Background: Phf14 is identified as a novel gene that regulates proliferation of mesenchymal cells.
Results: Phf14-null mice show interstitial pulmonary hyperplasia and mesenchymal fibroblasts exhibit increased proliferation rates accompanied by enhanced expression of PDGFRα.
Conclusion: Phf14 acts as a negative regulator of PDGFRα expression in mesenchymal cells.
Significance: Phf14 has potential as a target for the treatment of lung fibrosis.
Keywords: Cell Growth, Cell Proliferation, Development, Embryonic Stem Cell, Transcription Regulation, PDGFR, Mesenchyme
Abstract
The regulation of mesenchymal cell growth by signaling molecules plays an important role in maintaining tissue functions. Aberrant mesenchymal cell proliferation caused by disruption of this regulatory process leads to pathogenetic events such as fibrosis. In the current study we have identified a novel nuclear factor, Phf14, which controls the proliferation of mesenchymal cells by regulating PDGFRα expression. Phf14-null mice died just after birth due to respiratory failure. Histological analyses of the lungs of these mice showed interstitial hyperplasia with an increased number of PDGFRα+ mesenchymal cells. PDGFRα expression was elevated in Phf14-null mesenchymal fibroblasts, resulting in increased proliferation. We demonstrated that Phf14 acts as a transcription factor that directly represses PDGFRα expression. Based on these results, we used an antibody against PDGFRα to successfully treat mouse lung fibrosis. This study shows that Phf14 acts as a negative regulator of PDGFRα expression in mesenchymal cells undergoing normal and abnormal proliferation, and is a potential target for new treatments of lung fibrosis.
Introduction
The development and maintenance of adult and embryonic tissues requires fine control by cell surface receptors and extracellular ligands. Disruption of receptor-mediated signaling underlying these control processes is associated with developmental dysplasia, tissue degeneration, and tumorigenesis. Mesenchymal cells, which provide structural support for various types of tissues, can proliferate in response to extracellular stimuli, and aberrant growth of these cells causes pathogenetic fibrosis in adult tissues (1). PDGF and its receptor, PDGFRα, play a crucial role in the proliferation of mesenchymal cells (2). PDGFRα is widely expressed in mesenchyme during organogenesis and is critical to development of the palate (3), hair follicles (4), and lung tissues (5). It is also a marker of paraxial mesoderm cells at gastrulation, which is a germ layer committed to differentiate into bone, cartilage, and skeletal muscle (6). Gene disruption of Pdgfrα in mice results in abnormal patterning of somites, which are derived from paraxial mesoderm (7), and pulmonary hypoplasia (8). These findings indicate that PDGFRα signaling is required for both morphogenesis and organogenesis in mouse development. A knock-out mouse study showed that mesenchymal cell numbers in the intestine and testis markedly decreased in PDGFRα-null embryos (9, 10). In adult human tissues, widespread expression of PDGFRα is also observed in abnormal fibrotic and sclerosing processes associated with inflammation, during which expression of PDGFRα is induced by extracellular factors such as transforming growth factor β, interleukin-1α, basic fibroblast growth factor (bFGF), tumor necrosis factor α, and lipopolysaccharide (11). Taken together, these findings support the notion that PDGF/PDGFRα signaling is important for the normal development of mesenchymal tissues, but it also underlies the pathogenetic events triggered by abnormal mesenchyme proliferation. However, the mechanisms controlling PDGFRα expression in normal and abnormal growth of mesenchyme remain unclear. This study aimed to elucidate the role of PDGFRα expression and the regulatory mechanism governing PDGFRα expression in normal development and related diseases. We used an in vitro ES cell culture system using PDGFRα as a marker to identify progenitors of paraxial mesoderm (12–14). The advantage of this system is that gene expression in differentiating ES cells mimics that observed in embryonic development (15–17), enabling us to monitor the intermediate stages of embryonic development and differentiation in vitro (18, 19). Another advantage of this system is that cell surface markers such as PDGFRα can be used to purify cells of a specific lineage for analysis (20). In a previous study, we compared the gene expression profile of ES cells with those of ES cell-derived PDGFRα+ intermediate populations, and identified ARID3b, which is a transcription factor with an unknown function. We also showed that ARID3b plays an essential role in both mesenchymal development and tumorigenesis (21, 22). The expression pattern of ARID3b in various cells is very similar to that of PDGFRα. However, ARID3b do not regulate PDGFRα expression, implying that there are other regulatory factors that control proliferation of PDGFRα+ mesenchymal cells. For the sake of seeking those factors, we have conducted a screening for genes dominantly expressed in PDGFRα+ positive cell population in in vitro ES cell culture.
In the current study, we have identified a novel transcription factor, Phf14, which controls the proliferation of mesenchymal cells by regulating PDGFRα expression. Phf14 was annotated as a plant homology domain (PHD)2 finger transcription factor based on the amino acid sequence, although molecular function of the Phf14 protein has not been reported previously. PHD finger transcription factors are generally thought to be concerned with transcriptional regulation by interacting with modified histones (23), and dysregulation of several genes encoding PHD finger transcription factors, such as ING1 and RBP2, has been found in human cancer diseases suggesting these factors have an important role for cancer cell proliferation (24). However, it has not been studied whether PHD finger transcription factors regulate proliferation of mesenchymal cells in embryonic development.
Phf14-null mice died just after birth due to respiratory failure. Histological analyses revealed an abnormally hypertrophic pulmonary wall, indicating interstitial proliferation in alveolar tissues. Phf14-null mesenchymal fibroblasts show increased PDGFRα expression, which resulted in enhanced mesenchymal proliferation. Based on these results, we used an antibody against PDGFRα to successfully treat mouse lung fibrosis. We showed that Phf14 represses PDGFRα expression in mesenchymal cells undergoing normal and abnormal proliferation, and thus has potential as a target for the treatment of lung fibrosis.
EXPERIMENTAL PROCEDURES
Cell Culture
Murine CCE and TT2 ES cells were maintained as described previously (12, 21). Primary mouse embryonic fibroblasts (MEF) from each genotype were isolated from E14.5 embryos and cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS). Bone marrow mesenchymal cells were isolated from femurs and tibias of E18.5 embryos and cultured in α-minimum Eagle's medium (Invitrogen) supplemented with 10% FBS and 50 μm 2-mercaptoethanol as described previously (25). Cells were maintained in a humidified 5% CO2 atmosphere.
In Vitro ES Cell Differentiation and cDNA Subtraction
In vitro ES cell differentiation was performed as described previously (12). Briefly, 8 × 104 cells of CCE ES cells were seeded on collagen IV-coated 100-mm dish with α-minimum Eagle's medium (Invitrogen) supplemented with 10% FBS and 50 μm 2-mercaptoethanol. Four days later, the cells were harvested, and the total RNA of FACS-purified PSP or the VEGFR2 single positive cell population was isolated using Sepasol reagent (Nacalai tesque) according to the manufacturer's protocol. Purified poly(A)+ RNA was subjected to cDNA subtraction by using the PCR-Select cDNA Subtraction kit (Clontech Laboratories).
Phf14 Gene Targeting and Animal Studies
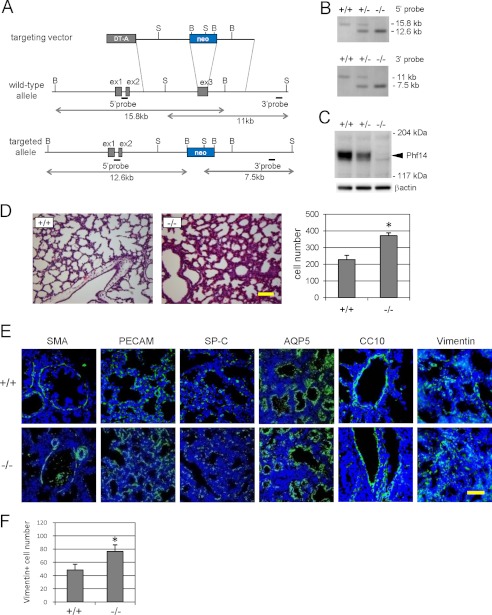
The 5′ and 3′ arms of the targeting construct homologous to Phf14 sequences were isolated from the B6 mouse strain BAC clone. Two correctly targeted clones were confirmed by Southern blot analysis with a 5′ upstream and a 3′ downstream genomic probe. BamHI-digested genomic DNA generated the wild-type 15.8-kb and targeted 12.6-kb fragments. SphI-digested genomic DNA generated the wild-type 11-kb and targeted 7.5-kb fragments (Fig. 2A). Targeted ES clones were microinjected into C57BL/6 blastocysts to generate chimeric mice and transfer the Phf14-neo allele to the germline. All mice strains were backcrossed into the C57BL/6 background. Phf14+/− (heterozygous) mice have been backcrossed more than 10 generations onto C57BL/6 and each of the experiments described herein were performed with mice that had been backcrossed at least 5 generations. For timed matings, all comparisons were made between siblings of the desired genotype, and noon of the vaginal plug day was considered E0.5.
FIGURE 2.
