hunter T. (1995) How do protein kinases discriminate between serine/threonine and tyrosine? Structural insights from the insulin receptor protein-tyrosine kinase. FASEB J. 9, 1255–1266 [DOI] [PubMed] [Google Scholar]
- 40. Goldberg J., Nairn A. C., Kuriyan J. (1996) Structural basis for the autoinhibition of calcium/calmodulin-dependent protein kinase I. Cell 84, 875–887 [DOI] [PubMed] [Google Scholar]
- 41. Hu S. H., Parker M. W., Lei J. Y., Wilce M. C., Benian G. M., Kemp B. E. (1994) Insights into autoregulation from the crystal structure of twitchin kinase. Nature 369, 581–584 [DOI] [PubMed] [Google Scholar]
- 42. Rosenberg O. S., Deindl S., Sung R. J., Nairn A. C., Kuriyan J. (2005) Structure of the autoinhibited kinase domain of CaMKII and SAXS analysis of the holoenzyme. Cell 123, 849–860 [DOI] [PubMed] [Google Scholar]
- 43. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 44. Bahadur R. P., Chakrabarti P., Rodier F., Janin J. (2004) A dissection of specific and non-specific protein-protein interfaces. J. Mol. Biol. 336, 943–955 [DOI] [PubMed] [Google Scholar]
- 45. Sun S. C. (2010) Controlling the fate of NIK: a central stage in non-canonical NF-κB signaling. Sci. Signal. 3, pe18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosebeck S., Madden L., Jin X., Gu S., Apel I. J., Appert A., Hamoudi R. A., Noels H., Sagaert X., Van Loo P., Baens M., Du M. Q., Lucas P. C., McAllister-Lucas L. M. (2011) Cleavage of NIK by the API2-MALT1 fusion oncoprotein leads to non-canonical NF-κB activation. Science 331, 468–472 [DOI] [PMC free article] [PubMed] [Google Scholar]