Phf14-null mice exhibit mesenchymal cell hyperplasia of embryonic lungs. A, strategy for producing Phf14-null mice. The third exon was replaced with a PGK-neomycin cassette. The flanking genomic probes used for Southern blot hybridization are indicated as bars. B, BamHI; S, SphI. B, Southern blot analysis of E18.5 embryos. Genomic DNA was digested with BamHI and SphI, and hybridized with 5′ and 3′ probes, respectively. Fragments obtained from wild-type and targeted alleles are indicated. C, immunoblot analysis confirmed the defective Phf14 expression in knock-out mice. Endogenous Phf14 protein (arrowhead) was observed in Phf14+/+ and Phf14+/− embryos, but not in Phf14−/− embryo. D, narrow airspace and hypertrophic pulmonary wall in lung of Phf14-null embryos. The sections of Phf14+/+ and Phf14−/− lungs of E18.5 embryos were stained with hematoxylin and eosin. The number of cells in a 200-μm square were calculated and the results are shown in the bar graph on the right. Scale bar, 100 μm. *, p < 0.05 versus Phf14+/+ (n = 3). E, immunofluorescence staining of E18.5 lung sections of Phf14+/+ and Phf14−/− mice for cell-lineage markers. Green fluorescence represents a positive immunoreactive signal for each marker protein as indicated at the top of the panels. Nuclei were counterstained with Hoechst 33258. Scale bar, 50 μm. F, increased vimentin+ cells in the lung of E18.5 Phf14−/− embryos, based on the number of vimentin+ cells in a 200-μm square. *, p < 0.05 versus Phf14+/+ (n = 3).
Polymerase Chain Reaction Analysis
RT-PCR and quantitative RT-PCR were performed as described previously (12, 26). Briefly, cDNA was synthesized from 5 μg of total RNA with Superscript III (Invitrogen) and random hexamers according to the manufacturer's protocol. Ten μl of cDNA (total, 50 ng) was mixed with a reaction buffer consisting of 2.5 μl of primer mix (2 mm each) and 12.5 μl of Thunderbird SYBR Green Reaction Mix (TOYOBO). Quantitative RT-PCR was performed using the Step One (Applied Biosystems) according to the manufacturer's instructions. The data were normalized to GAPDH expression and represented as mean ± S.D. from triplicate assays. The primers used for the analyses were listed in supplemental Table S1.
Gene Transfection and Cell Growth Assay with Various Treatments
Production and infection of retrovirus was done as described previously (27). For the analysis of MEF proliferation, 1 × 105 cells of MEFs were seeded into a 6-well plate and transferred every 3 days. The fold of cell growth was calculated by dividing the number of harvested MEFs with the number of inoculated. For the analysis of MEF proliferation with various growth factors, 1 × 105 cells MEFs were seeded into a 6-well plate and incubated for 12 h to allow the cells to attach to the plate, and the medium was changed to DMEM supplemented with 0.1% FBS. After a 24-h incubation, the medium was changed to DMEM supplemented with 0.1% FBS and each growth factor and/or APA5 monoclonal antibody at concentrations and times as indicated in figure legends. The proliferation ratio was represented as the ratio of cell number at each indicated time to 0 h treatment with growth factors. Growth factors, PDGF-BB, bFGF, and EGF were purchased from R&D Systems. For the analysis of MEF proliferation with MAPK inhibitors, 1 × 105 cells MEFs were seeded into a 6-well plate and incubated for 12 h to allow the cells to attach to the plate, and the medium was changed to DMEM supplemented with 10% FBS and each inhibitor at concentrations and times as indicated in figure legends. The proliferation ratio was represented as the ratio of cell number at each indicated time to 0 h treatment with inhibitors. All values were expressed as fold-change relative to the initial value, and are represented as mean ± S.D. from three independent cell lines of each genotype.
Construction of Plasmids
Phf14 cDNA fragments used in this study were amplified by RT-PCR from MEF total RNA. The full-length coding region of Phf14 was inserted into EcoRI/XhoI sites of pMSCV-IRES-EGFP (28), a retroviral vector that expresses full-length Phf14 and EGFP by using an internal ribosome entry site under the Murine Stem Cell Virus (MSCV) promoter. A fragment encoding the N-terminal domain of Phf14 (amino acids 1–227) into the pGEX-4T vector (Amersham Biosciences) to be expressed as GST-fused Phf14. PDGFRα genome DNA fragments used for the reporter assay were amplified by PCR from the BAC clone and inserted into pGL3-basic (Promega). For the reporter analysis of the promoter/enhancer activity, an enhancer fragment containing the 3′ downstream region from the PDGFRα transcription start site (from +76 to +440) was amplified by PCR, and inserted into BamHI/SalI site of pGL3-basic.
Immunoblot Analysis
For immunoblots, MEF cells or lung tissues were lysed with lysis buffer (50 mm Tris-HCl, pH 8.0, 2 mm EDTA, 250 mm NaCl, 0.1% Nonidet P-40, 10% glycerol), sonicated, and the supernatant was collected by centrifugation at 15,000 × g for 10 min at 4 °C. Ten μg of each supernatant was separated by SDS-PAGE, and the proteins were transferred to a PVDF membrane. The membrane was incubated with each antiserum, and the immunoreactive bands were visualized with Enhanced Chemiluminescence detection (PerkinElmer Life Sciences). The intensity of the immunoblotted bands were quantified using National Institutes of Health ImageJ software. All values were expressed as fold-change relative to control, and are represented as mean ± S.D. from three independent cell lines of each genotype. The antibodies used for the experiments are listed in supplemental Table S2.
Knockdown Assay by shRNA
A shRNA used for knockdown of Phf14 expression was designed and purchased as a plasmid of retroviral expression vector (HuSH-29) from OriGene Technologies, Inc. The sequence targeting Phf14 corresponds to coding regions 159 to 187 relative to the first nucleotide of the start codon. Scrambled shRNA was used as a control. Production and infection of retrovirus was done as described previously.
Promoter Analysis with Luciferase Reporter
For the luciferase assay, cells were plated on 24-well plates at a density of 4 × 104 cells/well and allowed to recover for 24 h; then, they were transfected with 10 ng of pRL-TK internal control plasmid, 250 ng of pGL3-basic-based reporter plasmid, and 250 ng of pMSCV-IRES-EGFP-based plasmid using FuGENE 6 transfection reagent (Roche Applied Sciences) as described by the manufacturer's protocol. After 48 h of incubation, the luciferase activities were assayed using the Dual Luciferase reporter assay system (Promega). The luminescence was quantified in a Lumat LB9506 luminometer (Berthold). Firefly luciferase activity was normalized for transfection efficiency based on Renilla luciferase activity. Values were expressed as fold-change relative to control, and are represented as mean ± S.D. from triplicate assays.
Antibody Generation
Polyclonal rabbit antibodies against Phf14 protein were prepared using GST-Phf14 (amino acid 1–227) fusion protein. The fusion protein was produced in Escherichia coli DH5α and purified by glutathione-Sepharose, thrombin digestion, and SDS-polyacrylamide gel electrophoresis. The purified Phf14 protein was inoculated into rabbits (Japanese white species) together with adjuvant. Anti-PDGFRα monoclonal antibody, APA5, was prepared as described previously (29).
Histological Analysis with Immunofluorescence Staining
Freshly dissected embryonic mouse lung tissues (E18.5) were fixed in paraformaldehyde and the tissues were sliced into 6-μm paraffin sections or 5-μm cryosections. The paraffin sections were used for hematoxylin and eosin staining. The cryosections were used for immunofluorescent staining with antibodies listed in supplemental Table S2. Alexa Fluor-conjugated secondary antibodies (Molecular Probes) were used for primary antibody detection. For quantification of immunofluorescent staining with PECAM, SP-C, AQP5, and vimentin, the immunofluorescent area in a 200-μm square of each specimen was measured in pixel value by ImageJ software omitting nuclear staining and areas of pulmonary lumen, and expressed as a ratio divided by the pixel value of total measurement area. For quantification of immunofluorescent staining with smooth muscle actin and CC10, the immunofluorescent area (in pixel) was measured from a transverse section containing a bronchiole by ImageJ and normalized by the length (in pixel) of the bronchial muscle layer (for smooth muscle actin) or surface epithelium (for CC10). The number of vimentin+ cells in Fig. 2F was calculated by counting immunohistochemically positive cells in a 200-μm square area. All values were represented as mean ± S.D. of three areas for each genotype or treatment.
Bleomycin-induced Pulmonary Fibrosis and Antibody Treatment
Male 6-week-old C57BL/6J mice (Crea Japan, Japan) were injected intranasally with 50 μl of 15 mg/ml of bleomycin hydrochloride (Nippon Kayaku, Japan) solution dissolved in sterile saline under anesthesia. Control mice were injected with saline. In the experiment for inhibition of the PDGFRα signal pathway, 200 μg of APA5 monoclonal antibody or control rat IgG was administered intraperitoneally to each mouse every day from day 1 to 6. At day 7, the lung tissues near terminal bronchioles were freshly dissected. For immunofluorescent staining of cryosections, dissected lung tissues were mounted with embedding compound (OCT compound, Sakura Finetek USA) and snap frozen in liquid nitrogen for preparation of 5-μm cryosections, and prepared sections were processed for immunostaining as described above. Lysates of dissected lung tissues were prepared and analyzed by immunoblot analysis as described in a previous section. The alveolar airspace area was measured as follows. Hematoxylin and eosin-stained sections of the lung were viewed using a stereomicroscope system (Leica, Switzerland) without of knowledge of drug treatment, and bright-field images were transferred via a CCD camera to a computer system using DP controller software (Olympus, Japan). The cell number in Fig. 7E was calculated by counting nuclei in a 200-μm square area of hematoxylin and eosin-stained sections. Large airways and bronchi were excluded from the analysis. Alveolar airspace area was calculated as a sum of the white areas in an image by the computerized process using ImageJ software and expressed as a lung tissue to airspace ratio divided by the measured tissue area. The values are represented as mean ± S.D. from three areas for each genotype.
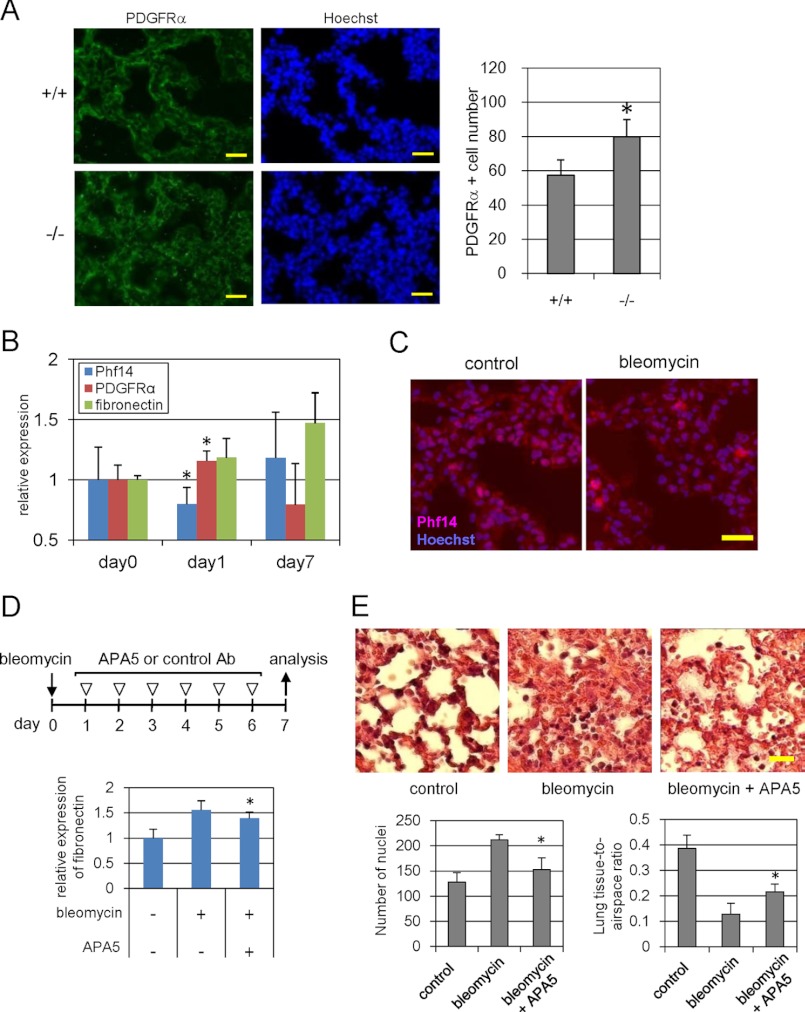
FIGURE 7.
Phf14 expression in bleomycin-induced pulmonary fibrosis. A, immunofluorescence for PDGFRα in E18.5 embryonic lungs. The number of PDGFRα+ cells significantly increased in E18.5 lung sections of Phf14−/− embryos. The number of PDGFRα+ cells in a 200-μm square of the immunofluorescence images is shown in the graph on the right. Nuclei were counterstained with Hoechst 33258. Scale bar, 20 μm. *, p < 0.05 versus Phf14+/+ (n = 3). B, expression of Phf14, PDGFRα, and fibronectin during bleomycin-induced lung fibrosis. After bleomycin treatment, the expressions of Phf14, PDGFRα, and fibronectin were measured on days 0, 1, and 7 by immunoblot analysis. The expression ratio of each protein was calculated relative to that on day 0 (no treatment). *, p < 0.05 versus day 0 (n = 3). C, immunofluorescence for Phf14 in bleomycin-treated lungs. The number of Phf14+ cells in the bleomycin-treated lung tissue decreased. Lung sections on day 1 after treatment were stained with the anti-Phf14 antibody. Nuclei were counterstained with Hoechst 33258. Scale bar, 20 μm. D, effect of anti-PDGFRα antibody administration on bleomycin-induced lung fibrosis. The expression of fibronectin, a fibroblastic marker, decreased after antibody treatment. After bleomycin or saline treatment, mice were treated with APA5, and anti-PDGFRα antibody that antagonizes PDGFRα signaling, or control IgG as indicated (upper panel). The expression of fibronectin in lung tissue was measured on day 7 by immunoblot analysis. The expression ratio was calculated relative to that of control (no treatment). *, p < 0.05 versus bleomycin treatment (n = 3). E, histological findings of lung tissues of D. The hypertrophy of alveolar walls that is strikingly observed in bleomycin-treated lung (center) was reversed by antibody treatment (right). Frozen sections of each lung tissue were also subjected to hematoxylin and eosin staining. Scale bar, 20 μm. The number of nuclei stained with hematoxylin in a 200-μm square was counted and expressed as mean ± S.D. in the graph on the left, below. The lung tissue-to-airspace ratio was calculated from the alveolar airspace area divided by the measured tissue area of a 200-μm square and expressed as mean ± S.D. in the graph on the right, below. *, p < 0.05 versus bleomycin treatment (n = 3).
FACS Analysis
The biotinylated rat monoclonal antibodies APA5 (anti-PDGFRα), APB5 (anti-PDGFRβ), and FITC conjugated-AVAS12 (anti-VEGFR-2) were purchased from eBioscience. Phycoerythrin-conjugated streptavidin (BD Biosciences) was used for detecting biotinylated antibodies. Cultured cells were harvested and collected using 0.05% trypsin-EDTA (Invitrogen). MEF suspensions were stained with APA5 or APB5 and analyzed by FACSCanto. Differentiated CCE ES cells were stained with APA5 and AVAS12, and a separate PSP population by FACS Aria II (BD Biosciences) as previously described (12). The positive ratio and mean value were calculated by using FlowJo software (Tree Star). Values are represented as mean ± S.D. from three independent cell lines of each genotype.
Chromatin Immunoprecipitation (ChIP) Analysis
ChIP assay was performed as described previously (30). In brief, MEFs were cross-linked with formaldehyde, and then the lysate was incubated with anti-Phf14 polyclonal antibodies or preimmune rabbit serum. Purified DNA pellets were used as templates for PCR amplification. The intensity of amplified DNA fragments was quantified using ImageJ software. All values are expressed as fold-change relative to input control, and are represented as mean ± S.D. from triplicate assays. The primers used for the PCR amplification were listed in supplemental Table S1.
Statistics
All data are presented as mean ± S.D. of three separate experiments (n = 3). The Student's t test was used to evaluate the statistical significance of difference and p value less than 0.05 was considered to be significant.
RESULTS
Phf14 Is a Novel Regulatory Gene of Mouse Embryonic Fibroblast Proliferation
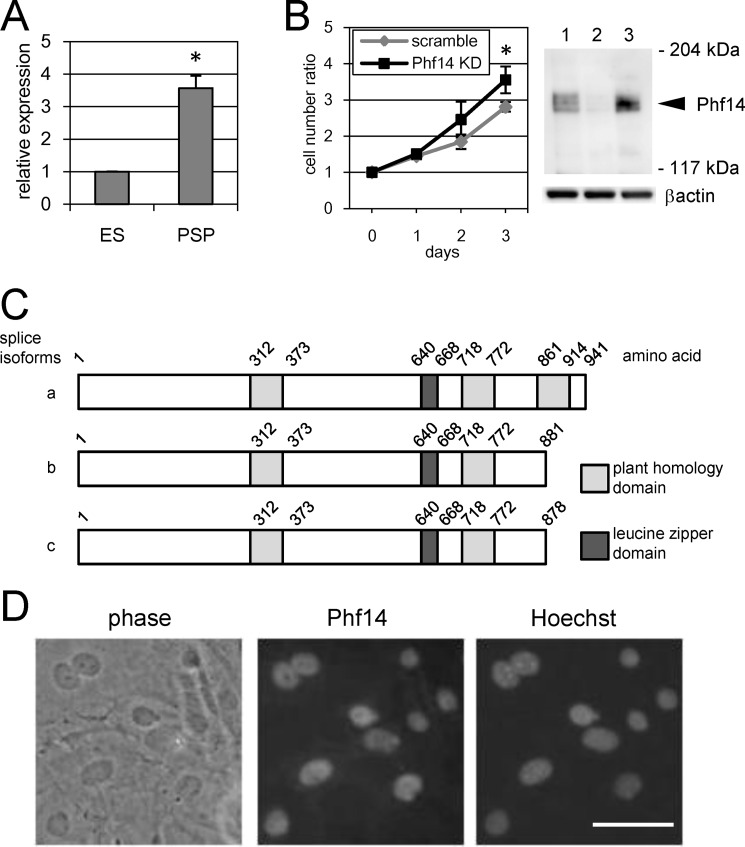
ES cells were differentiated under culture conditions selective for PSP and a vascular endothelial growth factor receptor 2 (VEGFR2) single positive cell population, which correspond to paraxial and lateral mesoderm cells, respectively (supplemental Fig. S1A) (12, 14). During this process, a remarkable change in gene expression is expected to underlie the differentiation and proliferation of ES cells into mesoderm cells. We carried out cDNA subtraction between PSP and VEGFR2 single positive cell population cells to detect any differentially expressed genes, identifying several genes specifically expressed in PSP (supplemental Fig. S1, B and C). Because PSP can generate mesenchymal cell lineages such as osteocytes and chondrocytes, we focused on those genes with high expression in PSP and identified several where expression was significantly different to that in ES control cells. To identify candidate genes with a role in the proliferation of mesenchymal cells, we investigated the effect of shRNA-induced knockdown of their expression on the proliferation of MEF, as a model of mesoderm-derived mesenchymal cells. Using this approach, we identified a novel gene, Phf14, which showed over 3-fold higher expression in PSP cells than in ES cells (Fig. 1A). The knockdown of Phf14 gene expression significantly increased (∼1.2-fold) MEF proliferation after 3 days (Fig. 1B). To confirm the knockdown of endogenous Phf14 protein, we raised an antiserum against the recombinant amino-terminal portion of mouse Phf14. The antiserum specifically detected bands of ∼160 kDa in MEF whole cell lysate that were not detectable with the knockdown of Phf14, indicating that these bands represent endogenous Phf14 proteins and that shRNA-induced knockdown of Phf14 was effective (Fig. 1B). The protein structure of Phf14 implicated it as a member of the PHD protein family because of the presence of tandem PHD structures, and we have identified three splice isoforms in the process of cDNA cloning (Fig. 1C). Quantitative RT-PCR analysis of three splice isoforms revealed that “type a” isoform was expressed over 100-fold higher than other isoforms in ES cells, PSP cells, and MEF cells (supplemental Fig. S2). Therefore, we used Phf14 type a for the following experiments as a major isoform of Phf14. In addition, Phf14 has a leucine-zipper (LZ) domain that is frequently observed in transcriptional factors involved in dimerization with other proteins carrying an LZ domain. According to several reports, most proteins of the PHD family are located in the nucleus and function in the regulation of gene expression (24). To confirm the notion of Phf14 as a PHD family protein, we examined the intercellular localization of the endogenous Phf14 protein by immunofluorescent staining in MEF cells and showed that it was localized in the nucleus (Fig. 1D). This is the first report describing a possible function of Phf14. Taken together, our findings suggested that Phf14 is a member of the PHD family of proteins and has an important role in regulating the expression of genes controlling MEF proliferation.
FIGURE 1.
Identification of Phf14 as a regulatory factor of MEF proliferation. A, elevation of Phf14 expression during ES cell differentiation. Expression of Phf14 in ES cells and ES cell-derived PDGFRα single positive population (PSP) cells was analyzed by quantitative RT-PCR (qRT-PCR). *, p < 0.05 versus ES cells (n = 3). B, suppression of Phf14 expression enhances MEF proliferation. Growth activity represents the relative ratio of cell number at the indicated days to that at day 0. Suppression of Phf14 expression was confirmed by immunoblot analysis with antiserum against Phf14 (right panel). MEF were transduced with either a scrambled control shRNA (lane 1) or Phf14 shRNA (lane 2). MEF with no transfectant was in lane 3. The arrowhead indicates the predicted size of endogenous Phf14. *, p < 0.05 versus scramble (n = 3). C, schematic diagram of the Phf14 protein structure. Alternative splice patterns produce three variants of the Phf14 protein. Numbers indicate the amino acid positions from the N terminus. Gray and dark gray boxes indicate the PHD and LZ domain, respectively. D, representative immunostaining of MEF showing Phf14 protein localized in the nuclei. Phase, a phase-contrast view of MEF. Hoechst, nuclei were counterstained with Hoechst 33258. Scale bar, 20 μm.
Phf14-null Mice Have a Perinatal Lethal Phenotype and Show Interstitial Hyperplasia in the Developing Lung
Phf14 expression was detected in all stages of embryogenesis (supplemental Fig. S3A), and was ubiquitously expressed in adult tissues, with increased expression in the intestine, colon, and lung (supplemental Fig. S3B). To elucidate the role of Phf14 during embryonic development, we generated Phf14-null mice according to the knock-out strategy described in Fig. 2, A and B. Immunoblot analysis confirmed the loss of Phf14 protein expression in knock-out mice (Fig. 2C). Phf14+/− mice were healthy and fertile, whereas Phf14−/− mice were perinatal lethal (supplemental Table S3). Although Phf14−/− embryos developed and survived up to E18.5 at the expected Mendelian ratio, Phf14−/− neonates died within 1 day after birth due to respiratory failure. Almost all Phf14−/− neonates exhibited cyanotic and labored breathing (supplemental Movie S1), indicating that Phf14−/− mice have respiratory impairment. The pattern of sternal ossification was partially impaired in Phf14-null mutants, although other skeletal structures were normal (supplemental Fig. S4, A and B). Histological analysis of lung tissue from E18.5 embryos revealed hypertrophy of the alveolar wall in Phf14−/− mice, but not in Phf14+/+ mice (Fig. 2D). Consistent with hypertrophy of the alveolar wall, the cell density in lung sections was significantly higher in Phf14−/− mice than in Phf14+/+ mice (Fig. 2D). These results implicated impaired alveolar formation as a likely cause of respiratory failure in the Phf14−/− neonate mice. To determine which type of cells had proliferated excessively in the lung tissue of Phf14−/− mice, we examined the expression of lung cell lineages markers in Phf14+/+ and Phf14−/− E18.5 embryo lungs (Fig. 2E). All types of alveolar and bronchial components were identified by immunofluorescent staining of the lung sections. Quantification of the immunofluorescent area for each marker protein revealed that the vimentin-positive area was significantly increased in Phf14−/− compared with Phf14+/+ (Fig. 2E and Table 1). In support of this observation, the number of vimentin+ mesenchymal cells was significantly higher in the thickened alveolar wall of Phf14−/− embryo lung sections than in the wild-type (Fig. 2, E and F). In contrast, there were no notable differences between Phf14+/+ and Phf14−/− embryo lungs in the tissue localization of CC10+ bronchial epithelial cells, SMA+ bronchial smooth muscle cells, PECAM+ vascular endothelial cells, AQP5+ type I alveolar epithelial cells, or SP-C+ type II alveolar epithelial cells (Table 1). These results indicated that the observed alveolar wall hypertrophy in Phf14−/− mice was due to excessive proliferation of mesenchymal cells.
TABLE 1.
Quantitative data of immunofluorescence analysis of wild-type and Phf14-null E18.5 lung sections
| Marker protein | Data of immunofluorescent area (mean ± S.D.) |
p value | |
|---|---|---|---|
| Phf14+/+ | Phf14−/− | ||
| SMA | 4.19 ± 0.72 | 4.12 ± 0.81 | 0.108 |
| PECAM | 0.112 ± 0.012 | 0.108 ± 0.007 | 0.264 |
| SP-C | 0.066 ± 0.020 | 0.057 ± 0.011 | 0.122 |
| AQP5 | 0.173 ± 0.022 | 0.189 ± 0.052 | 0.209 |
| CC10 | 5.15 ± 1.52 | 5.01 ± 0.23 | 0.448 |
| Vimentin | 0.158 ± 0.023 | 0.193 ± 0.031 | 0.045 |
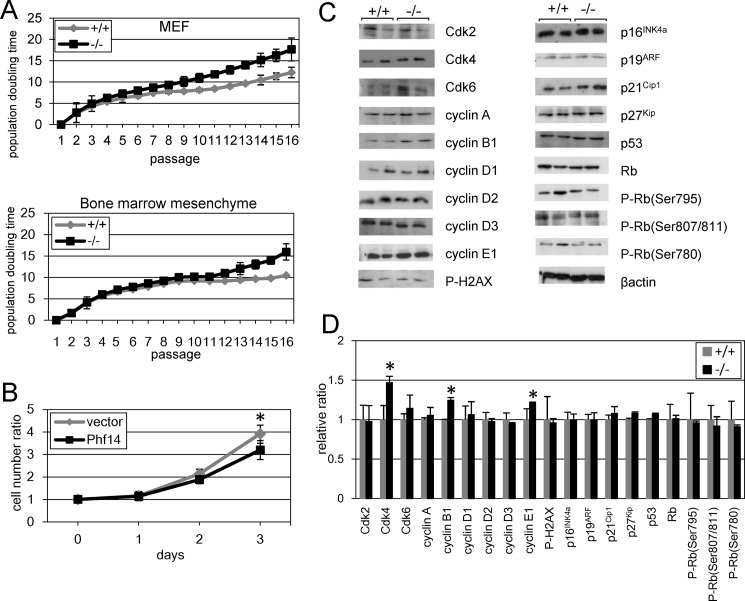
Lack of Phf14 Expression Enhances Proliferation of Mesenchymal Cells
We next examined the cell proliferation rate of mesenchymal cells derived from Phf14−/− embryos by measuring doubling times. MEFs and bone marrow-derived mesenchymal cells were prepared from Phf14+/+ and Phf14−/− embryos (Fig. 3A). Phf14−/− MEFs grew faster than Phf14+/+ MEFs from passage 3 and this higher growth rate was sustained for the following passages. Similarly, the proliferation rate of bone marrow-derived mesenchymal cells was higher than that of wild-type cells (Fig. 3A). Overexpression of Phf14 in Phf14−/− MEFs rescued the proliferation impairment of these cells (Fig. 3B). In contrast, overexpression of Phf14 in Phf14+/+ MEFs was not affected with the proliferation rate (supplemental Fig. S5A), suggesting the endogenous expression levels of Phf14 are enough for normal proliferation in MEF. These results indicated that lack of Phf14 expression enhances the proliferation rate of embryo-derived mesenchymal cells. We next investigated the molecular mechanism underlying the enhanced proliferation rate of Phf14−/− mesenchymal cells by analyzing the expression of key cell cycle regulatory molecules (Fig. 3, C and D). Immunoblot analyses revealed that expression of Cdk4, cyclin B1, and cyclin E1 was higher in Phf14−/− MEFs than in wild-type cells, whereas expression of the cyclin-dependent kinase inhibitors, p16INK4a, p19ARF, p21Cip1, and p27Kip, remained unchanged. These results suggested that lack of Phf14 expression increases the rate of cell cycle progression.
FIGURE 3.
Enhanced proliferation of MEF derived from Phf14-null mice. A, high proliferation rate of Phf14−/− mesenchymal cells. MEF and bone marrow mesenchymal cells were isolated from Phf14+/+ and Phf14−/− embryos and cultured continuously. The population doubling time was calculated from the cell number obtained at each passage. The average and standard deviations of three independent cultures are shown. B, normalized growth rate of Phf14−/− MEF subjected to overexpression of Phf14. Phf14−/− MEFs were infected with retroviral vector carrying Phf14 or control vector. The cell growth ratio at each day was determined. The ratio was significantly decreased in Phf14-expressing Phf14−/− MEF. *, p < 0.05 versus vector only (n = 3). C, the expression of several cell cycle regulatory factors was increased in Phf14−/− MEF. Whole cell lysates of Phf14+/+ and Phf14−/− MEF were prepared from cells at passage 6. The expression of cell cycle regulatory factors was examined by immunoblot analysis with antibodies against each factor. D, the intensity of each band was quantified, and the relative ratio of expression in Phf14−/− MEF to that in Phf14+/+ MEF was determined from three independent cell lines. The values are presented as the averages with standard deviation. The expressions of Cdk4, cyclin B1, and cyclin E1 were significantly increased in Phf14−/− MEF. *, p < 0.05 versus Phf14+/+ (n = 3).
Up-regulated PDGFRα Expression Is Associated with Enhanced Proliferation of Phf14−/− MEF
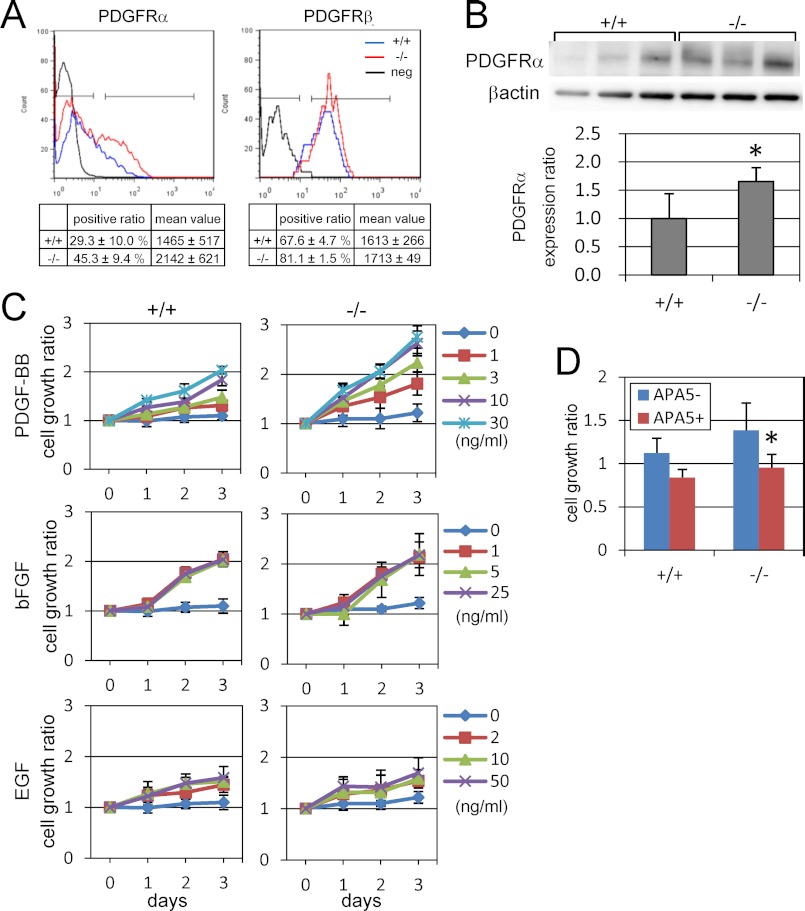
PDGF controls MEF proliferation via its receptors, PDGFRα and PDGFRβ, which are present on the cell surface as dimers. We therefore investigated whether PDGF signaling is involved in the enhanced growth of Phf14−/− mesenchymal cells by examining the abundance of these receptors. The proportion of PDGFRα+ cells was higher in Phf14−/− MEF than in Phf14+/+ MEF, indicating an increased number of PDGFRα+ cells in Phf14−/− MEF (Fig. 4A). This increase was confirmed by immunoblot analysis (Fig. 4B). In contrast, there was no notable difference in PDGFRβ expression between Phf14+/+ and Phf14−/− MEF (Fig. 4A and supplemental Fig. S6). We also found no differences in the expression levels of other receptors known to stimulate mesenchymal cell growth including epidermal growth factor receptor, and fibroblastic growth factor receptors 1, 2, and 3 (FGFR1, -2, and -3) (supplemental Fig. S6). It is possible that elevation of the PDGFRα level resulting from the increased number of PDGFRα+ cells in the Phf14−/− MEF population caused the enhanced cell proliferation rate. To test this hypothesis, we determined the cell proliferation rate of Phf14+/+ and Phf14−/− MEFs under serum-starved culture conditions supplemented with various concentrations of PDGF. In the presence of PDGF, Phf14−/− MEF proliferated at a higher rate than Phf14+/+ MEF, and in a dose-dependent manner (Fig. 4C). Even at low concentrations of PDGF (∼1 ng/ml), the growth rate of Phf14−/− MEF was markedly augmented compared to wild-type cells. In contrast, there were no significant differences between these cells in the presence of other growth factors such as EGF and bFGF (Fig. 4C). Blocking PDGF signaling by using APA5, a monoclonal antibody against PDGFRα, confirmed that the increased growth rate of Phf14−/− MEF is dependent on PDGF signaling in the presence of serum-containing medium (Fig. 4D).
FIGURE 4.
PDGFRα expression is enhanced in Phf14-null MEFs. A, expression of PDGFRα and PDGFRβ in Phf14+/+ and Phf14−/− MEF stained with fluorescence-conjugated anti-PDGFRα or PDGFRβ antibodies and analyzed by flow cytometry. The proportion and the average and S.D. of three independent cultures are shown in the table. The percentage of PDGFRα-positive cells in Phf14−/− MEF was higher than that in Phf14+/+ MEF. B, immunoblot analysis of PDGFRα expression in Phf14+/+ and Phf14−/− MEF. The data were normalized for β-actin expression. *, p < 0.05 versus Phf14+/+ MEF (n = 3). C, elevation of the sensitivity for PDGF signals in Phf14−/− MEF. Enhanced growth of Phf14−/− MEF was observed in cultures containing PDGF-BB (upper panels), but not with other factors including bFGF and EGF (middle and lower panels). Growth was represented as the ratio of cell counts to the initial seeded cell number. The average and standard deviations of three independent cultures are shown. D, inhibition of Phf14+/+ and Phf14−/− MEF growth by treatment with APA5, an anti-PDGFRα antibody. Phf14+/+ and Phf14−/− MEFs were grown in medium with and without APA5, which antagonizes PDGF signaling. The growth activity of MEFs is represented as the ratio of cell counts to the initial seeded cell number. *, p < 0.05 versus without APA5 (n = 3).
Growth Advantage of Phf14−/− Mesenchymal Cells Is Dependent on the Activation of ERK1/2 MAPK Kinases
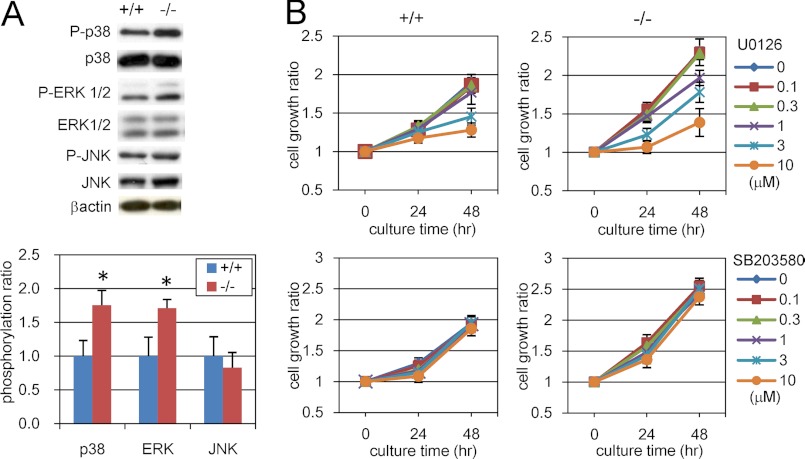
Next, we searched for downstream molecules of PDGF signaling in Phf14+/+ and Phf14−/− MEFs. PDGF/PDGFR signaling is mediated by MAPKs such as extracellular signal-regulated kinase (ERK) (31, 32). Accordingly, we observed enhanced phosphorylation of MAPKs, ERK, and p38, in Phf14−/− compared with Phf14+/+ MEF by immunoblot analysis (Fig. 5A). These results suggested that the increased expression of PDGFRα in Phf14−/− MEF leads to enhanced phosphorylation of ERK and p38 and in turn, a higher rate of proliferation compared with Phf14+/+ MEF. To test this possibility, we inhibited the activation of ERK and p38 by specific chemical inhibitors and calculated the growth rate of MEF. Treatment with U0126, an inhibitor of the MAPK/ERK signaling pathway, suppresses the proliferation of Phf14−/− MEF in a dose-dependent manner (Fig. 5B). In contrast, inhibition of p38 kinase activity with SB203580 had no effect on the rate of proliferation of Phf14+/+ or Phf14−/− MEF, suggesting that activation of p38 is not involved in growth, but has other functions in MEFs. The inhibitory activity of the compounds was confirmed by immunoblot analysis of the phosphorylation status of ERK1/2 and MAPKAPK-2, a downstream effector of p38 (supplemental Fig. S7). Together with previous evidence, these results indicated that the enhanced proliferation of Phf14−/− MEF was due to activation of PDGF/PDGFR signaling via activation of ERK1/2.
FIGURE 5.
Proliferation of Phf14-null MEF is dependent on ERK activation. A, phosphorylation of MAPKs in Phf14+/+ and Phf14−/− MEF. For each MAPK, the expression phosphorylation status was analyzed by immunoblot analysis (left panel). The data were normalized for the total expression of each MAPK (right panel). Phf14−/− MEF showed higher levels of p38 and ERK phosphorylation than Phf14+/+ MEF. *, p < 0.05 versus Phf14+/+ (n = 3). B, inhibition of ERK activity, but not p38 activity, can suppress the growth of Phf14−/− MEF treated with MEK/ERK and p38 kinase inhibitors, U0126 and SB203580, respectively. Growth is represented as the ratio of cell counts to the initial seeded cell number. The average and S.D. of three independent cultures are shown.
Phf14 Acts as a Transcription Factor to Control PDGFRα Expression
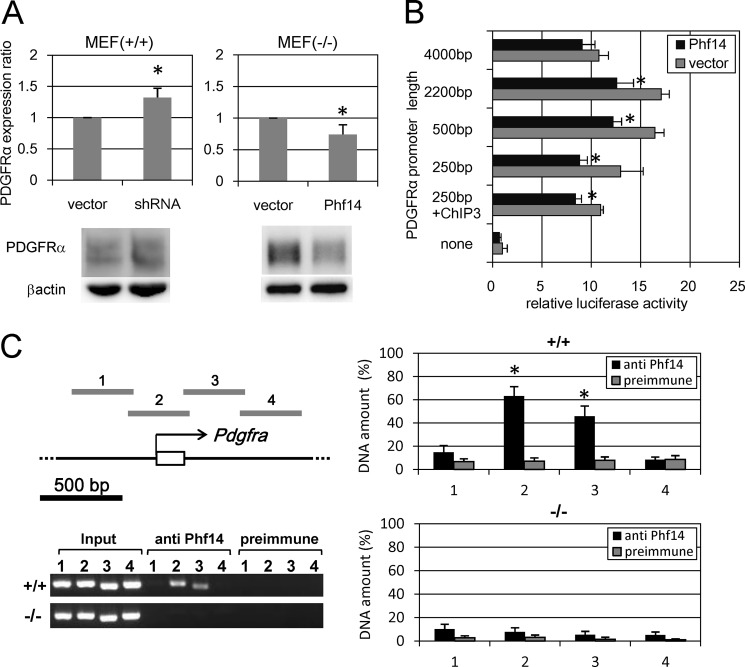
Because a lack of Phf14 increases the expression of PDGFRα in MEFs, Phf14 might regulate PDGFRα expression. To test this idea, we abolished Phf14 expression using RNA interference and exogenous overexpression. As expected, knockdown of endogenous Phf14 in Phf14+/+ MEF increased PDGFRα expression, whereas overexpression of Phf14 in Phf14−/− MEF decreased PDGFRα expression (Fig. 6A). Conversely, overexpression of Phf14 in Phf14+/+ MEFs was not affected with the expression of PDGFRα (supplemental Fig. S5B), suggesting that a certain expression level of Phf14 is enough for the regulation of PDGFRα expression in MEF.
FIGURE 6.
Phf14 directly represses the transcription of PDGFRα. A, regulation of PDGFRα expression by altering Phf14 expression. PDGFRα expression was measured by immunoblot analysis. In Phf14+/+ MEF, suppression of Phf14 expression by shRNA significantly enhanced the PDGFRα expression (left panel). In contrast, overexpression of Phf14 in Phf14−/− MEFs significantly suppressed PDGFRα expression (right panel). The data were normalized for β-actin expression. *, p < 0.05 versus vector only (n = 3). B, transcriptional activity of the PDGFRα promoter region in Phf14−/− MEF transfected with luciferase reporter vectors containing fragments 5′-upstream of the PDGFRα transcription start site as indicated together with the Phf14 expression vector (Phf14) or control vector (vector). ChIP3 represents a fragment from +76 to +440 corresponding to the region presented as 3 in C. The transcriptional activity of a 250-bp upstream region of the PDGFRα promoter was significantly repressed by exogenous expression of Phf14. *, p < 0.05 versus vector only (n = 3). C, chromatin immunoprecipitation of the PDGFRα promoter region with anti-Phf14 serum. The chromatin complex prepared from Phf14+/+ and Phf14−/− MEF was subjected to immunoprecipitation with anti-Phf14 or a preimmune serum. The regions shown in the upper left panel of the schematic diagram were amplified from the immunoprecipitated DNA fragments and electrophoresed. The amount of amplified DNA was quantified by densitometry and expressed as density relative to that of input control (right panels). Phf14 protein interacted with regions 2 and 3 of the PDGFRα promoter in Phf14+/+ MEF only. *, p < 0.05 versus preimmune serum (n = 3).
To explore the mechanism by which Phf14 affects PDGFRα expression, we examined whether Phf14 could act as a transcription factor using a luciferase reporter system to analyze function. We showed that the transcriptional activity of the upstream promoter region of PDGFRα in Phf14−/− MEF was partially repressed by exogenous expression of Phf14 (Fig. 6B), with a ∼250-bp region upstream of the transcription start site in PDGFRα implicated in the repressive activity of Phf14 (Fig. 6B). To confirm a direct association between Phf14 and this promoter region of PDGFRα, we performed ChIP assays on Phf14+/+ and Phf14−/− MEF using the anti-Phf14 antibody (Fig. 6C). Endogenous Phf14 protein bound to the proximal upstream region of the PDGFRα transcription start site as well as to the adjacent downstream region. This finding suggested the presence of multiple binding sites for Phf14 around the transcription start site or that Phf14 is a component of a transcription regulatory complex that recognizes different binding sites. It has been reported that the CCAAT/enhancer-binding protein could bind directly to several conserved binding elements in this ∼250-bp region, implying that this region may play an important role for transcriptional regulation of PDGFRα (33). To clarify whether Phf14 binds to any portion of this ∼250-bp region directly or not, we examined an in vitro DNA binding assay. However, no direct binding between in vitro translated Phf14 protein and oligonucleotides covering this ∼250-bp region was detected, suggesting Phf14 is associated with this region indirectly via other nuclear proteins that bind to this region directly (supplemental Fig. S8). We also tested the promoter/enhancer activity of the adjacent downstream region (from +76 to +440) identified by ChIP analysis and showed that the promoter activity was partially suppressed by Pfh14 overexpression in Phf14-null fibroblasts (250bp + ChIP3 in Fig. 6B). These results indicated that Phf14 acts as a transcription factor to suppress PDGFRα expression in mesenchymal cells.
Based on the LZ domain in Phf14, we speculated that Phf14 can interact with other LZ-containing transcription factors. However, we could not detect any interactions between the Phf14 protein and known LZ-containing transcription factors including Atf1, Jdp2, Fos, Jun, Creb, and Cebpa (supplemental Fig. S9).
PDGFRα Is a Target for the Treatment of Lung Fibrosis
We showed that Phf14 is expressed in adult lung at relatively high levels compared with other tissues (supplemental Fig. S3B). The splice isoform type a was the most highly expressed among the splice forms as observed in ES and MEF cells (supplemental Fig. S2). We also showed that Phf14-null mice die from respiratory failure immediately after birth, and that the lungs of these mice have hypertrophic enlargement of alveolar walls due to excess mesenchymal cells. We next examined the expression of PDGFRα in Phf14+/+ and Phf14−/− E18.5 embryo lungs to determine whether this hypertrophic alveolar phenotype is associated with increased PDGFRα expression (Fig. 7A). Compared with wild-type mice, the number of PDGFRα+ cells was significantly higher in Phf14−/− embryo lung sections. The phenotype of Phf14−/− lungs was similar to that seen in interstitial pneumonia and lung fibrosis, both of which are pathogenetic processes associated with mesenchymal hyperplasia and increased PDGFRα expression (34). To confirm the role of Phf14 in the hypertrophic process of drug-induced lung fibrosis, we utilized a mouse model of bleomycin-induced pulmonary fibrosis as previously described (35), and examined the role of Phf14 in these lungs by immunoblot analysis of PDGFRα and Phf14 expression. One day after bleomycin treatment, Phf14 expression was decreased by 30% and PDGFRα expression was increased by about 20% (Fig. 7B). This change in PDGFRα expression resembled that observed in Phf14-knockdown MEF (Fig. 6A). Quantitative RT-PCR analysis revealed that PDGFRα mRNA expression was increased in bleomycin-treated lung tissues (supplemental Fig. S10), indicating that transcription of the Pdgfrα gene was enhanced in response to this treatment. Consistent with the immunoblot analyses, immunostaining of lung sections with anti-Phf14 antibody revealed a decreased signal in the nuclei after bleomycin treatment (Fig. 7C). Although expression of Phf14 recovered to control levels, the progression of pulmonary fibrosis was confirmed by an increase in fibronectin expression 7 days after bleomycin treatment (Fig. 7B). Together with the increase in PDGFRα+ cells in Phf14-null lungs, these results suggested that dysregulation of Phf14 expression in the lung is related to the increased number of PDGFRα+ cells in bleomycin-induced lung fibrosis. It has been reported that both PDGF-A and PDGF-B mRNA levels were increased in bronchioalveolar lavage of the bleomycin-induced pulmonary fibrosis hamster model (36), and in alveolar macrophages obtained from human patients with idiopathic pulmonary fibrosis (37), indicating PDGF can act as a potent mitogen that drives the proliferation of fibroblasts in pulmonary fibrosis. In accordance with these reports, an increase for PDGF-A and PDGF-B mRNA was observed in bleomycin-treated lung tissues (supplemental Fig. S10), suggesting that the increase in ligands is sufficient to activate the PDGFRα-mediated signal pathway. The up-regulation of PDGFRα expression in drug-induced lung fibrosis and our finding that mesenchymal cell proliferation is due to PDGFRα up-regulation in Phf14-null mice highlight PDGFRα as a potential target for the treatment of lung fibrosis. To evaluate this possibility, we exposed the bleomycin-treated mice with APA5, an anti-PDGFRα monoclonal antibody known to inhibit the PDGFRα signaling pathway (29). We then evaluated the progression of pulmonary fibrosis by quantitatively analyzing the fibronectin expression and by histological analyses. Fig. 7D shows that up-regulation of fibronectin expression after bleomycin treatment was partially, but significantly, suppressed by APA5. Histological analyses of these lung tissues also revealed a reduction in the extent of fibrotic lesions in the alveolar wall following APA5 treatment (Fig. 7E). To assess this reduction quantitatively, we measured the alveolar airspace area and the number of nuclei for specimens for each treatment (Fig. 7E). As a result, the number of nuclei were significantly decreased in lung tissues treated with bleomycin and APA5 compared with those treated with bleomycin only. On the other hand, the alveolar airspace area was significantly increased. These results implicated that antagonizing PDGFRα signaling by antibodies relieved the progression of hyperplasia in bleomycin-treated lung tissues.
DISCUSSION
In this study, we identified a novel nuclear factor, Phf14, and its essential role in mesenchymal cell proliferation via the regulation of PDGFRα expression. We showed that the number of PDGFRα+ cells was higher among Phf14−/− MEF than wild-type cells, and that this increased responsiveness to PDGF increased the rate of cell proliferation.
PDGF signaling is initially mediated through its receptors, PDGFRα and PDGFRβ, to regulate cell proliferation and tissue organization in developmental processes such as angiogenesis, glomerulogenesis, alveologenesis, and hair morphogenesis (2). PDGF signaling controls both proliferation and differentiation in the development of mesodermal cells and their descendants, such as osteoblasts (38, 39). Transgenic mice overexpressing PDGF in lung epithelium show increased fetal lung mesenchymal cell density and perinatal lethality due to respiratory failure (40). A constitutively active form of PDGFRα expressed in knock-in mice also induced increased mesenchymal cell density in the fetal lung and perinatal lethality (41). Although these studies implicated PDGF and its receptor, PDGFRα, in the proliferation of lung mesenchymal cells, the molecular mechanism underlying PDGFRα expression and signaling in mesenchymal tissues remains unclear. This study has demonstrated that Phf14 that is newly isolated from ES cell culture negatively controls the cell proliferation of mesenchymal cells by suppressing PDGFRα expression. Phf14-null mouse mutants showed neonate lethality due to respiratory failure. The main pathological phenotype of these Phf14-null mutants was hyperplasia of the interstitial lung tissue, similar to that observed in other mutants with dysregulated PDGF signaling. We demonstrated that the interstitial hyperplasia was caused by an increased number of PDGFRα+ and vimentin+ mesenchymal cells. Thus, our results showed that Phf14 plays a role in the regulation of PDGFRα expression, the precise control of which is essential for lung mesenchymal cell proliferation during development.
The role of Phf14 in the regulation of PDGFRα expression suggests that PDGFRα and Phf14 interact directly in PDGFRα+ mesenchymal cells. Promoter-reporter and ChIP assays in the present study revealed binding of Phf14 to the proximal region of the PDGFRα transcription start site and the repressed transcription of PDGFRα in mesenchymal fibroblasts. Thus, Phf14 can act as a transcriptional factor to directly repress expression of PDGFRα by binding to the promoter. Although Phf14 has no conserved domains common to other transcription repressors, we expect that it functions in a complex by interacting with other nuclear factors via hydrophobic interactions through its LZ domain. The same domain is essential in ING2, a PHD-containing subunit of the Sin3 histone deacetylase complex, for it to function as a scaffold protein to mediate the interaction between p53 and p300, and for p53-dependent chromatin remodeling (42). We found some cis-elements in the Phf14-binding region of the PDGFRα gene, to which LZ-containing transcription factors such as C/EBP, CREB, c-Fos, and c-Jun could bind. However, we could not detect any direct associations between Phf14 and these factors. The repression of PDGFRα expression by Phf14 might therefore be mediated by interactions with as yet unknown factors.
The PHD structure occurs in a wide variety of eukaryotic proteins. To date, several mutations in PHD family proteins have been implicated in various human diseases such as immunodeficiency syndromes, neurological disorders, and cancer (24). Consistent with these findings, we showed down-regulation of Phf14 during bleomycin-induced fibrogenic inflammation in the adult lung that correlated with enhanced PDGFRα expression. Although the mechanism underlying bleomycin-induced repression of Phf14 expression is unknown, our results suggested that Phf14 is involved in the control of PDGFRα expression in the adult lung and in the pathological process leading to lung fibrosis. Bleomycin treatment induces DNA strand breaks through a series of complex oxidative reactions (43), and also generates reactive oxygen species such as superoxide and hydroxyl radicals (44). Such molecules can change the DNA binding activity of nuclear factor (NF)-κB, as well as Nrf-1 and -2 transcription factors, in pulmonary artery endothelial cells (45). Thus, it is possible that oxidative derivatives could influence the expression of Phf14 or the stability of the Phf14 protein.
This study revealed that up-regulation of PDGFRα expression induced by a lack of Phf14 enhanced the proliferation of lung mesenchymal cells, which in turn induced respiratory failure and perinatal death. Based on our results, we also examined the potential of antibody therapy to treat mouse lung fibrosis. We showed that a monoclonal antibody antagonizing PDGFRα signaling was effective in treating drug-induced lung fibrosis in mice. Perturbation of PDGFRα signaling is thought to be a key event in the mechanism underlying pulmonary inflammation and fibrosis (2), and several inflammatory agents induce pulmonary fibrosis by inducing an increase in PDGFRα expression (46, 47). It has been reported that bleomycin-induced pulmonary fibrosis in mice are accompanied by an increase in the expression of several extracellular matrix proteins including fibronectin and procollagens (48), and several histological findings in the mouse model resemble those in human pulmonary fibrosis (49). Although more detailed pathogenic studies are required to ensure that this mouse model of drug-induced pulmonary fibrosis accurately reflects human pulmonary fibrotic diseases, our results implicated antibodies that antagonize PDGFRα signaling as therapeutic candidates for lung fibrosis.
In summary, this study showed that Phf14 plays a role not only in the process of lung organogenesis, but also in the induction of pulmonary fibrosis. Phf14 negatively regulates mesenchymal cell proliferation by suppressing PDGFRα expression. Phf14-null mutants are neonatally lethal and show interstitial tissue hyperplasia in the lung. Studies of PHD family molecules in mammalian development and related diseases are limited, thus the present report highlights novel molecular mechanisms that regulates the growth of normal and pathogenetic mesenchyme.
Supplementary Material
Acknowledgments
We thank Megumi Goto, Mayumi Fukaura, Yasumitsu Fujie, and Genta Tsuzuki for technical support.
This work was supported in part by grants from the Ministry of Health, Labor, and Welfare of Japan and Core Research for Evolutional Science and Technology (CREST), Japan Science and Technology Agency.

This article contains supplemental Tables S1–S3, Figs. S1–S10, and Movie S1.
- PHD
- plant homology domain
- PSP
- PDGF receptor α single positive cell population
- MEF
- mouse embryonic fibroblasts
- LZ
- leucine zipper.
REFERENCES
- 1. Kalluri R., Neilson E. G. (2003) Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Invest. 112, 1776–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrae J., Gallini R., Betsholtz C. (2008) Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 22, 1276–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu X., Bringas P., Jr., Soriano P., Chai Y. (2005) PDGFR-α signaling is critical for tooth cusp and palate morphogenesis. Dev. Dyn. 232, 75–84 [DOI] [PubMed] [Google Scholar]
- 4. Karlsson L., Bondjers C., Betsholtz C. (1999) Roles for PDGF-A and sonic hedgehog in development of mesenchymal components of the hair follicle. Development 126, 2611–2621 [DOI] [PubMed] [Google Scholar]
- 5. Sun T., Jayatilake D., Afink G. B., Ataliotis P., Nistér M., Richardson W. D., Smith H. K. (2000) A human YAC transgene rescues craniofacial and neural tube development in PDGFRα knock-out mice and uncovers a role for PDGFR α in prenatal lung growth. Development 127, 4519–4529 [DOI] [PubMed] [Google Scholar]
- 6. Takakura N., Yoshida H., Ogura Y., Kataoka H., Nishikawa S. (1997) PDGFR α expression during mouse embryogenesis. Immunolocalization analyzed by whole-mount immunohistostaining using the monoclonal anti-mouse PDGFR α antibody APA5. J. Histochem. Cytochem. 45, 883–893 [DOI] [PubMed] [Google Scholar]
- 7. Soriano P. (1997) The PDGF α receptor is required for neural crest cell development and for normal patterning of the somites. Development 124, 2691–2700 [DOI] [PubMed] [Google Scholar]
- 8. Bleyl S. B., Moshrefi A., Shaw G. M., Saijoh Y., Schoenwolf G. C., Pennacchio L. A., Slavotinek A. M. (2007) Candidate genes for congenital diaphragmatic hernia from animal models: sequencing of FOG2 and PDGFRα reveals rare variants in diaphragmatic hernia patients. Eur. J. Hum. Genet. 15, 950–958 [DOI] [PubMed] [Google Scholar]
- 9. Karlsson L., Lindahl P., Heath J. K., Betsholtz C. (2000) Abnormal gastrointestinal development in PDGF-A and PDGFR-α-deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development 127, 3457–3466 [DOI] [PubMed] [Google Scholar]
- 10. Brennan J., Tilmann C., Capel B. (2003) Pdgfr-α mediates testis cord organization and fetal Leydig cell development in the XY gonad. Genes Dev. 17, 800–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heldin C. H., Westermark B. (1999) Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79, 1283–1316 [DOI] [PubMed] [Google Scholar]
- 12. Sakurai H., Era T., Jakt L. M., Okada M., Nakai S., Nishikawa S. (2006) In vitro modeling of paraxial and lateral mesoderm differentiation reveals early reversibility. Stem Cells 24, 575–586 [DOI] [PubMed] [Google Scholar]
- 13. Izumi N., Era T., Akimaru H., Yasunaga M., Nishikawa S. (2007) Dissecting the molecular hierarchy for mesendoderm differentiation through a combination of embryonic stem cell culture and RNA interference. Stem Cells 25, 1664–1674 [DOI] [PubMed] [Google Scholar]
- 14. Era T., Izumi N., Hayashi M., Tada S., Nishikawa S. (2008) Multiple mesoderm subsets give rise to endothelial cells, whereas hematopoietic cells are differentiated only from a restricted subset in embryonic stem cell differentiation culture. Stem Cells 26, 401–411 [DOI] [PubMed] [Google Scholar]
- 15. Wobus A. M., Boheler K. R. (2005) Embryonic stem cells. Prospects for developmental biology and cell therapy. Physiol. Rev. 85, 635–678 [DOI] [PubMed] [Google Scholar]
- 16. Nishikawa S., Jakt L. M., Era T. (2007) Embryonic stem-cell culture as a tool for developmental cell biology. Nat. Rev. Mol. Cell Biol. 8, 502–507 [DOI] [PubMed] [Google Scholar]
- 17. Kitagawa M., Era T. (2010) Differention of mesodermal cells from pluripotent stem cells. Int. J. Hematol. 91, 373–383 [DOI] [PubMed] [Google Scholar]
- 18. Tada S., Era T., Furusawa C., Sakurai H., Nishikawa S., Kinoshita M., Nakao K., Chiba T. (2005) Characterization of mesendoderm. A diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development 132, 4363–4374 [DOI] [PubMed] [Google Scholar]
- 19. Yasunaga M., Tada S., Torikai-Nishikawa S., Nakano Y., Okada M., Jakt L. M., Nishikawa S., Chiba T., Era T. (2005) Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat. Biotechnol. 23, 1542–1550 [DOI] [PubMed] [Google Scholar]
- 20. Takashima Y., Era T., Nakao K., Kondo S., Kasuga M., Smith A. G., Nishikawa S. (2007) Neuroepithelial cells supply an initial transient wave of MSC differentiation. Cell 129, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 21. Takebe A., Era T., Okada M., Martin Jakt L., Kuroda Y., Nishikawa S. (2006) Microarray analysis of PDGFR α+ populations in ES cell differentiation culture identifies genes involved in differentiation of mesoderm and mesenchyme including ARID3b that is essential for development of embryonic mesenchymal cells. Dev. Biol. 293, 25–37 [DOI] [PubMed] [Google Scholar]
- 22. Kobayashi K., Era T., Takebe A., Jakt L. M., Nishikawa S. (2006) ARID3B induces malignant transformation of mouse embryonic fibroblasts and is strongly associated with malignant neuroblastoma. Cancer Res. 66, 8331–8336 [DOI] [PubMed] [Google Scholar]
- 23. Soliman M. A., Riabowol K. (2007) After a decade of study, ING, a PHD for a versatile family of proteins. Trends Biochem. Sci. 32, 509–519 [DOI] [PubMed] [Google Scholar]
- 24. Baker L. A., Allis C. D., Wang G. G. (2008) PHD fingers in human diseases. Disorders arising from misinterpreting epigenetic marks. Mutat. Res. 647, 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang X., Hisha H., Taketani S., Adachi Y., Li Q., Cui W., Cui Y., Wang J., Song C., Mizokami T., Okazaki S., Li Q., Fan T., Fan H., Lian Z., Gershwin M. E., Ikehara S. (2006) Characterization of mesenchymal stem cells isolated from mouse fetal bone marrow. Stem Cells 24, 482–493 [DOI] [PubMed] [Google Scholar]
- 26. Era T., Asou N., Kunisada T., Yamasaki H., Asou H., Kamada N., Nishikawa S., Yamaguchi K., Takatsuki K. (1995) Identification of two transcripts of AML1/ETO-fused gene in t(8;21) leukemic cells and expression of wild-type ETO gene in hematopoietic cells. Genes Chromosomes Cancer 13, 25–33 [DOI] [PubMed] [Google Scholar]
- 27. Pear W. S., Nolan G. P., Scott M. L., Baltimore D. (1993) Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U.S.A. 90, 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hawley R. G., Lieu F. H., Fong A. Z., Hawley T. S. (1994) Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1, 136–138 [PubMed] [Google Scholar]
- 29. Takakura N., Yoshida H., Kunisada T., Nishikawa S., Nishikawa S. I. (1996) Involvement of platelet-derived growth factor receptor-α in hair canal formation. J. Invest. Dermatol. 107, 770–777 [DOI] [PubMed] [Google Scholar]
- 30. Loayza D., De Lange T. (2003) POT1 as a terminal transducer of TRF1 telomere length control. Nature 423, 1013–1018 [DOI] [PubMed] [Google Scholar]
- 31. Matsumoto T., Yokote K., Tamura K., Takemoto M., Ueno H., Saito Y., Mori S. (1999) Platelet-derived growth factor activates p38 mitogen-activated protein kinase through a Ras-dependent pathway that is important for actin reorganization and cell migration. J. Biol. Chem. 274, 13954–13960 [DOI] [PubMed] [Google Scholar]
- 32. Wu E., Palmer N., Tian Z., Moseman A. P., Galdzicki M., Wang X., Berger B., Zhang H., Kohane I. S. (2008) Comprehensive dissection of PDGF-PDGFR signaling pathways in PDGFR genetically defined cells. PLoS One 3, e3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Afink G., Westermark U. K., Lammerts E., Nistér M. (2004) C/EBP is an essential component of PDGFRA transcription in MG-63 cells. Biochem. Biophys. Res. Commun. 315, 313–318 [DOI] [PubMed] [Google Scholar]
- 34. Bonner J. C. (2004) Regulation of PDGF and its receptors in fibrotic diseases. Cytokine Growth Factor Rev. 15, 255–273 [DOI] [PubMed] [Google Scholar]
- 35. Thannickal V. J., Toews G. B., White E. S., Lynch J. P., 3rd, Martinez F. J. (2004) Mechanisms of pulmonary fibrosis. Annu. Rev. Med. 55, 395–417 [DOI] [PubMed] [Google Scholar]
- 36. Gurujeyalakshmi G., Hollinger M. A., Giri S. N. (1999) Pirfenidone inhibits PDGF isoforms in bleomycin hamster model of lung fibrosis at the translational level. Am. J. Physiol. 276, L311–318 [DOI] [PubMed] [Google Scholar]
- 37. Nagaoka I., Trapnell B. C., Crystal R. G. (1990) Up-regulation of platelet-derived growth factor-A and -B gene expression in alveolar macrophages of individuals with idiopathic pulmonary fibrosis. J. Clin. Invest. 85, 2023–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soriano P. (1994) Abnormal kidney development and hematological disorders in PDGF β-receptor mutant mice. Genes Dev. 8, 1888–1896 [DOI] [PubMed] [Google Scholar]
- 39. Lindahl P., Karlsson L., Hellström M., Gebre-Medhin S., Willetts K., Heath J. K., Betsholtz C. (1997) Alveogenesis failure in PDGF-A-deficient mice is coupled to lack of distal spreading of alveolar smooth muscle cell progenitors during lung development. Development 124, 3943–3953 [DOI] [PubMed] [Google Scholar]
- 40. Li J., Hoyle G. W. (2001) Overexpression of PDGF-A in the lung epithelium of transgenic mice produces a lethal phenotype associated with hyperplasia of mesenchymal cells. Dev. Biol. 239, 338–349 [DOI] [PubMed] [Google Scholar]
- 41. Olson L. E., Soriano P. (2009) Increased PDGFRα activation disrupts connective tissue development and drives systemic fibrosis. Dev. Cell 16, 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang Y., Wang J., Li G. (2006) Leucine zipper-like domain is required for tumor suppressor ING2-mediated nucleotide excision repair and apoptosis. FEBS Lett. 580, 3787–3793 [DOI] [PubMed] [Google Scholar]
- 43. Chen J., Stubbe J. (2005) Bleomycins. Towards better therapeutics. Nat. Rev. Cancer 5, 102–112 [DOI] [PubMed] [Google Scholar]
- 44. Oberley L. W., Buettner G. R. (1979) The production of hydroxyl radical by bleomycin and iron (II). FEBS Lett. 97, 47–49 [DOI] [PubMed] [Google Scholar]
- 45. Day R. M., Suzuki Y. J., Lum J. M., White A. C., Fanburg B. L. (2002) Bleomycin up-regulates expression of γ-glutamylcysteine synthetase in pulmonary artery endothelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 282, L1349-L1357 [DOI] [PubMed] [Google Scholar]
- 46. Bonner J. C., Lindroos P. M., Rice A. B., Moomaw C. R., Morgan D. L. (1998) Induction of PDGF receptor-α in rat myofibroblasts during pulmonary fibrogenesis in vivo. Am. J. Physiol. 274, L72–80 [DOI] [PubMed] [Google Scholar]
- 47. Lasky J. A., Tonthat B., Liu J. Y., Friedman M., Brody A. R. (1998) Up-regulation of the PDGF-α receptor precedes asbestos-induced lung fibrosis in rats. Am. J. Respir. Crit. Care Med. 157, 1652–1657 [DOI] [PubMed] [Google Scholar]
- 48. Kelley J., Chrin L., Shull S., Rowe D. W., Cutroneo K. R. (1985) Bleomycin selectively elevates mRNA levels for procollagen and fibronectin following acute lung injury. Biochem. Biophys. Res. Commun. 131, 836–843 [DOI] [PubMed] [Google Scholar]
- 49. Adamson I. Y., Bowden D. H. (1974) The pathogenesis of bloemycin-induced pulmonary fibrosis in mice. Am. J. Pathol. 77, 185–197 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.